Abstract
The optical properties of amyloid fibers are often distinct from those of the source protein in its non-fibrillar form. These differences can be utilized for label-free imaging or characterization of such structures, which is particularly important for understanding amyloid fiber related diseases such as Alzheimer’s and Parkinson’s disease. We demonstrate that two amyloid forming proteins, insulin and β-lactoglobulin (β-LG), show intrinsic fluorescence with emission spectra that are dependent on the excitation wavelength. Additionally, a new fluorescence peak at about 430 nm emerges for β-LG in its amyloid state. The shift in emission wavelength is related to the red edge excitation shift (REES), whereas the additional fluorescence peak is likely associated with charge delocalization along the fiber backbone. Furthermore, the spherulitic amyloid plaque-like superstructures formed from the respective proteins were imaged label-free with confocal fluorescence, multiphoton excitation fluorescence (MPEF), and second-harmonic generation (SHG) microscopy. The latter two techniques in particular yield images with a high contrast between the amyloid fiber regions and the core of amorphously structured protein. Strong multiphoton absorption (MPA) for the amyloid fibers is a likely contributor to the observed contrast in the MPEF images. The crystalline fibrillar region provides even higher contrast in the SHG images, due to the inherently ordered non-centrosymmetric structure of the fibers together with their non-isotropic arrangement. Finally, we show that MPEF from the insulin spherulites exhibits a spectral dependence on the excitation wavelength. This behavior is consistent with the REES phenomenon, which we hypothesize is the origin of this observation. The presented results suggest that amyloid deposits can be identified and structurally characterized based on their intrinsic optical properties, which is important for probe-less and label-free identification and characterization of amyloid fibers in vitro and in complex biological samples.
OCIS codes: (180.4315) Nonlinear microscopy, (190.4160) Multiharmonic generation, (190.4180) Multiphoton processes, (170.3880) Medical and biological imaging, (160.2540) Fluorescent and luminescent materials
1. Introduction
There are many proteins that misfold and aggregate into amyloid-like fibrillar structures upon mild denaturation. [1,2] Such fibers vary in size, but are typically 5 to 15 nm in diameter, have typical persistence lengths in the μm regime and are chemically and mechanically stable. [3–5] This family of fibers has backbones of intermolecular β-sheets that run along the fiber axis. [6] Amyloid fibers are found in a wide range of severe conditions, many of which are becoming increasingly prevalent. Some examples include: Alzheimer’s disease, diabetes mellitus type II, and Parkinson’s disease - for which the associated proteins are Aβ, islet amyloid polypeptide, and α-synuclein, respectively. [7–11] Other examples of amyloid-forming proteins are insulin and β-lactoglobulin (β-LG), [12–14] which to date have not been shown to induce degenerative conditions. As a result, these amyloids are well-suited to be used in basic research that aims to develop strategies for identification and characterization of amyloid structures. Additionally, the unique chemical, mechanical, and optical properties of amyloids make them interesting for practical use in engineering applications, such as organic light-emitting diodes, conducting nanowires, drug-delivery systems, among others. [15–21]
It has been shown that lysozyme and Aβ, as well as a few engineered protein sequences, develop intrinsic fluorescence in the visible regime upon aggregation into amyloid fibers. [22–25] In a few cases, electrical conduction through the amyloid fibers has been reported as well. [22,26] The origin and specific characteristics of the fluorescence and charge transport of these structures may be complex and multifaceted; [27,28] however, as these properties can arise even without any ring-structures in the peptide sequence, one mechanism that has been hypothesized for their emergence is charge delocalization along the fiber backbone due to long-range hydrogen bonding of the peptide units. [22,23,29,30] The double-bond form of the resonance structures in a peptide bond resembles a unit that one would expect to be conjugated in a long β-sheet sequence, which grants credibility to this hypothesis. [31] The fact that this resonance form would be highly discouraged in hydrophobic environments further corroborates this theory, since it has been shown that low humidity and vacuum conditions diminish both the fluorescence and charge transport of amyloid fibers. [22,23] A recent study demonstrated that the intrinsic fluorescence from amyloid fibers is also highly dependent on the pH. [32] This observation was attributed to proton transfer in the fibers, which may modulate the charge delocalization through them and affect the absorption and emission characteristics.
The amyloid deposits in brain tissue from patients with Alzheimer’s disease consist of various types of amyloid fiber aggregates, including a spherical superstructure called amyloid spherulites. [33–41] In these structures, amyloid fibers grow radially from the center, where one may find an amorphous core. The existence, size, and, morphology of the amorphous core is dependent on the source protein and the conditions under which they are formed. [43] It was previously demonstrated that the Aβ protein associated with Alzheimer’s disease can form spherulites in vitro, [41] but until recently [42] the procedure has not been time-efficient and the yield has been moderate. However, insulin and β-LG form both amyloid fibers and amyloid spherulites within hours and with high yields – therefore these proteins were used in this study. The assembly process depends on several factors, such as the protein concentration, temperature, pH and ion concentrations, and has been described elsewhere. [43–51] The microscopy efforts in this work focused on spherulites prepared from these proteins and the intrinsic optical properties of these amyloid structures were utilized in label-free imaging applications.
Most studies of amyloid structures by multiphoton excitation fluorescence (MPEF) and second-harmonic generation (SHG) imaging have relied on external probes, [52–57] while only a few have been label-free. [58–60] In this paper, we show that these techniques produce images with high contrast for the amyloids. This can be attributed to the previously demonstrated multiphoton absorption (MPA), [61] being as effective for insulin fibers as for typical two-photon dyes. Furthermore, the nonisotropic organisation and inherent noncentrosymmetry of the fibers in the crystalline fibrillar regions enhance their SHG susceptibility. In the following sections, we first briefly describe the preparation procedures that lead to either individual fibers or spherulites. The fibers were visualized by atomic force microscopy (AFM), while the spherulites were identified with cross-polarized microscopy. We then present the ultraviolet-visible (UV-Vis) absorption, as well as the fluorescence spectra and fluorescence lifetimes of the native proteins and the amyloid structures. Finally, we show images of the spherulites produced by confocal fluorescence, MPEF, and SHG microscopy without using any probes or labels.
2. Materials and methods
2.1. Chemicals
Bovine insulin (I5500) and β-LG from bovine milk (L0130) were purchased from Sigma Aldrich.
2.2. Preparation of the fibers and spherulites
Insulin fibers were prepared by dissolving 10 mg insulin in 4 mL 25 mM HCl, followed by 18h incubation at 70 °C while stirring with a magnetic stir bar at 300 rpm. β-LG fibers were prepared by dissolving 20 mg β-LG in 4 mL 25 mM HCl, followed by 42h incubation at 85 °C while stirring with a magnetic stir bar at 300 rpm. Insulin spherulites were prepared by dissolving 5 mg insulin in 1 mL 25 mM HCl and 5 mM NaCl, followed by 6h incubation at 70 °C without stirring. β-LG spherulites were prepared by dissolving 20 mg β-LG in 1 mL 25 mM HCl and 5 mM NaCl, followed by 18h incubation at 85 °C without stirring. MilliQ water (18.2 MΩ) was used in all solutions and the prepared fibers and spherulites were stored in 4 °C. Before use, each sample was centrifuged (5000g × 5 min for the fibers and 200g × 2 min for the spherulites) and resuspended twice in 25 mM HCl post-assembly, to reduce the amount of non-assembled protein.
2.3. AFM
The amyloid fiber solutions were diluted with 25 mM HCl to a final concentration of about 1 mg/mL followed by adsorption on clean Si substrates during 3 min. The surfaces were then rinsed by submersion into MilliQ water for 5 min. After drying with N2 gas, the surfaces were imaged with PeakForce AFM (Bruker-ICON).
