Abstract
It has been long established that hormones exert enduring influences on the developing brain that direct the reproductive response in adulthood, although the cellular mechanisms by which organisational effects are maintained have not been determined satisfactorily. Recent interest in epigenetic modifications to the nervous system has highlighted the potential for hormone-induced changes to the genome that could endure for the lifespan but not be transmitted to the next generation. Preliminary evidence suggests that this is indeed possible because sex differences in the histone code and in the methylation of CpGs in the promoters of specific genes have been identified and, at times, functionally correlated with behaviour. The present review provides an overview of epigenetic processes and discusses the current state-of-the-art, and also identifies future directions.
Keywords: oestrogens, steroids: neuroactive steroids, development, sex differences, preoptic area, hypothalamus
The developing brain is subject to intrinsic and tightly orchestrated temporally constrained directors of neuronal proliferation, differentiation and integration. These internal regulators are buffered by extrinsic variables that exert both subtle and profound effects on the final phenotype. Extrinsic variables range from aspects of the intrauterine environment affected by maternal diet, stress and infectious state, to postnatal parameters that include the newborns own stress, maternal behaviours and somatosensory stimulation. Males and females share common intrinsic variables that direct the development of fundamental parameters of brain development, although they diverge in important ways in response to extrinsic variables by showing differential responses to challenges, be they of psychological (i.e. maternal stress) or pathogenic (i.e. infection or inflammation) origin, and they are also on the receiving end of differences in maternal attention and care. One critical pivot point of the divergence between intrinsic and extrinsic influences on male versus female brain development is the difference in exposure to gonadal steroid hormones experienced by each sex, which has profound consequences on the brain as it develops and again in adulthood as part of a programme for ensuring maximum reproductive fitness in both sexes. A primary mechanism by which the early stage exposure to gonadal steroids ensures continuity in responsiveness in the adult brain is postulated to be via establishment of a cellular memory achieved by epigenetic alterations to the genome. The matching between early hormonal effects and adult hormonal responsiveness is codified as the Organisational/Activational Hypothesis of sexual differentiation of the brain (1). At the time it was established, over 50 years ago, this tenet was exclusive to differentiation of sexual behaviour. It has subsequently been widely applied to any early hormonal effect on the brain that impacts adult function, and these are many. It has become apparent, however, that the cellular processes, as well as the relevant parameters of a particular endpoint, are highly specific to that endpoint, if not unique, and that, although the general principles of the Organisational/Activational Hypothesis might apply, there are untold numbers of exceptions and variants on the theme. The complexity and diversity of sexually differentiated parameters prevents us from determining a coherent or unified theory of hormonal action on the developing brain at this time, although such a theory may emerge as our knowledge progresses. This requires that we understand at least some systems in great depth.
Epigenetics in the developing nervous system
A useful discussion of epigenetics begins with a definition because the term can mean different things in different fields. In the strictest sense, epigenetics refers to changes at the genome that are functionally relevant but do not change the DNA sequence, and, most importantly, are transmitted to the next generation. In other words, epigenetics is the same as genetics in that it is heritable. For this to be true, however, requires that any epigenetic changes in the periphery also occur in the germline, and that they survive the reprogramming of the genome that is a component of germ cell development. However, as noted above, this is the strictest and most confining definition of epigenetics. A second tier of rigour in the use of the term epigenetic is that associated with the programming of cell fate. Every cell in the body has the full complement of that individuals genome but only a subset of those genes will be expressed and will determine the identity of each cell. Many factors influence the profile of genes that will be expressed versus repressed, including nearest neighbours and cells of origin. The genes that are silenced must remain so to ensure their phenotype is not inappropriately changed or that they begin proliferating, which can have potentially catastrophic consequences. These are actually two separate epigenetic processes: one in which the daughter cell retains its phenotype (i.e. a dividing liver cell begets new liver cells) versus a cell maintaining its phenotype for life (i.e. once a neurone always a neurone). These are both accomplished via epigenetic silencing of the critical regulatory regions of each gene in a manner that is essentially irreversible. The third tier of epigenetic regulation is the one of relevance here; it is largely temporary and responsive to changes in the environment, drug exposure or experience. This has been referred to as context-dependent epigenetics to distinguish it from germ-line dependent epigenetics (2). The temporary component of context-dependent epigenetics is a relative term, meaning it can endure for extended periods, even throughout the lifespan, or be relatively short lived. This is the definition of epigenetics of interest to the mechanisms of sexual differentiation of the brain.
There are four generally agreed upon epigenetic modifiers. The two dominant and widely familiar forms are changes to the histone components of the chromatin and direct changes to the specific nucleotides of the DNA. Less well known are the microRNAs and transposed and transposable elements. MicroRNAs are small silencing RNAs generated via transcription, often at the same time as specific genes, with wide ranging pleiotrophic effects on a variety of gene targets (3). Transposons are the so called ‘jumping genes’ first identified in maize in the Noble prize winning work of Martha McClintock. By some estimates, as much as 75% of the mammalian genome consists of transposons and associated repeat elements, although the great majority are inactive, referred to as transposed elements (4,5). These have contributed to the expansion of the mammalian genome, as well as its diversity (6). Nonetheless, hundreds to thousands of transposons remain active, with substantial variation in the number and rate of retrotransposition between species, individuals, tissues and cells (6). The brain is emerging as a particularly active area of recombination, most notable in proliferating neural progenitor cells (7). Neither microRNAs, nor transposable elements have yet been implicated in hormonally-mediated sexual differentiation of the brain [although sex differences in microRNAs have been identified (3)] and so they are not discussed further, although they represent promising and exciting topics for investigation.
