Abstract
We describe the development and in-depth analysis of T4 DNA ligase-catalyzed DNA templated oligonucleotide polymerization toward the generation of diversely functionalized nucleic acid polymers. We demonstrate that the NNNNT codon set enables low codon bias, high fidelity, and high efficiency for the polymerization of ANNNN libraries comprising various functional groups. The robustness of the method was highlighted in the copolymerization of a 256-membered ANNNN library comprising 16 sub-libraries modified with different functional groups. This enabled the generation of diversely functionalized synthetic nucleic acid polymer libraries with 93.8% fidelity. This process should find ready application in DNA nanotechnology, DNA computing, and in vitro evolution of functional nucleic acid polymers.
Keywords: nucleic acids, molecular evolution, modified DNA, ligase, sequencing
Graphical abstract
Level playing field: A method to enzymatically generate diversely functionalized synthetic nucleic acid polymer libraries with chemical complexity similar to proteinogenic polymers is reported. The DNA-templated process enables the efficient sequence defined incorporation of 16 customizable functional groups in a modular codon library format at 93.8% fidelity and with low sequence bias.
Amongst nature’s sequence-defined biopolymers, the most diverse roles and functions are assigned to proteins. The hegemony of proteinogenic biopolymers as biological receptors and catalysts arises from their broad structural and chemical diversity, which enables them to effectively engage their molecular targets with high affinity and specificity.[1] Despite the ability of nucleic acid polymers to fold into complex three-dimensional structures, their functional group deficit has limited their ability to match the diverse activities of proteins.[2] Thus, technologies that enable the sequence-defined synthesis of nucleic acid polymers with diverse chemical functionality has received significant attention.[3] Indeed, DNA and RNA polymerases have been used to achieve the DNA-templated incorporation of modified nucleotides to facilitate the evolution of functionalized nucleic acid polymers with significantly improved activities over their unmodified counterparts.[4] Homomultivalent display of a hydrophobic functional group has been demonstrated to greatly increase the binding affinity and lower kinetic off-rates of nucleic acid aptamers raised against various protein targets.[5] Furthermore, the homomultivalent display of various functional groups has increased the catalytic potential of nucleic acid polymers for ribonuclease activity[6] and protease activity.[7] Expanding DNA into a heteromultivalent polymer comprising two or three functional groups has enabled the evolution of divalent metal-independent nuclease activity, thus opening the door to their application in vivo.[8]
Methods that permit the sequence-defined incorporation of a diverse library of functional groups along a library of ssDNA will allow the concomitant in vitro evolution of both the DNA architecture and the identities of the displayed functional groups. Therefore, the spatial optimization of the heteromultivalent ensemble of weak interactions between the functional groups and their molecular target can be achieved.[9] Current polymerase-based approaches using a standard genetic code are limited to generating heteromultivalent nucleic acid polymers comprising up to only four types of modifications.[10] As nature endows proteinogenic polymers with a much broader chemical repertoire to enable their function, expanding nucleic acid polymers beyond this limitation could greatly expand the functional activity of this class of polymer beyond their canonical form.