2.4. Cross-polarized microscopy
Droplets containing spherulites (undiluted) were put on microscope slides with coverslips on top. An in-house microscopy setup was used, with the sample between orthogonal linear polarizers. The images of the spherulites were captured with a charge-coupled device camera.
2.5. UV-Vis absorption, fluorescence spectroscopy and decay
Solutions with about 5 mg/mL protein in 25 mM HCl were prepared for all the amyloid samples and the native proteins. Absorption spectra from 240–500 nm were measured with a UV-Vis spectrometer (Varian Cary 5000) with a 25 mM HCl solution as the blank. Baseline correction was made with a model according to Eq. (1),
| (1) |
where Iobs is the measured intensity for the sample, I0 is the intensity for the blank, 1-A is the transmittance due to reflection/blocking at large structures, and 1-B/λ4 is the transmittance due to wavelength dependent scattering. The A and B factors were optimized to accomodate a flat baseline at zero at λ > 430 nm, for which little absorption was expected. All absorption spectra were normalized at 278 nm. The fluorescence spectra were acquired with a spectrofluorometer (Tecan, Infinite M1000), with λexc ranging from 310 nm to 450 nm. The relative signal intensities for the various λexc were noted for each sample and the spectra were normalized to their maximum values. The fluorescence decays were measured with a time-correlated single photon counting spectrometer (Picoquant, FluoTime 100) and the instrument response function was captured with 250 nm poly-L-lactic acid particles at low concentration. For all decay measurements, the integration time was adjusted so that the maximum count for each sample reached 10,000 and all results were fitted with double-exponential functions.
2.6. Confocal fluorescence, MPEF, and SHG microscopy
Droplets of the spherulites (undiluted) were placed on microscopy slides with cover slips on top, and then imaged. The fluorescence images were captured with a 40X/1.30 Plan-Neofluor oil immersion objective on a confocal microscope (Zeiss LSM 510 Meta) operating with λexc at 405 nm from a diode-pumped solid-state laser. The detection channel had a photon multiplier tube (PMT) and a longpass filter with a cut-on wavelength at 420 nm. The MPEF and SHG images were acquired with a multiphoton microscope (Olympus, FV1000 MPE BX61) pumped with a tunable nIR laser (Spectra-Physics Mai Tai HP) with 80 MHz repetition rate and 100 fs pulse width. Unless otherwise noted, λexc was 910 nm and the excitation laser power was 680 mW at the sample. No photodegradation was observed during acquisition for excitation powers below 1.2 W. Both detection arms had PMT detectors and included bandpass filters at 420–460 nm for the SHG channel and 495–540 nm for the MPEF channel. A 25X/1.05 XL Plan water immersion objective was used and the images were scanned at 100 μs/pixel, with pixel sizes <200 nm. For all the MPEF and SHG images in this work, the expected pump laser polarization orientation is 45° clockwise. When λexc was moved to 930 nm with the same pump power and detector settings, the intensity in the SHG channel decreased to about 5 % while over 90 % of the signal in the MPEF channel remained. This procedure was used as a confirmation that a majority of the signal detected in the SHG channel originates from SHG processes, while MPEF is detected in the MPEF channel. We also meassured MPEF in both channels with a range of λexc to observe how the relative intensities are dependent on the wavelength. For these studies we made additional measurements with another filter setup, allowing 460–500 nm in one detector arm and 520–560 nm in the other.
3. Results and discussion
3.1. Preparation and identification of fibers and spherulites
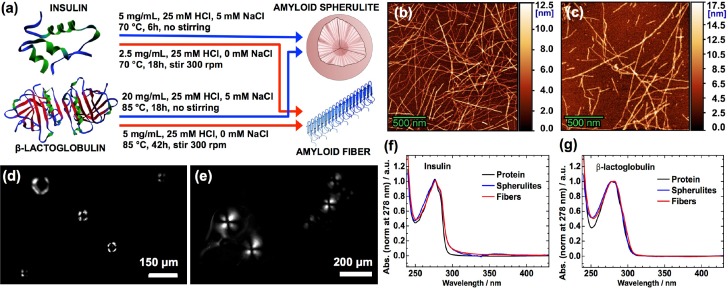
When proteins are brought to mild denaturation, they reach a flexible state that allows misfolding, reorganization and subsequent aggregation that may result in amyloid-like fibers. Insulin and β-LG are two proteins that form these fiber structures when heated close to their melting temperatures in acidic conditions. [62, 63] Thus, we dissolved bovine insulin and β-LG from bovine milk in 25 mM HCl water solutions and heated them to 70 °C and 85 °C, respectively. To promote fiber formation, lower concentrations (2.5 mg/mL for insulin and 5 mg/mL for β-LG) were used and 300 rpm stirring was applied to the solution. In contrast, higher concentrations (5 mg/mL for insulin and 20 mg/mL for β-LG) with additional 5 mM NaCl (not stirred) were the conditions used to favor spherulite formation (Fig. 1(a)). [48,49] Successful amyloid fiber formation was confirmed with AFM of the fibers adsorbed onto Si surfaces (Fig. 1(b),1(c)). When spherulites are imaged with cross-polarized microscopy, a characteristic Maltese-cross pattern appears due to the birefringent nature of amyloids, for which the optical axis is along the fiber. Therefore, the spherulites were imaged with a home-built cross-polarized microscopy setup to confirm their formation (Fig. 1(d),1(e)). At the center of the insulin spherulites, a dark region is observed in the structures, for which an associated large amorphous core has been suggested. [43–45,48] For β-LG spherulites, the corresponding dark region at the center is small, which implies a small or non-existent amorphous core. Finally, UV-Vis absorption spectra were captured and the expected absorption of the ring-structures in the proteins at about 278 nm was detected for all samples (Fig. 1(f),1(g)). A slight peak broadening for the fibers and spherulites was observed for both proteins, which may be related to either residual scattering that could not be compensated for, or a small change in the local environment for the amino-acids with ring-structures. At about 360 nm, a weak peak was observed for the insulin spherulites. This may be an effect from phonons being formed due to the rigid nature of the long amyloid fibers in the spherulites; however, further investigation of the details in this observation is needed and is beyond the scope if this paper.
Fig. 1.
Preparation and characterization of amyloid fibers and spherulites. (a) Insulin [64] and β-lactoglobulin [65] form fibers or spherulites, depending on the experimental conditions. AFM images of amyloid fibers from bovine insulin (b) and β-LG from bovine milk (c); bars are 500 nm for both images. Cross-polarized microscopy of the spherulites from insulin (d) and β-LG from bovine milk (e); bars are 150 μm for (d) and 200 μm for (e). Normalized absorption spectra of the native proteins (black), amyloid fibers (red), and amyloid spherulites (blue) from the bovine insulin (f) and the β-LG from bovine milk (g), in 25 mM HCl solutions at ∼5 mg/mL protein concentration.