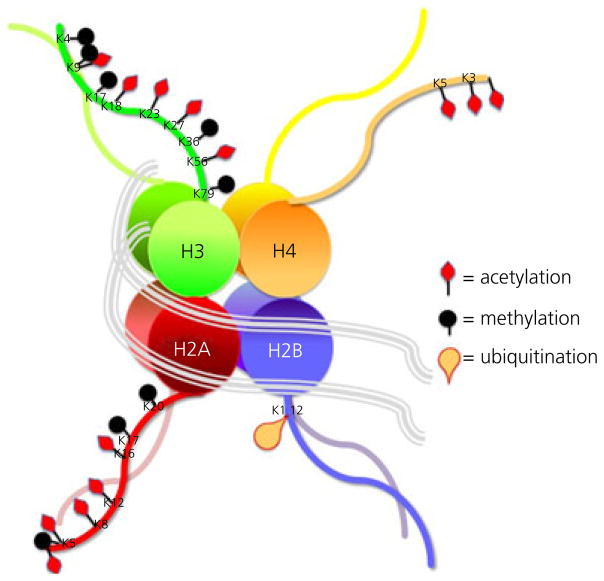
Changes to histones and DNA are emerging as essential components of the sexual differentiation process. We first review those involving the histones. DNA is tightly wound into structures called nucleosomes, which consist of an eight histone core that controls the access of transcription factors and polymerases to the DNA by altering the degree of compactness. Approximately 145–147 bp of genomic DNA wrapped around a histone is considered the basic unit of chromatin: the nucleosome. There are four core histones: H2, H3A, H3B and H4. H3 and H4 partner each other, as do the two H2 proteins. Within the nucleosome, each histone is found in duplicate. Another histone, H1, serves to link the cores, thus earning the name linker histone. The histones are built as octomers and are among the most highly conserved proteins across species and each has its own menu of possible changes that have highly predictable effects as either transcriptional activators or repressors (Fig. 1). For example, acetylation introduces a positive charge to the histone carbon tail that forces the molecule to relax from its tightly bound state, which in turn allows access of additional regulatory proteins and transcription factors. Referred to as the ‘histone code’, a combination of changes to the histones surrounding particular genes will largely determine the transcribability of that gene (8). Sometimes a combination of repressive and permissive histone marks is associated with the same gene, and this is referred to as bivalent domains (9). Histone changes associated with the epigenetic control of puberty are bivalent (10) and mirror the dual requirements of increased excitation and decreased inhibition also required for the transition into post-pubertal maturity.
Fig. 1.
The histone code. Nucleosomes are the fundamental unit of chromatin and consist of an octomer of histones of four varieties. Modifications to key amino acids in the carbon chain tails alter the electric charge which either repels or attracts the molecules and thereby gates the accessibility of transcription factors to the DNA. The dominant sites of modification are the lysines (K), which are either acetylated or methylated and sometimes both. Additional modifications include ubiquitination, as well as palmitylation and glycosylation (not shown here). Changes to the histone tail are achieved by specific enzymes, such as histone acetyl transferases and histone deactylases. The ‘histone code’ refers to the predictable effect of specific modifications on gene expression versus repression.
Changes to the histone tales are made enzymatically and faithfully, meaning that only specific amino acid residues are subject to specific types of modifications. In the case of acetylation, one of the more common histone modifications, acetyl groups are added only to lysines. Methylation can occur at the same lysine group and also have profound effects on transcription factor access (11,12). There are histone acetylation enzymes, called HATs for short, and histone deacetylation enzymes, called HDACs. Both types of enzymes come in a variety of forms and are readily, although sometimes nonspecifically, inhibited by various pharmacological agents (13–16). Many transcription factors associate with HATs, and this includes the steroid receptors, such as oestrogen receptor (ER), progesterone receptor (PR) and androgen receptor (AR) (17).
