The inability to reproduce key scientific results is a growing concern among scientists, funding agencies, academic journals and the public (Begley and Ellis 2012; Prinz et al. 2011). Studies examining research practices in neuroscience and other fields have highlighted problems with study design (Button et al. 2013), inadequate reporting of methods and results (Baker et al. 2014), errors in statistical analyses (Lazic 2010; Strasak et al. 2007) and data visualization (Weissgerber et al. 2015). Each of these factors may contribute to irreproducibility. These observations have sparked discussion about the respective roles and responsibilities of authors, peer reviewers and journals in improving the quality of scientific literature (2013; 2016; Collins and Tabak 2014; Landis et al. 2012).
In principle, one can distinguish between efforts to improve the quality of the research itself and initiatives aimed at improving the quality of scientific reporting. In practice, however, these two factors are often intertwined. Initiatives and guidelines designed to improve the quality of research often contain recommendations for investigators at all stages of the research process, including preparing manuscripts for publication. Without following these recommendations, authors might inadvertently omit critical details when describing their methods and results, which could render even an excellent study to appear as low quality.
The Journal of Neuroscience Research is introducing new policies, which are designed to ensure that papers contain essential information that reviewers and readers need to evaluate published studies. By having authors report complete, accurate and transparent scientific methods and statistical results; this policy will reduce inadequacies in experimental reporting and improve reproducibility and replication. These policy changes are in line with the National Institutes of Health Principles and Guidelines for Reporting Preclinical Research (NIHPG). This editorial provides an overview of these policies and highlights other resources that may help scientists to improve the quality of research and scientific data reporting.
Study Design
A reader’s ability to critically evaluate original research depends on clear reporting of the study design and methods. Reporting should include a general overview of the study design, including the objective of the research, an explanation of methods that were used to reduce the risk of bias and a detailed description of the experimental protocol that will allow others to replicate the study. Many papers, unfortunately, are missing crucial information (Landis et al. 2012). The ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines, for example, were designed to improve the quality of reporting in animal studies (Kilkenny et al. 2010). Despite their widespread endorsement, an analysis performed two years after their release revealed that many journals had not fully implemented the guidelines (Baker et al. 2014). Articles published after the guidelines became available were more likely to report the species, sex and age of animals. However, only 20% of studies provided information on blinding. Fewer than 10% of articles reported whether the study was randomized or presented a power calculation. Similar challenges in implementing the CONSORT (Consolidated Standards of Reporting Trials) guidelines for human clinical trials have led investigators to propose that reporting templates may be more effective than checklists (Altman 2015). This approach may improve the quality of reporting in animal studies and other types of basic science research.
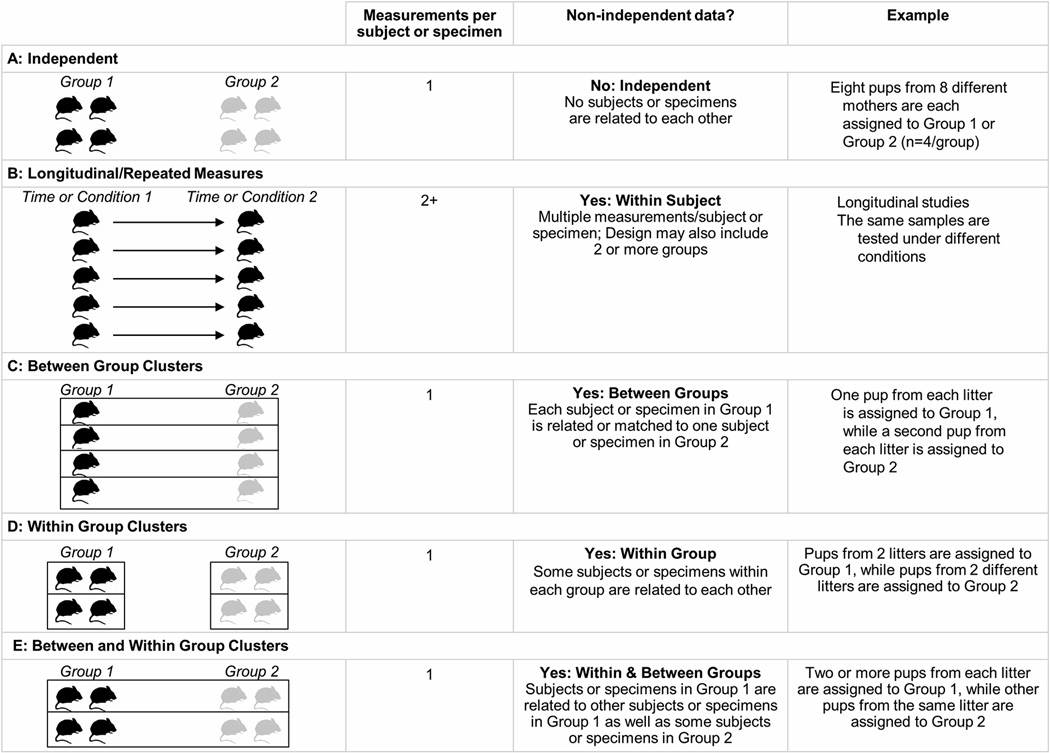
