Abstract
Cell-based assay based on time-stretch imaging is recognized to be well-suited for high-throughput phenotypic screening. However, this ultrafast imaging technique has primarily been limited to suspension-cell assay, leaving a wide range of solid-substrate assay formats uncharted. Moreover, time-stretch imaging is generally restricted to intrinsic biophysical phenotyping, but lacks the biomolecular signatures of the cells. To address these challenges, we develop a spinning time-stretch imaging assay platform based on the functionalized digital versatile disc (DVD). We demonstrate that adherent cell culture and biochemically-specific cell-capture can now be assayed with time-stretch microscopy, thanks to the high-speed DVD spinning motion that naturally enables on-the-fly cellular imaging at an ultrafast line-scan rate of >10MHz. As scanning the whole DVD at such a high speed enables ultra-large field-of-view imaging, it could be favorable for scaling both the assay throughput and content as demanded in many applications, e.g. drug discovery, and rare cancer cell screening.
OCIS codes: (110.0110) Imaging systems, (170.0180) Microscopy, (170.3880) Medical and biological imaging, (170.7160) Ultrafast technology
1. Introduction
Accessing detailed spatial information of cells, microscopy allows high-content cell-based phenotypic assay, but at a compromised measurement throughput [1,2]. Notable example is imaging flow cytometry in which streamlines cell interrogation by imaging single cells in suspension at a throughput of 1,000’s cells/sec is achieved [2]. However, this throughput is still at least two-orders-of-magnitude slower than classical non-imaging flow cytometry because of the inherent speed-versus-sensitivity limitation of the camera technologies. This pervasive problem hinders the acceptance of imaging cell-based assay in high-throughput screening (HTS) applications [3–5], e.g. phenotypic screening in the early drug discovery pipeline in which tens of thousands compounds per screen are involved; and circulating tumor cell (CTC) screening in which rare cell detection within enormous and heterogeneous population (some millions of blood cells) is mandated.
Optical time-stretch imaging has been demonstrated to overcome the speed limitation in conventional imaging by adopting an all-optical ultrafast image encoding concept [6–9]. The ultrafast continuous line-scan imaging (at a rate of tens of MHz) naturally favors imaging flow cytometry applications at a throughput which is impossible elsewhere, e.g. detecting rare cancer cells at 100,000 cells/sec [10]. Despite this uniquely fast imaging capability, two key challenges remain in the context of cell-based assay. First, time-stretch imaging predominantly generates intrinsic image contrasts from cells based on bright-field [10] or phase-contrast imaging [11,12]. Hence, the retrieved single-cell information mostly lacks chemical specificity. Second, assay of adherent cells, or fixed cells on planar substrates has been missing in the prior demonstrations of time-stretch imaging, which overwhelmingly focused on the suspension-cell assay format. Yet, planar platform is the prevalent format in cell-based assay, especially in the context of high-content imaging [1]. Cell-culture assay on the planar platform emulates the physiologically-relevant environments better than isolated cells in suspension, and thus are particularly effective in monitoring cell kinetics. In addition, planar platform allows space-multiplexed chemically specific cell-capture assays, e.g. array-based immunoassay. These assay types have yet been explored in time-stretch imaging.
To this end, we here demonstrate a spinning time-stretch imaging cell-based assay platform based on the current digital versatile disc (DVD) technology – a modified DVD drive for reconfigurable spinning control and modified DVDs as functionalized substrates for a variety of cell-based assay studies. The traditional rationale of adopting spinning assay platforms [13,14] is to leverage its centrifugal action for automated sample processing (e.g. fluid transport, mixing and valving) integrated with different types of assays (e.g. DNA microarray [15], and enzyme-linked immunosorbent assays/enzyme immunoassays (ELISA/EIA) [16]). In contrast, we here take advantage of the high-speed spinning motion, which naturally provides on-the-fly cellular imaging at an ultrafast rate that can only be made possible with time-stretch imaging. Indeed, spinning assays with imaging capability to-date remains scarce primarily because the conventional cameras or laser-/sample-scanning techniques run short of speed to provide blur-free high-resolution images under the high-speed spinning motion. Point-scanning cellular imaging on DVD has recently been demonstrated [17]. In contrast, the ultrafast line-scanning together with the all-optical encoding concept of our technique achieves almost 100-times higher in throughput. Moreover, we show that the modified DVD substrate can be flexibly functionalized for in-vitro cell culture experiments or chemically-specific cell-capture assay (i.e. binding assay), both can be assayed with time-stretch imaging at an ultrafast line-scan rate beyond 10MHz. Hence, scanning the whole disc with time-stretch imaging allows ultra-large field-of-view (FOV), thus high-throughput, imaging cell-based assay. More importantly, planar DVD platform provides a large footprint for highly multiplexed, and thus high-content, measurements by configuring the assay in an array format on the disc. Especially in the context of binding assay, the spinning-DVD time-stretch imaging platform provides not only biophysical phenotyping of cells derived from the morphology, but also extracts additional molecular signatures given by the chemically-specific cell binding. As a consequence, our platform could significantly expand the scope of ultrafast time-stretch imaging in cell-based assay beyond suspension assay.
2. Materials and methods
2.1 Substrate design and fabrication
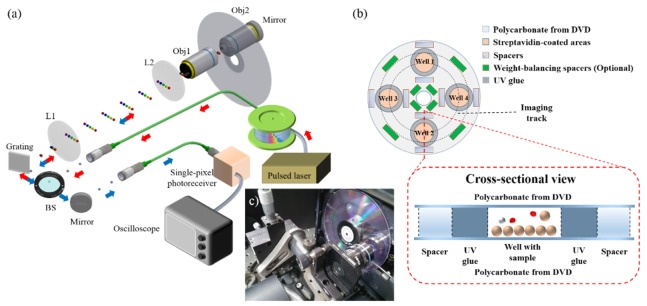
The substrates of commercial DVDs are typically made of polycarbonate, which is a popular choice of material in biomedical applications because of its biocompatibility and its superiormechanical strength. However, the reflective coatings on DVD generally forbid transmission imaging as adopted in this work (Fig. 1). To this end, our assay platform design was based on a double-layer polycarbonate substrate obtained from two separate DVDs. Specifically, each DVD (Maxell DVD-R) was split into two halves, each of which had the same disc shape but a reduced thickness, such that the originally sandwiched reflective layers can be removed. Only the transparent half (ca. 0.6 mm) was used for further surface functionalization.