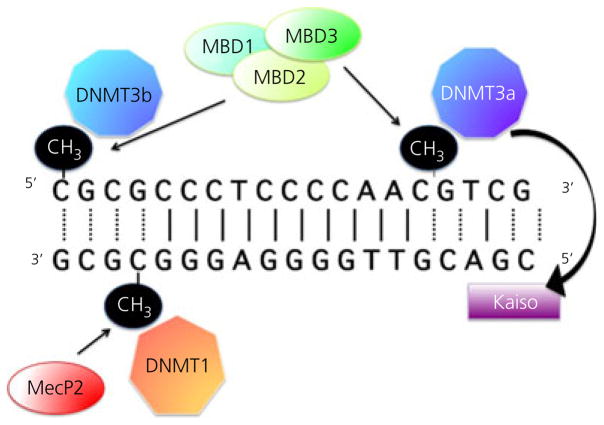
The second major mechanism of epigenetic modification are changes directly to the DNA and these occur largely in only one way, the addition of a methyl group to cytosines that are next to guanines in the genetic code, denoted as CpG (other bases can be methylated, although this occurs much less frequently and is of unknown significance). The ‘p’ denotes the phosphate bond between the two nucleotides and indicates these are contiguous to each other, and not base paired. The occurrence of cytosines next to guanines is not a frequent event, occurring far less than chance would predict. This is assumed to be a result of the propensity for cytosines next to guanines to undergo deamination and be converted to thymidines, thereby inadvertently introducing a site mutation. Strong selection pressure against such spontaneous mutations has presumably greatly reduced the occurrence of this nucleotide sequence. When CpGs do occur, they tend to be in clusters and are over-represented in the promoter regions of genes where they are referred to as CpG islands. A CpG island by definition is a stretch of DNA of at least 100 bp and they usually occur outside of nucleosomes. When they are found within a nucleosome, they tend to be in areas where the histone code is conducive to gene expression (18). Most importantly, when CpGs do occur, the configuration can be recognised by DNA methylating enzymes, called DNMTs, which add a methyl group to the 5′ carbon on the cytosine molecule (Fig. 2). There are two major types of DNMTs that are distinguished by the role they play in maintenance versus de novo methylation. DNMT-1 is the maintenance enzyme and is responsible for replication of the methylation pattern when a cell divides (19,20). This is essential both for retaining imprinted gene patterns, as well as for assurance that a cell’s phenotype is not altered. In other words, as noted above, when a liver cell divides, it should give rise to another liver cell and not a skin cell. Maintenance methylation is probably not of central importance to sexual differentiation of the brain since neurones are generally post-mitotic and hormonal affects are not transmitted transgenerationally. De novo methylation is the purview of DNMT 3a and 3b (21) and it is this form of methylation that likely impacts sexual differentiation by establishing the gene transcription patterns that endure.
Fig. 2.
Methylation of the DNA. Cytosines located proximal to guanines are the target of methylation by the DNA methyl transferase (DNMT) enzymes. Two classes of DNMT’s, DNMT1 and DNMT3, mediate maintenance versus de novo methylation. Increased methylation of cytosines can alter gene transcription directly by sterically hindering the access of transcription factors, or indirectly by recruiting methyl-binding domain (MBD) proteins, the most famous of which is MeCP2. Some MBDs, such as Kaiso, are also capable of binding to unmethylated DNA and recruiting DNMT activity. The relationship between cytosine methylation and gene expression is not as straightforward as that for changes to the histones.
Regardless of the ubiquitous presence of DNMTs, the majority of CpGs remain unmethylated, with, on average, only 5–20% of the cytosines within an island being methylated. However, it does not take much methylation to have a big impact on transcription. Several different nomenclatures are used to denote cytosine methylation, although 5mCyt is perhaps the most accurate. Repression of transcription following cytosine methylation can occur in two ways. First, there is the simple direct steric hindrance of transcription factors so that they cannot associate with the DNA. Second, there is the recruitment of an additional set of proteins called methyl-binding domain proteins, or MBDs. The most famous among these is MeCP2, which was identified because of its critical role in Rett syndrome, an X-linked disorder found predominantly in females and characterised by severe mental retardation (22,23). When the MeCP2 protein is missing or dysfunctional, there is inappropriate gene expression as a result of a lack of repression, including an mGluR receptor in the nervous system, which is assumed to be responsible for many of the deficits observed in Rett syndrome. The gene for MeCP2 is found on the X chromosome and is not subject to inactivation, hence explaining why Rett syndrome is more frequent and milder in girls because they have a second copy to compensate for a mutated copy on one X. In males, the Rett mutation is usually lethal.
Other MBDs are critical as well and identified as MBD2, 3 and 4 and Kaiso. These also impact transcription by either blocking access of transcription factors (24) or recruiting DNMTs to further methylate the region. Some MBDs are capable of binding to non-methylated stretches of DNA and initiating methylation via recruitment of DNMTs (25,26), providing yet another layer of regulation.
Although the canonical effect of DNA 5mC is gene repression, there is accumulating evidence that this is not always the case and that, in some instances, increased 5mC might reflect a past history of gene expression. Moreover, where in the gene the methylation occurs is important, with increased methylation in the first exon being more strongly associated with gene repression than increased methylation in the promoter, at least in some instances (27). When it comes to understanding the impact and functional significance of 5mC in the brain, it should be acknowledged that more studies are needed. Recent discoveries of highly dynamic patterns of 5mC are counter to the prevailing dogma that methylation is permanent (28). Changes to DNA methylation are being associated with the regulation of behaviour, stress responding and environmental insults, amongst other factors (29–33). This is resulting in a rethink of the regulation of DNA methylation and, more specifically of how demethylation might be achieved (34).
As noted above, the methylation of cytosines is considered permanent because it involves a covalent bond, one of the strongest bonds in nature. Yet quantification of 5mC levels in the brain unequivocally confirms that the amount of methylation changes, including a decrease in some cases. This has triggered the search for de-methylating enzymes that could actively remove the methyl group, a search that has been largely unsuccessful to date, although there is considerable controversy and disagreement on the topic. However, there is another way to remove a 5mC, and that involves removing the cytosine group altogether via DNA repair (34). The genome is constantly being subjected to potentially damaging insults. To protect against this onslaught, monitoring systems have evolved that constantly scan the DNA for sequence changes and correct them. This process can occur in one of two ways, either base-excision repair or nucleotide-excision repair, although both achieve the same end, which is to remove 5mCs and replace them with unmethylated Cs. This involves an intermediate step in which the cytosine is hydroxymethylated, denoted by 5hmC, and there are multiple enzymes and co-factors required. One is GADD-45 (growth-arrest-and-DNA-damage inducible enzyme). This co-factor is emerging as a potential mediator of demethylation in the brain and will no doubt be the subject of intense investigation (35,36).