Clear reporting of study design is essential, as it determines many aspects of the statistical analysis. Knowing whether the data are independent or non-independent is necessary to select statistical methods. Independent designs (Figure 1A) are analyzed using techniques such as unpaired t-tests or ANOVA, which assume independence. In contrast, repeated measures (Figure 1B) and clustered designs (Figure 1, Panels C-E) require analysis techniques that account for non-independence, such as paired t-tests or repeated measures ANOVA. Reporting of the study design is particularly important for animal and in vitro studies, as many experiments use different study designs for different outcome measures. Study design figures are valuable and underutilized tools for highlighting key features of the experimental design that affect the statistical methods.
Figure 1. Study designs for independent and non-independent data.
The figure illustrates study designs for independent (A), longitudinal or repeated measures (Panel B), Between group clustered (C), within group clustered (D) and between and within group clustered (E) designs.
Statistical Methods and Analysis
Inadequate reporting of statistical methods and misuse of statistical techniques are common in basic science research, even among articles published in top journals (Baker et al. 2014; Lazic 2010; Strasak et al. 2007). Problems with reporting may include not presenting a power calculation (Strasak et al. 2007), failing to state which statistical test was used (Strasak et al. 2007), providing adequate detail about the test (i.e. paired vs. unpaired t-test) (Strasak et al. 2007), not stating whether the assumptions of the statistical tests were examined (Strasak et al. 2007; Weissgerber et al. 2015), or not stating how replicates were analyzed (Lazic 2010). Problems with statistical analysis include using incorrect or suboptimal tests (Baker et al. 2014; Strasak et al. 2007), using mean and standard deviation or standard error to summarize data that were analyzed by non-parametric tests (Weissgerber et al. 2015), reporting p-values that do not match the test statistic (Garcia-Berthou and Alcaraz 2004; Nuijten et al. 2015), p-hacking (Head et al. 2015), and analyzing non-independent data as though they are independent (Lazic 2010). Along with addressing these problems, the scientific community is increasingly recognizing the need for better statistical education for basic scientists (Weissgerber et al. 2016a). This includes specific strategies for analysis and interpretation of small datasets, clustered data, and handling outliers, as well as learning to identify and avoid errors in data visualization and analysis that are common in the basic biomedical sciences.
The methods used to determine sample size are critical and should be addressed in the statistical methods section. A recent paper highlighted low statistical power and small sample sizes as major obstacle to reproducibility in the neurosciences (Button et al. 2013). While some types of experiments depend on statistical analyses and power calculations, others do not (Vaux 2012). Power calculations traditionally have focused on reducing the likelihood of false negatives (Halsey et al. 2015). This can lead to the misleading impression that an underpowered study is only problematic if no effect is found. This perspective is incomplete, as it ignores the potential for false positives. Additionally, when the sample size is too small, samples that yield significant differences are usually extreme in some way and will overestimate the magnitude of the difference between groups (Halsey et al. 2015).