Fig. 1.
(a) A schematic of the time-stretch imaging system. Laser pulses (from a fiber mode-locked laser) are first time-stretched within a dispersive fiber and thus form wavelength-swept waveforms. A holographic diffraction grating together the relay lenses (L1 and L2) and the objective lens (Obj1) is used to transform the wavelength-swept beam into 1D spectrally-encoded line-scan beam, which is projected onto the modified spinning DVD substrate. The encoded line-scan beam is returned along the same path, by placing a mirror at the entrance pupil of the back objective lens (Obj2), forming a double-pass configuration. Upon recombined back to the Gaussian beam profile, the imaged-encoded beam is eventually detected by a high-speed photo-receiver (bandwidth of 12GHz) and recorded by a high-speed real-time oscilloscope (bandwidth of 16GHz; sampling rate of 80GSa/s). (b) A design schematic of the modified DVD assay platform employed in this work (Four assay wells/sites are depicted in (b)). The substrate is composed of two polycarbonate layers (See the cross-sectional view), obtained from two separate DVDs, that are bonded together with UV-cured adhesive. This creates assay chambers with a height of ~100-120 µm (defined by the spacers). The spacers are also carefully aligned to stabilize the rapid spinning motion. (c) A photo of a spinning DVD mounted on a modified DVD drive.
For cell culture experiments (Fig. 2), the polycarbonate substrate was cleaned and sterilized with 70% ethanol and ultra-violet (UV) light. For chemically-specific cell-/microparticle-capture experiments (Figs. 3, 4, and 5), the substrate was further processed with streptavidin coating separately in four or eight pre-defined areas, which later formed the assay wells. Streptavidin coating was selected because of the strong binding affinity between streptavidin and biotin, as well as the wide availability of off-the-shelf biotinylated products (e.g. antibodies). Secondary biotinylated antibodies can be incubated in these areas for further specific capture. After the cell culture or specific cell-capture procedures (as detailed in Section 2.5), UV-cured adhesive was deposited surrounding the cell-specimen sites such that the cells under test were not in contact with the UV-cured adhesive before they were cured. Glass spacers with 100-120 μm in height were carefully positioned at various locations on the substrate (Fig. 1(b)) such that the weight was evenly distributed across the substrate in order to ensure stable spinning operation.
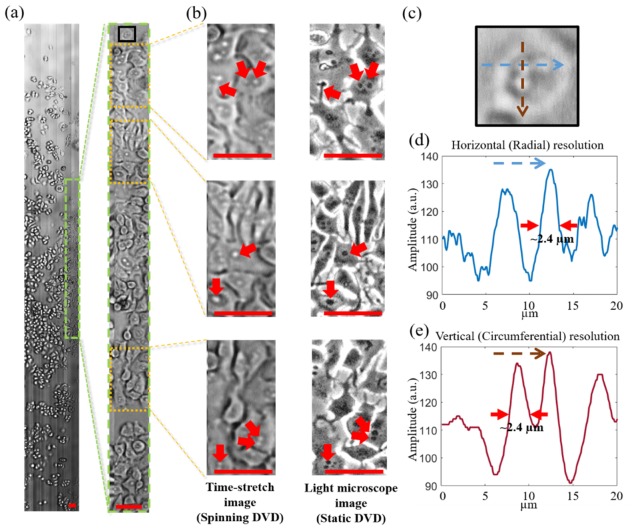
Fig. 2.
(a) Time-stretch image of MCF-7 (0.42 mm x 34 mm) cultured on the modified DVD captured at a single-shot line-scan of 11 MHz. The spinning speed is 900 rpm (linear speed of ~4 m/s). (b) zoom-in view of the area highlighted in (a). (c) (left column) further zoom-in view of the areas highlighted in (b). (Right column) Phase-contrast images of the same areas taken by commercial light microscope (Nikon Eclipse Ni-U). Arrows indicates the key cellular features identified in both time-stretch images and light microscope images. (c) Zoom-in view of a MCF-7 cell highlighted as the black box in (a). (d,e) The line-cuts along the fast axis (i.e. radial direction) and the slow axis (i.e. circumferential direction) of the selected cell in (c). The two line-cut plots clearly show that the ability of the system to resolve sub-cellular structure with a feature size better than 2.4 µm, consistent with the theoretical resolution of ~2 µm (see the text for detailed discussion). All scale bars (in red) represent 50 µm.
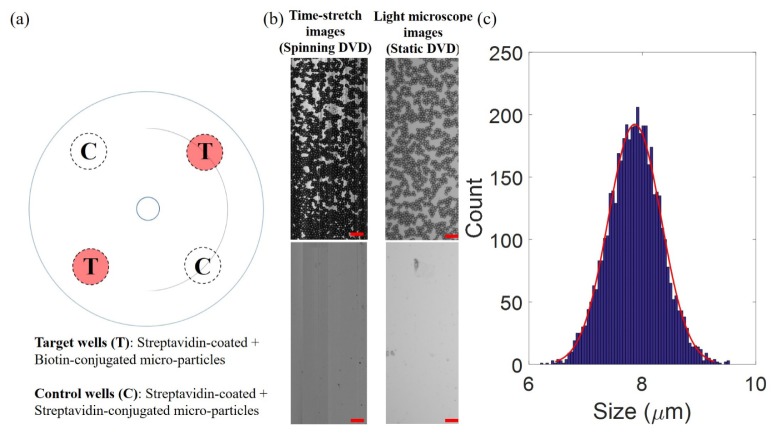
Fig. 3.