In summary, the epigenome of the brain is established by a combination of changes to the histones and the DNA, both of which are mediated enzymatically and which involve regulatory co-factors. Although presented as separate and distinct processes, there is in reality a close relationship between the two processes such that changes to the chromatin are required for DNA methylation and de-methylation and changes to the DNA can recruit enzymes and co-factors that modify the chromatin. The brain appears to be a place that is particularly dynamic in regard to epigenetic modifications, and many firmly established tenets are being rewritten as a result. The ability of epigenetic modifications within neurones, and potentially other cell types of the central nervous system, to transduce temporary signals such as hormonal fluctuations into enduring changes makes them a perfect candidate for mediating the long-term consequences of perinatal hormonally-directed sexual differentiation of the brain.
Sexual differentiation of the brain
The coin of the realm in nature is reproduction: the transference of ones genetic profile to the next generation. In sexually reproducing species, this is achieved by the packaging of an individuals genome in a haploid state into a single cell that can merge with a similarly haploid cell from another individual and generate a new diploid organism that merges the two genomes. Ovulation, spermatogenesis, fertilisation, gestation, parturition and lactation (in mammals) are all essential components of reproduction but none are of significance in the absence of mating between two individuals to allow fusion of haploid cells. The fusion must occur between a male and a female, which are operationally defined by the differential size and characteristics of the haploid cells: large ova in females and multiple small mobile sperm in males. The ova may be shed within the body of the female and fertilised there, as in most reptiles, birds and mammals, or may be expelled by the female and fertilised in the water, as in most fish and amphibians. However, males always expel their gametes because, by definition, it is the ova, or female cell, that will be fertilised. As a result, there is differential investment in the gametes because the consequences of fertilisation can be markedly different for the male versus female parent. An important way this differential investment is managed is behaviour, with males and females having different thresholds for sexual activity and/or receptivity and varying mate selection criteria, and with females generally having a higher receptivity threshold and more stringent mate selection criteria. These parameters are controlled by the brain and, athough influenced by the hormones emanating from the gonads, are essentially independent of peripheral reproductive structures.
Ensuring that brain and behaviour are coordinated with reproductive status is essential for the successful transference of genetic material to the next generation and, in mammals (and likely many other phyla as well), this is achieved via a two-step process referred to as the Organisational/Activational Hypothesis noted above. No longer a hypothesis, this tenet was first codified in a now iconic manuscript published in 1959 that puts forth the notion that androgen production by the developing male testis, embryonically and immediately postnatally, gains access to the brain and organises it to support the expression of male sexual behaviour in adulthood. However, the second phase is the requirement for adult androgen production, which then activates the organised neural substrate. In the absence of either phase of hormone production and action on the brain, male sexual behaviour will not be expressed. By contrast, the brain is organised by default to support the expression of female sexual behaviour but, nonetheless, requires activation in adulthood by ovarian hormones. Because the female organisation pathway, or feminisation, is the default, there is a third process by which this neural capacity for female sexual behaviour is eliminated in males, referred to as defeminisation. This too is an organisational process mediated by testicular hormones, although there is no activational component (37).
Today, we know a considerable amount about the neural substrates of masculinisation and defeminisation; less so for feminisation because, as the default process, it is difficult to gain a handle on, although progress is being made. One of the emerging principles is that, although the same steroid hormones, androgens or oestrogens, may mediate the organisation of many divergent endpoints (i.e. sex behaviour, maternal behaviour, social play behaviour, aggression, anxiety, learning strategies, etc.), in each case, the cellular mediators and endpoints are distinct. Differentiated cellular endpoints include sex differences in synaptogenesis, glial morphology, neurogenesis, glial genesis, apoptosis, cell migration and neurochemical phenotypic differentiation. Cellular mediators are equally varied and include prostaglandins, endocanbinoids, phosphoinositide 3-kinase, nuclear factor-κB and vasopressin, amongst others surely waiting to be discovered (38).
Combination of epigenetics and sexual differentiation
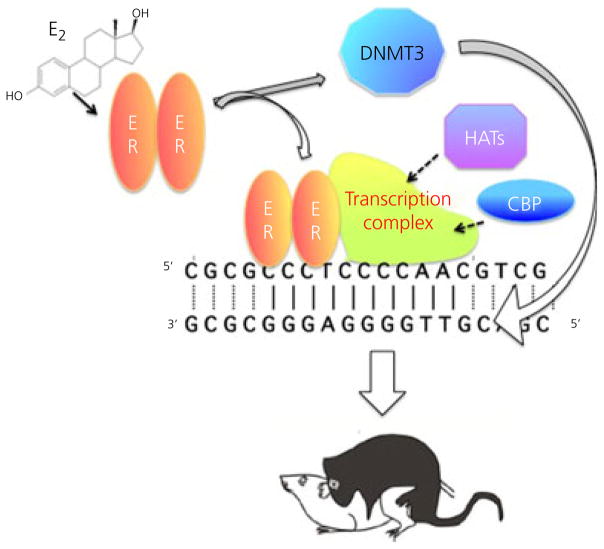
The combination of steroid hormone receptors that directly interact with both histone acetylation-modifying enzymes and the DNA, as well as the enduring nature of organisational hormone actions, makes the potential for epigenetic underpinnings of sexual differentiation appear to be a forgone conclusion (39,40). What are the details? In what way do steroids alter the epigenome? Where in the brain does this occur and what are the consequences? The answers to these questions are only beginning to emerge but it is already apparent that, as with the myriad of mechanisms of hormone action in the brain, there are also a myriad of epigenetic changes and these appear to vary by brain region, functional endpoint and likely by hormone (Fig. 3).