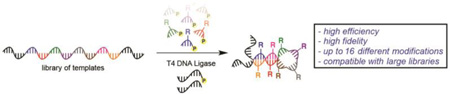
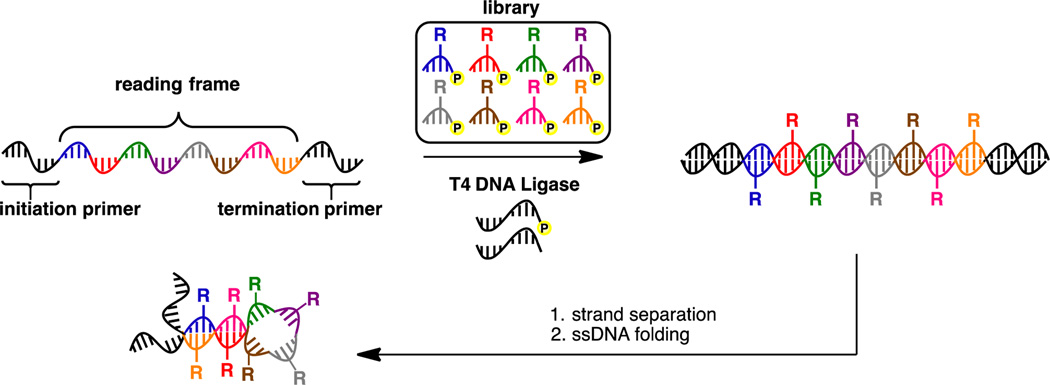
Our group has explored enzymatic methods to incorporate multiple instances of a diverse set of functional groups throughout a DNA polymer. Recently, we developed the Ligase-catalyzed OligOnucleotide PolymERization (LOOPER) method to access this class of sequence-defined synthetic biopolymers (Figure 1).[11] The method relies upon the T4 DNA ligase-catalyzed DNA-templated copolymerization of a library of modified 5’-phosphorylated pentanucleotides. As the method employs codons, rather than single nucleotides, the theoretical maximum number of unique modifications that can be incorporated for a pentanucleotide system is 1024.
Figure 1.
Synthesis of modified ssDNA polymers using LOOPER.
We recently developed a DNA duplex sequencing method to analyze the fidelity of LOOPER when polymerizing pentanucleotide libraries of various nucleic acid composition along large libraries of DNA templates.[12] The fidelity of the process was calculated as the percentage of pentanucleotides that were incorporated across from their cognate codon. Our initial analysis revealed that a reading frame comprising the NTNNN codon set generated the highest level of fidelity for pentanucleotide incorporation with 98.1% when using amine-modified building blocks (Table 1, entry 1). As this codon set comprises 256 unique codons, we sought to apply this set to the simultaneous incorporation of different modifications, with the ultimate goal of using a differentially modified pentanucleotide library toward the generation of heteromultivalent ssDNAs with diverse chemical functionality. However, amine-modified NNNAN library 1 (Figure 2) was found to polymerize with only modest efficiency, giving approximately 30% yield of full-length products. As attempts to improve the polymerization efficiency were unsuccessful, this prompted us to analyze the codon bias of the polymerization process as incomplete polymerization could potentially result in skewing of the codon distribution, which could convolute in vitro evolution efforts of this class of biopolymer. We defined the bias of the system by calculating the standard deviation of the enrichment of every codon within the codon set; whereby enrichment was calculated as the frequency of a codon in the polymer divided by the frequency of the same codon in the template.
Table 1.
Characterization of LOOPER with different pentanucleotide libraries[a]
| Entry | Pentanucleotide | Codon set | Yield[b] | Bias[c] | Fidelity[d] |
|---|---|---|---|---|---|
| 1 | 1 | NTNNN | 30% | 0.91 | 98.1% |
| 2 | 2 | NNNNT | 84% | 0.16 | 86.7% |
| 3 | 3 | NNNNT | 68% | 0.21 | 95.1% |
| 4 | 8 | NNNNT | 60% | 0.37 | 93.2% |
| 5 | 6 | NNNNT | 66% | 0.36 | 91.3% |
| 6 | 4 | NNNNT | 73% | 0.34 | 93.9% |
| 7 | 7 | NNNNT | 62% | 0.25 | 95.1% |
| 8 | 5 | NNNNT | 63% | 0.36 | 94.0% |
LOOPER and duplex barcoding process performed on 15 pmol of DNA template library. 35 amol of product was subjected to duplex DNA sequencing.
Determined by densitometry of full-length product by polyacrylamide gel electrophoresis.
Codon bias was calculated as σ(freq. polymer/freq. template).
Calculated for pentanucleotide incorporation.