The numbers of participants, animals or samples included in the original study should be clearly specified, with detailed explanations of the reasons for any attrition in the study. This includes providing information about how outliers were identified and handled in the analysis. A recent meta-analysis highlighted the problems with reporting of attrition in preclinical animal studies of stroke and cancer (Holman et al. 2016). In 64.2% of stroke studies and 72.9% of cancer studies, it was not possible to determine whether animals were excluded or did not complete the experiment. Among studies with clear evidence of attrition, most authors did not explain the reasons for attrition. This information is crucial and should be included to demonstrate that the results are not affected by systematic biases that may be introduced by the exclusion or loss of samples. Simulation studies indicate that biased exclusion of a few animals can inflate the estimated treatment effect (Holman et al. 2016).
The statistical methods section should also specify how non-independent replicates are handled in the analysis. Clustered data are frequently obtained in neuroscience research, but are often inappropriately analyzed or ignored (Lazic 2010). A cluster contains multiple observations, giving data a hierarchical structure (Galbraith et al., 2010). When analyzing these data, researchers often make the mistake of treating the data as if the observations were independent (i.e., pooling individual synapses to create a large dataset), which could increase statistical error. “Data reduction” is one of the most common strategies for analyzing clustered data (Galbraith et al. 2010). Replicates from a single subject or experiment (e.g., multiple synapses from the same slide, brain section, or animal) are converted into a single summary statistic (i.e. mean, median, etc.) and then analyzed using techniques that assume independence. While data reduction may be the only option for very small datasets, this approach reduces statistical power, as information is lost when the replicates are averaged. More sophisticated techniques, such as bootstrapping, permutation tests, and mixed models, may be valuable for investigators with larger datasets (Galbraith et al. 2010).
Sufficient details about statistical tests should be presented when reporting study results. For studies using traditional (frequentist) statistical analysis, this includes the test statistic, degrees of freedom, p-value, and whether the test was one-sided or two-sided. The description of the statistical methods should provide enough details to enable a reader with access to the data to verify the reported results (Bailar and Mosteller 1988). An analysis of papers published in psychology journals between 1985 and 2013 observed that approximately half of papers included at least one p-value that did not match the test statistic and degrees of freedom (Nuijten et al. 2015). Large errors that may have altered the conclusion of the test were found in 12.5% of papers (Nuijten et al. 2015).
Detailed discussion of these important topics is beyond the scope of this editorial. Many of these considerations have been incorporated in the updated Statistical and Data Reporting Guidelines within the Author Guidelines of the Journal of Neuroscience Research.
Data Visualization
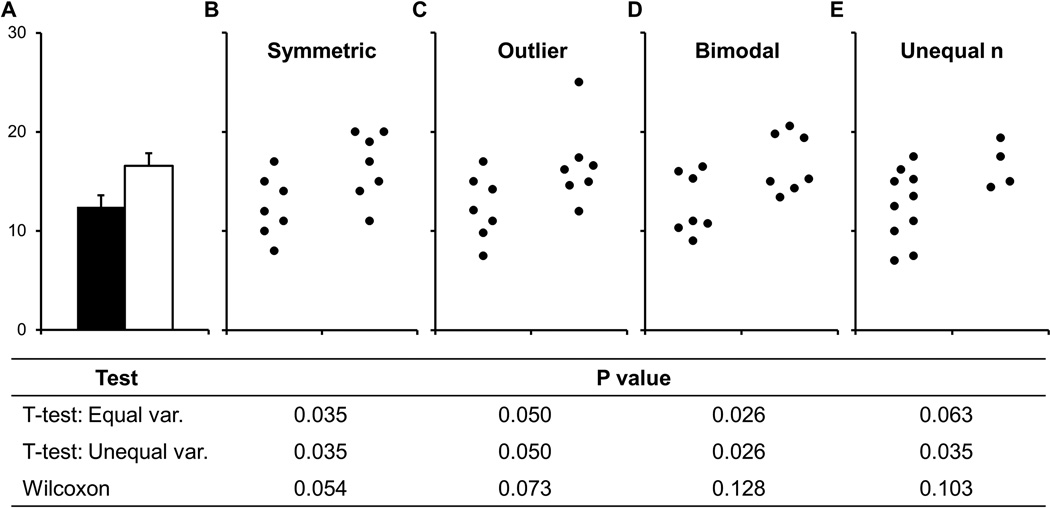
Figures are important, as they are often used to illustrate the most important findings from a study. A well-designed figure should convey information about the study design, in addition to showing the data. Selecting the right type of figure is essential (Table 1). Authors should consider the study design, type of outcome measure (i.e. categorical, continuous, etc.), and sample size when designing figures. Recent reports indicate that the common practice of using bar graphs to show continuous data for small sample size studies is a problem in the scientific literature (Saxon 2015; Weissgerber et al. 2015). Bar graphs are designed for categorical data; when used to display continuous data, bar graphs omit key information about the data distribution. This is problematic, as many different datasets can lead to the same bar graph and the actual data may suggest different conclusions from the summary statistics (Figure 2). The new guidelines recommend using figures that show the data distribution, such as univariate scatterplots (also called Cleveland dot plots (Cleveland 1993)), boxplots, violin plots, or kernel density plots when presenting continuous data. While line graphs are typically used to present longitudinal or repeated measures data, these figures provide limited information about data distribution and no information about whether responses vary among subjects. Showing univariate scatterplots or boxplots of differences for selected pairs of time points or conditions where important changes occur may alleviate this problem. Alternatively, recent publications have highlighted the importance of developing new techniques for the presentation of scientific results, such as interactive graphics (Krumholz 2015; Weissgerber et al. 2016b). We recently designed a free, web-based tool for creating interactive line graphs for scientific publications (Weissgerber et al. 2016b). This proof-of-concept tool shows how interactive alternatives to static graphics allow for additional exploration of published data. Tools such as this one have the potential to promote widespread use of interactive graphics and transform scientific publications from static papers into interactive datasets narrated by the authors.