Fig. 3.
Epigenetics and sexual differentiation. Oestradiol (E2) binds to and activates its nuclear transcription factor receptor (ER) which moves to the DNA and recruits a transcriptional complex. Included in this complex are enzymes with histone-acetylating ability to allow access to the DNA. Activated ER may also modify the activity of DNA methyl transferase (DNMT) enzymes and thereby alter the methylation status of the DNA. Taken together, these changes may provide the molecular basis for the organisational effects of early hormone exposure, which endure into adulthood and direct activational responses to sex-typic gonadal steroids. CBP, CREB-binding protein; HATs, histone acetyl transferases.
In taking the first steps towards unravelling the role of epigenetics in brain sexual differentiation, investigators have employed two approaches. The first is discovery-based, in essence taking a survey to determine whether there are changes in epigenetic marks during or as a consequence of sexual differentiation. The second approach is hypothesis-based, aiming to identify a particular sexually differentiated endpoint and then, via pharmacological or other manipulation, determine whether that endpoint is dependent upon epigenetic changes.
In taking the first approach, our laboratory focused on the primary transducers of steroid action: the two isoforms of the oestrogen receptor, ERα and ERβ, and PR (we did not examine AR but are not denying its critical role in sexual differentiation of the brain). Via pyrosequencing, we examined a region of the promoter for each gene in males, females and females treated with a masculinising dose of oestradiol. The advantage of pyrosequencing is that all of the target gene cDNA strands in a given sample are sequenced, resulting in hundreds of thousands of reads and therefore a high degree of accuracy and reliability for even small changes. The disadvantage of pyrosequencing is that only a limited portion of a CpG island can be sequenced on any given read and the primer design is constrained. With these advantages and disadvantages in mind, we compared the degree of methylation in one CpG island of each gene at three different time points; shortly after birth, at the time of weaning, and in full adulthood. We also compared across brain regions, examining the preoptic area (POA) and hypothalamus. Our first observation was a developmental increase in the POA and hypothalamus with respect to methylation of a portion of the ERα promoter located between exons 1 and 2 but upstream from the start site in Exon 2. There was an almost doubling between birth and 3 weeks of age, from approximately 5–10% methylation to 15–20% at most CpG sites (28). This would be predicted to cause a considerable decline in receptor expression between birth and post-natal 20, although there is no evidence that this is the case. Thus, in this instance, methylation marks may reflect past transcriptional history as opposed to future expression potential, or even changes in promoter usage and or post-transcriptional splicing. Interestingly, there were no changes in the percent methylation of the CpG islands assayed for ERβ or PR and, in the case of PR, there is a well established decline in expression with development (41).
In addition to the limited developmental shift in methylation, we also observed hormonal modulation and/or sex differences in the percent methylation of specific CpG residues in each of the genes promoters. What was most striking was the transient nature of those differences, with some disappearing over time and new ones emerging, suggesting a far higher degree of dynamism than previously anticipated (28). Examination of another portion of the ERα promoter found good correlation between methylation levels and ERα expression in the POA of 10-day-old rat pups, such that males had higher methylation and lower ER levels, and masculinisation of females with oestradiol treatment on the day of birth mimicked the profile of males (42). Thus, there are certainly instances where the degree of promoter methylation appears to be reflective of gene expression, although this is not always the case, and most certainly so when considering steroid receptors in the reproductively relevant regions, the POA and hypothalamus.
Although there are sex differences in ER expression in the POA and hypothalamus of the developing brain, they are by and large subtle. A far greater dynamic range of ER expression is found in the cortex where levels are quite high in the very young brain but decline with advancing age. The change in expression levels is neatly paralleled by changes to the methylation status of the promoter of ESR1 (gene for ERα) which increases with age as receptor expression declines. Both males and females show similar patterns of high to low ER expression over the course of development (43). A re-emergence of ERα expression following injury in the adult brain also appears to be the result of a removal of the repressive CpG methylations (44), perhaps as a manifestation of developmental processes being evoked following injury.
Focusing on the MBD genes is another approach for determining how and where epigenetics impacts the establishment of sex differences in the brain. The iconic MBD, MeCP2, is indeed higher in the ventromedial nucleus and amygdala of newborn female rats. These two brain regions are notable for their primary role in the control of female sexual behaviour and a panel of social and emotional behaviours, respectively, many of which differ between males and females. One particular behavior is social play, which is expressed during the juvenile period and occurs at a higher rate and with greater intensity in males in a wide range of species, from laboratory rodents to domesticated pets to wild animals, to humans and nonhuman primates. One of the more striking aspects of the sex difference in social play (also referred to as rough-and-tumble play) is that it peaks during a phase of life when steroid hormones are at their nadir: the juvenile hiatus between the high steroid levels required for the organisational phase of sexual differentiation and the return of steroids at puberty for activation. Thus, any sex differences that are hormonally determined and appear during the juvenile hiatus were ‘organised’ early in development and do not require ‘activation’, a scenario consistent with an epigenetic memory of earlier hormone exposure. Manipulation of MecP2 levels demonstrates that there is indeed an epigenetic programming of social play by hormones, and that the neural mechanism involves changes in vasopressin innervation (45,46). Vasopressin has emerged as a central neuropeptide in a variety of social behaviours, including pair bonding, social recognition and intra-individual aggression (47).