(a) Modified DVD assay design for time-stretch imaging of specifically-captured biotinated-polystyrene microparticles. The single-layer substrate in this experiment adopted a four-well design: two are target wells coated with streptavidin for specific capture of microparticles (marked as T) whereas another two are control wells in which no streptavidin coating is present (marked as C). The curve line on the DVD indicates the recording area. (b) (Left column) Time-stretch images of captured microparticles in wells T and C taken under a spinning speed of 3000 rpm (speed of 14 m/s). (Right column) Static images of the wells T and C taken by ordinary light microscope. (c) Statistical size distribution of the captured 4657 microparticles analysed from the time-stretch image (Red curve is the Gaussian fit). All scale bars (in red) represent 50 µm.
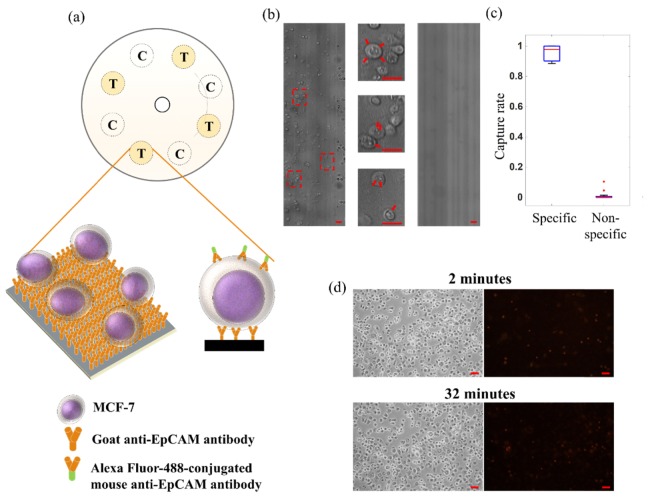
Fig. 4.
(a) Modified DVD assay design for time-stretch imaging of specifically-captured MCF-7. The substrate adopted an eight-well design: four are target wells coated with anti-EpCAM antibody (marked as T) whereas the other four are control wells with only streptavidin coating (marked as C). A schematic of the antibody-captured MCF-7 in the target wells is also shown. The curve line on the DVD indicates the recording area. (b) (Left) Time-stretch image of antibody-captured MCF-7 cells in the target well on the spinning DVD (at a spinning speed of 2400 rpm and a linear speed of 11 m/s). (Middle) Zoom-in views of the time-stretch image are also shown. (Right) Time-stretch image of control wells under the same spinning condition. (c) The analysis of the cell-capture specificity in the experiments. The percentages of specificity and non-specificity are calculated from the number of remaining cells upon rinsing out of the number of MCF-7 captured in the antibody-coated and streptavidin-coated wells respectively. (d) Static images of the captured cells (phase-contrast (left) and fluorescence (right)) treated with vital staining (propidium iodide) on the DVD substrate, taken after (top) 2-minutes and (bottom) 32-minute of spinning. All scale bars (in red) represent 50 µm.
Fig. 5.
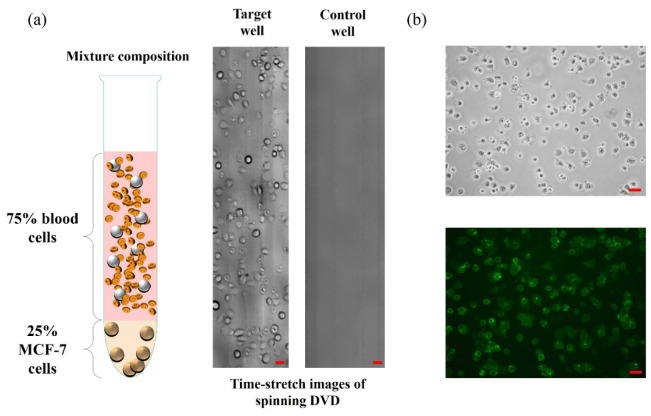
(a) (Left) Specimen of MCF-7 (25%) mixed with human buffy coat (75%) used for antibody-captured, and thus enrichment of, MCF-7. (Middle) Time-stretch image of the enriched MCF-7 (with anti-EpCAM antibody) in the target well. (Right) Time-stretch image of the control well in which only streptavidin is coated. Both the images in (a) were taken at a spinning speed of 900 rpm (linear speed of 4 m/s). Same substrate design as shown in Fig. 4. was adopted in this experiment. (b) Static images (Top: phase contrast; Bottom: fluorescence) of the enriched MCF-7 further stained with green fluorescent dye (Alexa Fluor-488 anti-EpCAM (RnD FAB9601G, 10 µg/mL)). This additional staining step was performed to further confirm the MCF-7 enriched on the DVD. All scale bars (in red) represent 50 µm.
Another identical but non-functionalized polycarbonate substrate was stacked and glued on top of the functionalized substrate with the spacers. The top substrate was further pressed to ensure complete contact with all the spacers. This resulted in a double-layer disc, with a thickness of ~1.3 mm, containing four to eight pre-defined assay compartments (Fig. 1(b)). The double-layer disc assembly was then exposed to the UV light (Thorlabs CS2010) for at least 30 seconds for further curing. The illumination was structured such that it avoided UV exposure, and thus phototoxicity of the cells within the compartments. We note that any disc-shaped substrates can in principle be adopted in the present time-stretch imaging system, given that the substrate material of choice is mechanically robust under high-speed spinning, biocompatible with cell culture or specific cell capture, and optically transparent for bright-field transmission illumination.