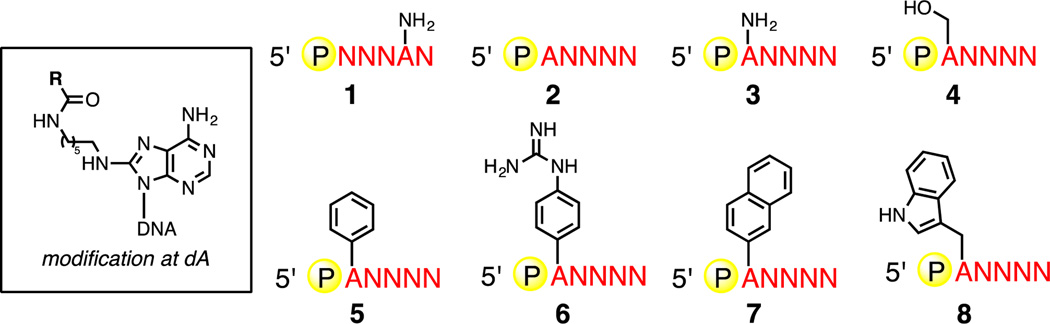
Figure 2.
Homofunctionalized 256-membered ANNNN libraries used to examine the influence of nucleobase modification on the fidelity of LOOPER. Inset: molecular architecture of functional group attachment site on dA.
Unfortunately, significant codon bias was observed for the polymerization of amine-modified NNNAN libraries along NTNNN codon sets (Figure 3a). To test whether the low polymerization yield resulted in higher levels of codon bias, we compared the bias of the NTNNN codon set against that of the NNNNT codon set, which had greater than 2-fold increase in yield, albeit with somewhat lower fidelity (Table 1, entry 3). The sequencing analysis revealed that the NNNNT codon set resulted in a significantly decreased codon bias in comparison to the NTNNN codon set (Figure 3a).
Figure 3.
Analysis of codon sets used during LOOPER. a) Codon bias observed for the NTNNN and the NNNNT codon sets during LOOPER with corresponding hexylamine-modified pentanucleotide libraries. b) DNA sequence logos for misincorporation at NTAGT codons within the NNNNT codon set. c) Error rate of different homofunctionalized ANNNN libraries parsed by 3’-nucleotide identity.
We concluded that codon sets which place the modified nucleobase in close proximity to the 3’-OH of the pentanucleotide resulted in decreased kinetics of phosphodiester bond formation by T4 DNA ligase and decreased efficiency of the polymerization process. Due to the higher yield and lower codon bias of the NNNNT codon set, we further pursued the optimization of this set for polymerization. Thus, we first surveyed temperature and ATP concentration to determine the optimal conditions with respect to yield, fidelity, and codon bias. We found that temperatures ranging from 10 °C to 30 °C, which are within the ideal range for T4 DNA ligase activity,[13] had no observable effect on fidelity (Table S1). This is most probably due to these temperatures being well above the Tm of the pentanucleotide libraries. The lowest sequence bias was observed at 25 °C, which correlated with polymerization efficiency (Table S1). ATP concentrations did influence the fidelity and efficiency of LOOPER. 1 mM ATP resulted in an error rate of 4.7%, which gradually increased to 5.9% with decreasing ATP concentrations to 25 µM (Table S2). However, 25 µM ATP was superior with respect to polymerization yield and codon bias (Table S2). Increased efficiency of polymerization at lower ATP concentrations are likely the result of decreased inhibition of DNA binding[14] and the minimization of an over-adenylated system.[11b]
In order to study the capacity of LOOPER to tolerate various functional groups on the pentanucleotide, we prepared several 5’-phosphorylated ANNNN pentanucleotide libraries containing different functional groups (Figure 2). The modifications were incorporated through amide bond-formation between the hexylamine appended to the C8 position of adenosine and various carboxylic acid derivatives (Figure 2, inset). This modification site has been previously shown to be the most permissive for this system.[11b] Duplex DNA sequencing was performed on the products of LOOPER for each monofunctionalized library, and the yields, biases, and fidelities for each library are reported in Table 1. Importantly, all monofunctionalized libraries polymerized with good yields and fidelities. On the whole, polymerization fidelities were not governed by the molecular size of the functional group modification. This is not surprising, as pentanucleotides modified with peptide fragments have been show to polymerize efficiency and with high fidelity.[11b] While codon biases were greater than either the unmodified or amine-modified ANNNN libraries, the biases remained considerably lower than the amine-modified NNNAN library, suggesting that this codon set could be applied to in vitro selection systems where strong levels of codon bias would be problematic, such as selections operating with poor enrichment levels.