A substantial body of literature highlights the enduring and profound impact of early maternal care on brain development, and that many of these effects are epigenetically mediated (48). However, there has been relatively little attention paid to sex differences in both how the dam cares for her pups and the sensitivity of the pups to variation in maternal care. Edelmann et al. (49) took advantage of the well established fact that rat dams perform more licking and grooming of their male pups than the females. Simulation of maternal grooming of pups with a paintbrush allowed for standardisation of care between males and females and increased the level of methylation in the promoter of the ERα gene and correspondingly decreased expression levels of the receptor in females to that of males (49). This neatly demonstrates a convergence between experiential variables (e.g. maternal care) and endogenous hormones (e.g. oestradiol) to mediate the establishment and perhaps maintenance of sex differences in the brain.
Taken, together these studies demonstrate a role for DNA methylation as a component of epigenetic programming of sexual differentiation. However, changes in the methylation status of DNA are only one way of achieving this goal because the histones are also critical regulatory sites that often work in concert with modifications to the DNA. Hyper-acetylated histones H3 and H4 are associated with activated gene transcription, whereas deacetylation results in repression and the levels of these have been measured in various brain regions, including those known to be subject to sexual differentiation. Tasi et al. (50) examined the activational histone marks H3K9/14ac and H3K9me3 in neonatal mouse pups and, contrary to expectation, there is no sex difference in the POA. However, there are sex differences in the cortex and hippocampus and, although the higher levels of H3K9/14ac in males could be attributed to gonadal hormones, the higher levels of H3K9me3 could not (50), suggesting a role for sex chromosome complement in establishing this particular sex difference, although this is an assertion that remains to be tested.
A central bedrock of brain sexual differentiation is the hormonal modulation of naturally-occurring cell death. Several hypothalamic and POA nuclei are larger in one sex and, in each case, this is a result of more cells being present in one sex as opposed to the density of cell packing. There are three ways to explain the existence of more cells in a brain region: (i) more cells are born there; (ii) more cells migrate into the region; or (iii) more cells die there. For the three nuclei of the POA that have been carefully examined in this regard, the sexually dimorphic nucleus (SDN), the anteroventral periventricular nucleus (AVPV) and the principal subdivision of the bed nucleus of the stria terminalis (pBNST), the dominant variable impacting a sex difference in size is greater cell death in one sex versus another and, in each case, the differential cell death is hormonally mediated (51). In other words, higher brain oestradiol levels promote cell survival in the SDN and pBNST of male rat pups but actually induce cell death in the AVPV. So, although the two sexes are born with the same number of neurones in these brain regions, more cells will live or die as a function of their different hormonal status and thereby sculpt a sex difference in size.
Noting that there is a delay of several days between the height of hormone exposure and changes in cell survival, Forger et al. (52) speculated this may be the result of epigenetic programming and used the second approach to determine epigenetic mechanisms: hypothesis-based interrogation. Valproic acid is a broad band pharmacological inhibitor of HDACs, and treatment of neonates with this agent increased H3 acetylation in the pBNST. Moreover, blocking brain HDAC activity in newborn male rats or females treated with a masculinising dose of testosterone also prevented the neuroprotective effects of both the endogenous (males) and exogenous (females) hormones, resulting in a mature pBNST that was the same size as that seen in control females (i.e. smaller). This is the first demonstration of hormonally-mediated epigenetic programming of a structural change in the brain, and includes changes to vasopressin innervation, not all of which involve the pBNST (52), suggesting it is epigenetic programming of innervation that extends more broadly.
A combination of the two experimental approaches has been used to good effect by Matsuda et al. (53) by interrogating the POA for its histone code at specific gene loci at the same time as using pharmacological inhibition of the HDACs and assessing the impact of neonatal treatment on adult behaviour. Immunoprecipitated chromatin from the POA of embryonic and neonatal males and females was assessed for H3 and H4 acetylation in the dominant brain promoter for ERα (1b) and aromatase (If), as well as the gonadal promoter for aromatase which is active in brain (II). A varied profile of relative acetylation between the two histones, the two ages and the two sexes emerged, again highlighting the dynamic nature of epigenetic modifications in the brain. Treatment of neonates with the HDAC inhibitor trichostatin A infused directly into the brain on the first 2 days of life markedly reduced adult male sexual performance. The HDAC inhibitors available to date are not specific to subtypes but antisense oligonucleotide-mediated inhibition indicated that both HDAC2 and HDAC4 are required for normal masculinisation of behaviour. Further support for this view is the observation that both enzymes bound to the ERα promoter, although only HDAC2 was found to be associated with the aromatase (II) promoter (53).