2.2 Spinning platform design
We modified a commercial DVD drive (Sony NEC Optiarc Inc. ND-3570A) by routing the stepper motor control signal from a custom-made Arduino-based proportional–integral–derivative (PID) controller. In this way, the spinning rate can be arbitrarily controlled with high stability. Specifically, the controller first acquired the reading of the spinning rate through the built-in Hall sensors of the DVD motor. Using a calibrated algorithm for precise damping-torque control, we converted the difference value between the current and targeted spinning speed (in revolutions per minute, rpm) into a control voltage that was fed back to the DVD driver. This feedback-control was operated in real-time and continuously to compensate the fluctuating air-resistance encountered during high-speed spinning (900-14000 rpm). To configure transmission time-stretch imaging, we removed the original laser source and photodetector in the DVD drive. We also created windows on both sides of the DVD drive chassis such that imaging light path could access through the double-layer disc loaded on the tray of the DVD drive.
2.3 Time-stretch imaging system
The central concept of time-stretch imaging is to achieve ultrafast optical imaging by an all-optical image encoding concept which combines spectral encoding (by the use of diffraction grating) and wavelength-time mapping (through group velocity dispersion (GVD)), all within a single-shot broadband laser pulse. In this way, the imaging scan-rate is determined by the laser repetition rate, which can typically be MHz or even GHz [18]. The light source was a home-built broadband (~10 nm) mode-locked fiber laser centered at the wavelength of 1060 nm. The laser repetition rate, or equivalently the line-scan rate, is 11MHz with a high pulse-to-pulse stability (~1% fluctuation in pulse intensity) [18]. The laser pulses from the mode-locked laser were amplified by a custom ytterbium-doped fiber amplifier module and then time-stretched by a 20-km long single-mode fiber (with an average GVD of 0.54 ns/nm). The time-stretched laser pulses were spatially transformed into one-dimensional (1D) spectral showers, by a diffraction grating (groove density = 1200 lines/mm), and were projected onto the spinning disc by an objective lens (numerical aperture (NA) of 0.6). It essentially provides a line-scan illumination with a field-of-view (FOV) of ~70 μm wide. The spatial resolution was estimated as 2 μm by considering the space-time mapping in this system, which is currently limited by diffraction limit of objective lens. To be specific, the image resolution along the radial (δx) direction, i.e. the fast axis, is governed by three limiting regimes [19]: (i) the spatial-dispersion-limited resolution, which is linked to the spectral resolution of the diffraction grating, denoted as δxspatial, (ii) the stationary-phase approximation limit, which describes the ambiguity of the wavelength-time mapping process δxSPA, which inversely scales with the square root of GVD and (iii) the photodetector-limited resolution δxdet, which is related to the finite bandwidth of the photodetector. In general, the largest value among these three parameters determines the overall resolution in a time-stretch image δx, i.e. δx = max {δxspatial, δxSPA, δxdet}. In our case δx = δxspatial = 1.7 µm. On the other hand, the image resolution along the spinning (circumferential) direction (δy), i.e. the slow axis, is limited by the diffraction limit (governed by the NA of the objective lens). In our case, δyobj = 1.98 µm. Note that the linear-motion speed (F, in m/s) and the repetition rate of the laser (R, in Hz) should be carefully configured to ensure Nyquist sampling condition along the slow axis, i.e. 2F/R < δx. This condition is satisfied in our case (R = 11.8 MHz) for the when the spinning speed is below ~5000 rpm.
This spectral-shower or line illumination, which was orthogonal to the spinning direction and double-passed the disc by another objective lens and a mirror (Fig. 1). The returned image-encoded spectral shower was converted back to a single Gaussian pulsed beam by the same grating, and was then detected by a single-pixel photodetector (bandwidth of 12 GHz, New Focus). The serial-time waveform data was digitized by a real-time oscilloscope (bandwidth of 16 GHz; sampling rate of 80 GSa/s) for subsequent custom image reconstruction routine, which involved background subtraction, line-scan cropping and stacking for forming a two-dimensional (2D) image. Note that 2D image reconstruction can be done in real-time. The image focus position can thus be adjusted in real-time as well.
2.4 Image stitching for large FOV visualization
Apart from controlling the spinning rate of the DVD motor, the PID controller was integrated with a custom-design triggering circuit module, which consists of a frequency divider and another Arduino-based microprocessor. It played a critical role for robust image stitching which enables large FOV imaging for ultra-large-scale, and thus high-throughput imaging cell-based assay. Specifically, this integrated module controlled the oscilloscope for capturing different segments on the spinning disc in real-time by continuously updating the imaged segment positions. The final large FOV images were obtained through off-line digital image-segment stitching in both the radial and circumferential directions after acquiring multiple images at known segment position. For radial stitching, the modified DVD drive was also scanned transversely.
2.5 Sample preparations
The human breast adenocarcinoma cell lines (MCF-7) were trypsinized from the culture dish and centrifuged before mixing with standard cell culture medium formulated with 90% Dulbecco's Modified Eagle Medium (DMEM), 10% Fetal Bovine Serum (FBS) and 1% Penicillin-Streptomycin (Pen Strep). Cells were cultured in a CO2 incubator and the medium was renewed two times per week. A portion of cells were labelled with a vital stain (trypan blue) and were counted manually using a standard hemocytometer to ensure the viability of cells.
For the experiments of cell culture (Fig. 2), about 30,000 MCF-7 cells were mixed with 300 μL standard cell culture medium and were then loaded on the pre-defined areas on the half-disc substrate. The mixture was spatially confined within the area by surface tension on the hydrophobic polycarbonate surface. This substrate was then incubated for two days before bonded with another non-functionalized polycarbonate half-disc, as described in Section 2.1.
