Genomic‐driven cancer medicine is increasingly becoming a part of routine clinical practice. For successful implementation of precision cancer medicine, strategically organized molecular tumor boards are critical to provide objective evidence‐based translation of observed molecular alterations into patient‐centered clinical action. Molecular tumor board implementation models along with clinical and economic outcomes will define future treatment standards.
Keywords: Cancer, Molecular tumor board, Precision medicine, Personalized medicine, Lessons learned
Abstract
Background.
The increasing practicality of genomic sequencing technology has led to its incorporation into routine clinical practice. Successful identification and targeting of driver genomic alterations that provide proliferative and survival advantages to tumor cells have led to approval and ongoing development of several targeted cancer therapies. Within many major cancer centers, molecular tumor boards are constituted to shepherd precision medicine into clinical practice.
Materials and Methods.
In July 2014, the Clinical Genomics Action Committee (CGAC) was established as the molecular tumor board companion to the Personalized Medicine Clinical Service (PMCS) at Moffitt Cancer Center in Tampa, Florida. The processes and outcomes of the program were assessed in order to help others move into the practice of precision medicine.
Results.
Through the establishment and initial 1,400 patients of the PMCS and its associated molecular tumor board at a major cancer center, five practical lessons of broad applicability have been learned: transdisciplinary engagement, the use of the molecular report as an aid to clinical management, clinical actionability, getting therapeutic options to patients, and financial considerations. Value to patients includes access to cutting‐edge practice merged with individualized preferences in treatment and care.
Conclusions.
Genomic‐driven cancer medicine is increasingly becoming a part of routine clinical practice. For successful implementation of precision cancer medicine, strategically organized molecular tumor boards are critical to provide objective evidence‐based translation of observed molecular alterations into patient‐centered clinical action. Molecular tumor board implementation models along with clinical and economic outcomes will define future treatment standards.
Implications for Practice.
It is clear that the increasing practicality of genetic tumor sequencing technology has led to its incorporation as part of routine clinical practice. Subsequently, many cancer centers are seeking to develop a personalized medicine services and/or molecular tumor board to shepherd precision medicine into clinical practice. This article discusses the key lessons learned through the establishment and development of a molecular tumor board and personalized medicine clinical service. This article highlights practical issues and can serve as an important guide to other centers as they conceive and develop their own personalized medicine services and molecular tumor boards.
Introduction
Advances in genomic technology have opened new options for cancer treatment [1], [2], [3], [4]. Successful identification and targeting of the driver genomic alterations that provide proliferative and survival advantages to tumor cells have led to approval of several targeted cancer therapies, such as imatinib for BCR‐ABL‐positive chronic myelogenous leukemia [5], vemurafenib and dabrafenib for BRAF V600‐mutated melanoma [6], [7], and crizotinib and ceritinib for ALK‐rearranged non‐small cell lung cancer (NSCLC) [8], [9]. As genomic sequencing and targeted therapies have demonstrated clinical efficacy, the current pharmaceutical pipeline contains several agents targeting altered cancer genes across many cancer types. The increasing practicality of genomic sequencing technology has spurred investigators to further understand the clinical impact of these mutations, and analysis of the cancer genome is increasingly becoming routine clinical practice [10], [11], [12], [13].
The disease courses of many patients progress beyond U.S. Food and Drug Administration (FDA)‐approved therapies or National Comprehensive Cancer Network guidelines on the basis of alterations detected in their tumor [14]. This has led to identification of clinical trials, “off‐label” treatment, or compassionate‐use protocols in attempts to objectively provide options to prolong survival and increase quality of life. Within many major cancer centers, molecular tumor boards are constituted to shepherd precision medicine into clinical practice [15], [16], [17].
Materials and Methods
In July 2014, the Clinical Genomics Action Committee (CGAC) was established as the molecular tumor board companion to the Personalized Medicine Clinical Service (PMCS) at Moffitt Cancer Center in Tampa, Florida. CGAC was conceived to aid in the rational implementation of cancer genomics (and other ways of individualizing treatment) by providing a multidisciplinary assessment of advanced diagnostic strategies and complex clinical results. The committee provides oversight and guidance to the PMCS and discusses patients with all types of cancer to develop consensus on therapeutic recommendations, enabling the translation of scientific findings into evidence‐based recommendations for individualized treatments. Specific responsibilities of CGAC include the following: (a) providing a consensus forum for determining objective patient management recommendations when multiple therapy options are being considered or when a variant of unclear therapeutic significance is identified, (b) performing multidisciplinary assessment of requests for introduction of precision medicine assays at Moffitt Cancer Center, and (c) assisting in the development of Moffitt electronic health record clinical decision support rules that alert clinicians to actionable variants.