Further analysis of the NNNNT codon sets revealed several trends with respect to codon sequence and fidelity. We analyzed the consensus sequence for the misincorporations of each codon during LOOPER with amine-modified and phenyl-modified ANNNN libraries (Figure S1 and S2). A representative example of the four sequence logos generated for the NTAGT codon set is shown in Figure 3b. One of the most striking trends is that single-nucleotide errors arising from misincorporation most frequently occur at the first two nucleotides at the 5’-end of the codon (3’-end of the pentanucleotide building block). The only exception is when the 5’-nucleotide of the codon is a dC, which is largely conserved during misincorporations. These trends are witnessed across the entire 256-membered codon set (Figure S1 and S2). These data are consistent with our observation that the rate of LOOPER is faster when extending from the 3’-OH of a primer, rather than the 5’-phosphate (Figure S3). This extension preference results in a primarily unidirectional polymerization process. Thus, errors are most likely to occur distal to the site of ligation, namely the 3’-end of the incoming pentanucleotide. The different monofunctionlized ANNNN libraries were also parsed by nucleotide identity at each position to determine if specific nucleotides influenced the fidelity of incorporation. Indeed, we observed higher error rates for pentanucleotide building blocks that contained either a dA or dT at the 3’-position. Other positions were less sensitive to the nucleotide identity (Figure S4). It is possible that a 128-membered codon set lacking a dA or dT at the 5’-position, namely SNNNT, could result in higher fidelity, albeit at the cost of sequence diversity. A somewhat similar approach has been used in the successful molecular evolution of TNA aptamers with a reduced genetic code.[15] Alternatively, noncanonical bases such as 2,6-diaminopurines, could be used in place of dA to modulate the fidelity.[16]
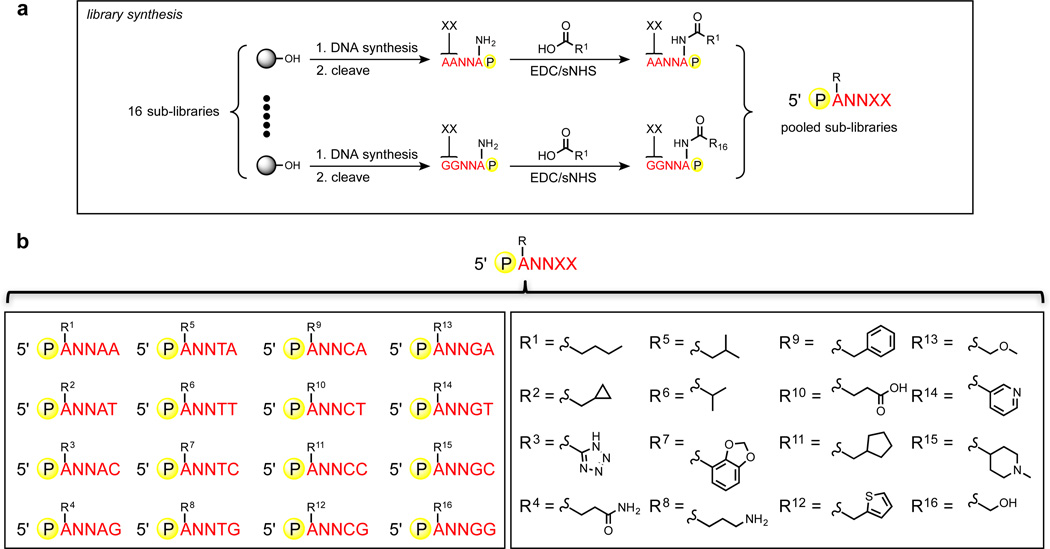
Inspired by the trinucleotide code used during ribosomal translation of mRNA into proteins, we sought to organize the 256-membered NNNNT codon set into degenerate sub-libraries, where each sub-library would encode a unique modification. Thus, evolution of the DNA scaffold and functional group can occur concomitantly. Namely, errors that arise during LOOPER could either result in synonymous mutations that change the ssDNA scaffold, while maintaining the displayed functional group identity, or missense mutations that concomitantly change the identity of the displayed functional group and the ssDNA scaffold. We explored the use of a codon set derived from XXNNT, where XX represents a dinucleotide sequence that specifies the functional group modification on the pentanucleotide. Thus, from the 256-membered codon set, 16 