For the chemically-specific microparticle-capture experiments (Fig. 3), biotin polystyrene microspheres (Spherotech, 7.79 μm) were employed. 20 μL of the stock supplied microsphere solution was incubated on all pre-defined capture (target) wells of the disc for 30 minutes (see the disc schematic in Fig. 3(a)). All wells are washed with 1 × PBS for 5 times to prevent non-specific microsphere capture. Here, we adopted only a single-layer half-disc substrate structure, bypassing the need for covering the disc substrate with another disc. To prevent crystallization of PBS upon drying, we gently washed the cell sites with reverse osmosis (RO) water once for 5 seconds.
For the experiments of chemically-specific cell capture (Fig. 4), four or eight target wells were defined on the single-layer polycarbonate substrate and were treated with streptavidin coating (following the protocol provided with BioteZ Polystreptavidin R Coating Kit) (Fig. 4(a)). Biontinylated horse anti-goat antibody (Vector Labs BA-9500, 10 μg/mL) was incubated in the streptavidin-coated sites for 30 minutes, followed by rinsing for 5 times with 1 × phosphate-buffered saline (PBS). Then, goat anti-EpCAM antibody (RnD AF960, 10 μg/mL) was further incubated only in the target wells for 30 minutes and followed by rinsing for 5 times with 1 × PBS, such that the target wells can capture MCF-7, which has epithelial cell adhesion molecules (EpCAM) as surface markers [3]. 10 μL MCF-7 in 1 × PBS was loaded to all the wells on the substrate for 30 minutes that allowed binding between the antibodies and EpCAM, and thus capture of MCF-7 (Fig. 4(a)). The samples on the wells were pipetted up and down every 15 minutes to reduce non-specific adhesion. Followed by rinsing with 1 × PBS for 5 times, it reduced non-specific binding. We also separately tested and verified all the coating layers by standard biochemical methods. The streptavidin layer was tested with 3,3′-Diaminobenzidine (DAB) staining using biotinylated horseradish peroxidase H (Vector Labs PK-6100) such that the streptavidin-coated substrate appeared to be brown in color upon staining. The biotinylated horse anti-goat antibody (Vector Labs BA-9500) layer on top of streptavidin layer was verified by fluorescence imaging after incubation with the extra Alexa Fluor 488 goat anti-mouse antibody (Life Technologies A-11001) on the substrate (Fig. 4(a)). For every layer, control experiments were conducted to verify minimal non-specific binding.
For the experiment of MCF-7 enrichment/screening (Fig. 5), the human buffy coat was extracted and provided by Hong Kong Red Cross. All the blood donors have given written consent for clinical care and research purposes. The human buffy coat was then mixed with MCF-7 cells for the experiments shown in Fig. 5(a). The buffy coat samples were obtained one day prior to the experiments and stored under the room temperature overnight. On the other hand, MCF-7 cells were trypsinized from the culture dish and was counted such that ~6×105 MCF-7 was then mixed with ~3×106 cells from human buffy coat (220 μL in total). 10 μL of the mixture solution was then incubated in each target site for 30 minutes, with pipetting up-and-down every 15 minutes to reduce non-specific adhesion. We note that the human buffy coat samples obtained from Hong Kong Red Cross were originally mixed with a portion of red blood cells and serum. Therefore, the binding sites were rinsed with 1x PBS for 7 times after the capturing process of MCF-7 to minimize the unspecific binding, especially due to the blood clot.
3. Results and discussion
3.1 Time-stretch imaging of cell culture on spinning disc
We performed time-stretch imaging of adherent cells on the modified DVD at a spinning speed of 900 rpm, which is equivalent to a linear speed of 4 m/s within the line-scan region. Not only can the system capture a large FOV, (as large as 34 mm × 420 μm (Fig. 2(a))) at a line-scan rate of 11MHz, but can also deliver high-resolution cellular imaging that reveal sub-cellular structures without motion-blur (see Fig. 2(b)). Note that the image contrast can be further enhanced by accessing the phase contrast through the use of interferometry [11,12], or phase gradient contrast by an asymmetric-detection technique [20]. Notably, interferometric time-stretch imaging can further quantify the phase information of cells, from which a set of biophysical phenotypes can be extracted, e.g. cell size, mass, and density [7,11].
It was found that the spinning speed regime adopted in this experiment can ensure no observable change in morphology of the adherent cells under the ultrafast spinning action. This can be verified by the static images of the same area taken by a conventional light microscope using a 10× objective lens (Nikon Eclipse Ni-U). The cellular morphology visualized in the static images and the time-stretch spinning images are generally consistent with each other. In the current system, the FOV of the time-stretch image is limited by the finite memory depth provided by oscilloscope. When integrated with high-throughput data acquisition platform, e.g. graphic processing unit (GPU) or field programmable gated array (FPGA), continuous whole disc imaging in real-time is feasible. It is noteworthy that FPGA has the flexible reconfigurability in terms of handling enormous data acquisition, processing, storage and even higher-level data analytics in real-time.
3.2 Time-stretch imaging of microspheres captured on spinning disc
Four sites/wells were symmetrically defined on the disc substrate. Two of them were coated with streptavidin for biotinylated-microsphere binding whereas the other two wells without streptavidin coating were defined as control wells (Fig. 3(a)). For the sake of imaging demonstration and simplicity, we adopted a single-layer substrate design and reflection imaging, i.e. we removed the back objective lens and the mirror such that only the reflected and back-scattered light was collected. It is because, in contrast to biological cells, polystyrene microspheres give sufficiently high back-scattered light contrast and can be exposed to air during imaging without introducing any detrimental effect to the microspheres. The substrate was dried inside a desiccator prior to high-speed spinning for time-stretch imaging. A stitched image with large FOV of 0.384 mm × 140 mm was transformed into a curved image, with an arc of 180° (See the overlaid image on the disc schematic shown in Fig. 3(a)). Specifically captured microspheres (4657 microspheres) are clearly visualized under a high rotational speed of 3000 rpm, or a linear speed of ~14 m/s (Fig. 3(b)), in comparison with the image taken in the control sites, i.e. no microspheres are observed (Fig. 3(b)). Note that the time-stretch images of the spinning substrate are highly consistent with the static images of the same regions, captured by an ordinary light microscope (Fig. 3(b)). To further exemplify the capability of quantitative analysis derived from this high-throughput imaging technique, we digitally segmented individual microspheres in the image and quantified the statistical distribution of the size (Fig. 3(c)). The measured mean diameter, i.e. 7.85 μm (a standard deviation of 0.68 μm), is consistent with the specification provided by the supplier.