Process
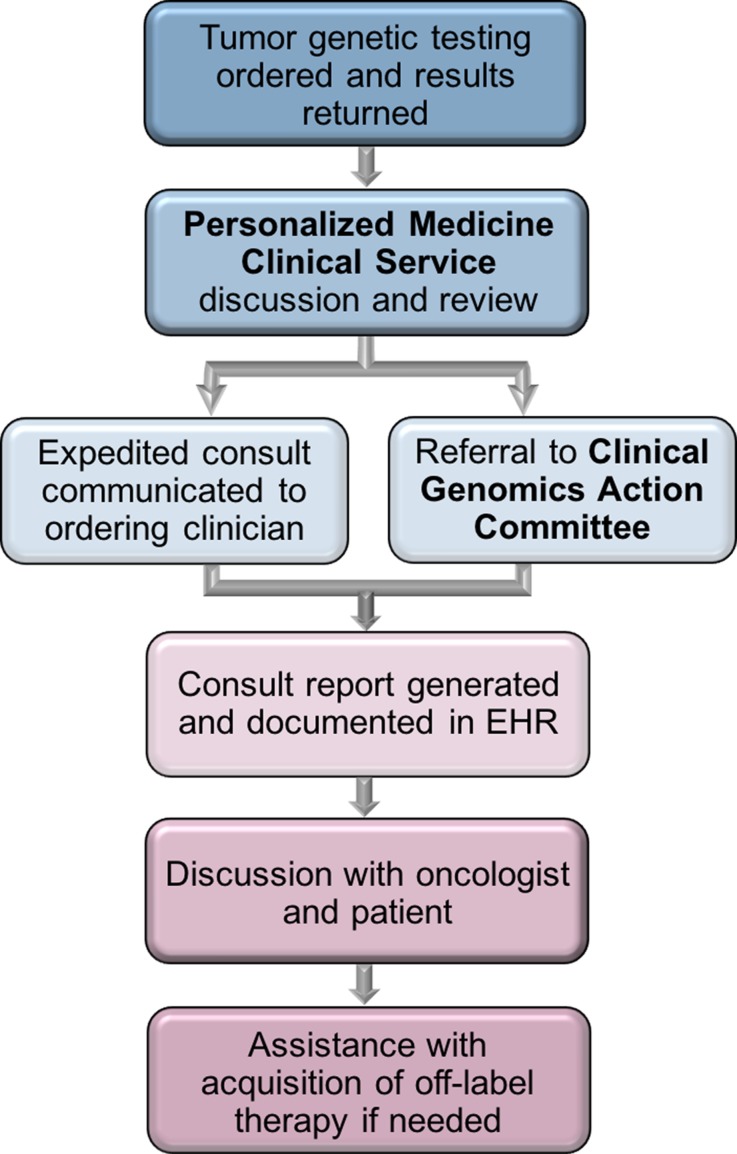
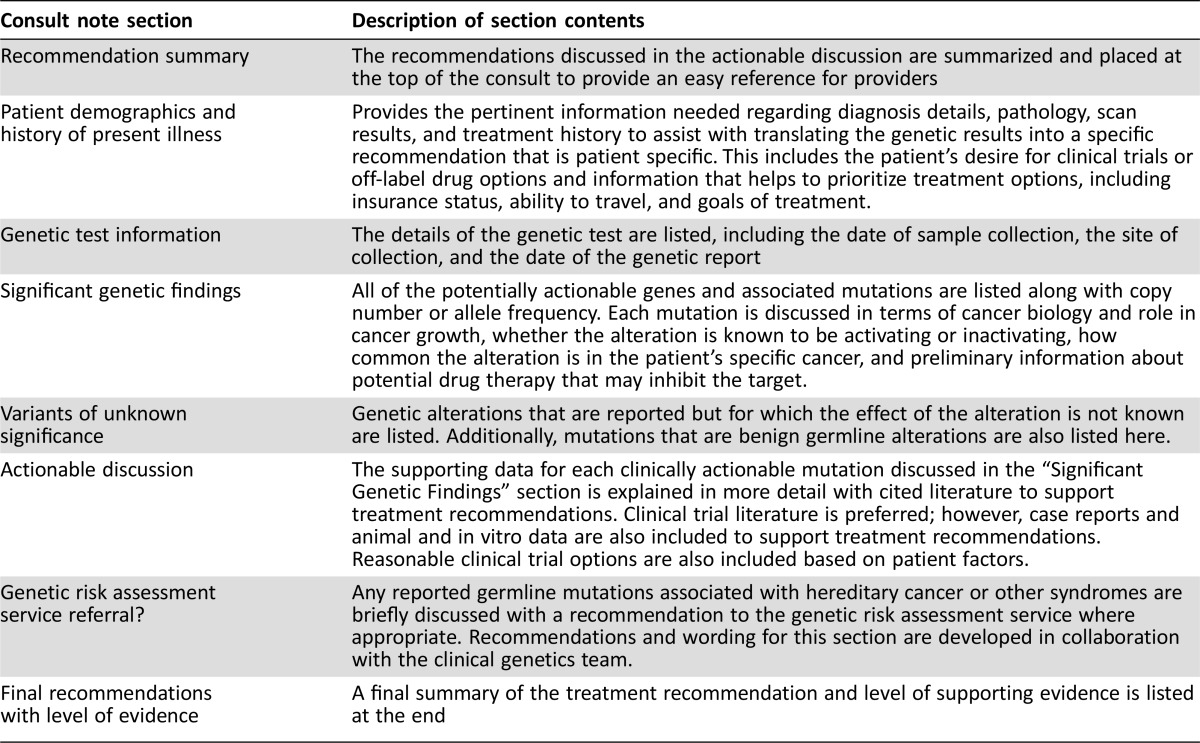
The PMCS reflexively receives results of all next‐generation tumor sequencing (NGS) panels ordered as part of clinical care at Moffitt. These include in‐house developed NGS panels in addition to those sent to reference laboratories. NGS results are reviewed at a weekly PMCS meeting, with PMCS interpretation and recommendation provided to the ordering clinician through email and a clinical consult note in the electronic health record (EHR) for all cases (Table 1). Controversial or challenging cases are discussed at the monthly CGAC meeting (n = 59; mean, 2.3 cases per meeting) (Table 2) to establish a group consensus related to the significance of alterations detected, possible therapeutic options, and the recommended procession of therapy. Once consensus has been achieved, consultation reports, including the key points of the CGAC discussion, are generated, entered into the EHR, and communicated to the ordering clinician and subsequently the patient. Clinical trial enrollment is facilitated, or if off‐label therapy or compassionate use is pursued, the PMCS offers assistance in obtaining the medication and insurance approval (Fig. 1).