Table 1.
Quick guide to figures that basic scientists often use
| Study Type | Type of Outcome Variable |
Objective | Type of Figure |
|---|---|---|---|
| Cross- sectional |
Categorical | Compare values for two or more groups |
Bar graph |
| Continuous | Compare values for two or more groups |
Small n Univariate scatterplot Medium n Box plot with data points overlaid Large n Box plot, violin plot or kernel density plot |
|
| Continuous | Examine relationship between two continuous variables |
Scatterplot | |
| Longitudinal /repeated measures |
Categorical | Examine changes over time, for one or more groups |
Line graph |
| Continuous | Examine changes over time, for one or more groups |
Small datasets with 2 time points or conditions spaghetti plot and univariate scatterplot showing change scores* Larger datasets, or datasets with >2 time points or conditions: Interactive line graph or line graph and univariate scatterplot(s) showing change scores for time points where important changes occur |
When working with paired data, it is critically important to show change scores to allow the reader to assess the direction and magnitude of changes and determine whether responses vary among subjects.
Figure 2. Many different datasets can lead to the same bar graph.
The full data may suggest different conclusions from the summary statistics. The means and standard errors for the four example datasets show in Panels B-E are all within 0.5 units of the means and standard errors shown in the bar graph (A). P-values were calculated in R statistical software (version 3.0.3) using an unpaired t-test, an unpaired t-test with Welch’s correction for unequal variances or a Wilcoxon rank sum test. In Panel B, the distribution in both groups appears symmetric. Although the data suggest a small difference between groups, there is substantial overlap between groups. In Panel C, the apparent difference between groups is driven by an outlier. Panel D suggests a possible bimodal distribution. Additional data are needed to confirm that the distribution is bimodal and to determine whether this effect is explained by a covariate. In Panel E, the smaller range of values for group 2 may simply be due to the fact that there are only three observations. Additional data for group 2 would be needed to determine whether the groups are actually different. Abbreviations: var, variance. Reprinted from Weissgerber et al. 2015 under a creative commons license.
A recent article provides a valuable overview of graphic design principles that investigators should consider when creating figures for scientific publications (Duke et al. 2015). Numerous investigators have released free templates, tools or code that allows scientists to create graphics for continuous data. Resources for creating univariate scatterplots include Excel templates (Weissgerber et al. 2015), instructions for GraphPad PRISM (Weissgerber et al. 2015) and code for creating these graphics in R (Ashander 2015; Marwick 2015) or SPSS (https://www.ctspedia.org/do/view/CTSpedia/TemplateTesting). R code is also available for investigators who want to create boxplots, with or without the data points overlaid (Ashander 2015). Authors who have clustered data can use the web-based tool provided by Pallmann and Hothorn to create customized boxplots with data points overlaid (Pallmann and Hothorn 2015) (https://lancs.shinyapps.io/ToxBox/). This application is easy to use and does not require any programming skills. Investigators can also use this tool to create box plots that show data points for studies with independent designs. A newly published article provides a detailed discussion about the advantages and disadvantages of different types of graphics that are frequently used to present categorical and continuous data in scientific publications (Rice and Lumley 2016). Code and links to resources for creating these graphics in Excel, R and Stata are available on the authors’ website (http://faculty.washington.edu/kenrice/heartgraphs/). Other recent papers have provided detailed overviews of different strategies for visualizing differences in effect size (Wilcox 2006) and MATLAB scripts for visualizing fMRI data (Allen et al. 2012).