3.3 Time-stretch imaging of antibody-captured cells on spinning disc
Under the rotational speed of 2400 rpm (i.e. a linear speed as high as 11 m/s), our time-stretch imaging system was able to acquire high-resolution images of individual captured MCF-7 cells in the target wells (Fig. 4(b)). Note that the final stitched image has a FOV of 0.55 mm × 70 mm, covering both the control and target wells (across an arc of 90° as shown in Fig. 4(a)). DVD surface modification procedure and the binding specificity are clearly validatedby the comparison between the images of the target and control wells (Fig. 4(b)). The specific capture rate was determined to be ~95.8% whereas the non-specific capture rate was ~1.4% (Fig. 4(c)). It should be noted that the specific capture rate is in principle limited by the available binding area. The significance of this demonstration is that time-stretch imaging integrated with this spinning cell-based assay format can reveal not only the morphological information of the cells, but also the biomolecular signature of the cells through biochemically-specific binding (e.g. the surface markers EpCAM of MCF-7 in this case) – providing additional information for enhancing the assay accuracy and specificity.
We also assessed the viability of the cells influenced by high-speed spinning operation. Vital staining was performed by incubating captured cells with propidium iodide (PI) on the substrate. Orange-red fluorescence emission from PI serves as the indicator for dead cells. No noticeable change in the viability of the cells after the 2-min and 32-min spinning operations was observed (only 0.5% increase in the dead cell counts). It verifies that the high-speed spinning during time-stretch imaging introduces minimal detrimental effect to the cells. In addition, the vast majority of the captured cells remains unchanged in their position on disc over the spinning duration of 30-min (Fig. 4(d)). It demonstrates the superior binding strength, and thus robustness of this cell-capture assay format.
3.4 Cancer cell screening: Mixture of human blood and MCF-7
Instead of using pure population of MCF-7 (as shown in Fig. 4), we further conducted an experiment involving a mixed population of human blood cells and MCF-7. Specifically, MCF-7 cells were mixed with human buffy coat (extracted from human whole blood), followed by MCF-7 capture and screening on the spinning disc. The same substrate design as shown in Fig. 4(a) was employed. Similar to the experiment with the pure MCF-7 population (Fig. 4), four out of eight sites on disc were designated as target wells, which were coated with streptavidin, biotinylated horse anti-goat antibody and finally goat anti-EpCAM antibody. The screening process demonstrated a highly efficient specific cell-capture, which was again visualized by time-stretch imaging, under the spinning speed of 900 rpm (Fig. 5(a)). The specificity of MCF-7 capture was further validated by conjugating additional green fluorescent probe (Alexa Fluor-488) with anti-EpCAM antibody and detecting the corresponding fluorescence emission after the time-stretch spinning imaging operation (Fig. 5(b)) – confirming the captured cells are not the white blood cells.
This experiment is particularly relevant to the applications of CTCs enrichment, detection and enumeration. In this example, we utilized EpCAM, a cell adhesion molecule commonly expressed on epithelial cells, as a biomarker to distinguish MCF-7 cells from the white blood cells through an immunological binding approach. Therefore, our system not only can perform EpCAM-based CTC enrichment, similar to the existing techniques [3,21,22], but also allows in situ quantitative image analysis of the captured cells with the single-cell precision, thanks to the high-resolution and high-throughput imaging capability. Notably, coupled with quantitative phase time-stretch imaging, this high-throughput spinning imaging cell-based assay could allow single-cell biophysical phenotyping, e.g. cell size, mass, density and other cellular mechanical properties. These intrinsic phenotypes are known to be closely correlated with the malignancy transformation and are thus effective biomarkers of cancer screening as well as drug development [23–25]. Especially regarding the drug development process, large-format spinning disc also favors highly-multiplexed imaging assay and can potentially be used for efficient (cancer) drug screening against hundreds to tens of thousands compounds.
4. Conclusion
We have developed a high-throughput imaging cell-based assay platform based on integration of time-stretch imaging and high-speed spinning DVD. Leveraging the commercial DVD technology, which inherently provides high-speed unidirectional sample scanning, our system is capable of delivering real-time, continuous cellular imaging on disc at a line-scan rate beyond 10 MHz. Note that the actual imaging throughput depends on the cell density and the scanning FOV. Assuming the cells with the diameter of 10 - 30 µm and occupying 10-20% of the total disc space, we can estimate the overall throughput to be as high as 100,000 – 1,000,000 cells/sec. In practice, such an enormous size of image data can only be handled when the imaging system is integrated with a high-throughput data acquisition platform, e.g. GPU or FPGA, in order to support continuous whole disc image acquisition and processing in real-time. Compared to time-stretch imaging, automated microscopy, which is a widely adopted tool for image-based high-content screening [1], runs short of sufficient imaging throughput. This is primarily limited by the frame rate achieved by the current camera technologies, which hinders fast automated specimen stage scanning. Moreover, essentially all classical optical microscopes have a fundamental space-bandwidth-product limit of optical imaging, in which high resolution imaging compromises the FOV, and thus throughput [26]. These factors generally explain the relatively lower throughput, 1,000’s cells/sec, compared to the spinning-disc time-stretch imaging system.