Table 1. Demographics of all Personalized Medicine Clinical Service cases as of June 27, 2016 (n = 1,402).
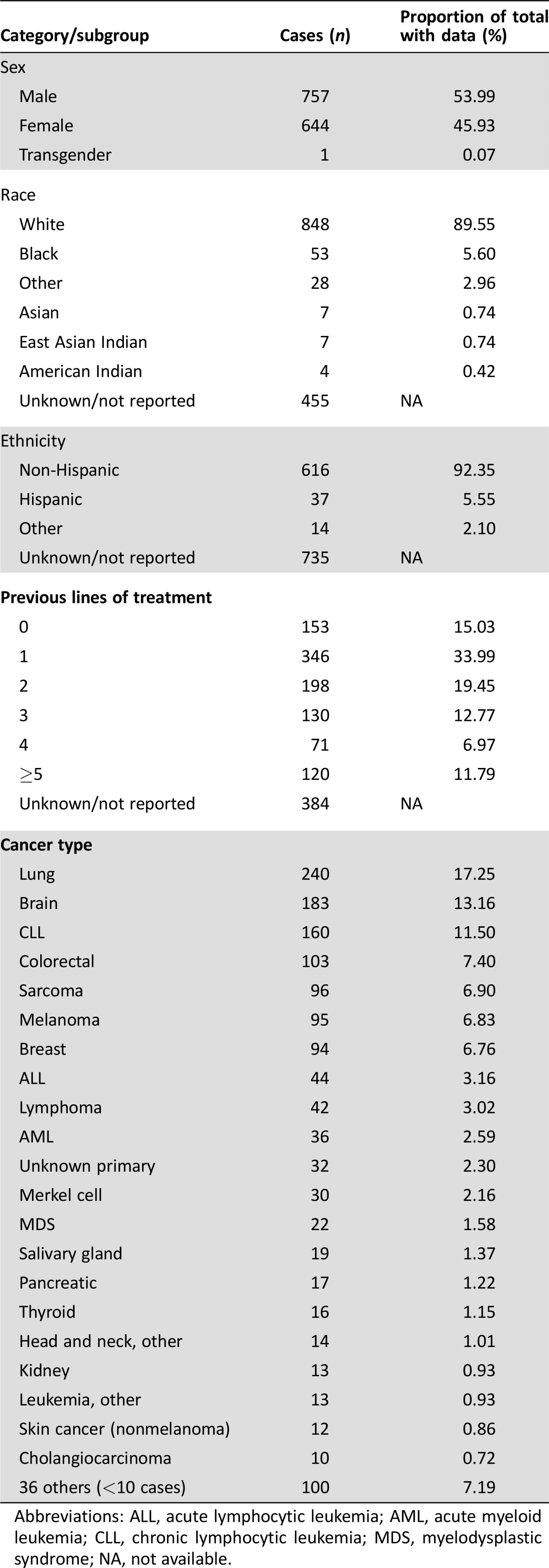
Abbreviations: ALL, acute lymphocytic leukemia; AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia; MDS, myelodysplastic syndrome; NA, not available.
Table 2. Cases presented at Clinical Genomics Action Committee meetings (n = 58).
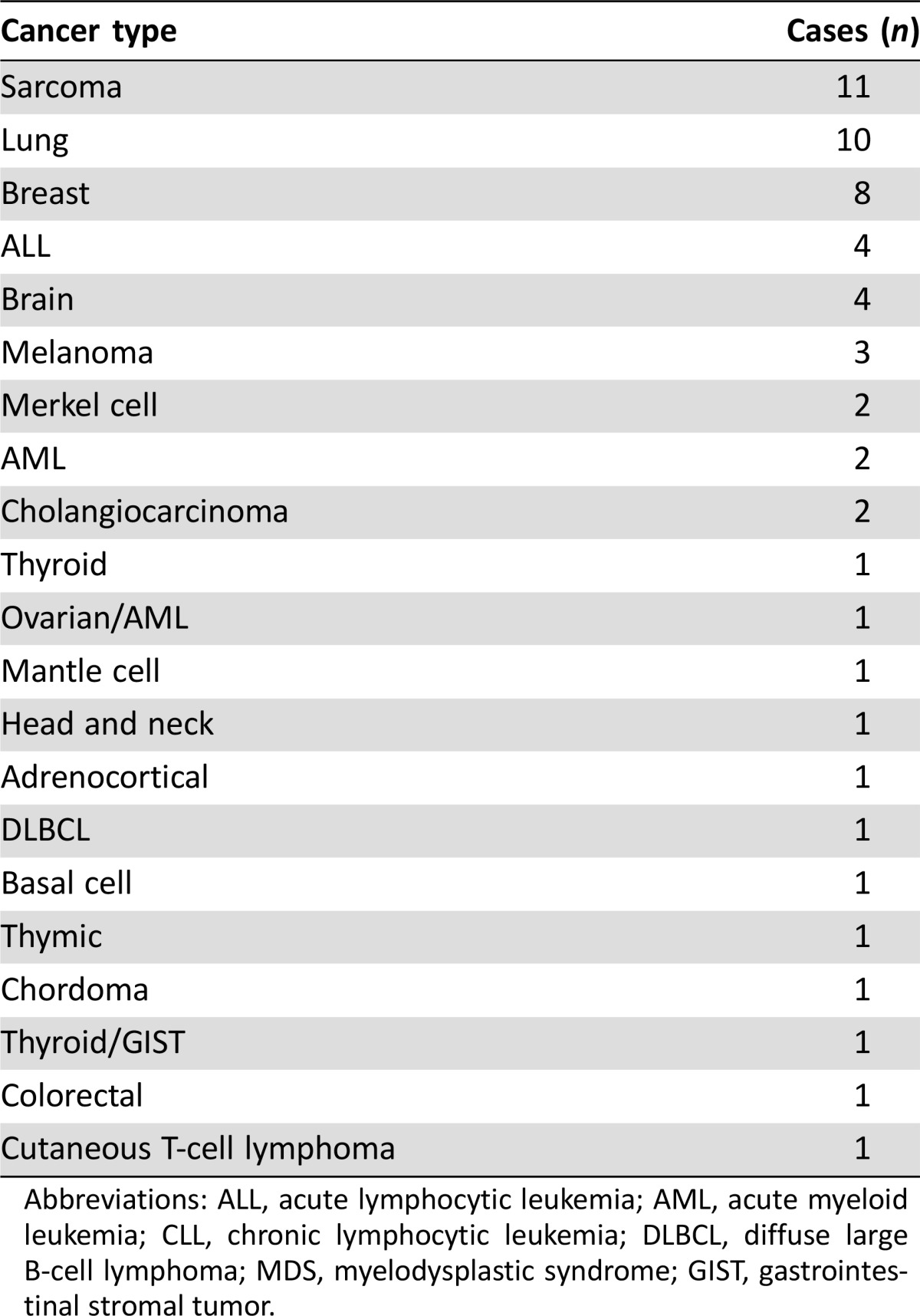
Abbreviations: ALL, acute lymphocytic leukemia; AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia; DLBCL, diffuse large B‐cell lymphoma; MDS, myelodysplastic syndrome; GIST, gastrointestinal stromal tumor.
Figure 1.
Tumor genome analysis workflow.
Abbreviation: EHR, electronic health record.
Several practical lessons of broad applicability have been learned through the establishment and initial 1,400 patients of the PMCS and its associated molecular tumor board at a major cancer center.
Key Lessons Learned
Transdisciplinary Engagement
To accommodate the diverse results generated by broad tumor sequencing, a comprehensive, collaborative approach is needed. CGAC includes members from pathology, medical genetics, bioinformatics, translational research, laboratory science, pharmacy, patient representatives, nursing, social work, and physicians from across oncologic and hematologic diseases. Engagement from all of these groups is crucial to having a true transdisciplinary evaluation of the patient and their genomic information, and all are encouraged to stimulate discussion on potentially relevant findings even if those findings are outside of their designated specialty. Cases are typically presented by a member of the PMCS or a rotating fellow/resident, with clinical details supported by the treating physician. All experts in the room bring a unique perspective, and committee meetings present a unique opportunity to have representation from all of these groups in one place at a given time united to further the personalization of cancer care. While a particular patient case being discussed may revolve around a specific tumor type, lessons and insight from other clinicians in the room have proven helpful in describing their experience with the molecular aberrations and corresponding targeted therapy or evidence and developments in their field. The presence of both researchers and clinicians has enabled a bidirectional flow of information. Not only does cutting‐edge research inform discussion of therapeutic options, but clinician practice of medicine and realities of the multipayer health care system help determine which information is of greatest utility.