Conclusions
A number of studies examining the research process have provided valuable insight into deficiencies in the study design, analysis and reporting of published research. Journal policy changes, such as the one being implemented by the Journal of Neuroscience Research, are essential components of a comprehensive strategy to promote transparent reporting and improve the quality and reproducibility of scientific studies. Initiatives designed to raise awareness about common problems in the literature, improve adherence to reporting guidelines and develop tools and templates to improve data visualization should all be a part of the solution.
Acknowledgments
Funding
TLW was supported by the Office of Research on Women’s Health (BIRCWH K12HD065987). This work was supported by P-50 AG44170 (V.D. Garovic) from the National Institute on Aging.
Footnotes
Conflict of Interest Statement
The authors have nothing to disclose.
TLW drafted the editorial. All authors revised the manuscript for intellectually important content.
References
- Reproducing our irreproducibility. Nature. 2013;496:398. [Google Scholar]
- Take the long view. Nat Med. 2016;22(1):1. doi: 10.1038/nm.4033. [DOI] [PubMed] [Google Scholar]
- Allen EA, Erhardt EB, Calhoun VD. Data visualization in the neurosciences: overcoming the curse of dimensionality. Neuron. 2012;74(4):603–608. doi: 10.1016/j.neuron.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman DG. Making research articles fit for purpose: structured reporting of key methods and findings. Trials. 2015;16:53. doi: 10.1186/s13063-015-0575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashander J. Easy alternatives to bar charts in native R graphics, Rapid evolution: Theory, computation and inference. 2015 Apr 28; 2015. http://www.ashander.info/posts/2015/04/barchart-alternatives-in-base-r/
- Bailar JC, 3rd, Mosteller F. Guidelines for statistical reporting in articles for medical journals. Amplifications and explanations. Ann Intern Med. 1988;108(2):266–273. doi: 10.7326/0003-4819-108-2-266. [DOI] [PubMed] [Google Scholar]
- Baker D, Lidster K, Sottomayor A, Amor S. Two years later: journals are not yet enforcing the ARRIVE guidelines on reporting standards for pre-clinical animal studies. PLoS Biol. 2014;12(1):e1001756. doi: 10.1371/journal.pbio.1001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483(7391):531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES, Munafo MR. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci. 2013;14(5):365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- Cleveland WS. Visualizing Data. Hobart Press; 1993. [Google Scholar]
- Collins FS, Tabak LA. Policy: NIH plans to enhance reproducibility. Nature. 2014;505(7485):612–613. doi: 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke SP, Bancken F, Crowe B, Soukup M, Botsis T, Forshee R. Seeing is believing: good graphic design principles for medical research. Statistics in medicine. 2015;34(22):3040–3059. doi: 10.1002/sim.6549. [DOI] [PubMed] [Google Scholar]
- Galbraith S, Daniel JA, Vissel B. A study of clustered data and approaches to its analysis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(32):10601–10608. doi: 10.1523/JNEUROSCI.0362-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Berthou E, Alcaraz C. Incongruence between test statistics and P values in medical papers. BMC medical research methodology. 2004;4:13. doi: 10.1186/1471-2288-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsey LG, Curran-Everett D, Vowler SL, Drummond GB. The fickle P value generates irreproducible results. Nat Methods. 2015;12(3):179–185. doi: 10.1038/nmeth.3288. [DOI] [PubMed] [Google Scholar]
