Rare oncogenic aberrations, such as the ALK fusion and RRAS mutation, may drive pancreatic carcinogenesis independent of the KRAS mutation.
Keywords: ALK, RRAS, KRAS, Pancreatic adenocarcinoma, Gene fusion
Abstract
Purpose.
Oncogenic mutations in the KRAS gene are a well‐known driver event, occurring in >95% of pancreatic cancers. The objective of this study was to identify driver oncogene aberrations in pancreatic cancers without the KRAS mutation.
Methods.
Whole‐exome and transcriptome sequencing was performed on four cases of KRAS mutation‐negative pancreatic ductal adenocarcinoma, which were identified in a cohort of 100 cases.
Results.
One case harbored an oncogenic DCTN1‐ALK fusion. The fusion gene enabled interleukin‐3‐independent growth of Ba/F3 cells and rendered them susceptible to the anaplastic lymphoma kinase tyrosine kinase inhibitors crizotinib and alectinib. The structure of the breakpoint junction indicated that the fusion was generated by nonhomologous end joining between a segment of DCTN1 exon DNA and a segment of ALK intron DNA, resulting in the generation of a cryptic splicing site. Another case harbored an oncogenic RRAS mutation that activated the GTPase of the RRAS protein.
Conclusion.
Rare oncogenic aberrations, such as the ALK fusion and RRAS mutation, may drive pancreatic carcinogenesis independent of the KRAS mutation.
Implications for Practice.
The oncogenic DCTN1‐ALK fusion and the RRAS mutation were associated with the development of pancreatic ductal adenocarcinoma (PDAC) in the absence of the KRAS mutation. Constitutional activation of DCTN1‐ALK fusion protein was suppressed by the anaplastic lymphoma kinase tyrosine kinase inhibitors crizotinib and alectinib. Thus, a small subset of PDAC patients might benefit from therapy using these inhibitors.
摘要
目的. KRAS基因致癌性突变是众所周知的胰腺癌驱动事件, 95%以上的胰腺癌病例均携带这一突变。本研究的目的是在不携带KRAS突变的胰腺癌患者中鉴定驱动致癌基因畸变。
方法. 从一个含100例病例的队列中鉴别出了4例KRAS突变阴性胰腺导管腺癌病例, 随后对其进行全外显子组和转录组测序。
结果. 1例病例携带致癌性DCTN1‐ALK融合。该融合基因可使Ba/F3细胞的生长不依赖于白细胞介素‐3, 从而使其对间变性淋巴瘤激酶酪氨酸激酶抑制剂克唑替尼和Alectinib敏感。断点连接处的结构表明, DCTN1外显子DNA片段与ALK内含子DNA片段的非同源末端彼此连接后产生了一个隐蔽的剪接位点, 从而引起基因融合。另一病例携带致癌性RRAS突变, 该突变激活了RRAS蛋白的GTP酶。
结论. 诸如ALK融合和RRAS突变等罕见的致癌性畸变可能引发不依赖于KRAS突变的胰腺癌变。 The Oncologist 2017;22:158–164
对临床实践的提示:致癌性DCTN1‐ALK融合和RRAS突变可在不伴KRAS突变的情况下引发胰腺导管腺癌(PADC)。间变性淋巴瘤激酶酪氨酸激酶抑制剂克唑替尼和Alectinib可抑制DCTN1‐ALK融合蛋白的结构性活化。因此, 少部分PDAC患者可从上述抑制剂治疗中获益。
Introduction
Pancreatic adenocarcinoma is one of the deadliest malignancies; indeed, it is the seventh leading cause of cancer‐related death worldwide [1]. Approximately 80% of patients are diagnosed with unresectable disease [2]; therefore, efficient chemotherapeutic methods, including molecular targeting, are being sought on the basis of examination of pancreatic cancer genomes. Oncogenic mutations in the KRAS gene are a well‐known driver event in >95% of pancreatic cancers [3]. Despite the low druggability of the KRAS protein, several studies indicate that oncogenic KRAS mutants are a promising therapeutic target; thus, treatments may include suppression of enzyme activity by mutation‐specific inhibitors [4], or targeting vulnerabilities in cancer cells harboring a KRAS mutation [5]. In addition, several genome‐wide sequencing studies identified additional genetic aberrations that occur alongside the KRAS mutation in pancreatic cancer genomes and proposed new therapeutic approaches based on these data [6], [7], [8], [9], [10], [11].