sub-libraries can be generated for the sequence-specific incorporation of 16 unique modifications along a ssDNA scaffold – a chemical complexity similar to that seen in proteins. As most errors from misincorporation occur at the two nucleotide positions at the 5’-end of the codon, this codon set would have a higher level of missense mutations than other codon sets, such as the NNXXT codon set, which would be more prone to synonymous mutations. Using the data garnered from the homofunctionalized ANNNN libraries, along with lessons learnt from homofunctionalized and heterofunctionalized aptamer and nucleic acid enzyme selections,[4] we selected 16 functional groups and synthesized the corresponding 16 sub-libraries (Figure 4). The functional group set comprised various Brønsted acids and bases, as well as a broad representation of polar and hydrophobic groups to enable the biopolymer to engage in molecular recognition and catalysis.
Figure 4.
Assembly of the 256-membered heterofunctionalized pentanucleotide library for the generation of heteromultivalent DNA by LOOPER. a) Synthesis of each 16 ANNXX libraries. b) Assignment of the 16×16 library.
The 16 sub-libraries were mixed in equimolar ratios to generate the 256-membered heterofunctionalized pentanucleotide library, which was used in LOOPER with a library of templates comprising eight repeats of the NNNNT codon set within the reading frame. The full-length product was isolated in 59% yield, and was subjected to duplex DNA sequencing.[12] The aggregated fidelity of the polymerization was determined to be 93.8% (table 2, entry 1), which is comparable to the level of fidelity observed with the homofunctionalized libraries, suggesting that this method could be applied to the generation of diversely functionalized ssDNA and dsDNA. The bias of the codon set was somewhat increased over the homofunctionlized libraries, which prompted us to evaluate each separate 16-membered sub-library within the 256-membered pentanucleotide library. The enrichments, biases, and fidelities of each codon sub-library are summarized in table 2. As anticipated, there is variability amongst sub-libraries, and indeed, variability within each sub-library (see Table S3). The lowest fidelity sub-library was from the 5’P-ANNAC library modified with tetrazole (table 2, entry 6), while the highest fidelity sub-library was from the 5’P-ANNTG modified with the hexylamine (table 2, entry 13). As a statistically significant trend, codons that were enriched in the polymerization were also higher fidelity (Figure S5). As the polymerization could not be pushed to completion, this is likely due to the fact that higher GC content pentanucleotides polymerize more efficiently during LOOPER, and that higher GC content has consistently led to higher fidelities of incorporation in pentanucleotide libraries (Figure S6)
Table 2.