3.2. Characterization of the intrinsic fluorescence
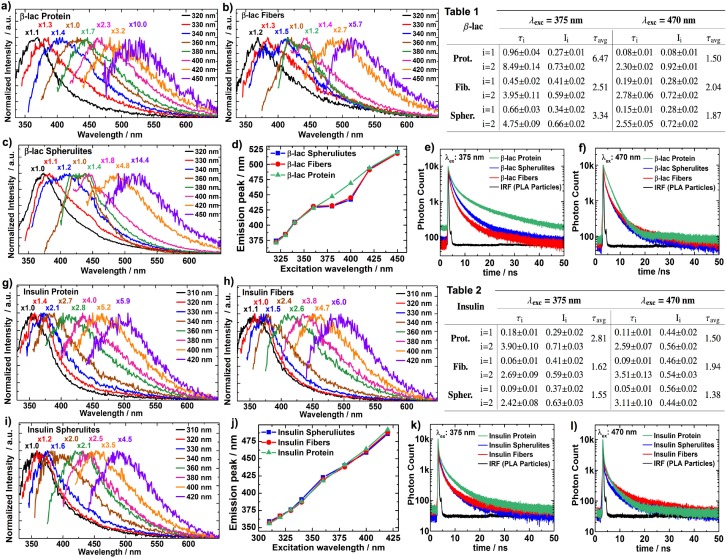
Before proceeding to imaging the spherulite structures, we thoroughly investigated the intrinsic fluorescence of the fibers, spherulites and native proteins of the insulin and β-LG. For all these samples, intrinsic fluorescence was observed and the emission spectra were dependent on the excitation wavelength (Fig. 2(a)–2(d),2(g)–2(j)), which is in conflict with Kasha’s rule. Raman scattering moves with the excitation wavelength, but cannot account for all the observed features and Raman microscopy of the spherulites could conclusively exclude this as a viable explanation. However, this effect is based on another well-documented phenomenon - the red edge excitation shift (REES). [66,67] Solvent interactions result in a statistical distribution of the ground state (Eg) and the first excited state (Ee) energies for the fluorophores. Of this distribution, only the fluorophores having Eg above the mean (Ēg) and Ee below the mean (Ēe) will be probed when they are excited with light at the red edge of the absorption spectra. If the solvent relaxation (reorientation) is slower than the fluorescence decay, this will lead to a red-shifted emission spectrum, compared to the scenario when the excitation wavelength is sufficient for transitions beyond the gap between Ēg and Ēe. Usually, REES is not observed for fluorophores in liquid solvents at room temperature, since the solvent relaxation in such cases is sufficiently rapid to consistently yield a transition to the same Ee distribution before radiative decay occurs. However, the dynamics of water and other molecules surrounding the intrinsic fluorophores within proteins are slow enough to allow a substantial wavelength-dependent REES. The absorption peaks for the amino-acids with ring-structured side-chains (which are typically responsible for the intrinsic fluorescence in proteins) are at about 280 nm. The λexc used in Fig. 2 (310 – 450 nm) is therefore far out on the red edge. Conforming with the REES discussed above, an excitation wavelength dependence of the emission was observed for the native proteins for both the β-LG and insulin. However, when λexc is decreased to 300 nm and below, the emission obeys Kasha’s rule. When we made similar measurements for free tyrosine amino acids in 25 mM HCl solutions, the emission peaks were at no point dependent on λexc (data not shown). This further corroborates that the mechanism for the observed results is REES, since the solvent relaxation for the free tyrosines is expected to be fast. REES is also largely observed for the fiber and spherulite structures; however, in the case of the amyloids from β-LG, a static weak emission peak appeared at about 430 nm with λexc at 360 – 400 nm (Fig. 2(b)–2(d)). This observation is most likely associated with additional fluorescence arising from charge delocalization along the backbone of the fibers in the amyloid structures, as discussed in the introduction.
Fig. 2.
Intrinsic fluorescence. The data for the intrinsic fluorescence from the β-LG from bovine milk (a–f) and bovine insulin (g–l), at ∼5 mg/mL in 25 mM HCl water solutions. Fluorescence emission spectra are shown for the native proteins (a, g), the amyloid fibers (b, h), and the amyloid spherulites (c, i), when λexc ranges from 310 to 450 nm. The emission peaks are plotted versus the λexc for both β-LG (d) and insulin (j) for the respective types of samples: native proteins (green, ▵), amyloid fibers (red, ○), and amyloid spherulites (blue, □). The fluorescence decays for λexc at 375 nm (e, k) and 470 nm (f, l) are presented for the native proteins (green), the amyloid spherulites (blue), and the amyloid fibers (red) – as well as the instrument response function (black) measured with 250 nm poly-L-lactic acid particles. The lifetimes (ns) and fractional intensities for double-exponential fits of the decays are shown for both the β-LG (Table 1) and insulin (Table 2) samples with λexc at 375 nm and 470 nm. The 95 % confidence intervals for the fitted parameters are included.
To confirm that this is indeed a new fluorescence not present for the native protein, the fluorescence decays were measured for the native protein and the amyloid structures for β-LG. When λexc was 470 nm, similar decays for the native proteins and the amyloid analogs were observed (Fig. 2(f)), while faster decays were measured for the amyloids when λexc was 375 nm (Fig. 2(e)). This shows that the faster decay is not an omnipresent feature for the fibers and spherulites throughout the excitation spectra, but is rather indicative of a specific and additional fluorescence pathway when λexc is 375 nm. Interestingly, a corresponding fluorescence was not readily observed for insulin amyloids, for which the emission spectra (Fig. 2(g)–2(j)) exhibited features similar as for the native protein throughout the range of excitation wavelengths used in this work. However, the fluorescence decays (Fig. 2(k),2(l)) show a slightly faster decay for the insulin amyloids compared to the native protein when excited at 375 nm. This may indicate that insulin also develops intrinsic fluorescence in the amyloid state, but with a very low absorption cross-section and/or quantum yield. It is not easy to conclusively determine the origin of the distinctions in the intrinsic optical properties for the amyloid states of β-LG and insulin. However, it is likely that the detailed organizations in these superstructures affect the prospects of charge delocalization in them, for instance via their proton transfer capability. [32] It is therefore not surprising that amyloids from insulin do not exhibit the exact same optical properties as those from β-LG, even though the specific details for the origin of this observation are currently unknown.
It has been shown that the intrinsic fluorescence decays from the ring-structures in proteins often require at least double-exponential functions to produce sensible fits. The origin of this feature is non-trival and depends on the specific positioning of the fluorescent amino acids, possible energy transfers between them, as well as transitions to more than one excited electronic state. [67–69] Accordingly, double-exponential fits were used for all samples, which yielded short (τ1) and long (τ2) decays (Table 1 and 2 in Fig. 2). With λexc at 470 nm, both τ1 and τ2 were quite similar across the samples for each protein and no clear and interpretable trend is observed. For insulin, the fluorescence lifetimes for the amyloid structures were slightly shorter compared to the native protein when λexc was 375 nm. It seems like τ2, with the range 2–4 ns, is the main reason for this. A larger distinction in the decay times between the amyloids and the native protein for the same λexc was observed for β-LG. Both τ1 and τ2 were longer for the native protein, however the main difference was again measured for τ2, which was 4.75 ns and 3.95 ns for the amyloid spherulites and fibers, while it was 8.5 ns for the native protein. The fluorescence lifetimes measured for the β-LG amyloid structures conform with the corresponding values measured for amyloid-specific intrinsic fluorescence of other proteins. [24] The fluorescence with longer lifetime for the native protein is likely from tryptophan, which has been shown to exhibit a wide range of decay times that depend on the environment. For insulin, the main origin of the observed fluorescence for all samples is likely from the tyrosine residues in the protein sequence, which lacks tryptophanes.