However, it is unclear whether oncogene aberrations drive KRAS mutation‐negative pancreatic cancer, which comprises approximately 5% of all cases. Here, we performed whole‐exome and transcriptome sequencing on four cases of pancreatic ductal adenocarcinoma (PDAC) in which the absence of a KRAS mutation was revealed by a targeted deep‐sequencing analysis approach with high depth coverage. The results indicate that ALK fusion and RRAS mutation function as rare oncogenic aberrations that possibly drive pancreatic carcinogenesis independent of the KRAS mutation.
Materials and Methods
Samples
One hundred pancreatic cancer patients who underwent pancreatectomy with curative resection at the National Cancer Center Hospital in Tokyo, Japan, between March 2005 and June 2012 were enrolled. All the tumors were examined pathologically and classified as PDAC, but not as duodenal, biliary, or ampullary carcinoma, by using the World Health Organization classification [12] and the UICC TNM classification [13]. The tumors included 98 conventional adenocarcinomas and two adenosquamous carcinomas (supplemental online Table 1). Fresh frozen cancer and noncancerous tissue samples from the patients were obtained from the National Cancer Center Biobank, which collected tissue samples after receiving written informed consent. Pathologists then macroscopically confirmed the presence of tumor cells in all tumor tissue samples. The Ethics Committee of the National Cancer Center approved the study protocol.
Selection of KRAS Mutation‐Negative Cases
Genomic DNAs from cancer tissues taken from the 100 cases were extracted by using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany, https://www.qiagen.com). Ten nanograms of genomic DNA was then subjected to targeted deep‐sequencing analysis by using an Ion Ampliseq Cancer Hotspot Panel v2 system (Thermo Fisher Scientific Life Sciences, Waltham, MA, http://www.thermofisher.com), which examined 190 hot spots in 50 cancer‐related genes, and an Ion Proton sequencer (Thermo Fisher Scientific Life Sciences). KRAS and other mutations were detected by using Torrent Suite software (Thermo Fisher Scientific Life Sciences), and the lack of mutations in hot spot regions of KRAS in four negative cases was further validated by viewing the sequencing reads using the Integrative Genomics Viewer (IGV), a high‐performance visualization tool for next‐generation sequencer datasets (Broad Institute, Cambridge, MA, https://www.broadinstitute.org/igv/).
Whole‐Exome Sequencing
Exome sequencing was conducted by using 2.5 μg of cancerous and noncancerous DNA isolated from snap‐frozen tissues taken from the four KRAS mutation‐negative patients. Exome capture was performed by using the SureSelect Human All Exon V5 (Agilent Technologies, Santa Clara, CA, http://www.agilent.com), according to the manufacturer's instructions. Exome sequencing was performed on the HiSeq 2000 platform by using 75 bp paired‐end reads (Illumina, San Diego, CA, http://www.illumina.com/). Basic alignment and sequence quality control were undertaken by using the Picard and Firehose pipelines. The reads were aligned against the reference human genome from UCSC human genome 19 (Hg19) using the Burrows‐Wheeler Aligner Multi‐Vision software package (http://bio-bwa.sourceforge.net). Because duplicate reads were generated during the polymerase chain reaction (PCR) amplification process, paired‐end reads that aligned at the same genomic positions were removed by using SAMtools (http://samtools.sourceforge.net/). Somatic single nucleotide variants (SNVs) were called by the MuTect program (Broad Institute), which applies a Bayesian classifier to allow the detection of somatic mutations with a low allele frequency [14]. Somatic insertion/deletion (InDel) mutations were called by the GATK Somatic IndelDetector (http://archive.broadinstitute.org/cancer/cga/indelocator). SNV and InDel detection was supported by visual examination using IGV software.
Whole‐Transcriptome Sequencing
The TruSeq RNA Sample Prep Kit (Illumina) was used to prepare RNA sequencing libraries from 1 µg of total RNA. The resultant libraries were subjected to paired‐end sequencing of 75‐base pair reads on a HiSeq 2000 system (Illumina). Fusion transcripts were detected by using the TopHat‐Fusion algorithm [15]. Expression of the DCTN1‐ALK fusion transcripts in pancreatic cancer cells was validated by reverse transcriptase PCR (RT‐PCR). Briefly, total RNA (500 ng) was reverse‐transcribed into cDNA by using Superscript III Reverse Transcriptase (Thermo Fisher Scientific Life Sciences). cDNA (corresponding to 10 ng of total RNA) was then subjected to PCR amplification using Kapa Taq DNA Polymerase (Kapa Biosystems, Woburn, MA, https://www.kapabiosystems.com). The reactions were carried out in a thermal cycler under the following conditions: 30 cycles at 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 1 minute, with a final extension at 72°C for 1 minute. The gene encoding glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) was amplified to estimate the efficiency of cDNA synthesis. PCR products were directly sequenced in both directions by using an ABI 3130xl DNA Sequencer (Thermo Fisher Scientific Life Sciences) and the BigDye Terminator kit. The following PCR primers were used for RT‐PCR and Sanger sequencing: 5′‐ATCGGGAA CTGACAAACCAG‐3′ and 5′‐TGCCAGCAAAGCAGTAGTTG‐3′.
