Abstract
The mechanisms underlying the neurodevelopmental deficits associated with CHARGE syndrome, which include cerebellar hypoplasia, developmental delay, coordination problems, and autistic features, have not been identified. CHARGE syndrome has been associated with mutations in the gene encoding the ATP-dependent chromatin remodeler CHD7. CHD7 is expressed in neural stem and progenitor cells, but its role in neurogenesis during brain development remains unknown. Here we have shown that deletion of Chd7 from cerebellar granule cell progenitors (GCps) results in reduced GCp proliferation, cerebellar hypoplasia, developmental delay, and motor deficits in mice. Genome-wide expression profiling revealed downregulated expression of the gene encoding the glycoprotein reelin (Reln) in Chd7-deficient GCps. Recessive RELN mutations have been associated with severe cerebellar hypoplasia in humans. We found molecular and genetic evidence that reductions in Reln expression contribute to GCp proliferative defects and cerebellar hypoplasia in GCp-specific Chd7 mouse mutants. Finally, we showed that CHD7 is necessary for maintaining an open, accessible chromatin state at the Reln locus. Taken together, this study shows that Reln gene expression is regulated by chromatin remodeling, identifies CHD7 as a previously unrecognized upstream regulator of Reln, and provides direct in vivo evidence that a mammalian CHD protein can control brain development by modulating chromatin accessibility in neuronal progenitors.
Introduction
Mutations in genes encoding chromatin remodeling factors have emerged as a major cause of neurodevelopmental disorders (1). The mechanisms employed by these factors to ensure precise regulation of developmental gene expression remain largely unexplored. Furthermore, the neuroanatomical abnormalities that result from these mutations and the behavioral and psychiatric features associated with specific brain defects remain unidentified for most of these conditions. Chromodomain–helicase–DNA-binding (CHD) proteins are ATP-dependent chromatin remodeling factors that use ATP to catalyze nucleosome translocation along chromatin, presumably to modulate access of transcriptional regulators (2). Mutations in CHD7 cause CHARGE syndrome, a complex developmental syndrome defined by a constellation of birth defects, which include coloboma, heart defects, atresia of the choanae, retarded growth and development, and genital and ear abnormalities (3–5). Although neurodevelopmental abnormalities are not considered for clinical diagnosis, 99% of patients exhibit developmental delay and 74% suffer from intellectual disability (6). The alterations in chromatin and brain structure that underlie these deficits have not been identified.
ChIP sequencing (ChIP-Seq) experiments in a variety of cell lines have identified widespread CHD7 recruitment across the genome (7–9). CHD7 appears to localize primarily to distal regulatory elements marked by lysine 4 monomethylation on histone 3 (H3K4me1). In vitro nucleosome remodeling assays have confirmed the ability of CHD7 to translocate nucleosomes along a chromatin template and demonstrated a loss or reduction in this activity in CHARGE syndrome–associated CHD7 mutants (10). These findings point to chromatin remodeling defects as a central pathogenic mechanism in CHARGE syndrome. However, whether this holds true in vivo and the effect of such changes on developmental gene expression have not been investigated.
As a first step toward identifying the neuroanatomical alterations that may underlie the neurological deficits in CHARGE syndrome, we recently reported cerebellar hypoplasia in 35% of patients with CHD7 mutations (11). It remains unknown to what extent cerebellar defects contribute to the developmental delay and intellectual disability associated with CHARGE syndrome.
The cerebellum develops over several weeks in mice and several months in humans. The cerebellar territory is established during early embryogenesis by the action of secreted signaling molecules from a secondary organizer located at the mid-hindbrain boundary (reviewed in ref. 12). We previously reported that biallelic expression of Chd7 was essential for maintaining appropriate levels of Fgf8 expression at this early stage of cerebellar development (11). As deregulated Fgf8 expression and signaling from this embryonic organizer selectively disrupt the formation of the cerebellar vermis, these findings identified a role for CHD7 in early embryonic cerebellar development (13, 14). Toward the end of embryogenesis, the cerebellum initiates a period of rapid growth, primarily driven by the proliferation of granule cell progenitors (GCps) in the external granule layer (EGL) on the surface of the cerebellar anlage. The primary mitogen driving GCp proliferation is Sonic Hedgehog (SHH), which is produced by postmigratory Purkinje cells (PCs) that become organized in a layer beneath the EGL during late embryonic stages (15–17). This process is associated with the formation of cerebellar folia. Intriguingly, we have also detected cerebellar foliation anomalies in CHARGE syndrome patients (11), implying a role for CHD7 in GCp development.
Several genes associated with cerebellar hypoplasia, developmental delay, ataxia, and intellectual disability in humans are expressed in cerebellar GCps (reviewed in ref. 18). Homozygous mutations in RELN, which encodes the secreted glycoprotein reelin, or the gene encoding its receptor VLDLR are responsible for severe cerebellar hypoplasia and intellectual disability in humans (19, 20). Studies in mouse models have localized Reln expression to GCps and have reported essential roles for RELN signaling in PC migration, maturation, and GCp proliferation (21–23). RELN has also been associated with psychiatric disease in several contexts. For example, RELN expression has been reported to be reduced in postnatal cerebella from autism patients (24, 25).
In this article, we report a role for CHD7 in controlling GCp proliferation and survival, and a striking downregulation of Reln gene expression in Chd7-deficient GCps. Increasing Reln expression partially rescued GCp proliferation and cerebellar hypoplasia, providing functional genetic evidence that Reln downregulation contributes to the cerebellar hypoplasia phenotype. Finally, we show that DNA accessibility is reduced at the Reln locus, and throughout the genome in CHD7-deficient GCps, consistent with a role for CHD7 in maintaining DNA accessibility through nucleosome remodeling.
Results
Chd7 deletion from neuronal progenitors results in cerebellar hypoplasia.