As the molecular treatment of cancer increasingly becomes a part of standard care, the expectation is that molecular tumor boards will become essential much in the same way that disease‐specific tumor boards are today. Community clinicians who may lack the resources to meet the forthcoming challenges can partner with large academic medical centers or use centralized molecular tumor boards. A side benefit of the multidisciplinary discussions is a greater appreciation for the perspectives of complementary disciplines. Molecular tumor boards should bring together a varied group of experts and function in a way that builds on existing structures and processes within the health system while maintaining the flexibility to adapt to new challenges and support the community in which they serve.
Following are some tips for the community practice. Most cancer patients in the United States are treated at community‐based practices [18] and thus are unlikely to have access to the molecular tumor boards concentrated at large academic medical centers. Self‐contained, single‐site molecular tumor boards in the community are typically not feasible because molecular tumor boards rely on a breadth of experts who are not always integrated into community practices (bioinformaticians and translational researchers, for example). Community oncologists seeking to provide highest‐level care to their patients are faced with the challenge of developing and maintaining high‐level expertise in interpreting and acting upon molecular reports, combined with mechanisms for clinical trial enrollment or finding alternate solutions.
Large registry trials, such as the American Society of Clinical Oncology's Targeted Agent and Profiling Utilization Registry (TAPUR), which offer a molecular tumor board component, may represent a current pathway for obtaining external advice on patients who have received NGS [19]. Other community practices may pursue an outsourced molecular tumor board from an academic partner institution, which would gain reciprocal value though increased genomic data and potential clinical trial participants. Alternatively, private molecular tumor boards composed of national experts may be developed to provide such services to community practices. Although community practices may not have the resources to support full‐time in‐house molecular tumor boards, alternatives, including partnerships with academic centers and regional and private tumor boards, allow for broader patient access to multidisciplinary expertise.
The Molecular Report as an Aid to Clinical Management
To promote implementation of tumor genomic data, alterations identified by NGS are communicated to clinicians by the test providers as a summarized molecular report. This molecular report serves as an aid to clinical management and represents the start of the process of molecularly targeted precision cancer therapy. However, there is a gap between the content of the lab reports and the clinical action that should result from the data. There are practical reasons for this gap (liability, customer autonomy, insufficient clinical context), but it is a major issue in getting the most out of the test.