Analysis of Breakpoint Junction for DCTN1‐ALK Fusion
Genomic DNA (10 ng) was subjected to PCR amplification by using Kapa Taq DNA Polymerase (Kapa Biosystems). The reactions were performed in a thermal cycler under the following conditions: 30 cycles at 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 2 minutes, with a final extension at 72°C for 1 minute. The following PCR primers were used for RT‐PCR and Sanger sequencing: 5′‐ATCGGGAACTGACAAACCAG‐3′ and 5′‐TGCCAGCAAAGCAGTAGTTG‐3′. The PCR primers used for genomic PCR and Sanger sequencing were the same as those used for RT‐PCR.
Immunohistochemistry
Immunohistochemical staining was performed on formalin‐fixed, paraffin‐embedded tissues using the avidin‐biotin complex method. Briefly, 4‐µm‐thick sections cut from representative blocks were deparaffinized and rehydrated in an alcohol gradient. After blocking endogenous peroxidase with methanol containing 0.3% H2O2 and blocking biotin with a biotin‐blocking system (Agilent Technologies), the sections were autoclaved at 121°C for 10 minutes in citrate buffer (10 mM sodium citrate, pH 6.0) for antigen retrieval. After blocking with normal goat serum, the sections were reacted overnight at 4°C with an anti‐ALK antibody (5A4, 1:200; Santa Cruz Biotechnology, Dallas, TX, https://www.scbt.com). The sections were then sequentially reacted with biotin‐conjugated anti‐mouse IgG antibodies (Vector Laboratories, Burlingame, CA, https://vectorlabs.com) and Vectastain Elite ABC reagent (Vector Laboratories). DAB Enhancer (Agilent Technologies) was used as the chromogen, and nuclei were counterstained with hematoxylin.
Cell Lines and Reagents
NCI‐H1299 cells and Ba/F3 cells were provided by Dr. J. D. Minna (UT Southwestern Medical Center) and Dr. Hiroyuki Mano (University of Tokyo), respectively. 293FT cells and WEHI‐3B cells were obtained from Thermo Fisher Scientific Life Sciences and RIKEN BioResource Center (Ibaraki, Japan, http://en.brc.riken.jp/), respectively. NCI‐H1299 and WEHI‐3B cells were cultured in RPMI medium containing 10% fetal bovine serum (FBS). Ba/F3 cells were cultured in RPMI medium containing 10% FBS and 10% WEHI‐3B‐conditioned medium (a source of interleukin [IL]‐3). 293FT cells were cultured in DMEM containing 10% FBS.
Alectinib and crizotinib were purchased from Selleck Chemicals (Houston, TX, http://www.selleckchem.com). Antibodies against ALK (catalog no. 3633), phosphor‐ALK pTyr1604 (catalog no. 3341), and β‐actin (catalog no. 3700) were purchased from Cell Signaling Technology (Danvers, MA, https://www.cellsignal.com).
Examination of Properties of ALK Fusion Products
To construct the lentiviral vectors used to express the DCTN1‐ALK fusion proteins, full‐length cDNAs were PCR‐amplified from tumor cDNA and inserted into pLenti‐6/V5‐DEST plasmids (Thermo Fisher Scientific Life Sciences). The procedure followed the National Institutes of Health guidelines for recombinant DNA research. The integrity of each cDNA insert was verified by Sanger sequencing.