Chd7 is expressed in neural stem cells, where it has been reported to regulate the expression of developmental and disease-associated genes (7). Chd7 expression localizes to regions of ongoing neurogenesis in the developing brain: the ventricular zone (VZ) and hippocampus (Figure 1, A and C), as well as the rhombic lip stream, VZ, and EGL of the cerebellum (Supplemental Figure 1, A–E; supplemental material available online with this article; doi:10.1172/JCI83408DS1). Chd7 expression is maintained in postmitotic, differentiating, and migrating neural progenitors in the rostral migratory stream of the forebrain, and upregulated in the differentiating inner EGL and internal granule cell layer of the cerebellum (Supplemental Figure 1E and Figure 1, A and C), implicating Chd7 in neural differentiation. Several recent studies have identified roles for Chd7 in adult neural stem cell populations in the forebrain (26–28); yet Chd7 appears to have a relatively minor role in regulating neural stem cell expansion in the embryonic and perinatal forebrain (26). To determine whether Chd7 has a function in embryonic neurogenesis, we inactivated Chd7 in neuronal progenitors using a nestin-Cre line (29). In agreement with previous demonstrations of robust gene recombination in the cerebellar anlage by E11 (14), Chd7 mRNA was not detectable in the nestin-Cre Chd7fl/fl conditional knockout (cko) cerebellum at E12.5 (Supplemental Figure 1, F and G), and CHD7 protein, present in cell nuclei in the VZ and migratory cells emerging from the rhombic lip in control embryos, was absent from these cells (Supplemental Figure 1, H and I). The expression of Fgf8 from the mid-hindbrain organizer was maintained and the expression patterns of the Otx2 and Gbx2 homeobox genes were not altered in these mutants (Supplemental Figure 1, J–O), indicating that the deletion of Chd7 after the establishment of this embryonic organizer had little effect on the expression of genes linked to organizer function (11).
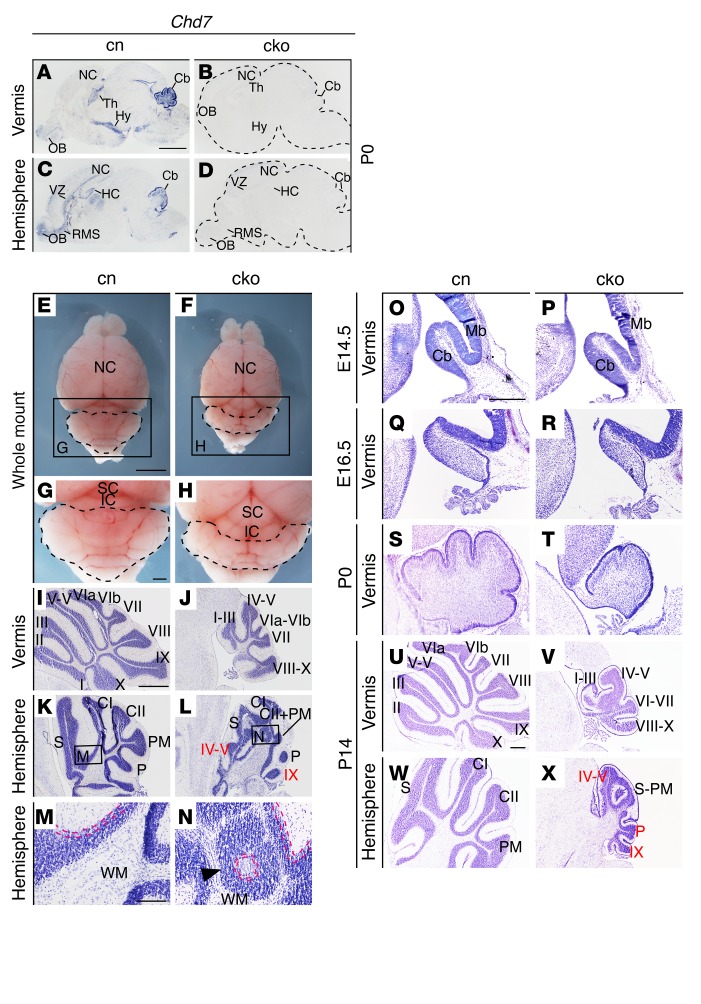
Figure 1. Chd7 deletion from embryonic neural progenitors causes cerebellar hypoplasia and foliation abnormalities.
(A–D) In situ hybridization for Chd7 exon 3 transcripts (blue) in sagittal sections through P0 Chd7fl/fl (control [cn]) and nestin-Cre Chd7fl/fl (cko) brains. Sections through the cerebellar vermis (A and B) and hemispheres (C and D) are shown. Note Chd7 expression in the olfactory bulb (OB), neocortex (NC), thalamus (Th), hypothalamus (Hy), hippocampus (HC), ventricular zone (VZ), rostral migratory stream (RMS), and cerebellum (Cb) in cn sections (A and C), absent in the cko (B and D). (E and F) Whole-mount images (anterior to the top) with cerebellum outlined. (G and H) High-power images with cerebella outlined. SC, superior colliculus; IC, inferior colliculus. (I–N) Cresyl violet–stained sagittal sections through the cerebellum, anterior to the left, with vermis folia labeled with Roman numerals according to Inouye and Oda (77). The simplex (S), Crus I (CI), Crus II (CII), paramedian (PM), and pyramidis (P) folia are labeled in hemisphere sections. Note cerebellar hypoplasia (J and L) and disorganized folia in cko hemispheres (L, boxed area). Note the expansion of lobules IV–V and IX from vermis into the hemispheres (L, red text). (M and N) Magnified view of the boxed areas in K and L with PC layers outlined in broken red lines. Ectopic granule cells organized around a circular cluster of PCs are indicated with a black arrowhead. WM, white matter. (O–X) Cresyl violet–stained sections through the developing cerebellum at the time points indicated, anterior to the left, with cerebellar folia labeled as above. Note the vermis hypoplasia at E16.5 and P0, and striking hypoplasia of both vermis and hemispheres at P14 (V and X) in the cko. Mb, midbrain. Scale bars: 1 mm (A, E, I, and O), 300 μm (G and U), 100 μm (M).
Newborn cko animals lacked Chd7 expression throughout the entire brain (Figure 1, B and D). These mice were born at the expected Mendelian ratios, but postnatal survival rate was reduced (Supplemental Table 1). Two mutants survived to P21; these were smaller than their littermates, overall brain size was reduced accordingly, but cerebellar size was disproportionally reduced (compare cerebella outlined in Figure 1, E with F, and G with H). Histological examination confirmed pronounced hypoplasia of all cerebellar lobules in the vermis (Figure 1, I and J). The cerebellar hemispheres were also hypoplastic and displayed highly irregular cerebellar foliation with apparently misdirected folia (Figure 1, K and L) and ectopic clusters of granule cells around clusters of PCs (Figure 1N). Following specific folia in serial sections (Supplemental Figure 2) revealed that vermis folia IV–V and IX extended abnormally into the lateral hemispheres (Supplemental Figure 2, I and J).