The availability of dedicated personalized medicine experts, such as a personalized medicine consult service or a molecular tumor board, can help oncologists navigate the nuances of the report. To support a practitioner who orders the test or to aid patients who bring a large test report to their clinician, there needs to be a mechanism for external molecular reports to be assessed by the personalized medicine service or molecular tumor board. It is common for radiologists to reveal the relevance of a T1‐ versus T2‐weighted magnetic resonance imaging examination or for pathologists to understand the role of specific stains when reviewing a biopsy specimen. This same principle can apply to the evaluation of the cancer genome.
Variability across reports from reference laboratories is also often not recognized by oncologists applying new tests. Understanding and interpreting results across the breadth of genes evaluated (from a single gene to the whole genome), the types of alterations detected (e.g., mutations, copy number alterations, rearrangements, translocations, fusions), and other factors (e.g., read depth, sequence coverage, the effect of the subclonal detection of mutant variants or equivocal levels of copy number variation) are critical. Evaluation of a genomic report, as with other specialty reports, requires an understanding of the capabilities and elements of the report.
Most reports include information curated from medical literature describing the function of the gene, the frequency with which it has been reported in the patient's cancer type and other cancers, any known prognostic role, and possible therapeutic strategies. This information is dynamic, and thus it is important to consider how it is being curated, from what sources, and how often. Within the report, clinical recommendations of therapies, including those for off‐label use or clinical trials to be considered, can be broad and abundant. These recommendations are written to apply to a generalized patient population and logistically cannot account for patient‐specific factors that are not shared with the sequencing lab. This becomes evident when the report supplies a recommendation for a clinical trial that the patient is ineligible for because of having received too many previous lines of therapy or multiple comorbidities, for example, or is not recruiting locally. The ordering physician and the supportive personalized medicine team should consider the relevance of the treatment options in a way that is personalized to individual patients, their clinical history, and their treatment preferences. The molecular tumor board therefore offers not only an opportunity to harmonize (or at least provide greater context) to the different test platforms but enables appropriate dialogue and personalization of results in order to use them in the context of patient history and preferences.
Clinical Actionability
The goal of the PMCS/CGAC is to assist the treating oncologist in the translation of molecular variants into clinical action for the individual patient. The clinical actionability of these variants includes providing the rationale for potential therapeutic options, contributing to diagnostic evidence, or helping prognosticate disease course. For a growing list of genomic variants there is clear impact on specific cancer therapies, often with corresponding presence in the FDA drug label (e.g., KRAS, EGFR, ALK fusions, and ROS‐1). These variants are handled in the pathology report, with little added value from a PMCS or CGAC. However, most variants (>80% in our experience) do not have well‐defined clinical consequence. The availability of a tumor genomics assessment supported by bioinformatics is a key tool for addressing the need for time, expertise, and resources.
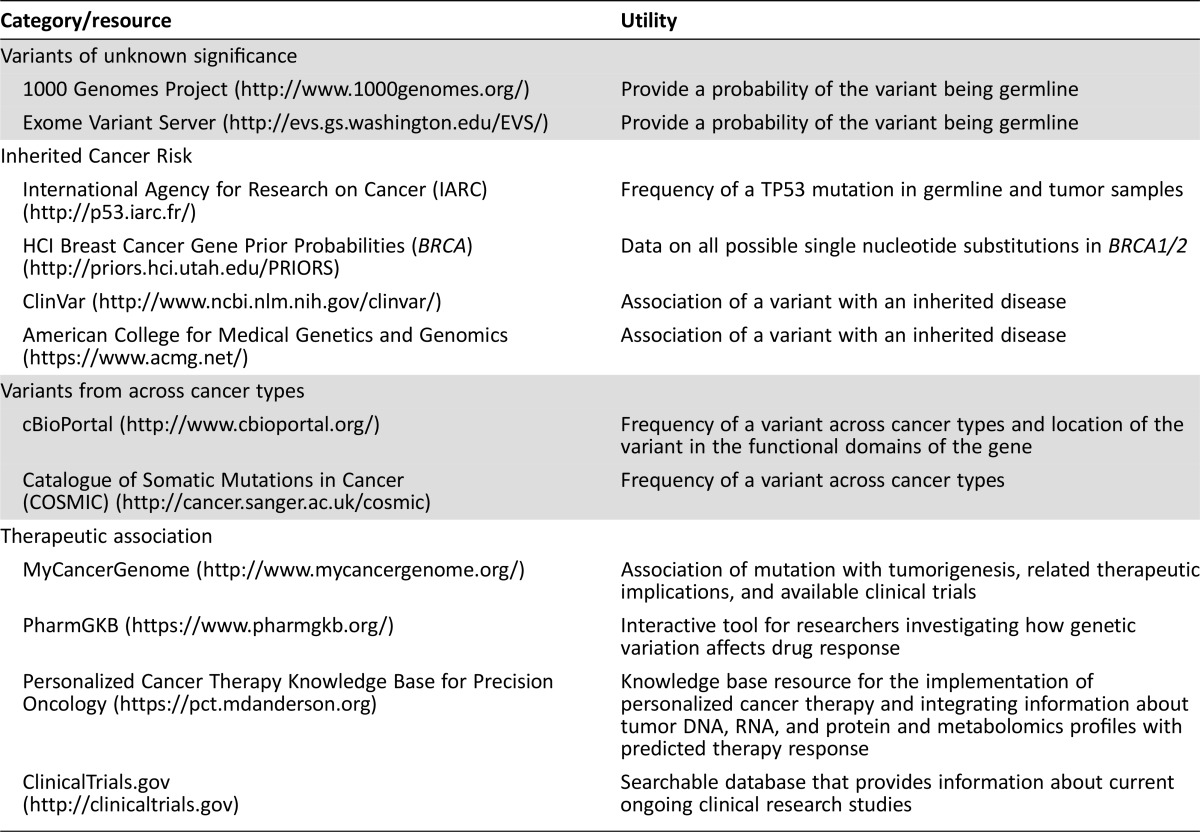
Many commercial sequencing strategies focus on tumor tissue and therefore identify both somatic and germline variants. It has been reported that the absence of a matched germline control results in false‐positive somatic mutation determinations [20]. There is a need to understand whether the variant is likely to be germline (and potentially associated with inherited cancer syndromes), somatic (and whether it has been observed before and in what types of cancer), and in a location in the gene that includes a biologically relevant domain on the resulting protein. Use of databases, such as 1000 Genomes Project or Exome Variant Server, will provide support of germline inheritance; ClinVar or the International Agency for Research on Cancer will indicate whether the variant has been associated with inherited disease.