Conclusions
Sexual differentiation of reproductive behaviour is a two-step process that involves early organisational actions induced by steroids followed by adult activational hormonal effects that manifest the process. The long intervening gap between developmental hormonal exposure, much of which occurs in utero, and the onset of the behaviour that is being regulated suggests there is a form of cellular memory that must be maintained throughout that period. The limited studies reviewed here suggest that the memory is at least in part, epigenetic, and involves changes at both the DNA and the associated histones that constitute the chromatin. However, research is still in its early days and these studies reflect the promise of what is to come rather than comprising a genuine determination of how epigenetic changes impact or maintain sexual differentiation. Many of the observations made to date are not functionally connected to outcome or understood in sufficient depth to reveal the cellular mechanisms at play. Barriers to progress include the enormous heterogeneity in the brain, with neurones of varying neurochemical phenotype and distinct afferent and efferent partners. Moreover, there is no reason to assume that all epigenetic changes in the brain are restricted to neurones given the essential roles for astrocytes (54,55) and, most recently, microglia (56) in the sexual differentiation of male sexual behaviour. A second major barrier is one common to all studies of brain epigenetics: the challenge of obtaining quantitative data that are broadly representative and highly accurate. Studies with sufficient numbers of subjects to provide confidence are limited by the use of sampling procedures that are incomplete and/or have a random component (28). Alternatively, deep sequencing approaches, such as RNA-Seq or Methyl-Seq, are so prohibitive in terms of cost and the amount of data generated that relatively few animals are surveyed. At this time, a trade-off remains with respect to accuracy versus reliability. It is anticipated that these problems will be solved soon, and with them will come major advances in our understanding of how steroids acting during a sensitive period in brain development induce the epigenetic changes that endure for a lifetime.
References
- 1.Phoenix CH, Goy RW, Gerall AA, Young WC. Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology. 1959;65:369–382. doi: 10.1210/endo-65-3-369. [DOI] [PubMed] [Google Scholar]
- 2.Crews D. Epigenetics and its implications for behavioral neuroendocrinology. Fron Neuroendocrinol. 2008;29:344–357. doi: 10.1016/j.yfrne.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan CP, Bale TL. Sex differences in microRNA regulation of gene expression: no smoke, just miRs. Biol Sex Differ. 2012;3:22. doi: 10.1186/2042-6410-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey R, Mukerji M. From ‘JUNK’ to just unexplored noncoding knowledge: the case of transcribed Alus. Brief Funct Genomics. 2011;10:294–311. doi: 10.1093/bfgp/elr029. [DOI] [PubMed] [Google Scholar]
- 5.Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 10.Lomniczi A, Loche A, Castellano JM, Ronnekleiv OK, Bosch M, Kaidar G, Knoll JG, Wright H, Pfeifer GP, Ojeda SR. Epigenetic control of female puberty. Nat Neurosci. 2013;16:281–289. doi: 10.1038/nn.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nature reviews Nat Rev Mol cell biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 13.Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discovery Today. 2009;14:942–948. doi: 10.1016/j.drudis.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Lau OD, Kundu TK, Soccio RE, Ait-Si-Ali S, Khalil EM, Vassilev A, Wolffe AP, Nakatani Y, Roeder RG, Cole PA. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell. 2000;5:589–595. doi: 10.1016/s1097-2765(00)80452-9. [DOI] [PubMed] [Google Scholar]
- 15.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 16.Wang DF, Helquist P, Wiech NL, Wiest O. Toward selective histone deacetylase inhibitor design: homology modeling, docking studies, and molecular dynamics simulations of human class I histone deacetylases. J Med Chem. 2005;48:6936–6947. doi: 10.1021/jm0505011. [DOI] [PubMed] [Google Scholar]
- 17.Kim MY, Hsiao SJ, Kraus WL. A role for coactivators and histone acetylation in estrogen receptor alpha-mediated transcription initiation. EMBO J. 2001;20:6084–6094. doi: 10.1093/emboj/20.21.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cirio MC, Ratnam S, Ding F, Reinhart B, Navara C, Chaillet JR. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev Biol. 2008;8:9. doi: 10.1186/1471-213X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 22.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 23.Honda S, Satomura S, Hayashi S, Imoto I, Nakagawa E, Goto Y, Inazawa J. Concomitant microduplications of MECP2 and ATRX in male patients with severe mental retardation. J Hum Genet. 2012;57:73–77. doi: 10.1038/jhg.2011.131. [DOI] [PubMed] [Google Scholar]
- 24.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buck-Koehntop BA, Martinez-Yamout MA, Dyson HJ, Wright PE. Kaiso uses all three zinc fingers and adjacent sequence motifs for high affinity binding to sequence-specific and methyl-CpG DNA targets. FEBS Lett. 2012;586:734–739. doi: 10.1016/j.febslet.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buck-Koehntop BA, Stanfield RL, Ekiert DC, Martinez-Yamout MA, Dyson HJ, Wilson IA, Wright PE. Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso. Proc Natl Acad Sci USA. 2012;109:15229–15234. doi: 10.1073/pnas.1213726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz JM, Nugent BM, McCarthy MM. Developmental and hormone-induced epigenetic changes to estrogen and progesterone receptor genes in brain are dynamic across the life span. Endocrinology. 2010;151:4871–4881. doi: 10.1210/en.