To identify the developmental stage at which these phenotypes first emerge, sagittal sections from E12.5 to P14 cerebella were examined. Cerebellar size was normal at E12.5 (Supplemental Figure 1, F–I) and E14.5 (Figure 1, O and P). Clear cerebellar growth retardation became evident at E16.5 (Figure 1, Q and R), a time point shortly after the initiation of GCp proliferation. At birth (P0), cerebellar hypoplasia was most clearly present in the cerebellar vermis, where it was associated with a failure to initiate the formation of most of the cardinal cerebellar fissures (Figure 1, S and T). By P14, the cerebellum was reduced in size, with irregular foliation that included expansion of vermis lobules IV–V and IX into the hemispheres (Figure 1, U–X). Taken together, these experiments identified a critical role for Chd7 in cerebellar growth after E14.5, implying a function for CHD7 in the expansion of cerebellar GCps.
CHD7 regulates the proliferation, differentiation, and survival of cerebellar GCps.
To specifically determine the function of Chd7 within the granule cell lineage, we deleted Chd7 from these cells from the time of their specification in the rhombic lip using a Math1-Cre transgene (30). Transgenic expression resulted in the recombination of the Chd7 conditional allele from the early stages of EGL formation at E14.5 (Supplemental Figure 3, C and D). By postnatal stages, efficient Chd7 deletion was evident in GCps in the anterior and central cerebellar vermis, with Chd7 expression reduced somewhat in anterior lobule VIII and spared in the most posterior (posterior IX and X) lobules, in agreement with previous reports on the activity of this transgene (Supplemental Figure 3, E–H, and ref. 31). Efficient Chd7 deletion was evident throughout the entire cerebellar hemispheres (Supplemental Figure 3, I and J). Immunostaining showed the same pattern of CHD7 protein depletion in the cerebellar vermis (Supplemental Figure 3, K–R). Together, these experiments identified the granule cell lineage as the predominant cell type in the postnatal cerebellum that express CHD7, with only faint expression remaining in cells in the white matter after granule cell–specific Chd7 deletion (Supplemental Figure 3, H and J). Efficient Chd7 deletion in GCps was further confirmed by quantitative reverse transcriptase PCR (RT-PCR) analysis of purified GCps from P7 animals (Supplemental Figure 3S).
The GCp-specific conditional mutants, from here on referred to as cko animals, survived to adulthood. Examination of whole-mount P21 cerebella revealed significant cerebellar hypoplasia (Figure 2, A and B). Histological sections confirmed hypoplasia of lobules I–VIII (Figure 2, C and D), with the most striking hypoplasia presenting in central lobules. All lobules in the hemispheres were smaller, and vermis folia IV–V and IX were again found to extend abnormally into the hemispheres (Supplemental Figure 4 and Figure 2, E and F) with associated foliation irregularities (Figure 2, G and H). Cerebellar development was followed from E13.5 to reveal the first signs of reduced cerebellar growth. Surface area measurements to estimate cerebellar size were performed on Cresyl violet–stained sagittal sections through the most medial aspect of the cerebellar vermis. Delayed initiation of cerebellar foliation in the vermis became evident at E17.5 (Supplemental Figure 5) before cerebellar size was significantly altered (mean area ± SEM = 0.26 ± 0.068 mm2 in controls, 0.26 ± 0.026 mm2 in cko). Hypoplasia became evident at E18.5 (mean ± SEM = 0.50 ± 0.014 mm2 in controls, 0.30 ± 0.026 mm2 in cko). To visualize the trajectory of cerebellar growth, representative sections (vermis and hemisphere) from control and cko cerebella were traced and overlaid (Figure 2I). This analysis revealed interesting differences: whereas vermis growth was clearly reduced from E18.5 onward, hypoplasia of the hemispheres became obvious only from P7 (Figure 2I).
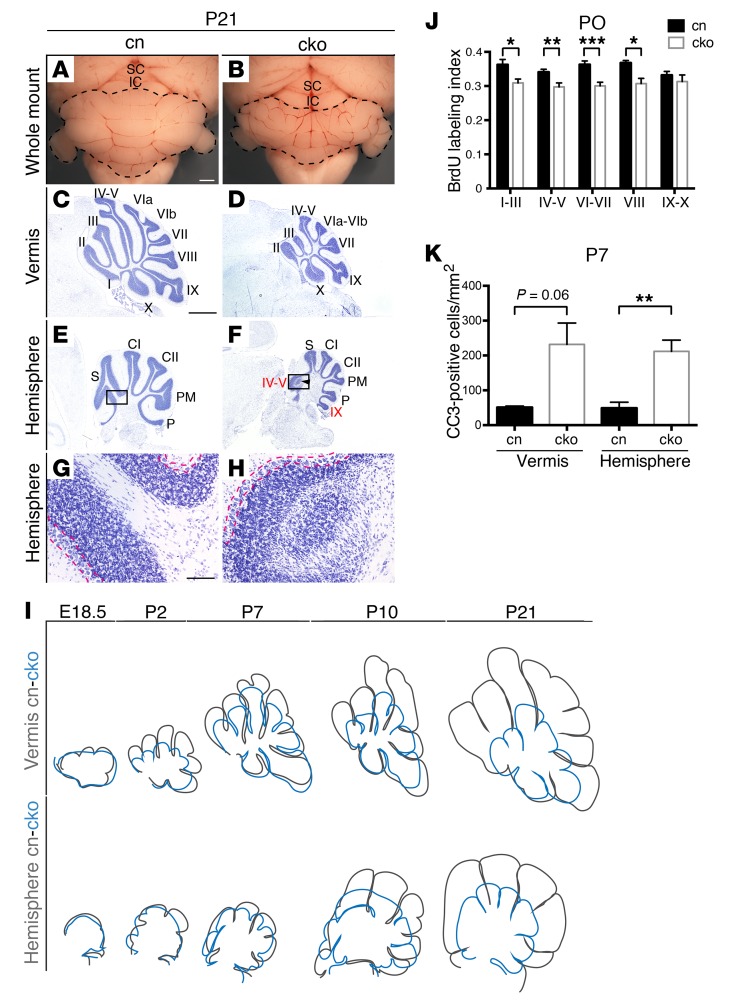
Figure 2. Chd7 regulates the proliferation and survival of cerebellar GCps.