Suppression of phosphorylation of the ALK kinase domain in the DCTN1‐ALK fusion polypeptide was examined in NCI‐H1299 lung cancer cells, which do not express endogenous ALK proteins. NCI‐H1299 cells transiently transfected with the DCTN1‐ALK expression plasmid or with an empty (control) plasmid for 12 hours were treated with dimethyl sulfoxide (DMSO; Sigma‐Aldrich, St. Louis, MO, http://www.sigmaaldrich.com/) or with the indicated inhibitor (dissolved in DMSO) for 24 hours. Whole‐cell lysates were subjected to immunoblot analysis. Briefly, cells were lysed in NETN (100 mM NaCl, 1 mM EDTA, 20 mM Tris‐HCl, and 0.5% NP‐40) buffer containing Complete Protease and PhosSTOP Phosphatase Inhibitor Cocktail (Roche, Mannheim, Germany, http://www.roche.com). Proteins were then subjected to SDS polyacrylamide gel electrophoresis, followed by immunoblotting onto polyvinylidene difluoride membranes. The membranes were blocked for 1 hour with Tris‐buffered saline (TBS) containing 0.1% Tween 20/1% bovine serum albumin and then probed with primary antibodies. After washing with TBS containing 0.1% Tween 20, the membranes were incubated with horseradish peroxidase‐conjugated anti‐mouse or anti‐rabbit secondary antibodies and visualized by using an enhanced chemiluminescence reagent (PerkinElmer, Waltham, MA, http://www.perkinelmer.com). Signal intensity was measured by using an LAS3000 imaging system (Quansys Biosciences, West Logan, UT, http://www.quansysbio.com).
Ba/F3 Cell Viability Assays
Lentiviruses were generated in 293FT cells (6 × 106 cells per 10‐cm plate) transfected with ViraPower packaging mix (Thermo Fisher Scientific Life Sciences) and a pLenti‐6/V5‐DEST plasmid containing DCTN1‐ALK or EML4‐ALK cDNA using the Lipofectamine 3000 reagent (Thermo Fisher Scientific Life Sciences). Viral supernatants were collected 42 hours after medium exchange and used to infect 4.0 × 105 Ba/F3 cells in the presence of 10 μg/ml Polybrene (Sigma‐Aldrich). After overnight incubation at 37°C/5% CO2, the cells were distributed into 24‐well plates and selected for 1 week in medium containing IL‐3 and 8 μg/ml blasticidin (Thermo Fisher Scientific Life Sciences). Blasticidin‐resistant cells were grown in IL‐3‐free medium for 2 weeks, and expression of exogenous DCTN1‐ALK proteins was confirmed by immunoblot analysis coupled with Sanger sequencing of RT‐PCR products derived from the cells.
Twenty‐four hours before inhibitor treatment, 2,000 Ba/F3 cells were plated in 96‐well plates (in quadruplicate). Serially diluted inhibitors were then added to the wells. Cell viability was measured 72 hours after drug treatment using the CellTiter‐Glo luminescent cell viability reagent (Promega, Madison, WI, https://www.promega.com) and EnVision (PerkinElmer). Cell viability was calculated as the cell count in drug‐treated samples relative to that in untreated samples. Data were displayed graphically by using GraphPad Prism software, version 6.0 (GraphPad Software Inc., San Diego, CA, http://www.graphpad.com).
Results and Discussion
Clinicopathological Characteristics of the Four KRAS Mutation‐Negative Patients
We examined a cohort of 100 pancreatic cancer cases (96 cases with a KRAS mutation and 4 without) who underwent radical resection (supplemental online Table 1). In the original cohort of 100, we detected mutations in three other genes (TP53 [42%, 42/100], SMAD4 [13%, 13/100], and CDKN2A [7%, 7/100], all of which are common in pancreatic cancer) at frequencies similar to those reported previously [6], [7], [8], [9], [10], [11]. The clinicopathological characteristics of the four PDAC cases that did not harbor KRAS mutations are shown in Table 1. All cases were primary tumors and were negative for mutations in other druggable oncogenes, such as BRAF and PIK3CA, mutations in which were detected recently in KRAS mutation‐negative PDACs [8]. Interestingly, three of these cases did not recur after surgery (Table 1), whereas 74 (77%) of the other 96 cases, excluding 10 cases with metastatic lesions, did. Sufficient RNA and genomic DNA for next‐generation sequencing were available for 4 and 3 of the four cases without KRAS mutations, respectively; therefore, these cases were subjected to whole RNA (n = 4) and exome (DNA) sequencing (n = 3).
Table 1. Characteristics of four pancreatic cancer cases without KRAS mutation.
UICC TNM Classification of Malignant Tumours, 7th edition [13].
Abbreviation: ND, not detected.