Another assessment is the frequency of a somatic variant in such databases as cBioPortal or Catalogue of Somatic Mutations in Cancer (COSMIC) (Table 3). Understanding how often and where (i.e., which type of cancer) a variant has been observed will provide a level of confidence in calling it a somatic mutation and can broaden the search for impact (e.g., a variant that is rare in sarcoma but observed in 20% of NSCLCs gives guidance on where to look for gene‐effect relationships). Many of the resulting variants are germline in nature, and most are not of direct relevance to therapeutic care, although focusing on smaller panels of cancer‐related genes reduces this. To ensure that incidental germline findings receive appropriate follow‐up, the PMCS has worked with Moffitt's Genetic Risk Assessment Service to develop a list of genes based on the American College of Medical Genetics and Genomics genes associated with inherited cancer syndromes [21] and other potentially actionable genes found on the somatic genomic panels but are supported by clinical literature as associated with inherited cancer syndromes that warrant patient referral if a mutation is reported. Clinicians and centers must be prepared for incidental findings, be able to recognize them, and know when and how to refer to genetic counselors and medical geneticists [22].
Table 3. Informatics resources.
A common challenge in precision medicine is genomic variants, which are located in a functional protein domain and have possible functional consequences (e.g., nonsynonymous mutation, stop codon, frame shift) but have not been biologically characterized to the point of definitive recommendations. In the context of a patient case, variants of almost known significance (VAKS) are triaged differently than variants of known significance or variants of unknown significance and are a frequent subject of discussion at CGAC meetings because they may make the patient eligible for certain targeted therapies. Input from basic scientists helps to clarify the potential effect of the mutation and subsequently the affected pathways, triggering discussion of mechanisms of drug response. In the absence of other options, these data are tempting treatment targets, but this temptation has to be carefully weighed against therapeutic options available to the patient in question and the patient's prognosis. Molecular tumor boards should be prepared to face these molecular dilemmas and consider processes for handling them, ranging from withholding action until guideline consensus is established to developing a research enterprise to evaluate the variants. The presence of these variants as potential therapeutic targets can also feed back to researchers who may have the resources and interest to test VAKS for functional activity.
Getting Therapeutic Options to Our Patients
The PMCS is involved in the interpretation of a variety of different somatic genomic assays from different laboratories, each with its own unique clinical reports. An added value provided by the PMCS is operating beyond the more generic information provided and reporting it in a consistent, individualized format based on each patient's prior therapies and unique clinical characteristics (Table 4). This helps contextualize results, improving the clinical utility of each consult and enhancing implementation. An additional patient‐centered goal of the clinical consult is anticipation of a patient's needs to consider future treatments beyond the next line of therapy. This is also an important consideration when weighing possible clinical trials. Reviewing the inclusion criteria for a particular trial may optimize the number of alternatives a patient may have. For example, if options A and B are both equally acceptable to the patient and supported by the medical team, but giving off‐label option A before clinical trial option B would exclude the patient from option B, then a more favorable order for therapy should be considered.
Table 4. Personalized medicine consult note description.
Additional value of the clinical consult note may include facilitating off‐label therapy by using the consult report as a summary of evidence‐based justification to payers. Capturing the discussion of a diverse panel of clinicians, health care professionals, and scientists through the CGAC helps create strong, literature‐based recommendations that allow for clear and concise letters of medical necessity and assist with peer‐to‐peer discussions with payers. Experience with this appeal process also allows the PMCS to help secure insurance approval for off‐label use.
Finally, although the clinical consult notes are written primarily for the medical team, they also facilitate discussions directly with patients in the clinic. Rather than having a separate “personalized medicine clinic,” PMCS has integrated these discussions into the routine care and clinic visits of each patient. A PMCS team member will coordinate with the attending oncologist to meet with the patient in the disease‐specific clinic where the patient is usually seen.
Proactive Financial Considerations
Although the cost of genomic sequencing has declined rapidly in recent years, NGS is much more expensive than companion diagnostics for targeted therapies [23]. To provide large‐scale somatic mutation testing that informs treatment decisions, hospitals must either make a substantial investment in equipment and Clinical Laboratory Improvement Amendments test development or contract with third‐party providers. Still, the list price of sequencing can be as much as $7,200 per sample [24]. Many laboratories have decided not to bill patients or have charged significantly reduced prices for testing services, with the goal of generating clinical utility data and demonstrating sufficient value for payers to make favorable coverage and reimbursement decisions. This economic model is not sustainable in the long term, eventually requiring health systems, payers, patient, or some combination to be willing to pay for testing. Although sequencing is becoming a routine part of clinical care, research efforts to further drive down costs and increase quality will continue to be important.