(A and B) Dorsal whole-mount images of P21 Chd7fl/fl cn and Math1-Cre Chd7fl/fl GCp-specific conditional knockout (cko) mouse cerebella, anterior to the top. Note hypoplasia and abnormal foliation in the cko. (C–F) Cresyl violet–stained sagittal sections through the cerebellum, anterior to the left. Folia are numbered as in Figure 1. Note hypoplasia and abnormal folia indicated by an arrowhead and labeled in red in the cko. (G and H) Magnified view of the boxed areas in E and F with PC layers outlined by red broken lines. (I) Traced outlines of representative cerebellar sections from cn (black) and cko (blue) mice at the indicated time points. Foliation defects are first noted at E18.5 in the vermis and P2 in the hemispheres. Hypoplasia becomes evident in the vermis by E18.5 and hemispheres by P10 (see also Supplemental Figure 5). (J) Quantification of GCp proliferation in the P0 EGL by BrdU incorporation within a 1-hour time window. The BrdU labeling index for the indicated regions representing the fraction of BrdU-positive GCps in the EGL as counted in 100-by-100-μm areas. (K) The number of cleaved caspase-3–positive (CC3-positive) cells in the EGL per square millimeter of granule cell area at P7 is shown for vermis and hemispheres (n = 3 per genotype). *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test. Scale bars: 300 μm (A), 1 mm (C), 100 μm (G).
To determine the underlying cause of cerebellar hypoplasia, GCp proliferation was quantified by BrdU incorporation between E18.5 and P14. This analysis revealed a significant reduction in GCp proliferation in vermis lobules I–VIII at early postnatal stages (Supplemental Figure 6, A and B), most prominent at P0 (Figure 2J). No significant reduction in GCp proliferation was seen in hemispheres (Supplemental Figure 6D). These results indicated that reduced GCp proliferation likely contributed to reduced growth of the vermis, but not the hemispheres. To assess the contribution of apoptosis, sections were stained with an antibody against activated cleaved caspase-3. The number of apoptotic GCps was increased in both vermis and hemispheres at P7 (Supplemental Figure 6, E and F), but reached statistical significance only in the hemispheres (Figure 2K). These data suggested that the reduced postnatal growth of the hemispheres was largely caused by increased cell death, whereas both proliferative and apoptotic changes contributed to vermis hypoplasia.
Reduced PC numbers in Chd7 cko cerebella.
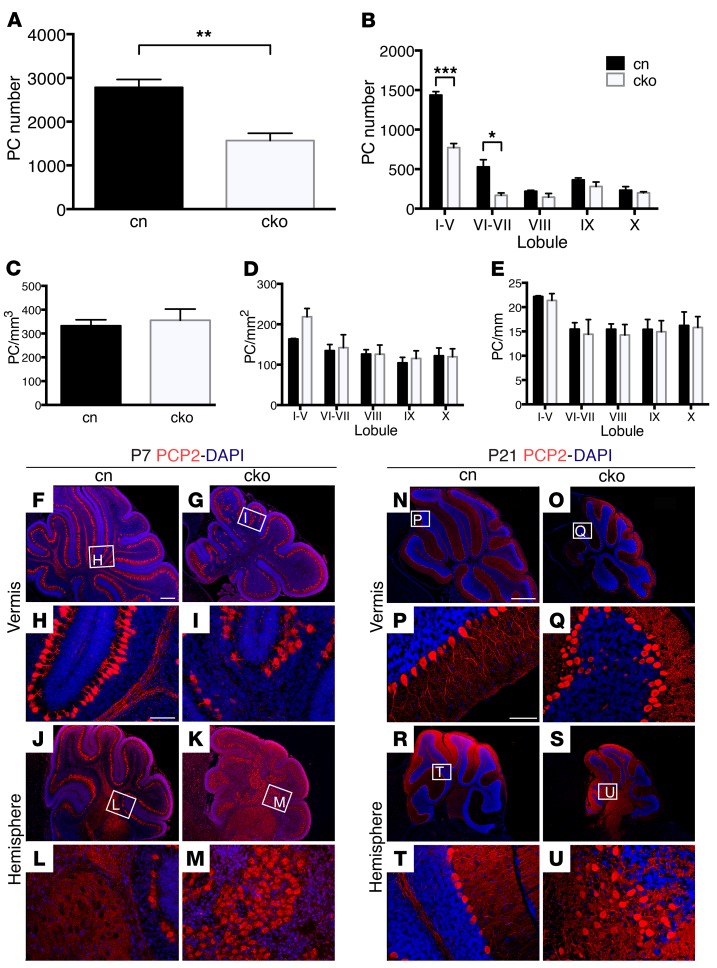
To determine the impact of the diminished production of granule neurons in the cerebellum on PCs, PC number and distribution were examined. Total PC numbers were reduced in cko cerebella at P21 (Figure 3A). A finer analysis of PC numbers across different cerebellar regions revealed that the reduction in PC number was due to fewer PCs in lobules I–VII of cko animals (Figure 3B), in agreement with the pronounced hypoplasia of anterior and central cerebellar lobules in the mutants (Figure 2D). PC density was not altered, indicating that the reduction in PC numbers remained proportional to the reduced cerebellar size (Figure 3, C–E). PCs in the cko cerebellar vermis were organized in monolayers for most of the cerebellum, with small regions of slightly disorganized cells observed at P7 (Figure 3, F–I) and P21 (Figure 3, N–Q). By contrast, large patches of disorganized PCs were present in the hemispheres at P7 (Figure 3, J–M) and P21 (Figure 3, R–U).
Figure 3. Deletion of Chd7 from GCps results in PC abnormalities.