- Head ML, Holman L, Lanfear R, Kahn AT, Jennions MD. The extent and consequences of p-hacking in science. PLoS Biol. 2015;13(3):e1002106. doi: 10.1371/journal.pbio.1002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman C, Piper SK, Grittner U, Diamantaras AA, Kimmelman J, Siegerink B, Dirnagl U. Where Have All the Rodents Gone? The Effects of Attrition in Experimental Research on Cancer and Stroke. PLoS Biol. 2016;14(1):e1002331. doi: 10.1371/journal.pbio.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8(6):e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumholz HM. The End of Journals. Circ Cardiovasc Qual Outcomes. 2015;8(6):533–534. doi: 10.1161/CIRCOUTCOMES.115.002415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis SC, Amara SG, Asadullah K, Austin CP, Blumenstein R, Bradley EW, Crystal RG, Darnell RB, Ferrante RJ, Fillit H, Finkelstein R, Fisher M, Gendelman HE, Golub RM, Goudreau JL, Gross RA, Gubitz AK, Hesterlee SE, Howells DW, Huguenard J, Kelner K, Koroshetz W, Krainc D, Lazic SE, Levine MS, Macleod MR, McCall JM, Moxley RT, 3rd, Narasimhan K, Noble LJ, Perrin S, Porter JD, Steward O, Unger E, Utz U, Silberberg SD. A call for transparent reporting to optimize the predictive value of preclinical research. Nature. 2012;490(7419):187–191. doi: 10.1038/nature11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazic SE. The problem of pseudoreplication in neuroscientific studies: is it affecting your analysis? BMC neuroscience. 2010;11:5. doi: 10.1186/1471-2202-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwick B. Using R for the examples in “Beyond Bar and Line Graphs: Time for a New Data Presentation Paradigm’. 2015 doi: 10.1371/journal.pbio.1002128. https://cdn.rawgit.com/benmarwick/new-data-presentation-paradigm-using-r/582a80eaba654237231fe4b06d3eda5a61587d73/Weissgerber_et_al_supplementary_plots.html. [DOI] [PMC free article] [PubMed]
- Nuijten MB, Hartgerink CH, van Assen MA, Epskamp S, Wicherts JM. The prevalence of statistical reporting errors in psychology (1985–2013) Behav Res Methods. 2015 doi: 10.3758/s13428-015-0664-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallmann P, Hothorn LA. Boxplots for grouped and clustered data in toxicology. Archives of Toxicology. 2015;90(7):1631–1638. doi: 10.1007/s00204-015-1608-4. [DOI] [PubMed] [Google Scholar]
- Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nat Rev Drug Discov. 2011;10(9):712. doi: 10.1038/nrd3439-c1. [DOI] [PubMed] [Google Scholar]
- Rice K, Lumley T. Graphics and statistics for cardiology: comparing categorical and continuous variables. Heart. 2016;102(5):349–355. doi: 10.1136/heartjnl-2015-308104. [DOI] [PubMed] [Google Scholar]
- Saxon E. Beyond bar charts. BMC Biol. 2015;13(1):60. doi: 10.1186/s12915-015-0169-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasak AM, Zaman Q, Marinell G, Pfeiffer KP, Ulmer H. The use of statistics in medical research: A comparison of the New England Journal of Medicine and Nature Medicine. The American Statistician. 2007;61(1):47–55. [Google Scholar]
- Vaux DL. Research methods: Know when your numbers are significant. Nature. 2012;492(7428):180–181. doi: 10.1038/492180a. [DOI] [PubMed] [Google Scholar]
- Weissgerber T, Garovic VD, Milin-Lazovic J, Winham S, Obradovic Z, Trzeciakowski JP, Milic N. Re-inventing Biostatistics Education for Basic Scientists. PLoS Biol. 2016a;14(4):e1002430. doi: 10.1371/journal.pbio.1002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissgerber T, Milic N, Winham S, Garovic VD. Beyond Bar and Line Graphs: Time for a New Data Presentation Paradigm. PLoS Biol. 2015;13(4):e1002128. doi: 10.1371/journal.pbio.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissgerber TL, Garovic VD, Savic M, Winham SJ, Milic NM. From Static to Interactive: Transforming Data Visualization to Improve Transparency. PLoS Biol. 2016b;14(6):e1002484. doi: 10.1371/journal.pbio.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox R. Graphical Methods for Assessing Effect Size: Some Alternatives to Cohen's d. The Journal of Experimental Education. 2006;74(4):353–367. [Google Scholar]