The key to success for reference laboratories, patients, and payers is to accurately estimate the value provided by each test. In some cases, a more targeted genotyping approach may cost significantly less while still providing the majority of clinically actionable data compared with broader, whole‐exome/genome testing. Patients with new diagnoses or in early stages of treatment may benefit most from smaller panels that would indicate appropriate targeted therapies or inform decisions between standard‐of‐care options. Broad tumor sequencing is unlikely to alter first‐line treatment regimens. Once patients have exhausted most or all standard‐of‐care options, large panels are then more likely to provide value by directing them to clinical trials or possible off‐label use of approved drugs. However, this should be weighed against the availability of tissue. A reasonable argument for broader profiling at diagnosis can be made for cancers with tumors that are difficult to biopsy or have low tissue yields. Variants that are not actionable or informative may become so during the patient's natural history. Early use of NGS may require retesting as relevant mutations may have arisen during multiple cycles of therapy. Because of the high cost of these broad panels, serial retesting should be avoided when possible.
The emergence of so‐called liquid biopsies allows for less invasive interrogation of patients’ tumor mutation profiles by taking advantage of circulating tumor DNA in readily available body fluids such as plasma [25]. Liquid biopsies are poised to provide a sequencing option with distinct advantages to some of the challenges discussed previously. By using more abundant media, such as plasma or urine, as opposed to scarce tumor tissue, the issue of tissue availability is mitigated. However, the relative ease of procurement and clinical niche as a platform for detecting resistance mutations through serial sequencing at disease progression or therapeutic resistance increases the number of times a given patient may have their tumor sequenced, bringing a commensurate increase in sequencing cost.
Unnecessary costs should be contained to maximize patient benefit per dollar spent. However, it is important to recognize that the costs of genomic testing are relatively small compared with the total cost of treatment, particularly in complex diseases such as cancer. The tangible benefits of consumer‐directed genomic testing can be debated, but the success of companies such as 23andMe demonstrates a willingness to pay for personal genomic information. Patients may thus be willing to bear the cost of testing to help direct therapy and make difficult treatment choices. Of course, economic analyses should be performed to objectively assess the value delivered for the cost based on the perspective of multiple payers.
Conclusion
As the treatment of cancer increasingly transcends the boundary between distinct site‐of‐origin based care and the shared genomic origins of disease [26], there has been great investment in bringing targeted molecular strategies to the patient. This transformation has spurred the molecular tumor board, a transdisciplinary approach that facilitates both the sharing of disease‐specific expertise and the engagement of translational experts to shepherd precision cancer medicine into clinical practice. These teams will face complex challenges for which traditional evidence‐based medicine decisions are not feasible. The tumor board must be prepared to objectively weigh evidence, while simultaneously accounting for patient‐specific factors, to reach consensus decisions on the tumor genomic data. In the best of cases, these approaches will provide patients options where there were none and, done correctly, can lead to individual and societal advances in overall survival.
A critical need is the development of a relational clinical genomics database that can provide mechanisms to answer many of the questions posed above. By building collections of objective data on treatment selection and therapeutic response and the impact of particular variants, these databases possess the power to eventually turn the unknown into anecdotes, and anecdotes into verifiable data [27], [28], [29]. This will also provide objective data for payers to make reimbursement decisions. In the meantime, publicly available informatics resources are proving invaluable tools for assessment and translation of the novel into the familiar.
Automated systems will need to be developed to support teams as the volume of NGS results surpasses the burden that can be manually handled. These systems will identify cases in need of manual review, generate automated consults for cases that meet predefined criteria where manual review is not required, match patients to appropriate therapies and clinical trials, and aid in the curation of detected variants. Other information technology challenges, such as EHR integration and clinical decision support, exist and will require solutions that fit into the institutional information technology configuration.
Precision medicine approaches to cancer and immunotherapy have joined traditional chemotherapy, radiation therapy, and surgery as the pillars of cancer therapy. To successfully bring precision cancer medicine to patients, molecular tumor boards are critical tools capable of translating observed molecular alteration into clinical action and ultimately creating the pool of data for which future treatment standards will be set.
Acknowledgments
We thank the residents and fellows who have rotated on the clinical service. Research support was received from the DeBartolo Family Personalized Medicine Institute, the State of Florida Cancer Research Endowed Chair, and the Collins Charitable Foundation.