(A) PC counts in cn and cko mouse P21 cerebella. Note the significant reduction in total PC number in the cko. (B) Lobule-specific PC counts. Note the significant reduction in PC number in the anterior and central lobules of the cko. (C–E) PC density in relation to the total vermis volume (mm3) (C), lobule granule cell area (mm2) (D), and length of the PC layer (mm) (E) (n ≥ 5 per genotype). *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test. (F–U) Examples of Purkinje cell protein 2 (PCP2) immunostains to visualize PCs in P7 cerebellar sections through the vermis (F–I) or hemispheres (J–M), with nuclear DAPI counterstain, anterior to the left. Magnified views of PC layers indicated by white boxed areas in corresponding low-power images are depicted in H, I, L, and M. Note the occasional disorganized PC distribution in the cko vermis (I), and more pronounced clusters of mislocalized PCs in the cko cerebellar hemispheres (M). (N–U) Examples of PCP2 immunostains to visualize PCs in P21 cerebellar sections through the vermis (N–Q) or hemispheres (R–U), with nuclear DAPI counterstain, anterior to the left. Magnified views of PC layers indicated by white boxed areas in corresponding low-power images are depicted in P, Q, T, and U. Note the occasional multilayered PC distribution in the cko vermis (Q), and more pronounced clusters of mislocalized PCs in the cko cerebellar hemispheres (U). Scale bars: 300 μm (F), 100 μm (H and P), 1 mm (N).
To understand the cause of these abnormalities, we investigated the distribution of LHX1/5+ PC progenitors over time as they migrate from their site of origin, the VZ toward the pial surface (23). This analysis revealed a relatively normal PC distribution, including formation of the PC plate under the pial surface at E16.5 (Supplemental Figure 7, A–H). Abnormal PC organization was evident by E18.5, with more dispersed PC progenitors in the central vermis (Supplemental Figure 7Q) and apparently mislocalized progenitors in the hemispheres (Supplemental Figure 7, S and T), consistent with the altered PC distributions seen in these regions at later stages. As CHD7 is also expressed in cells in the VZ, we asked whether nestin-Cre cko mice, in which Chd7 has been deleted from these progenitors in addition to GCps, exhibited more pronounced PC developmental defects. Indeed, although a clear PC plate still formed in the medial cerebellum by E16.5 (Supplemental Figure 7I), PC distribution was clearly irregular in the rest of the cerebellum (Supplemental Figure 7, J–L). Mislocalized cells were evident in the vermis and hemispheres by E18.5 (Supplemental Figure 7, U–X). Together, these findings suggested that the deletion of Chd7 from the early VZ progenitors contributed to PC defects in nestin-Cre cko mice, although we cannot rule out the possibility that the deletion of Chd7 from the rhombic lip stream and EGL earlier than in Math1-Cre cko mutants also contributed to these defects.
GCp-specific Chd7 conditional mouse mutants exhibit motor delay and coordination deficits.
Next, we asked whether the cerebellar defects were sufficient to cause behavioral abnormalities in these mice. Mutant mouse pups exhibited normal growth (Supplemental Figure 8A). Pups were assessed for the acquisition of developmental milestones, which revealed a delay in acquiring the righting reflex (Figure 4A), the ability to turn around when placed head facing downward on a sloping platform (negative geotaxis; Figure 4B), and the ability to reach out toward an object (Figure 4C). To determine whether adult cko animals had any motor deficits, their motor coordination on a revolving rotarod was examined. Male cko mice performed significantly worse on this test than their sex-matched control littermates (Figure 4D), whereas female cko mice showed no difference compared with controls (Figure 4E). There were no significant differences in body weight or grip strength between control and cko animals (Supplemental Figure 8, B and C), excluding general muscle weakness as a potential cause of this motor phenotype. The performance of cko animals of both sexes improved to a similar extent over the 3 days of testing, indicating that these mutants had no deficit in motor learning. Mutant animals showed no signs of repetitive behaviors (Figure 4F) or anxiety (Supplemental Figure 8, D and E).
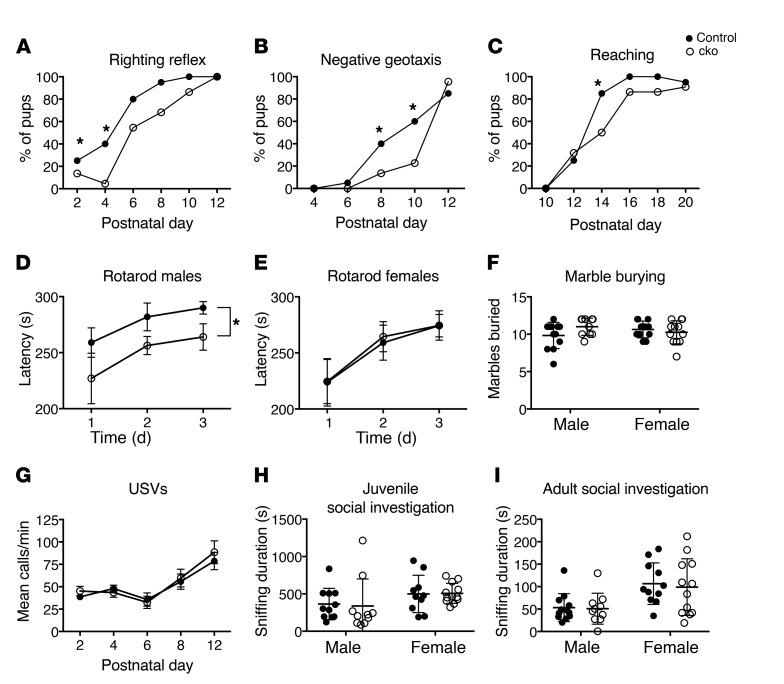
Figure 4. Deletion of Chd7 from GCps leads to developmental delay and motor coordination deficits.