3.3. Label-free imaging of amyloid spherulites
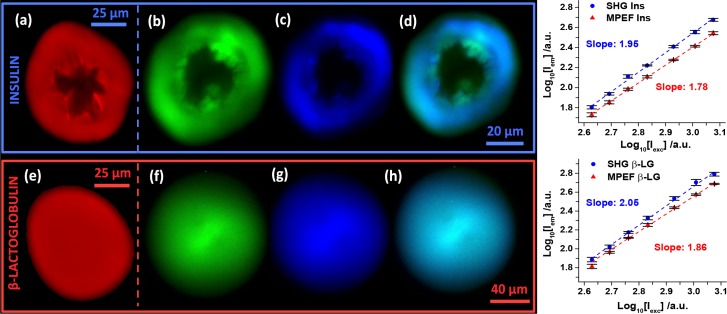
The intrinsic fluorescence investigated in the previous section can be utilized for label-free imaging of the amyloid spherulites in solution. Scanning confocal fluorescence microscopy with λexc at 405 nm was used to image the insulin and β-LG spherulites (Fig. 3(a),3(e)). Also MPEF and SHG microscopy with λexc at 910 nm, produced clear images of the spherulites (Fig. 3(b)–3(d),3(f)–3(h)). Power dependence plots for the average pixel intensity of the spherulite structures revealed a quadratic relationship (1.95 and 2.05 for insulin and β-LG, respectively) for the SHG images, which was expected. A nearly quadratic relationship (1.78 and 1.86 for insulin and β-LG, respectively) for the MPEF images was measured. The slightly lower power dependence for the MPEF images indicates that a larger relative contribution of linear processes (e.g. linear fluorescence or scattering) is present in this channel. The amorphous cores for the insulin spherulites were large and had quite irregular shapes, while they were non-existent for the β-LG spherulites. Another distinction was that many heterogenous features can be discerned in detail for the insulin spherulites, whereas a homogenous morphology was obserev for the β-LG spherulites. A clear contrast between the amorphous core and the surrounding crystalline fiber region in the insulin spherulites was observed for all three techniques. The line-intensities for representative regions of interests are plotted for comparable insulin spherulites to quantify these contrasts (Fig. 4(a)–4(c)).
Fig. 3.
Label-free imaging of amyloid spherulites. Spherulites from bovine insulin (a–d) and β-LG from bovine milk (e–h) imaged with confocal fluorescence (a,e - red), MPEF (b,f - green), and SHG (c,g - blue). MPEF and SHG were imaged simultaneously in different channels (420–460 nm and 495–540 nm, respectively) and overlays of the two are shown (d,h). Scale bars: 25 μm (a,e), 20 μm (b–d), and 40 μm (f–h). The λexc were 405 nm (a,e) and 910 nm (b–d,f–h). The power dependences for SHG and MPEF were measured for four replicates of both structures with powers ranging from 425–1190 mW. The error bars represent the St. Dev. and the slopes were obtained with least square fits.
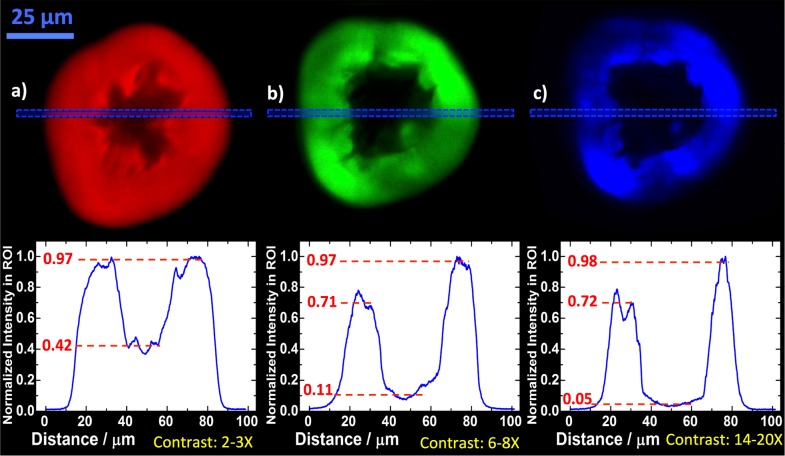
Fig. 4.
Contrast between the amorphous core and fibrillar region. For the bovine insulin spherulites, the relative intensities for the core and the fibrillar regions were evaluated by taking regions of interest (dashed blue boxes) linescans for the confocal fluorescence (a - red), MPEF (b - green) and SHG (c - blue). The scale bar is 25 μm for all spherulites and the λexc were 405 nm (a) and 910 nm (b, c), respectively.
The fiber regions had a 2 – 3× higher intensity for the confocal fluorescence. Based on the analysis in the previous section, the origin of this contrast is likely not due to new fluorescence pathways for the fibers. One contributing factor could be a distinction in protein density, however, it is unlikely that this alone would yield a 2 – 3 times higher intensity. However, it is possible that the packing and rigidity of the fiber regions discourage non-radiative decay pathways in favor of radiative pathways. This would lead to a higher quantum yield for the fiber region without affecting the overall fluorescence decay. Additional contrast was obtained for the MPEF, which had a 6 – 8× intensity difference. After excluding the possibility of hyper-Raman contributions, it was concluded that this is likely due to a strong MPA for insulin amyloids. It has been shown that insulin amyloid fibers exhibit a MPA cross-section comparable to those for good two-photon dyes. [61] In that study, the MPA order exhibited a strong wavelength dependence and was >3 for insulin fibers when λexc = 910 nm, which deviates from the order we have measured for MPEF at this wavelength. This indicates that the features that give rise to the wavelength dependence in MPA are not coupled to emissive decay pathways. However, despite this distinction between the previous study and our findings, a strong nonlinear absorption cross-section exclusively for the fibrillar structures is a likely explanation for the higher contrast in MPEF compared to the confocal fluorescence. As a 40 nm window is relatively large for SHG imaging, there is a risk that some MPEF could emerge in the channel for SHG detection (with a bandpass filter at 420 – 460 nm). Therefore, λexc was shifted from 910 nm to 930 nm as a control, which would have a limited effect on the observed MPEF, while the SHG signal would vanish completely as it shifts out from the bandpass region for the filter setup. Indeed, the vast majority of the signal disappeared (results not shown), demonstrating the contribution of SHG processes to the SHG images. For the core in the insulin spherulites, one would likely find proteins that are isotropically arranged with a random coil conformation, [42] and the SHG susceptibility would thus be low there. Therefore, the 14 – 20× intensity difference in the SHG image is a clear indication that the cores of the insulin spherulites are indeed amorphous, as has been previously hypothesized. A final comment to make is that the confocal fluorescence response seems uniform for the fiber region in the insulin spherulites, while the responses for the MPEF and SHG are more heterogeneous. This is because the excitation light is polarized in the latter two techniques, which favors fluorescence from fibers with the optical axis aligned with the polarization of the light. Such artifacts can be alleviated by the use of circularly polarized light. However, the polarization dependence can also be utilized to make conclusions about the detailed structure of the sample under study. In our case, the results indicate that the fibers mainly grow radially outward from the spherulite cores, which agrees with previous studies.
These imaging techniques can also be used to sample various cross-sections of the spherulites, which provides information on their 3D-structure (Fig. 5). A z-stack of cross-sections from an insulin spherulite was collected, from which four 3D animations were made. The results can be found in the supplementary material: Visualization 1 (557.7KB, MOV) and Visualization 2 (274.3KB, MOV) show half of a 37 μm spherulite imaged with SHG and MPEF respectively, while Visualization 3 (356.7KB, MOV) and Visualization 4 (270.8KB, MOV) show the corresponding SHG and MPEF images of the entire spherulite. The amorphous core can easily be identified as a “cavity” in the structure. Finally, to further demonstrate the ability of these techniques to detect detailed features in the spherulite structures, some pressure was gently applied on the spherulites to induce cracks in them. The samples were then placed in the microscope and imaged with MPEF and SHG (Fig. 6), with λexc at 910 nm. The SHG approach in particular was able to display the cracked features with high contrast. This is due to the high contrast for SHG demonstrated above, in combination with the fact that the microscope was used in back-scattering mode. The SHG signals are mainly produced in the forward direction (due to the conservation of momentum), which means that the signals need to be back-scattered to reach the detector. Consequently, enhanced scattering at cracks and defects make these features more pronounced. However, also the MPEF image exhibit enhanced signals close to some of the defects. This is also reasonable, because when the focus of the excitation light is close to defect sites, on would epect to get the regular MPEF signal and potentially additional signals from light that travels in the direction of the defects that subsequently scatter it to the detector.