Detection of DCTN1‐ALK Fusion in a Case of PDAC
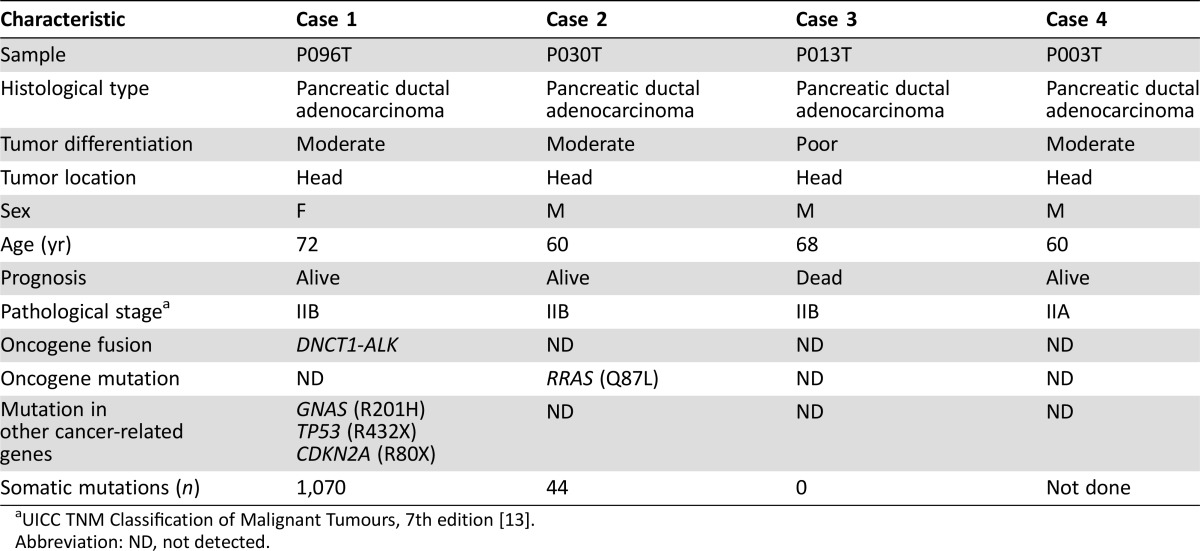
Analysis of RNA sequencing data and subsequent validation of RT‐PCR products by Sanger sequencing revealed a DCTN1‐ALK oncogene‐fusion transcript in one (P096T) of the four PDACs lacking the KRAS mutation (Fig. 1A). Sanger sequencing of genomic‐PCR products encompassing the breakpoint junction validated the tumor‐specific occurrence of this oncogene fusion (Fig. 1B). The structure of the breakpoint junction indicated that fusion was generated by nonhomologous end joining between DCTN1 and ALK intron DNAs, resulting in the generation of a cryptic splicing site 3′ to breakpoint junction.
Figure 1.
DCTN1‐ALK fusion in a case of KRAS mutation‐negative pancreatic ductal adenocarcinoma (PDAC). (A): Detection of gene‐fusion transcripts by RT‐PCR. RT‐PCR products for glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) are shown (left). A tumor (P096T) positive for gene fusion is shown alongside its corresponding noncancerous tissue (P096N). Sanger sequencing of RT‐PCR products derived from the tumor reveals expression of DCTN1‐ALK fusion transcripts, in which exon 12 of DCTN1 is joined to exon 20 of ALK. A “G” nucleotide, which comes from ALK intron 19, is boxed (Fig. 1B). (B): Detection of a gene‐fusion DNA by genomic‐PCR. A tumor (P096T) positive for the gene fusion is shown alongside its corresponding noncancerous tissue (P096N). Sanger sequencing of a genomic‐PCR product showed that the fusion was generated by nonhomologous end joining between DCTN1 and ALK intron DNAs, resulting in the generation of a cryptic splice donor site 3′ to breakpoint junction. A “G” nucleotide, which comes from ALK intron 19, was included in the transcripts by splicing at the cryptic splicing site (boxed). (C): Schematic representations of the dynactin 1 (DCTN1)‐anaplastic lymphoma kinase (ALK) proteins in PDAC and other tumors. (D): Histologic image of the DCTN1‐ALK fusion‐positive PDAC (middle power view). Left, hematoxylin and eosin staining. Moderately differentiated tubular adenocarcinoma cells with fusiform or cribriform features infiltrate the desmoplastic stroma. Right, ALK staining. Granular cytoplasmic staining is visible in the adenocarcinoma component. Magnification x100.
Abbreviations: PCR, polymerase chain reaction; RT‐PCR, reverse transcriptase polymerase chain reaction.
The DCTN1‐ALK fusion resulted in a polypeptide in which the ALK kinase domain was fused to a coiled‐coil domain of dynactin 1 (DCTN1), as reported in several human tumors, such as atypical Spitz tumor [16], [17], non‐small cell lung carcinoma [18], and inflammatory myofibroblastic tumor [19], [20] (Fig. 1C). DCTN1 is a subunit of a macromolecular complex, dynactin, which is involved in various aspects of intercellular transport and organelle movement [21]. The fusion protein contained the DCTN1 coiled‐coil domain, which functions during protein dimerization, and also retained the full ALK kinase domain. This indicated that the fusion protein is likely to form a homodimer via the coiled‐coil domain of DCTN1, causing constitutive activation of the kinase function of ALK (a situation similar to that observed for the EML4‐ALK fusion in non‐small cell lung cancer) [22].