Behavioral assessments of separate cohorts of adult (D–F, H, and I) cn (male n = 12, female n = 11) and cko (male n = 10, female n = 12) mice and pups for analysis of developmental milestones (cn n = 20, cko n = 22) or ultrasonic vocalizations (USVs) (cn n = 21, cko n = 23). (A–C) The percentage of pups at the indicated postnatal ages demonstrating the full righting reflex (A), a score of 1 or higher for negative geotaxis (B), or the full reaching response (C). Note the delay in acquiring these motor milestones in cko compared with cn animals (*P ≤ 0.05, χ2 tests). (D and E) The mean latency of male (D) and female (E) mice to fall from the rotarod. Note the significant difference between the male cko and cn animals (*P ≤ 0.05, repeated-measures ANOVA, with Student’s t test as post hoc analysis). (F) The mean number of ultrasonic vocalizations per minute at the indicated postnatal ages for cn and cko mice (repeated-measures ANOVA). (G) Average number of marbles buried within 30 minutes. Note no significant difference between the groups (between-subjects ANOVA). (H and I) The mean duration of total social investigation of an age- and sex-matched novel conspecific at P21 (H) and in adulthood (I). No significant difference between the cn and the cko groups was observed (repeated-measures ANOVA).
Cerebellar defects and dysfunction have also been proposed to be associated with autism spectrum disorders (reviewed in ref. 32). We therefore also performed tests to assess social interactions and communication. Mice communicate via vocalizations in the ultrasonic and sonic ranges, particularly in social situations such as when pups are separated from the mother (33). Mutant pups exhibited no difference in ultrasonic vocalizations (Figure 4G). Social investigation of conspecific age- and sex-matched mice by juvenile cko mice was normal (Figure 4H), as were reciprocal social investigations (Figure 4I) and sociability in the 3-chamber social approach test (Supplemental Figure 8F) in adult mice. In contrast to the Chd7 heterozygous mouse models (34, 35), no differences were observed in olfactory discrimination tests (Supplemental Figure 8, G and H), consistent with the cerebellar-specific Chd7 deletion in these mutants.
In addition to the well-established role of the cerebellum in sensorimotor function, structural and functional studies have also implicated the cerebellum in cognition and spatial processing (36). In humans, cognitive tasks tend to activate central cerebellar lobules VI, VII, and Crus I/II (reviewed in refs. 37, 38). Given the particularly strong phenotypes seen in the central cerebellar lobules of the cko animals, we also evaluated the performance of these mutants in the Morris water maze test, a cognitive task that assesses visual-spatial processing and memory. These conditional mutants exhibited no deficits in this task compared with control animals (Supplemental Figure 8I).
In conclusion, our behavioral tests indicate that cerebellar hypoplasia and PC hypocellularity in Chd7 cko mutants are associated with developmental delay and motor coordination. By contrast, these defects are not sufficient to cause deficits in social behaviors or cognition.
CHD7 controls gene expression levels in GCps.
Chd7 encodes a chromatin remodeling factor that has been postulated to function as a “transcriptional rheostat” to maintain developmental gene expression at physiological levels (9). Thus, to identify CHD7-regulated genes in GCps, a direct comparison of the Chd7-deficient and control transcriptome was performed by RNA sequencing (RNA-Seq). RNA was isolated from purified GCps at P7, when sufficient numbers of pure, primary GCps can be isolated ex vivo. A total of 881 coding transcripts with significantly changed expression (FDR < 0.05) were identified in Chd7-deficient GCps, compared with controls. Nearly equal numbers of genes were downregulated (n = 435) or upregulated (n = 446) (Figure 5A and Supplemental Table 2).
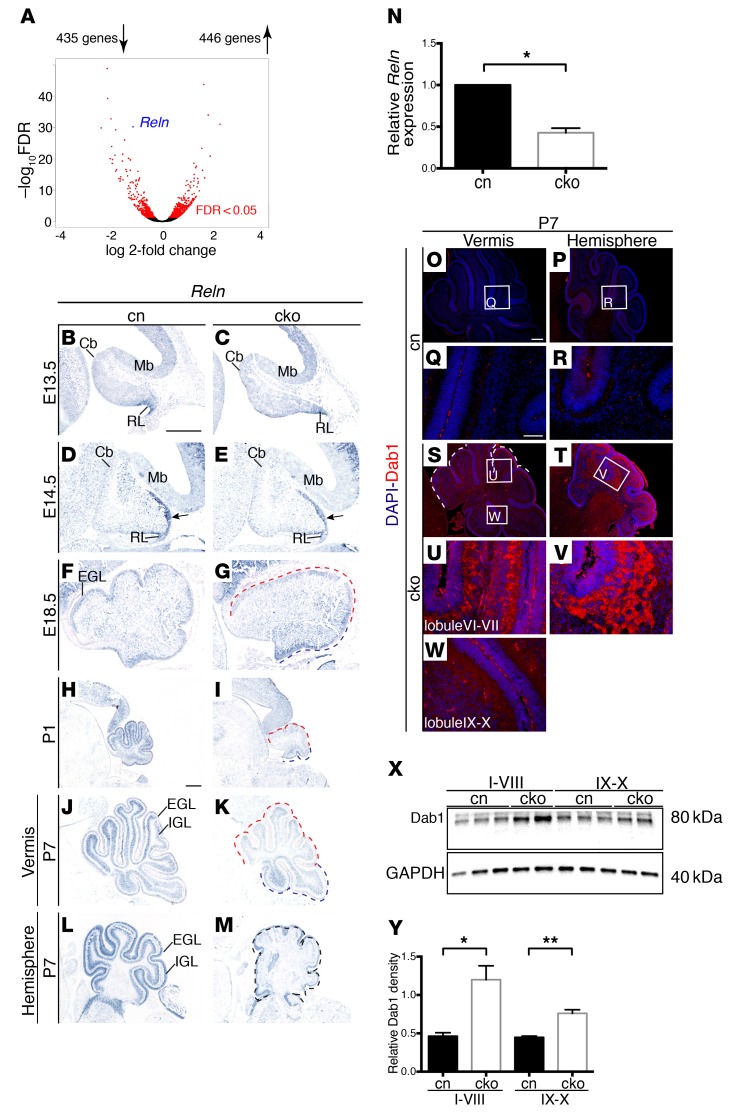
Figure 5. CHD7 regulates Reln gene expression in GCps.