More significantly, we have demonstrated that this system is compatible with both adherent cell culture and biochemically-specific cell capture assay formats – further enlarging the scope of time-stretch imaging, which has predominately been limited to the suspension-cell format [7,10–12,20]. Furthermore, the single-cell phenotypes extracted from the time-stretch images are no longer restricted primarily to biophysical properties, but also the molecular signatures (e.g. surface markers of cells), thanks to the biocompatibility of the DVD with the standard chemically-specific binding assay strategies. We also note that this spinning-disc assay platform can be compatible with not only bright-field and quantitative phase imaging, but also fluorescence imaging. For instance, a recent demonstration of ultrafast all-optical laser-scanning fluorescence imaging at a line-scan rate of > 1 MHz, based on a concept of free-space angular-chirp-enhanced delay (FACED) [27], could be a viable option to be combined with the current high-speed spinning assay platform to enrich fluorescence-image-based single-cell analysis, such as CTC detection with multi-color fluorescence staining [28].
For the sake of demonstration, up to 8 assay sites have been incorporated on the DVD-based substrate in the current work. Nonetheless, high-density assay array/matrix on DVD can readily be designed and fabricated with the existing microfabrication technologies and thus allows highly multiplexed cell-based assay at high-throughput, enabled by time-stretch imaging. The planar platform also makes it straightforward to combine the ultrafast imaging capability with the established centrifugal microfluidic technologies for active fluid control, e.g. sampling, mixing and valving, on the same disc. Such assay integration could allow more advanced assay functionalities and applications. As a result, time-stretch spinning imaging cell-based assay could hold promise for addressing the unmet need for scaling both the measurement throughput as well as content in many applications, e.g. HTS in drug development, and rare cancer cell screening.
Acknowledgment
We thank Shing Chan for providing the human buffy coat. This work was partially supported by Research Grants Council of the Hong Kong Special Administrative Region of China (HKU 17207714, HKU 17207715 HKU 720112E), Innovation and Technology Fund (ITS/090/14) and the University Development Funds of HKU.
References and links
- 1.Boutros M., Heigwer F., Laufer C., “Microscopy-Based High-Content Screening,” Cell 163(6), 1314–1325 (2015). 10.1016/j.cell.2015.11.007 [DOI] [PubMed] [Google Scholar]
- 2.Natasha I. A. V., Barteneva S., Imaging Flow Cytometry: Methods and Protocols, 1st ed., Methods in Molecular Biology (Humana Press, 2016). [Google Scholar]
- 3.Alix-Panabières C., Pantel K., “Challenges in circulating tumour cell research,” Nat. Rev. Cancer 14(9), 623–631 (2014). 10.1038/nrc3820 [DOI] [PubMed] [Google Scholar]
- 4.Feng Y., Mitchison T. J., Bender A., Young D. W., Tallarico J. A., “Multi-parameter phenotypic profiling: using cellular effects to characterize small-molecule compounds,” Nat. Rev. Drug Discov. 8(7), 567–578 (2009). 10.1038/nrd2876 [DOI] [PubMed] [Google Scholar]
- 5.Lang P., Yeow K., Nichols A., Scheer A., “Cellular imaging in drug discovery,” Nat. Rev. Drug Discov. 5(4), 343–356 (2006). 10.1038/nrd2008 [DOI] [PubMed] [Google Scholar]
- 6.Goda K., Jalali B., “Dispersive Fourier transformation for fast continuous single-shot measurements,” Nat. Photonics 7(2), 102 (2013). 10.1038/nphoton.2012.359 [DOI] [Google Scholar]
- 7.Lau A. K., Shum H. C., Wong K. K., Tsia K. K., “Optofluidic time-stretch imaging - an emerging tool for high-throughput imaging flow cytometry,” Lab Chip 16(10), 1743–1756 (2016). 10.1039/C5LC01458A [DOI] [PubMed] [Google Scholar]
- 8.Goda K., Tsia K. K., Jalali B., “Serial time-encoded amplified imaging for real-time observation of fast dynamic phenomena,” Nature 458(7242), 1145–1149 (2009). 10.1038/nature07980 [DOI] [PubMed] [Google Scholar]
- 9.Lei C., Guo B., Cheng Z., Goda K., “Optical time-stretch imaging: Principles and applications,” Appl. Phys. Rev. 3(1), 011102 (2016). 10.1063/1.4941050 [DOI] [Google Scholar]
- 10.Goda K., Ayazi A., Gossett D. R., Sadasivam J., Lonappan C. K., Sollier E., Fard A. M., Hur S. C., Adam J., Murray C., Wang C., Brackbill N., Di Carlo D., Jalali B., “High-throughput single-microparticle imaging flow analyzer,” Proc. Natl. Acad. Sci. U.S.A. 109(29), 11630–11635 (2012). 10.1073/pnas.1204718109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C. L., Mahjoubfar A., Tai L. C., Blaby I. K., Huang A., Niazi K. R., Jalali B., “Deep Learning in Label-free Cell Classification,” Sci. Rep. 6, 21471 (2016). 10.1038/srep21471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau A. K., Wong T. T., Ho K. K., Tang M. T., Chan A. C., Wei X., Lam E. Y., Shum H. C., Wong K. K., Tsia K. K., “Interferometric time-stretch microscopy for ultrafast quantitative cellular and tissue imaging at 1 μm,” J. Biomed. Opt. 19(7), 076001 (2014). 10.1117/1.JBO.19.7.076001 [DOI] [PubMed] [Google Scholar]