Fidelity of LOOPER with a heterofunctionalized library[a]
| Entry | Pentanucleotide | R group[b] | Enrich[c] | Bias[d] | Fidelity[e] |
|---|---|---|---|---|---|
| 1 | 5’P-A*NNNN | R1-R16 | 0.99 | 0.41 | 93.8% |
| 2 | 5’P-A*NNAA | 0.71 | 0.75 | 93.9% | |
| 3 | 5’P-A*NNCA | 1.01 | 1.03 | 93.8% | |
| 4 | 5’P-A*NNGA | 1.04 | 1.07 | 94.5% | |
| 5 | 5’P-A*NNTA | 0.66 | 0.69 | 89.3% | |
| 6 | 5’P-A*NNAC | 0.91 | 0.94 | 87.3% | |
| 7 | 5’P-A*NNCC | 1.48 | 1.49 | 96.6% | |
| 8 | 5’P-A*NNGC | 1.70 | 1.71 | 96.6% | |
| 9 | 5’P-A*NNTC |  |
0.78 | 0.78 | 91.4% |
| 10 | 5’P-A*NNAG | 1.03 | 1.08 | 93.9% | |
| 11 | 5’P-A*NNCG | 1.18 | 1.20 | 96.2% | |
| 12 | 5’P-A*NNGG | 1.46 | 1.50 | 97.4% | |
| 13 | 5’P-A*NNTG | 1.41 | 1.46 | 97.5% | |
| 14 | 5’P-A*NNAT | 0.74 | 0.77 | 89.2% | |
| 15 | 5’P-A*NNCT | 0.69 | 0.70 | 93.0% | |
| 16 | 5’P-A*NNGT | 1.09 | 1.12 | 94.8% | |
| 17 | 5’P-A*NNTT | 0.69 | 0.71 | 89.5% |
LOOPER and duplex barcoding process performed on 15 pmol of DNA template library. 35 amol of product was subjected to duplex DNA sequencing.
Functional group derived from carboxylic acid used in coupling to C6 amino dA.
Enrichment calculated as (freq. polymer/freq. template)
Codon bias was calculated as σ(freq. polymer/freq. template).
Calculated for pentanucleotide incorporation.
In summary, our findings reported herein demonstrate that the LOOPER process represents a viable approach toward the library generation of sequence-defined nucleic acid polymers with diverse chemical functionality. We show that LOOPER can operate with high fidelity and low sequence bias along an NNNNT codon set, enabling the sequence-defined copolymerization of a 256-membered ANNNN pentanucleotide library. Furthermore, we demonstrate that this library can be divided into 16 sub-libraries, each comprising 16 pentanucleotides adorned with a unique functional group. This heterofunctionalized pentanucleotide library was shown to polymerize with excellent fidelity and minimal bias. The high fidelity of the process and customizability of the functional groups gives promise to its application to modified nucleic acid polymer synthesis for DNA nanotechnology;[17] in vitro selection of nucleic acid polymers for desired molecular function;[4] and DNA computing.[18] Efforts are currently being directed toward the study of how LOOPER can enable the molecular evolution of modified nucleic acid polymers.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R21CA207711) and the Office for the Vice President of Research, University of Georgia. We would like to thank Dr. Saravanaraj Ayyampalayam for valuable help with the analysis of DNA sequencing data, the Georgia Genomics Facility for DNA sequencing, and the PAMS core facility at the University of Georgia for their help in the characterization of oligonucleotides.