Fig. 5.
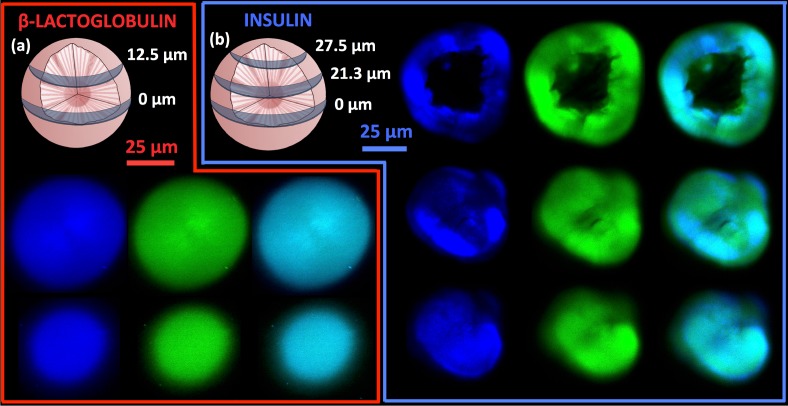
Various cross-sections for the spherulites. Cross-sections of a spherulite from β-LG from bovine milk (a) at 0 μm (top row) and 12.5 μm (bottom row) from the center with SHG (blue/left), MPEF (green/middle), and overlays (right); the scale bar is 25 μm. Cross-sections of a spherulite from bovine insulin (b) at 0 μm (top row), 21.3 μm (middle row), and 27.5 μm (bottom row) from the center with SHG (blue/left), MPEF (green/middle), and overlays (right); the scale bar is 25 μm. λexc was 910 nm in all cases. 3D-animations created from a z-stack of a 37 μm insulin spherulite are provided in the supplementary material. Both half the spherulite imaged with SHG ( Visualization 1 (557.7KB, MOV) ) and MPEF ( Visualization 3 (356.7KB, MOV) ), as well as the entire spherulite imaged with SHG ( Visualization 2 (274.3KB, MOV) ) and MPEF ( Visualization 4 (270.8KB, MOV) ) are shown.
Fig. 6.
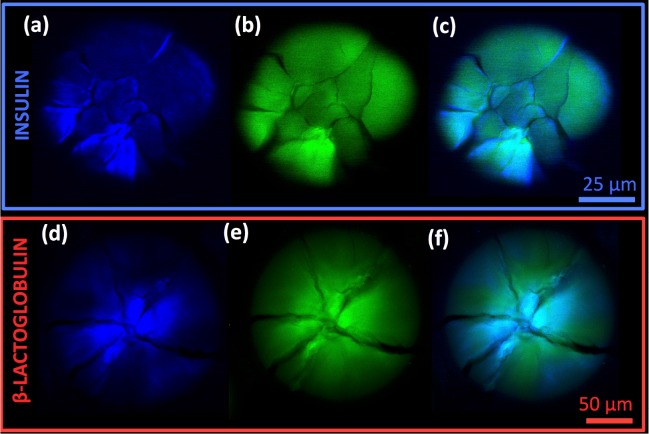
Imaging of cracked spherulites. SHG images (a, d - blue), MPEF images (b, e - green), and overlays (c,f) of cracked spherulites from bovine insulin (a–c) and β-LG from bovine milk (d–f); the scale bars are 25 μm (a–c) and 50 μm (d–f), and λexc was 910 nm. Note that the images of the insulin spherulite were not recorded at its center and, therefore, lack the amorphous core.
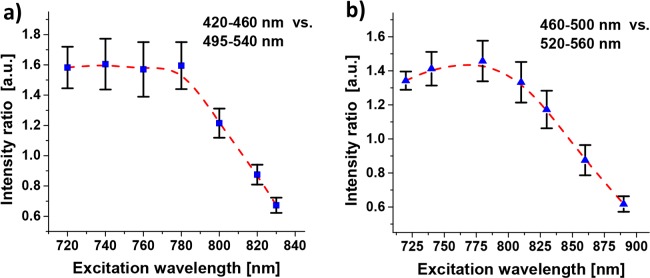
Lastly, REES in linear fluorescence has been readily demonstrated for these structures above. As far as we know, there are no conceptual reasons why REESs would not be possible to observe for MPEF as well. However, little research has been devoted to this and we are not aware of any studies of this kind. In order to investigate if REES may be found in the MPEF from our insulin spherulite structures, a range of different excitation wavelengths that would yield only MPEF and no SHG in both the detection arms were used. Thereafter, ratios of the average intensities of the images in the respective channels were formed. The results show that up to about 790 nm, the excitation wavelength has limited effect on the relative intensities in the two channels. Thereafter, the signal in the channel for the lower wavelength starts to decrease relative the one for longer wavelengths (Fig. 7(a)). This is consistent with the REES phenomenon. However, since 790 nm is relatively close to the lower window boundary, the experiment was repeated with a second filter setup, with 460–500 nm and 520–560 nm windows (Fig. 7(b)). Again, the decline started at about 790 nm, which shows that it cannot be an effect from hyper Raman scattering. To conclusively determine that the observed behavior really is REES, it would be necessary to measure the MPA spectra as well as complete MPEF spectra for several excitation wavelengths. That is beyond the scope of this work, but we hypothesize that the obtained results are due to REES in MPEF. In such a case, a wavelength of 910 nm (the typical λexc in this work) is far out on the red-edge of the MPA. This would explain why this wavelength allowed very high excitation powers without photodegradation, while the higher MPA cross-sections for shorter wavelengths required lower excitation powers (e.g. <100 mW for λexc<800 nm).
Fig. 7.
Excitation wavelength dependence in MPEF images. Ratio of average intensities for the insulin spherulites in the two detector channels. The bandpass filters windows compared are (a) 420–460 nm vs. 495–540 nm and (b) 460–500 nm vs. 520–560 nm. The error bars are St. Dev. from measurements on four spherulites and the excitation laser power was 80–400 mW, depending on the wavelength. The dashed red lines are splines intended as visual guide.
4. Conclusion
In this paper, the intrinsic fluorescence of amyloid structures from insulin and β-LG was investigated in detail. Fibers and spherulites from both proteins exhibit enhanced fluorescence. However, only the β-LG structures develop an additional fluorescence decay pathway in the visible regime that could be readily detected with fluorescence spectroscopy. The origin of the new intrinsic fluorescence for β-LG is likely due to charge delocalization, while the origin of the enhanced signal for the insulin amyloid structures in confocal fluorescence microscopy is likely due to a transition from non-radiative to radiative pathways for the tyrosines in the peptide sequence. The ability to image amyloid spherulites with confocal fluorescence, MPEF, and SHG microscopy without the use of any probes or labels was also demonstrated. In particular, the nonlinear techniques provide images with excellent contrast for the fibrillar regions within the spherulites. This work provides a first demonstration of the virtue of multiphoton absorption in MPEF microscopy of amyloid structures. Additional contrast was obtained in the SHG images, thanks to a high SHG susceptibility exclusively for the ordered fibrillar regions. Also, it was demonstrated that these techniques can be used to obtain 3D-images of amyloid spherulites and display detailed information about their structure, including cracks and defects. Finally, we showed that the MPEF exhibits a spectral dependence on the excitation wavelength. We hypothesize that the REES phenomenon is responsible for this effect, since the observed behavior is consistent with this mechanism. As far as we know, REES in MPEF has not previously been reported, but additional investigations are needed at this time to confirm the existence and use of REES in nonlinear optical spectroscopy and imaging. Enhanced intrinsic fluorescence, strong MPA and intrinsic molecular ordering are a combination of features that likely distinguishes amyloids from most other structures in normal tissue. [58–60] A visualization strategy including all these features would be a helpful tool to selectively find and characterize amyloid deposits in, for example, brain tissue from patients with Alzheimer’s disease, without the use of any labels or probes. As a result, non-destructive identification of amyloids in such tissues becomes possible without introduction of any contaminating agents. This subsequently allows chemical analyses of the identified amyloid structures by vibrational sum-frequency scattering spectroscopy, [70] coherent anti-stokes Raman scattering microscopy, [60, 71] and time-of-flight secondary ion mass spectrometry, [72] among other techniques. The fundamental optical properties of amyloids discussed and presented in this paper are essential for continuing efforts with the goal of providing innovative strategies for the study of amyloid fiber structures. An improved pathway for making amyloid spherulites from Aβ1–40 was recently developed. [42] As the presence of such structures has been demonstrated in the brain tissue of patients with Alzheimer’s disease, they have a potential to become important in vitro models for Alzheimer research. This emphasizes the need in biomedical research for noninvasive techniques that can provide detailed information on amyloid spherulites.