(A) Volcano plot of RNA-Seq data comparing gene expression of purified GCps from P7 cko and cn mice. Red points indicate genes with statistically significant (FDR < 0.05, n = 2) differences in expression. Reln is highlighted in blue. (B–M) In situ hybridization with an antisense Reln probe on sagittal cerebellar sections at the indicated time points. Note reduced Reln expression in the rhombic lip stream at E14.5 (arrow in E), anterior vermis of E18.5, P1, and P7 cerebella (red dashed line in G, I, and K), and all lobules of the hemispheres at P7 (M), with normal expression in the posterior vermis (navy dashed line in G, I, and K). IGL, internal granule layer. (N) Quantification of Reln transcript levels in purified P7 GCps confirms significant reduction in Reln expression in cko GCps (3 samples of purified GCps extracted from pooled cerebellar samples of each genotype). (O–W) Immunostaining for DAB-1 on sagittal sections of P7 mouse cerebella. High-magnification views of indicated regions in O, P, S, and T are shown in Q, R, and U–W, respectively. Increased DAB-1 protein levels in the anterior vermis lobules and hemisphere of cko mice are indicative of reduced RELN signaling. (X) Western blot analysis of DAB-1 protein levels in lobules I–VIII and IX–X. Note the increase in protein levels in cko samples (n = 3) compared with cn (n = 2), with more marked increases identified in lobules I–VIII of the cko. (Y) DAB-1 protein levels quantified relative to GAPDH. *P < 0.05, **P < 0.01, Student’s t test. Scale bars: 300 μm (B, H, and O), 100 μm (Q).
This experiment revealed significant downregulation of the Reln gene (labeled in blue in Figure 5A). Homozygous mutation of the RELN gene is associated with pronounced cerebellar hypoplasia in humans and mice (19, 39), suggesting that Reln downregulation might contribute to cerebellar hypoplasia in Chd7 cko mutants. In situ hybridization studies revealed a subtle reduction of Reln expression in the EGL already at E14.5 (Figure 5, D and E), E18.5 (Figure 5, F and G), and substantial downregulation in Chd7-deficient GCps in the anterior and central cerebellum at P1 (Figure 5, H and I) and P7 (Figure 5, J and K). In agreement with the efficient deletion of Chd7 throughout the lateral cerebellum (Supplemental Figure 3J), Reln expression was strongly downregulated in cerebellar hemispheres (Figure 5, L and M). Quantitative RT-PCR analysis showed that Reln expression was reduced by over 50% in cko GCps, compared with control GCps (Figure 5N). RELN signaling is associated with the degradation of the intracellular protein DAB-1 (21, 40–42), and therefore, reduced RELN signaling should result in increased levels of DAB-1 protein. Immunostaining with a DAB-1–specific antibody revealed a clear upregulation of DAB-1 protein in PCs in the anterior and central vermis lobules and hemispheres of Chd7 cko cerebella (Figure 5, S–V), compared with lobule X (Figure 5W) and control cerebella (Figure 5, O–R). These findings were corroborated by immunoblotting, which also revealed increased DAB-1 protein in the cko cerebellum, compared with the control cerebellum (Figure 5, X and Y). These observations confirmed that Reln downregulation was sufficient to cause a substantial reduction in RELN signaling in the developing cerebellum and identify CHD7 as an important regulator of Reln gene expression and signaling levels in the developing cerebellum.
Reduced RELN signaling contributes to cerebellar hypoplasia in Chd7 cko mice.
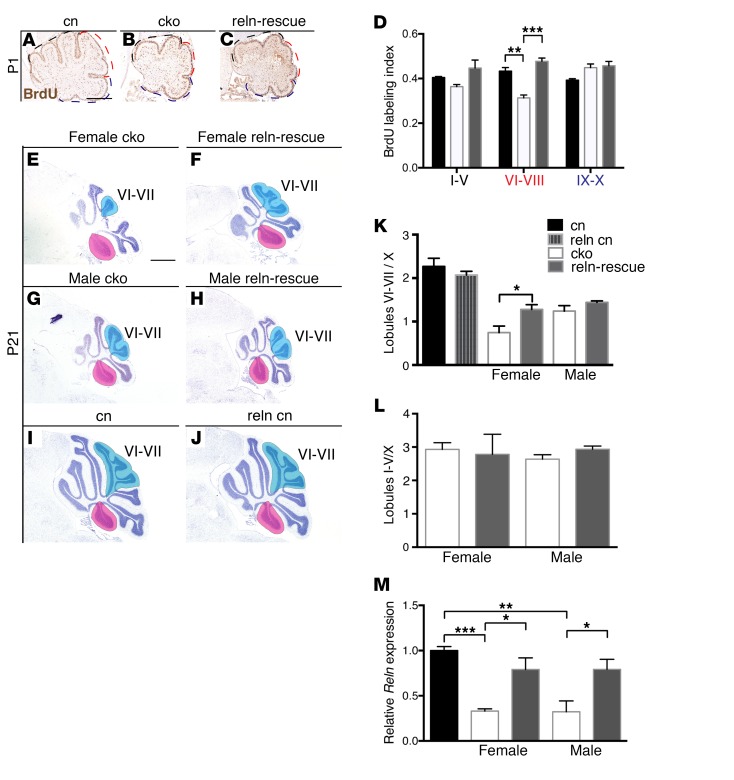
To test whether reduced RELN signaling was responsible for cerebellar hypoplasia in Chd7 cko mice, we sought to rescue this phenotype by increasing Reln expression in these mutants in vivo. For this purpose, we used a nestin-Reln transgenic mouse (RelnTG) reported to misexpress Reln throughout the developing cerebellum at 10%–20% of WT levels until P5 (43). Despite its low, transient, and ectopic expression, this transgene was capable of partially rescuing the cerebellar hypoplasia in homozygous reeler mice (43). We therefore generated Math1-Cre Chd7fl/fl RelnTG (referred to as reln-rescue) mice. We first asked whether the transgene could normalize the GCp proliferation defect observed in Chd7 cko mutants at P1 (Figure 6, A–D). The defect was fully corrected in reln-rescue mice (Figure 6D). To examine whether transient transgenic Reln expression had any lasting effect on the size of these central cerebellar lobules, P21 cerebella were examined. As Chd7 is not deleted (Supplemental Figure 3, H and R) and Reln expression is not altered (Figure 5K) in lobule X as a result of the inefficiency of the Math1-Cre transgene in the posterior cerebellum, we measured the size of the central lobules as a fraction of lobule X as internal control. This analysis revealed a significant increase in the relative size of the central lobules VI–VII in female reln-rescue mice, compared with Chd7 cko mice (Figure 6, E, F, and K). By contrast, RelnTG expression did not significantly increase the size of the central lobules in male mice (Figure 6, G, H, and K). Control RelnTG mice had a normal cerebellar size and morphology (Figure 6, I–K). The relative size of anterior lobules (I–V) was not rescued by RelnTG expression (Figure 6L), consistent with the lack of significant effect on GCp proliferation here (Figure 6D). Quantification of Reln transcript levels in E18.5 cerebella confirmed an almost 2-fold upregulation of Reln transcripts in RelnTG mice compared with mutants (Figure 6M), consistent with the near-complete rescue of proliferation at P1 (Figure 6D). Importantly, there was no difference in Reln levels between male and female cko cerebella, indicating that sex differences in Reln expression levels could not explain the sex-specific rescue effects seen (Figure 6M). In summary, our genetic studies confirmed that reduced Reln expression significantly contributes to the proliferation deficit of Chd7-deficient GCps in central cerebellar lobules and may affect cerebellar growth in a sex-specific manner.