References
- 1.Cech TR. Cell. 2009;136:599–602. doi: 10.1016/j.cell.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Wilson DS, Szostak JW. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 3.a) Badi N, Lutz JF. Chem Soc Rev. 2009;38:3383–3390. doi: 10.1039/b806413j. [DOI] [PubMed] [Google Scholar]; b) Lutz JF, Ouchi M, Liu DR, Sawamoto M. Science. 2013;341:1238149. doi: 10.1126/science.1238149. [DOI] [PubMed] [Google Scholar]
- 4.a) Meek KN, Rangel AE, Heemstra JM. Methods. 2016 doi: 10.1016/j.ymeth.2016.03.008. [DOI] [PubMed] [Google Scholar]; b) Kong D, Yeung W, Hili R. ACS Comb Sci. 2016;18:355–370. doi: 10.1021/acscombsci.6b00059. [DOI] [PubMed] [Google Scholar]
- 5.a) Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, Janjic N. Mol Ther Nucleic Acids. 2014;3:e201. doi: 10.1038/mtna.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Davies DR, Gelinas AD, Zhang C, Rohloff JC, Carter JD, O'Connell D, Waugh SM, Wolk SK, Mayfield WS, Burgin AB, Edwards TE, Stewart LJ, Gold L, Janjic N, Jarvis TC. Proc Natl Acad Sci U S A. 2012;109:19971–19976. doi: 10.1073/pnas.1213933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF. J Am Chem Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- 7.Zhou C, Avins JL, Klauser PC, Brandsen BM, Lee Y, Silverman SK. J Am Chem Soc. 2016;138:2106–2109. doi: 10.1021/jacs.5b12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Perrin DM, Garestier T, Helene C. J Am Chem Soc. 2001;123:1556–1563. doi: 10.1021/ja003290s. [DOI] [PubMed] [Google Scholar]; b) Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. Nucleic Acids Res. 2009;37:1638–1649. doi: 10.1093/nar/gkn1070. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hollenstein M, Hipolito CJ, Lam CH, Perrin DM. ACS Comb Sci. 2013;15:174–182. doi: 10.1021/co3001378. [DOI] [PubMed] [Google Scholar]; d) Thomas JM, Yoon JK, Perrin DM. J Am Chem Soc. 2009;131:5648–5658. doi: 10.1021/ja900125n. [DOI] [PubMed] [Google Scholar]
- 9.Mammen M, Choi S-K, Whitesides GM. Angewandte Chemie International Edition. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 10.Jager S, Rasched G, Kornreich-Leshem H, Engeser M, Thum O, Famulok M. J Am Chem Soc. 2005;127:15071–15082. doi: 10.1021/ja051725b. [DOI] [PubMed] [Google Scholar]
- 11.a) Hili R, Niu J, Liu DR. J Am Chem Soc. 2013;135:98–101. doi: 10.1021/ja311331m. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Guo C, Watkins CP, Hili R. J Am Chem Soc. 2015;137:11191–11196. doi: 10.1021/jacs.5b07675. [DOI] [PubMed] [Google Scholar]; c) Lei Y, Kong D, Hili R. ACS Comb. Sci. 2015;17:716–721. doi: 10.1021/acscombsci.5b00119. [DOI] [PubMed] [Google Scholar]
- 12.Lei Y, Kong D, Hili R. ACS Comb Sci. 2015;17:716–721. doi: 10.1021/acscombsci.5b00119. [DOI] [PubMed] [Google Scholar]
- 13.Wu DY, Wallace RB. Gene. 1989;76:245–254. doi: 10.1016/0378-1119(89)90165-0. [DOI] [PubMed] [Google Scholar]
- 14.Cherepanov AV, de Vries S. Eur J Biochem. 2003;270:4315–4325. doi: 10.1046/j.1432-1033.2003.03824.x. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Zhang S, Chaput JC. Nat Chem. 2012;4:183–187. doi: 10.1038/nchem.1241. [DOI] [PubMed] [Google Scholar]
- 16.Wu XL, Delgado G, Krishnamurthy R, Eschenmoser A. Org Lett. 2002;4:1283–1286. doi: 10.1021/ol020016p. [DOI] [PubMed] [Google Scholar]
- 17.a) Wang F, Lu CH, Willner I. Chem Rev. 2014;114:2881–2941. doi: 10.1021/cr400354z. [DOI] [PubMed] [Google Scholar]; b) Pinheiro AV, Han D, Shih WM, Yan H. Nat Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) McLaughlin CK, Hamblin GD, Sleiman HF. Chem Soc Rev. 2011;40:5647–5656. doi: 10.1039/c1cs15253j. [DOI] [PubMed] [Google Scholar]
- 18.Benenson Y. Nat Rev Genet. 2012;13:455–468. doi: 10.1038/nrg3197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.