Acknowledgments
We thank Dr. Paul Wiggins for generously lending equipment for the cross-polarized microscope and for sample preparation suggestions for imaging. We also acknowledge the Lynn and Mike Garvey Cell Imaging Lab and Dr. Ron Seifert for multiphoton microscope instrument time and supervision. Finally, we thank Dr. Niclas Solin for valuable discussions on the preparation and structure of amyloids.
Funding
This research was supported by NIH Grant No. EB-002027 to the National ESCA and Surface Analysis Center for Biomedical Problems.
References and links
- 1.Chiti F., Dobson C. M., “Protein misfolding, functional amyloid, and human disease,” Annu. Rev. Biochem. 75, 333–366 (2006). 10.1146/annurev.biochem.75.101304.123901 [DOI] [PubMed] [Google Scholar]
- 2.Charych D. H., Venema P., van der Linden E., “Fibrillar structures in food,” Food Funct. 3(3), 221–227 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Shirahama T., Cohen A. S., “High-resolution electron microscopic analysis of the amyloid fibril,” J. CellBiol. 33(3), 679–708 (1967). 10.1083/jcb.33.3.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makin O. S., Atkins E., Sikorski P., Johansson J., Serpell L. C., “Molecular basis for amyloid fibril formation and stability,” Proc. Natl. Acad. Sci. U. S. A. 102(2), 315–320 (2005). 10.1073/pnas.0406847102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith J. F., Knowles T. P. J., Dobson C.M., MacPhee C. E., Welland M. E., “Characterization of the nanoscale properties of individual amyloid fibrils,” Proc. Natl. Acad. Sci. U. S. A. 103(43), 15806–15811 (2006). 10.1073/pnas.0604035103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sunde M., Serpell L. C., Bartlam M., Fraser P. E., Pepys M. B., Blake C. C. F., “Common core structure of amyloid fibrils by synchrotron X-ray diffraction,” J. Mol. Biol. 273(3), 729–739 (1997). 10.1006/jmbi.1997.1348 [DOI] [PubMed] [Google Scholar]
- 7.Masters C. L., Simms G., Weinman N. A., Multhaup G., McDonald B. L., Beyreuther K., “Amyloid plaque core protein in Alzheimer disease and Down syndrome,” Proc. Natl. Acad. Sci. U. S. A. 82(12), 4245–4249 (1985). 10.1073/pnas.82.12.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serpell L. C., “Alzheimer’s amyloid fibrils: structure and assembly,” Biochim. Biophys. Acta 1502(1), 16–30 (2000). 10.1016/S0925-4439(00)00029-6 [DOI] [PubMed] [Google Scholar]
- 9.Westermark P., Wernstedt C., Wilander E., Hayden D. W., O’Brien T. D., Johnson K. H., “Amyloid fibrils in human insulinoma and islets of Langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells,” Proc. Natl. Acad. Sci. U. S. A. 84(11), 3881–3885 (1987). 10.1073/pnas.84.11.3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westermark P., Engström U., Johnson K. H., Westermark G. T., Betsholtz C., “Islet amyloid polypeptide: pinpointing amino acid residues linked to amyloid fibril formation,” Proc. Natl. Acad. Sci. U. S. A. 87(13), 5036–5040 (1990). 10.1073/pnas.87.13.5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spillantini M. G., Schmidt M. L., Lee V. M., Trojanowski J. Q., Jakes R., Goedert M., “α-synuclein in Leqy bodies,” Nature 388(6645), 839–840 (1997). 10.1038/42166 [DOI] [PubMed] [Google Scholar]
- 12.Jiménez J. L., Nettleton E. J., Bouchard M., Robinson C. V., Dobson C. M., Saibil H. R., “The protofilament structure of insulin amyloid fibrils,” Proc. Natl. Acad. Sci. U. S. A. 99(4), 9196–9201 (2002). 10.1073/pnas.142459399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bromley E. H. C., Krebs M. R. H., Donald A. M., “Aggregation across the length-scales in β-lactoglobulin,” Faraday Discuss. 128(4), 13–27 (2005). 10.1039/B403014A [DOI] [PubMed] [Google Scholar]
- 14.Hamada D., Dobson C. M., “A kinetic study of β-lactoglobulin amyloid fibril formation promoted by urea,” Protein Sci. 11(10), 2417–2426 (2002). 10.1110/ps.0217702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mankar S., Anoop A., Sen S., Maji S. K., “Nanomaterials: amyloids reflect their brighter side,” Nano Rev. 2, 6032 (2011). 10.3402/nano.v2i0.6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka H., Herland A., Lindgren L. J., Tsutsui T., Andersson M. R., Inganäs O., “Enhanced current efficiency from bio-organic light-emitting diodes using decorated amyloid fibrils with conjugated polymer,” Nano Lett. 8(9), 2858–2861 (2008). 10.1021/nl801510z [DOI] [PubMed] [Google Scholar]
- 17.Rizzo A., Solin N., Lindgren L. J., Andersson M. R., Inganäs O., “White light with phosphorescent protein fibrils in OLEDs,” Nano Lett. 10(6), 2225–2230 (2010). 10.1021/nl1012008 [DOI] [PubMed] [Google Scholar]
- 18.Schiebel T., Parthasarathy R., Sawicki G., Lin X-M., Jaeger H., Lindquist S. L., “Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition,” Proc. Natl. Acad. Sci. U. S. A. 100(8), 4527–4532 (2003). 10.1073/pnas.0431081100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elfwing A., Bäcklund F. G., Musumeci C., Inganäs O., Solin N., “Protein nanowires with conductive properties,” J. Mater. Chem. C 3(25), 6499–6504 (2015). 10.1039/C5TC00896D [DOI] [Google Scholar]
- 20.Mains J., Lamprou D. A., McIntosh L., Oswald I. D., Urquhart A. J., “Beta-adrenoceptor antagonists affect amyloid nanostructure; amyloid hydrogels as drug delivery vehicles,” Chem. Commun. (Camb.) 49(44), 5082–5084 (2013). 10.1039/c3cc41583j [DOI] [PubMed] [Google Scholar]
- 21.Hamedi M., Herland A., Karlsson R. H., Inganäs O., “Electrochemical devices made from conducting nanowire networks self-assembled from amyloid fibrils and alkoxysulfonate PEDOT,” Nano Lett. 8(6), 1736–1740 (2008). 10.1021/nl0808233 [DOI] [PubMed] [Google Scholar]