The targeted deep‐sequencing method used to select KRAS mutation‐negative PDAC revealed that the PDAC case harboring the DCTN1‐ALK fusion also carried a known activating GNAS mutation, R201H. Activating GNAS mutations drive the development of intraductal papillary mucinous neoplasm (IPMN) [23], and the frequent co‐occurrence of GNAS and KRAS mutations in PDACs derived from IPMN suggests that both GNAS and KRAS mutations contribute to its genesis [8], [24], [25]. The case harboring the DCTN1‐ALK fusion had no features consistent with IPMN; however, some were suggestive of IPMN rather than conventional PDAC. Morphology did not rule out a possible diagnosis of invasive carcinoma derived from IPMN, although it was not diagnostic for this type of tumor (Fig. 1D). Therefore, we diagnosed PDAC; however, the PDAC might have originated from IPMN.
The PDAC case harboring the DCTN1‐ALK fusion showed strong staining for the ALK protein, similar to ALK fusion‐positive lung cancer (Fig. 1D). Further immunostaining for the ALK protein in another 150 surgical and 90 biopsy specimens from pancreatic cancers, including 70 cases with coexisting IPMN and 20 cases of IPMN without invasive cancer, did not detect additional cases showing ALK expression. This is consistent with the results of recent immune‐histochemical and genome‐wide screening showing that pancreatic cancers lack ALK fusion [6], [9], [26]. Thus, ALK fusion is likely a rare event in pancreatic carcinogenesis.
Characterization of DCTN1‐ALK Fusion Protein
Despite recurrent observation of the DCTN1‐ALK fusion in human tumors [16], [18], [20], to our knowledge functional analysis of the fusion product has not been undertaken. Therefore, we asked whether the DCTN1‐ALK fusion protein is linked to carcinogenesis and therapy, as observed for the EML4‐ALK fusion protein.
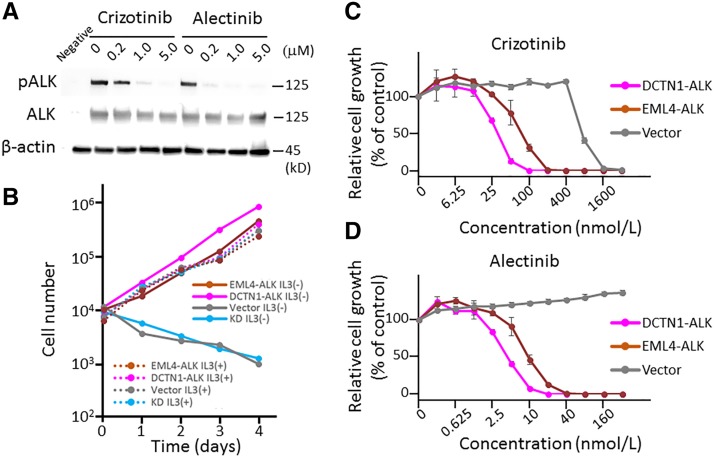
Phosphorylation (i.e., activation of ALK kinase) of the DCTN1‐ALK protein was suppressed by crizotinib and alectinib, ALK tyrosine kinase inhibitors used to treat non‐small cell lung cancer harboring the ALK fusion (Fig. 2A). Exogenous expression of DCTN1‐ALK fusion gene cDNA induced IL‐3‐independent growth of Ba/F3 cells (as does the EML4‐ALK fusion), supporting the constitutive activation of ALK kinase (Fig. 2B). This growth was suppressed by crizotinib and alectinib when used at similar concentrations (Fig. 2C): The concentrations of inhibitor that cause 50% growth inhibition for crizotinib and alectinib for the DCTN1‐ALK and EML4‐ALK fusions were 31.5 nM and 75.8 nM and 4.3 nM and 8.8 nM, respectively. These results indicate that the DCTN1‐ALK fusion acts as a driver oncogene and can be used as a target for ALK tyrosine kinase inhibitors. The result is also consistent with the finding that inflammatory myofibroblastic tumors treated with crizotinib and a multikinase vascular endothelial growth factor inhibitor, pazopanib, showed partial regression [19].
Figure 2.