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker-Andresen D, Ratnu VS, Bredy TW. Dynamic DNA methylation: a prime candidate for genomic metaplasticity and behavioral adaptation. Trends Neurosci. 2013;36:3–13. doi: 10.1016/j.tins.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Hodes GE. Sex, stress, and epigenetics: regulation of behavior in animal models of mood disorders. Biol Sex Differ. 2013;4:1. doi: 10.1186/2042-6410-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nugent BM, McCarthy MM. Epigenetic underpinnings of developmental sex differences in the brain. Neuroendocrinology. 2011;93:150–158. doi: 10.1159/000325264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zovkic IB, Guzman-Karlsson MC, Sweatt JD. Epigenetic regulation of memory formation and maintenance. Learn Mem. 2013;20:61–74. doi: 10.1101/lm.026575.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zovkic IB, Sweatt JD. Epigenetic mechanisms in learned fear: implications for PTSD. Neuropsychopharmacology. 2013;38:77–93. doi: 10.1038/npp.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niehrs C, Schafer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012;22:220–227. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology. 2012;37:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic deletion of Gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. J Neurosci. 2012;32:17059–17066. doi: 10.1523/JNEUROSCI.1747-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Todd BJ, Schwarz JM, McCarthy MM. Prostaglandin-E2: a point of divergence in estradiol-mediated sexual differentiation. Horm Behav. 2005;48:512–521. doi: 10.1016/j.yhbeh.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy MM, De Vries G, Forger N. Sexual differentiation of the brain: mode, mechanisms and meaning. In: Pfaff D, Arnold AP, Etgen AM, Fahrbach SE, Rubin RT, editors. Hormones, Brain and Behavior. San Diego, CA: Academic Press; 2009. pp. 1707–1744. [Google Scholar]
- 39.McCarthy MM, Auger AP, Bale TL, De Vries GJ, Dunn GA, Forger NG, Murray EK, Nugent BM, Schwarz JM, Wilson ME. The epigenetics of sex differences in the brain. J Neurosci. 2009;29:12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menger Y, Bettscheider M, Murgatroyd C, Spengler D. Sex differences in brain epigenetics. Epigenomics. 2010;2:807–821. doi: 10.2217/epi.10.60. [DOI] [PubMed] [Google Scholar]
- 41.Wagner CK. The many faces of progesterone: a role in adult and developing male brain. Front Neuroendocrinol. 2006;27:340–359. doi: 10.1016/j.yfrne.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Kurian JR, Olesen KM, Auger AP. Sex differences in epigenetic regulation of the estrogen receptor-alpha promoter within the developing preoptic area. Endocrinology. 2010;151:2297–2305. doi: 10.1210/en.2009-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westberry JM, Trout AL, Wilson ME. Epigenetic regulation of estrogen receptor alpha gene expression in the mouse cortex during early postnatal development. Endocrinology. 2010;151:731–740. doi: 10.1210/en.2009-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westberry JM, Prewitt AK, Wilson ME. Epigenetic regulation of the estrogen receptor alpha promoter in the cerebral cortex following ischemia in male and female rats. Neuroscience. 2008;152:982–989. doi: 10.1016/j.neuroscience.2008.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auger AP, Jessen HM, Edelmann MN. Epigenetic organization of brain sex differences and juvenile social play behavior. Horm Behav. 2010;59:658–663. doi: 10.1016/j.yhbeh.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurian JR, Olesen KM, Auger AP. Mecp2 organizes juvenile social behavior in a sex-specific manner. J Neurosci. 2008;28:7137–7142. doi: 10.1523/JNEUROSCI.1345-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Vries GJ. Sex differences in vasopressin and oxytocin innervation of the brain. Prog Brain Res. 2008;170:17–27. doi: 10.1016/S0079-6123(08)00402-0. [DOI] [PubMed] [Google Scholar]
- 48.Curley JP, Mashoodh R, Champagne FA. Epigenetics and the origins of paternal effects. Horm Behav. 2011;59:306–314. doi: 10.1016/j.yhbeh.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edelmann MN, Auger AP. Epigenetic impact of simulated maternal grooming on estrogen receptor alpha within the developing amygdala. Brain Behav Immun. 2011;25:1299–1304. doi: 10.1016/j.bbi.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai HW, Grant PA, Rissman EF. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 2009;4:47–53. doi: 10.4161/epi.4.1.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCarthy MM. Estradiol and the developing brain. Physiol Rev. 2008;88:91–124. doi: 10.1152/physrev.00010.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray EK, Hien A, de Vries GJ, Forger NG. Epigenetic control of sexual differentiation of the bed nucleus of the stria terminalis. Endocrinology. 2009;150:4241–4247. doi: 10.1210/en.2009-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsuda KI, Mori H, Nugent BM, Pfaff DW, McCarthy MM, Kawata M. Histone deacetylation during brain development is essential for permanent masculinization of sexual behavior. Endocrinology. 2011;152:2760–2767. doi: 10.1210/en.2011-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mong JA, Kurzweil RL, Davis AM, Rocca MS, McCarthy MM. Evidence for sexual differentiation of glia in rat brain. Horm Behav. 1996;30:553–562. doi: 10.1006/hbeh.1996.0058. [DOI] [PubMed] [Google Scholar]
- 55.Mong JA, McCarthy MM. Steroid-induced developmental plasticity in hypothalamic astrocytes: implications for synaptic patterning. J Neurobiol. 1999;40:602–619. doi: 10.1002/(sici)1097-4695(19990915)40:4<602::aid-neu14>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 56.Lenz KM, Nugent BM, Haliyur R, McCarthy MM. Microglia are essential to masculinization of brain and behavior. J Neurosci. 2013;33:2761–2772. doi: 10.1523/JNEUROSCI.1268-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]