- 22.del Mercato L. L., Pompa P. P., Maruccio G., Della Torre A., Sabella S., Tamburro A. M., Cingolani R., Rinaldi R., “Charge transport and intrinsic fluorescence in amyloid-like fibrils,” Proc. Natl. Acad. Sci. U. S. A. 104(46), 18019–18024 (2007). 10.1073/pnas.0702843104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharpe S., Simonetti K., Yau J., Walsh P., “Solid-State NMR characterization of autofluorescent fibrils formed by the elastin-derived peptide GVGVAGVG,” Biomacromolecules 12(5), 1546–1555 (2010). 10.1021/bm101486s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan F. T., Schierle G. S. K., Kumita J. R., Bertoncini C. W., Dobson C. M., Klaminski C. F., “Protein amyloids develop an intrinsic fluorescence signature during aggregation,” Analyst 138(7), 2156–2162 (2013). 10.1039/c3an36798c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinotsi D., Buell A. K., Dobson C. M., Kaminski Schierle G. S., Kaminski C. F., “A label-free, quantitative assay of amyloid fibril growth based on intrinsic fluorescence,” ChemBioChem 14(7), 846–850 (2013). 10.1002/cbic.201300103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amit M., Cheng G., Hamley I. W., Ashkenasy N., “Conductance of amyloid β based peptide filaments: structure-function relations,” Soft Matter 8(33), 8690–8696 (2012). 10.1039/c2sm26017d [DOI] [Google Scholar]
- 27.Rosenberg B., “Electrical conductivity of proteins,” Nature 193(4813), 364–365 (1962). 10.1038/193364a0 [DOI] [PubMed] [Google Scholar]
- 28.Amit M., Appel S., Cohen R., Cheng G., Hamley I. W., Ashkenasy N., “Hybrid proton and electron transport in peptide fibrils,” Adv. Funct. Mater. 24(37), 5873–5880 (2014). 10.1002/adfm.201401111 [DOI] [Google Scholar]
- 29.Perutz M. F., Finch J. T., Berriman J., Lesk A., “Amyloid fibers are water-filled nanotubes,” Proc. Natl. Acad. Sci. U. S. A. 99(8), 5591–5595 (2002). 10.1073/pnas.042681399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shukla A., Mukherjee S., Sharma S., Agrawal V., Kishan K. V. R., Guptasarma P., “A novel UV laser-induced visible blue radiation from protein crystals and aggregates: scattering artifacts or fluorescence transitions of peptide electrons delocalized through hydrogen bonding?” Arch. Biochem. Biophys. 428(2), 144–153 (2004). 10.1016/j.abb.2004.05.007 [DOI] [PubMed] [Google Scholar]
- 31.Novakovskaya Y. V., “Conjugation in hydrogen-bonded systems,” Struct. Chem. 23(4), 1253–1266 (2012). 10.1007/s11224-012-0029-8 [DOI] [Google Scholar]
- 32.Pinotsi D., Grisanti L., Mahou P., Gebauer R., Kaminski C. F., Hassanali A., Schierle G. S. K., “Proton transfer and structure-specific fluorescence in hydrogen bond-rich protein structures,” J. Am. Chem. Soc. 138(4), 3046–3057 (2016). 10.1021/jacs.5b11012 [DOI] [PubMed] [Google Scholar]
- 33.Jin L-W., Claborn K. A., Kurimoto M., Geday M. A., Maezawa I., Sohraby F., Estrada M., Kaminsky W., Kahr B., “Imaging linear birefringence and dichroism in cerebral amyloid pathologies,” Proc. Natl. Acad. Sci. U. S. A. 100(26), 15294–15298 (2003). 10.1073/pnas.2534647100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lührs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B., Doeli H., Schubert D., Riek R., “3D structure of Alzheimer’s amyloid-β(1–42) fibrils,” Proc. Natl. Acad. Sci. U. S. A. 102(48), 17342–17347 (2005). 10.1073/pnas.0506723102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ban T., Morigaki K., Yagi H., Kawasaki T., Kobayashi A., Yuba S., Naiki H., Goto Y., “Real-time and single fibril observation of the formation of amyloid β spherulitic structures,” J. Biol. Chem. 281(44), 33677–33683 (2006). 10.1074/jbc.M606072200 [DOI] [PubMed] [Google Scholar]
- 36.Yagi H., Ban T., Morigaki K., Naiki H., Goto Y., “Visualization and classification of amyloid β supramolecular assemblies,” Biochemistry 46(51), 15009–15017 (2007). 10.1021/bi701842n [DOI] [PubMed] [Google Scholar]
- 37.Gellerman G. P., Byrnes H., Striebinger A., Ullrich K., Mueller R., Hillen H., Barghorn S., “Aβ-globulomers are formed independently of the fibril pathway,” Neurobiol. Dis. 30(2), 212–220 (2008). 10.1016/j.nbd.2008.01.010 [DOI] [PubMed] [Google Scholar]
- 38.Fändrich M., Schmidt M., Grigorieff N., “Recent progress in understanding Alzheimer’s β-amyloid structures,” Trends Biochem. Sci. 36(6), 338–345 (2011). 10.1016/j.tibs.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.House E., Jones K., Exley C., “Spherulites in human brain tissue are composed of β sheets of amyloid and resemble senile plaques,” J. Alzheimers Dis. 25(1), 43–46 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Shimanouchi T., Shimauchi N., Ohnishi R., Kitaura N., Yagi H., Goto Y., Umakoshi H., Kuboi R., “Formation of spherulitic amyloid β aggregate by anionic liposomes,” Biochem. Biophys. Res. Commun. 426(2), 165–171 (2012). 10.1016/j.bbrc.2012.07.107 [DOI] [PubMed] [Google Scholar]
- 41.Exley C., House E., Collingwood J. F., Davidson M. R., Cannon D., Donald A. M., “Spherulites of Aβ42 in vitro and in Alzheimer’s disease,” J. Alzheimers Dis. 20(4), 1159–1165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cannon D., Eichhorn S. J., Donald A. M., “Structure of spherulites in insulin, β-lactoglobulin, and Amyloid β,” ACS Omega. 1(5), 915–922 (2016). 10.1021/acsomega.6b00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Domike K. R., Donald A. M., “Thermal dependence of thermally induced protein spherulite formation and growth: kinetics of β-lactoglobulin and insulin,” Biomacromolecules 8(12), 3930–3937 (2007). 10.1021/bm7009224 [DOI] [PubMed] [Google Scholar]
- 44.Krebs M. R. H., MacPhee C. E., Miller A. F., Dunlop I. E., Dobson C. M., Donald A. M., “The formation of spherulites by amyloid fibrils of bovine insulin,” Proc. Natl. Acad. Sci. U. S. A. 101(40), 14420–14424 (2004). 10.1073/pnas.0405933101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krebs M. R. H., Bromley E. H. C., Rogers S. S., Donald A. M., “The mechanism of amyloid spherulite formation by bovine insulin,” Biophys. J. 88(3), 2013–2021 (2005). 10.1529/biophysj.104.051896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogers S. S., Krebs M. R. H., Bromley E. H. C., van der Linden E., Donald A. M., “Optical microscopy of growing insulin amyloid spherulites on surfaces in vitro,” Biophys. J. 90(3), 1043–1054 (2006). 10.1529/biophysj.105.072660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Domike K. R., Hardin E., Armstead D. N., Donald A. M., “Investigating the inner structure of irregular beta-lactoglobulin spherulites,” Eur. Phys. J. E: Soft Matter Biol. Phys. 29(2), 173–182 (2009). 10.1140/epje/i2009-10465-y [DOI] [PubMed] [Google Scholar]
- 48.Domike K. R., Donald A. M., “Kinetics of spherulite formation and growth: salt and protein concentration dependence on proteins beta-lactoglobulin and insulin,” Int. J. Biol. Macromol. 44(4), 301–310 (2009). 10.1016/j.ijbiomac.2008.12.014 [DOI] [PubMed] [Google Scholar]