Character of DCTN1‐ALK protein linked to carcinogenesis and therapy. (A): Immunoblot analysis of full‐length DCTN1‐ALK proteins. An expression vector encoding DCTN1‐ALK cDNA was introduced into H1299 lung cancer cells, which do not express endogenous ALK [28]. The transfectants were then exposed to crizotinib and alectinib. Levels of phosphorylation at tyrosine 1604 were determined 24 hours after treatment with 0, 0.2, 1.0, or 5.0 μM of each drug. (B): Interleukin (IL)‐3‐independent growth of Ba/F3 cells stably expressing DCTN1‐ALK or EML4‐ALK cDNAs. Parental cells and Ba/F3 cells transduced with lentiviral cDNA or empty vector were cultured in medium with and without IL‐3, and the number of cells was counted. (C and D): Viability of Ba/F3 cells stably expressing DCTN1‐ALK or EML4‐ALK cDNAs after treatment with crizotinib (C) or alectinib (D). Ba/F3 cells transduced with lentiviral cDNA or empty vector were subjected to the assay, and the number of cells was counted at 72 hours.
Abbreviations: ALK, anaplastic lymphoma kinase; DCTN1, dynactin 1; pALK, phosphorylated ALK.
Detection of the RRAS Mutation in a Case of PDAC
Analysis of exome sequencing data revealed somatic mutations in two of the three KRAS‐negative PDACs (supplemental online Table 2). The P096T tumor harboring a DCTN1‐ALK fusion carried 990 somatic mutations, while the other tumor, P030T, carried 41 somatic mutations. Notably, the latter carried a missense mutation, Q87L, in the RRAS gene. The same RRAS mutation occurs in lung cancer (http://cancer.sanger.ac.uk/cosmic) and juvenile myelomonocytic leukemia [27]; therefore, it is a recurrent mutation in human tumors. The amino acid residue, Gln87, in the RRAS protein corresponds to the Gln61 residue in the HRAS and KRAS proteins [27]: the oncogenic substitution, Gln61Leu, is observed in several human cancers. Therefore, it is likely that this PDAC is driven by the RRAS mutation rather than by the KRAS mutation. Histologically, the P096T tumor was a tubular adenocarcinoma with a cribriform figure, the cells of which invaded dense fibrous stroma but showed relatively weak vascular and neural invasion.
Conclusion
We found that development of a small subset of pancreatic cancers is likely to be driven by oncogenic aberrations other than the KRAS mutation. To our knowledge, DCTN1‐ALK is the first oncogenic ALK fusion identified in pancreatic cancer, and it may be druggable. We examined only four cases of KRAS mutation‐negative pancreatic cancer; however, another study analyzing six cases of KRAS mutation‐negative pancreatic cancer identified PIK3CA and BRAF mutations in one and three cases, respectively [8]. Analysis of a larger number of KRAS mutation‐negative pancreatic cancers might help identify more genes involved in pancreatic carcinogenesis and improve existing therapies for advanced pancreatic cancer.
See http://www.TheOncologist.com for supplemental material available online.
Acknowledgments
The National Cancer Center Biobank is supported by the National Cancer Center Research and Development Fund. This work was supported in part by a MEXT KAKENHI grant (no. 26461039) to H.U., and by the Foundation for the Promotion of Cancer Research in Japan to H.U.