Author Contributions
Conception/Design: Todd C. Knepper, Gillian C. Bell, J. Kevin Hicks, Eric Padron, Jamie K. Teer, Teresa T. Vo, Nancy K. Gillis, Neil T. Mason, Howard L. McLeod, Christine M. Walko
Provision of study material or patients: Todd C. Knepper, Gillian C. Bell, J. Kevin Hicks, Eric Padron, Jamie K. Teer, Teresa T. Vo, Nancy K. Gillis, Neil T. Mason, Howard L. McLeod, Christine M. Walko
Collection and/or assembly of data: Todd C. Knepper, Gillian C. Bell, J. Kevin Hicks, Eric Padron, Jamie K. Teer, Teresa T. Vo, Nancy K. Gillis, Neil T. Mason, Howard L. McLeod, Christine M. Walko
Data analysis and interpretation: Todd C. Knepper, Gillian C. Bell, J. Kevin Hicks, Eric Padron, Jamie K. Teer, Teresa T. Vo, Nancy K. Gillis, Neil T. Mason, Howard L. McLeod, Christine M. Walko
Manuscript writing: Todd C. Knepper, Gillian C. Bell, J. Kevin Hicks, Eric Padron, Jamie K. Teer, Teresa T. Vo, Nancy K. Gillis, Neil T. Mason, Howard L. McLeod, Christine M. Walko
Final approval of manuscript: Todd C. Knepper, Gillian C. Bell, J. Kevin Hicks, Eric Padron, Jamie K. Teer, Teresa T. Vo, Nancy K. Gillis, Neil T. Mason, Howard L. McLeod, Christine M. Walko
Disclosures
Eric Padron: Incyte, Novartis (H), Incyte, Cell Therapeutics, Forma Therapeutics (RF); Jamie K. Teer: Patent filed on a genetic variation storage model, not currently licensed (IP); Howard L. McLeod: Cancer Genetics Inc., Kew Corp. (C/A), Cancer Genetics Inc. (O). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1. Roychowdhury S, Iyer MK, Robinson DR et al. Personalized oncology through integrative high‐throughput sequencing: a pilot study. Sci Transl Med 2011;3:111ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dancey JE, Bedard PL, Onetto N et al. The genetic basis for cancer treatment decisions. Cell 2012;148:409–420. [DOI] [PubMed] [Google Scholar]
- 3. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–674. [DOI] [PubMed] [Google Scholar]
- 4. Wong KM, Hudson TJ, McPherson JD. Unraveling the genetics of cancer: Genome sequencing and beyond. Annu Rev Genomics Hum Genet 2011;12:407–430. [DOI] [PubMed] [Google Scholar]
- 5. Druker BJ, Talpaz M, Resta DJ et al. Efficacy and safety of a specific inhibitor of the BCR‐ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med 2001;344:1031–1037. [DOI] [PubMed] [Google Scholar]
- 6. Chapman PB, Hauschild A, Robert C et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Flaherty KT, Infante JR, Daud A et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367:1694–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shaw AT, Kim DW, Nakagawa K et al. Crizotinib versus chemotherapy in advanced ALK‐positive lung cancer. N Engl J Med 2013;368:2385–2394. [DOI] [PubMed] [Google Scholar]
- 9. Shaw AT, Kim DW, Mehra R et al. Ceritinib in ALK‐rearranged non‐small‐cell lung cancer. N Engl J Med 2014;370:1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson DB, Dahlman KH, Knol J et al. Enabling a genetically informed approach to cancer medicine: A retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next‐generation sequencing panel. The Oncologist 2014;19:616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beltran H, Eng K, Mosquera JM et al. Whole‐exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol 2015;1:466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwaederle M, Daniels GA, Piccioni DE et al. On the road to precision cancer medicine: Analysis of genomic biomarker actionability in 439 patients. Mol Cancer Ther 2015;14:1488–1494. [DOI] [PubMed] [Google Scholar]
- 13. Mantripragada KC, Olszewski AJ, Schumacher A et al. ReCAP: Clinical trial accrual targeting genomic alterations after next‐generation sequencing at a non‐National Cancer Institute‐designated cancer program. J Oncol Pract 2016;12:e396–e404. [DOI] [PubMed] [Google Scholar]
- 14. Meric‐Bernstam F, Brusco L, Shaw K et al. Feasibility of large‐scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J Clin Oncol 2015;33:2753–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwaederle M, Parker BA, Schwab RB et al. Molecular tumor board: The University of California‐San Diego Moores Cancer Center experience. The Oncologist 2014;19:631–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Erdmann J. All aboard: Will molecular tumor boards help cancer patients? Nat Med 2015;21:655–656. [DOI] [PubMed] [Google Scholar]
- 17. Tafe LJ, Gorlov IP, de Abreu FB et al. Implementation of a molecular tumor board: The impact on treatment decisions for 35 patients evaluated at Dartmouth‐Hitchcock Medical Center. The Oncologist 2015;20:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simone JV. Understanding cancer centers. J Clin Oncol 2002;20:4503–4507. [DOI] [PubMed] [Google Scholar]
- 19. Dizon DS, Krilov L, Cohen E et al. Clinical cancer advances 2016: Annual report on progress against cancer from the American Society of Clinical Oncology. J Clin Oncol 2016;34:987–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones S, Anagnostou V, Lytle K et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med 2015;7:283ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Richards S, Aziz N, Bale S et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robson ME, Bradbury AR, Arun B et al. American Society of Clinical Oncology policy statement update: Genetic and genomic testing for cancer susceptibility. J Clin Oncol 2015;33:3660–3667. [DOI] [PubMed] [Google Scholar]
- 23.National Human Genome Research Institute . The cost of sequencing a human genome. Available at: https://www.genome.gov/sequencingcosts/. Accessed October 2, 2015.
- 24.Foundation Medicine . Billing and reimbursement: Answers for patients and caregivers. Available at: http://foundationone.com/docs/ONE-F-003-20131115%20Billing%20Guide%20Patients.pdf. Accessed September 12, 2015.
- 25. Crowley E, Di Nicolantonio F, Loupakis F et al. Liquid biopsy: Monitoring cancer—genetics in the blood. Nat Rev Clin Oncol 2013;10:472–484. [DOI] [PubMed] [Google Scholar]
- 26. Hyman DM, Puzanov I, Subbiah V et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med 2015;373:726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fenstermacher DA, Wenham RM, Rollison DE et al. Implementing personalized medicine in a cancer center. Cancer J 2011;17:528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hunter DJ, D'Agostino RB Sr. Let's not put all our eggs in one basket. N Engl J Med 2015;373:691–693. [DOI] [PubMed] [Google Scholar]
- 29. Rose S. Huge data‐sharing project launched. Cancer Discov 2016;6:4–5. [DOI] [PubMed] [Google Scholar]