- 13.Gorkin R., Park J., Siegrist J., Amasia M., Lee B. S., Park J. M., Kim J., Kim H., Madou M., Cho Y. K., “Centrifugal microfluidics for biomedical applications,” Lab Chip 10(14), 1758–1773 (2010). 10.1039/b924109d [DOI] [PubMed] [Google Scholar]
- 14.Strohmeier O., Keller M., Schwemmer F., Zehnle S., Mark D., von Stetten F., Zengerle R., Paust N., “Centrifugal microfluidic platforms: advanced unit operations and applications,” Chem. Soc. Rev. 44(17), 6187–6229 (2015). 10.1039/C4CS00371C [DOI] [PubMed] [Google Scholar]
- 15.Wang L., Li P. C., “Microfluidic DNA microarray analysis: a review,” Anal. Chim. Acta 687(1), 12–27 (2011). 10.1016/j.aca.2010.11.056 [DOI] [PubMed] [Google Scholar]
- 16.Park J., Sunkara V., Kim T. H., Hwang H., Cho Y. K., “Lab-on-a-disc for fully integrated multiplex immunoassays,” Anal. Chem. 84(5), 2133–2140 (2012). 10.1021/ac203163u [DOI] [PubMed] [Google Scholar]
- 17.Ramachandraiah H., Amasia M., Cole J., Sheard P., Pickhaver S., Walker C., Wirta V., Lexow P., Lione R., Russom A., “Lab-on-DVD: standard DVD drives as a novel laser scanning microscope for image based point of care diagnostics,” Lab Chip 13(8), 1578–1585 (2013). 10.1039/c3lc41360h [DOI] [PubMed] [Google Scholar]
- 18.Lau A. K. S., Tang A. H. L., Xu X. W. J., Wong K. K. Y., Tsia K. K., “Optical Time Stretch for High-Speed and High-Throughput Imaging — From Single-Cell to Tissue-Wide Scales,” IEEE J. Sel. Top. Quantum Electron. 22, 6803115 (2016). [Google Scholar]
- 19.Tsia K. K., Goda K., Capewell D., Jalali B., “Performance of serial time-encoded amplified microscope,” Opt. Express 18(10), 10016–10028 (2010). 10.1364/OE.18.010016 [DOI] [PubMed] [Google Scholar]
- 20.Wong T. T., Lau A. K., Ho K. K., Tang M. Y., Robles J. D., Wei X., Chan A. C., Tang A. H., Lam E. Y., Wong K. K., Chan G. C., Shum H. C., Tsia K. K., “Asymmetric-detection time-stretch optical microscopy (ATOM) for ultrafast high-contrast cellular imaging in flow,” Sci. Rep. 4, 3656 (2014). 10.1038/srep03656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozkumur E., Shah A. M., Ciciliano J. C., Emmink B. L., Miyamoto D. T., Brachtel E., Yu M., Chen P. I., Morgan B., Trautwein J., Kimura A., Sengupta S., Stott S. L., Karabacak N. M., Barber T. A., Walsh J. R., Smith K., Spuhler P. S., Sullivan J. P., Lee R. J., Ting D. T., Luo X., Shaw A. T., Bardia A., Sequist L. V., Louis D. N., Maheswaran S., Kapur R., Haber D. A., Toner M., “Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells,” Sci. Transl. Med. 5(179), 179ra47 (2013). 10.1126/scitranslmed.3005616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riethdorf S., Fritsche H., Müller V., Rau T., Schindlbeck C., Rack B., Janni W., Coith C., Beck K., Jänicke F., Jackson S., Gornet T., Cristofanilli M., Pantel K., “Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system,” Clin. Cancer Res. 13(3), 920–928 (2007). 10.1158/1078-0432.CCR-06-1695 [DOI] [PubMed] [Google Scholar]
- 23.Otto O., Rosendahl P., Mietke A., Golfier S., Herold C., Klaue D., Girardo S., Pagliara S., Ekpenyong A., Jacobi A., Wobus M., Töpfner N., Keyser U. F., Mansfeld J., Fischer-Friedrich E., Guck J., “Real-time deformability cytometry: on-the-fly cell mechanical phenotyping,” Nat. Methods 12(3), 199–202 (2015). 10.1038/nmeth.3281 [DOI] [PubMed] [Google Scholar]
- 24.Zangle T. A., Teitell M. A., “Live-cell mass profiling: an emerging approach in quantitative biophysics,” Nat. Methods 11(12), 1221–1228 (2014). 10.1038/nmeth.3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tse H. T., Gossett D. R., Moon Y. S., Masaeli M., Sohsman M., Ying Y., Mislick K., Adams R. P., Rao J., Di Carlo D., “Quantitative diagnosis of malignant pleural effusions by single-cell mechanophenotyping,” Sci. Transl. Med. 5(212), 212ra163 (2013). 10.1126/scitranslmed.3006559 [DOI] [PubMed] [Google Scholar]
- 26.Lohmann A. W., Dorsch R. G., Mendlovic D., Ferreira C., Zalevsky Z., “Space–bandwidth product of optical signals and systems,” J. Opt. Soc. Am. A 13(3), 470–473 (1996). 10.1364/JOSAA.13.000470 [DOI] [Google Scholar]
- 27.J. Wu, Y. Xu, J. Xu, X. Wei, A. C. Chan, A. H. Tang, A. K. Lau, B. M. Chung, H. C. Shum, E. Y. Lam, K. K. Wong and K. K. Tsia, Ultrafast Laser-Scanning Time-Stretch Imaging at Visible Wavelengths. Light: Science & Applications accepted article preview 12 August 2016. [DOI] [PMC free article] [PubMed]
- 28.Lawson D. A., Bhakta N. R., Kessenbrock K., Prummel K. D., Yu Y., Takai K., Zhou A., Eyob H., Balakrishnan S., Wang C.-Y., Yaswen P., Goga A., Werb Z., “Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells,” Nature 526(7571), 131–135 (2015). 10.1038/nature15260 [DOI] [PMC free article] [PubMed] [Google Scholar]