Author Contributions
Conception/Design: Takashi Kohno, Nobuyoshi Hiraoka
Provision of study material or patients: Hideki Ueno, Hideyuki Hayashi, Yasunari Sakamoto, Shunsuke Kondo, Chigusa Morizane, Kazuaki Shimada, Takuji Okusaka
Collection and/or assembly of data: Yoko Shimada, Yoshinori Ino, Takashi Nakaoku, Nobuyoshi Hiraoka
Data analysis and interpretation: Yoko Shimada, Nobuyoshi Hiraoka
Manuscript writing: Takashi Kohno, Nobuyoshi Hiraoka
Final approval of manuscript: Yoko Shimada, Takashi Kohno, Hideki Ueno, Yoshinori Ino, Hideyuki Hayashi, Takashi Nakaoku, Yasunari Sakamoto, Shunsuke Kondo, Chigusa Morizane, Kazuaki Shimada, Takuji Okusaka, Nobuyoshi Hiraoka
Disclosures
Chigusa Morizane: Yakult Honsha, Novartis (C/A), Pfizer, Novartis, Yakult Honsha, Eli Lilly (H), GlaxoSmithKline, Pfizer, Nobelpharma, Ono Pharmaceutical, Taiho Pharmaceutical (RF). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supplementary Information
References
- 1. Torre LA, Bray F, Siegel RL et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2. Von Hoff DD, Ervin T, Arena FP et al. Increased survival in pancreatic cancer with nab‐paclitaxel plus gemcitabine. N Engl J Med 2013;369:1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iacobuzio‐Donahue CA, Velculescu VE, Wolfgang CL et al. Genetic basis of pancreas cancer development and progression: Insights from whole‐exome and whole‐genome sequencing. Clin Cancer Res 2012;18:4257–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ostrem JM, Peters U, Sos ML et al. K‐ras(g12c) inhibitors allosterically control gtp affinity and effector interactions. Nature 2013;503:548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McLornan DP, List A, Mufti GJ. Applying synthetic lethality for the selective targeting of cancer. N Engl J Med 2014;371:1725–1735. [DOI] [PubMed] [Google Scholar]
- 6. Murphy SJ, Hart SN, Halling GC et al. Integrated genomic analysis of pancreatic ductal adenocarcinomas reveals genomic rearrangement events as significant drivers of disease. Cancer Res 2016;76:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sausen M, Phallen J, Adleff V et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 2015;6:7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Witkiewicz AK, McMillan EA, Balaji U et al. Whole‐exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Biankin AV, Waddell N, Kassahn KS et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491:399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jones S, Zhang X, Parsons DW et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yachida S, White CM, Naito Y et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long‐term survivors. Clin Cancer Res 2012;18:6339–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hruban RH, Boffetta P, Hiraoka N. Pancreas In: WHO Classification of Tumours of the Digestive System. Lyon: IARC, 2010:279–291. [Google Scholar]
- 13. Sobin LH, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumours. 7th ed. New York: Wiley‐Blackwell, 2009:1–336. [Google Scholar]
- 14. Cibulskis K, Lawrence MS, Carter SL et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature Biotechnol 2013;31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim D, Salzberg SL. TopHat‐fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol 2011;12:R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wiesner T, He J, Yelensky R et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun 2014;5:3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Busam KJ, Kutzner H, Cerroni L et al. Clinical and pathologic findings of spitz nevi and atypical spitz tumors with alk fusions. Am J Surg Pathol 2014;38:925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iyevleva AG, Raskin GA, Tiurin VI et al. Novel ALK fusion partners in lung cancer. Cancer Lett 2015;362:116–121. [DOI] [PubMed] [Google Scholar]
- 19. Subbiah V, McMahon C, Patel S et al. Stump un“stumped”: Anti‐tumor response to anaplastic lymphoma kinase (ALK) inhibitor based targeted therapy in uterine inflammatory myofibroblastic tumor with myxoid features harboring DCTN1‐ALK fusion. J Hematol Oncol 2015;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X, Krishnan C, Nguyen EP et al. Fusion of dynactin 1 to anaplastic lymphoma kinase in inflammatory myofibroblastic tumor. Hum Pathol 2012;43:2047–2052. [DOI] [PubMed] [Google Scholar]
- 21. Schroer TA. Dynactin. Annu Rev Cell Dev Biol 2004;20:759–779. [DOI] [PubMed] [Google Scholar]
- 22. Takeuchi K, Soda M, Togashi Y et al. RET, ROS1 and ALK fusions in lung cancer. Nature Med 2012;18:378–381. [DOI] [PubMed] [Google Scholar]
- 23. Cooper CL, O'Toole SA, Kench JG. Classification, morphology and molecular pathology of premalignant lesions of the pancreas. Pathology 2013;45:286–304. [DOI] [PubMed] [Google Scholar]
- 24. Furukawa T, Kuboki Y, Tanji E et al. Whole‐exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep 2011;1:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takano S, Fukasawa M, Maekawa S et al. Deep sequencing of cancer‐related genes revealed gnas mutations to be associated with intraductal papillary mucinous neoplasms and its main pancreatic duct dilation. PLoS One 2014;9:e98718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Graham RP, Oliveira AM, Zhang L. Rare ALK expression but no ALK rearrangement in pancreatic ductal adenocarcinoma and neuroendocrine tumors. Pancreas 2013;42:949–951. [DOI] [PubMed] [Google Scholar]
- 27. Flex E, Jaiswal M, Pantaleoni F et al. Activating mutations in rras underlie a phenotype within the rasopathy spectrum and contribute to leukaemogenesis. Hum Mol Genet 2014;23:4315–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kohno T, Ichikawa H, Totoki Y et al. KIF5B‐RET fusions in lung adenocarcinoma. Nature Med 2012;18:375–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.