- 49.Smith M. I., Foderà V., Sharp J. S., Roberts C. J., Donald A. M., “Factors affecting the formation of insulin amyloid spherulites,” Colloids Surf. B Biointerfaces 89, 216–222 (2012). 10.1016/j.colsurfb.2011.09.018 [DOI] [PubMed] [Google Scholar]
- 50.Cannon D., Donald A. M., “Control of liquid crystallinity of amyloid-forming systems,” Soft Matter 9(10), 2852–2857 (2013). 10.1039/c2sm26946e [DOI] [Google Scholar]
- 51.Backlund F. G., Pallbo J., Solin N., “Controlling amyloid fibril formation by partial stirring,” Biopolymers 105(5), 249–259 (2016). 10.1002/bip.22803 [DOI] [PubMed] [Google Scholar]
- 52.Bacskai B. J., Kajdasz S. T., Christie R. H., Carter C., Games D., Seubert P., Schenk D., Hyman B. T., “Imaging of amyloid-beta deposits in brains of living mice permits direct observation of clearance of plaques with immunotherapy,” Nat. Med. 7(3), 369–372 (2001). 10.1038/85525 [DOI] [PubMed] [Google Scholar]
- 53.Christie R. H., Bacskai B. J., Zipfel W. R., Williams R. M., Kajdasz S. T., Webb W. W., Hyman B. T., “Growth arrest of individual senile plaques in a model of Alzheimer’s disease observed by in vivo multiphoton microscopy,” J. Neurosci. 21(3), 858–864 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bacskai B. J., Klunk W. E., Mathis C. A., Hyman B. T., “Imaging amyloid-β deposits in vivo,” J. Cereb. Blood Flow Metab. 22(9), 1035–1041 (2002). 10.1097/00004647-200209000-00001 [DOI] [PubMed] [Google Scholar]
- 55.Heo C. H., Kim K. H., Kim H. J., Baik S. H., Song H., Kim Y. S., Lee J., Mook-Jung I., Kim H.M, “A two-photon fluorescent probe for amyloid-β plaques in living mice,” Chem. Commun. (Camb.) 49(13), 1303–1305 (2013). 10.1039/c2cc38570h [DOI] [PubMed] [Google Scholar]
- 56.Murugan N. A., Zalesny R., Kongsted J., Nordberg A., Agren H., “Promising two-photon probes for in vivo detection of β amyloid deposits,” Chem. Commun. (Camb.) 50(79), 11694–11697 (2014). 10.1039/C4CC03897E [DOI] [PubMed] [Google Scholar]
- 57.Kim D., Moon H., Baik S. H., Singha S., Jun Y. W., Wang T., Kim K. H., Park B. S., Jung J., Mook-Jung I., Ahn K. H., “Two-photon absorbing dyes with minimal autofluorescence in tissue imaging: application to in vivo imaging of amyloid-β plaques with a negligible background signal,” J. Am. Chem. Soc. 137(21), 6781–6789 (2015). 10.1021/jacs.5b03548 [DOI] [PubMed] [Google Scholar]
- 58.Zipfel W. R., Williams R. M., Christie R., Nikitin A. Y., Hyman B. T., Webb W. W., “Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation,” Proc. Natl. Acad. Sci. U. S. A. 100(12), 7075–7080 (2003). 10.1073/pnas.0832308100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwan A. C., Duff K., Gouras G. K., Webb W. W., “Optical visualization of Alzheimers pathology via multiphoton-excited intrinsic fluorescence and second harmonic generation,” Opt. Express 17(5), 3679–3689 (2009). 10.1364/OE.17.003679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee J. H., Kim D. H., Song W. K., Oh M. K., Ko D. K., “Label-free imaging and quantitative chemical analysis of Alzheimer’s disease brain samples with multimodal multiphoton nonlinear optical microspectroscopy,” J. Biomed. Opt. 20(5), 56013 (2015). 10.1117/1.JBO.20.5.056013 [DOI] [PubMed] [Google Scholar]
- 61.Hankzyc P., Samoc M., Nordén B., “Multiphoton absorption in amyloid protein fibres,” Nat. Photonics 7(12), 969–972 (2013). 10.1038/nphoton.2013.282 [DOI] [Google Scholar]
- 62.Wittingham J. L., Scott D. J., Chance K., Wilson A., Finch J., Brange J., Dodson G. G., “Insulin at pH 2: structural analysis of the conditions promoting insulin fibre formation,” J. Mol. Biol. 318(2), 479–490 (2002). 10.1016/S0022-2836(02)00021-9 [DOI] [PubMed] [Google Scholar]
- 63.Gosal W. S., Clark A. H., Pudney P. D. A., Ross-Murphy S. B., “Novel amyloid fibrillar networks derived from a globular protein: β-lactoglobulin,” Langmuir 18(19), 7174–7181 (2002). 10.1021/la025531a [DOI] [Google Scholar]
- 64.Pechkova E., Bragazzi N., Bozdaganyan M., Belmonte L., Nicolini C., “A review of the strategies for obtaining high-quality crystals utilizing nanotechnologies and microgravity,” Crit. Rev. Eukaryot. Gene Expr. 24(4), 325–339 (2014). 10.1615/CritRevEukaryotGeneExpr.2014008275 [DOI] [PubMed] [Google Scholar]
- 65.Brownlow S., Cabral J. H. M., Cooper R., Flower D. R., Yewdall S. J., Polikarpov I., North A. C. T., Sawyer L., “Bovine beta-lactoglobulin at 1.8 Å resolution – still an enigmatic lipocalin,” Structure 5(4), 481–495 (1997). 10.1016/S0969-2126(97)00205-0 [DOI] [PubMed] [Google Scholar]
- 66.Demchenko A. P., “The red-edge effect: 30 years of exploration,” Luminescence 17(1), 19–42 (2002). 10.1002/bio.671 [DOI] [PubMed] [Google Scholar]
- 67.Chattopadhyay A., Sourav H., “Dynamic insight into protein structure utilizing red edge excitation shift,” Acc. Chem. Res. 47(1), 12–19 (2014). 10.1021/ar400006z [DOI] [PubMed] [Google Scholar]
- 68.Lakowicz J. R., Sourav H., “On spectral relaxation in proteins,” Photochem. Photobiol. 72(4), 421–437 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wlodarczyk J., Kierdaszuk B., “Interpretation of fluorescence decays using a power-like model,” Biophys. J. 85(1), 589–598 (2003). 10.1016/S0006-3495(03)74503-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johansson P. K., Koelsch P., “Vibrational sum-frequency scattering for detailed studies of collagen fibers in aqueous environments,” J. Am. Chem. Soc. 136(39), 13598–13601 (2014). 10.1021/ja508190d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kiskis J., Fink H., Nyberg L., Thyr J., Li J-Y., Enejder A., “Plaque-associated lipids in Alzheimer’s diseased brain tissue visualized by nonlinear microscopy,” Sci. Rep. 5, 13489 (2015). 10.1038/srep13489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lazar A. N., Lazar A. N., Bich C., Panchal M., Desbenoit N., Petit V. W., Touboul D., Dauphinot L., Marquer C., Laprevote O., Brunelle A., Duyckaerts C., “Time-of-flight secondary ion mass spectrometry (TOF-SIMS) imaging reveals cholesterol overload in the cerebral cortex of Alzheimer disease patients,” Acta Neuropathol. 125(1), 133–144 (2013). 10.1007/s00401-012-1041-1 [DOI] [PubMed] [Google Scholar]