Figure 6. Reln downregulation is partly responsible for cerebellar hypoplasia in GCp-specific Chd7 cko mice.
(A–C) Sagittal sections of BrdU-treated cn, cko, and reln-rescue cerebella at P1, immunostained with anti-BrdU antibody (brown). Dashed lines indicate anterior (black), central (red), and posterior (navy) cerebellum used for BrdU counts. (D) Quantification of the BrdU labeling indices in regions indicated in A–C. Note reduced GCp proliferation in the central lobules (VI–VIII) of the cko mice and restoration in reln-rescue mice (n = 3 per genotype). (E–J) Cresyl violet–stained sagittal sections of the cerebellar vermis at P21; sex-genotype combinations are indicated, anterior to the left. Blue shading indicates the central lobules (VI–VII); lobule X is highlighted in pink. (K) Quantification of the volume of lobules VI–VII relative to lobule X calculated from serial sections through the cerebellar vermis. Note the rescue of the central lobule hypoplasia in female reln-rescue mice (n = 3 per genotype). (L) Quantification of the volume of lobules I–V in the cko and reln-rescue mice relative to lobule X calculated from serial sections through the cerebellar vermis. (M) Quantification of Reln transcript levels in E18.5 whole cerebella. Note that Reln expression is reduced in the cko and restored in the reln-rescue irrespective of sex (n = 5 per genotype). *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test.
CHD7 is required for the maintenance of DNA accessibility at the Reln promoter.
CHD7 is thought to function primarily by modulating DNA accessibility at developmental regulatory elements through its ATP-dependent chromatin remodeling activity. We hypothesized that CHD7 assists in the maintenance of an “open,” accessible chromatin structure at CHD7-regulated regulatory elements in primary GCps. To address this question, DNA accessibility in control and Chd7 cko GCps was compared using an assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-Seq) (44). In total, 142,047 ATAC-Seq peaks, i.e., regions where the DNA is accessible to transposase, were identified in the control cells (Supplemental Table 3). We compared the ability of ATAC-Seq to identify regions of accessible DNA with DNase I hypersensitive sites sequencing (DNase-Seq) data recently published from GCps (45). Our analysis of DNase-Seq data identified 50,022 accessible peaks in P7 GCps, 88% (43,782) of which were within 100 bp of an ATAC-Seq peak, indicating that ATAC-Seq can reliably identify accessible regions.
Having validated the ATAC-Seq approach, we asked whether DNA accessibility was significantly altered at any of these regions in Chd7-deficient GCps. We identified 4,921 regions that showed significantly reduced DNA accessibility in Chd7-deficient GCps (CTRL/KO, Figure 7A), and only 210 regions with increased accessibility (KO/CTRL, Figure 7A). This finding suggested that CHD7 primarily functions in the maintenance of open, accessible chromatin states in the genome.
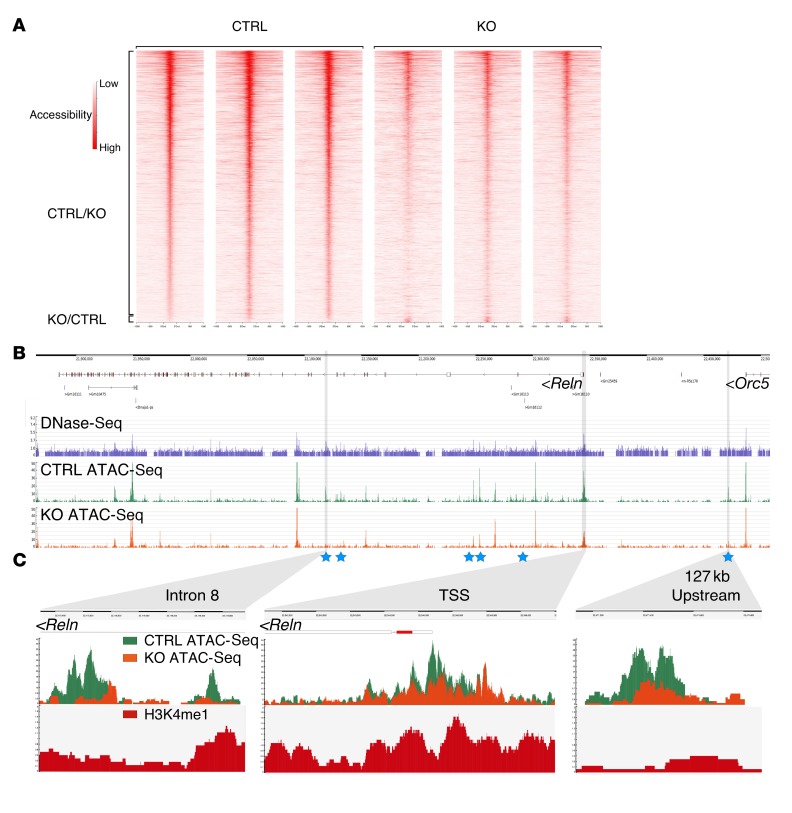
Figure 7. CHD7 deletion alters chromatin organization and reduces DNA accessibility at the Reln locus.
(A) Heatmaps representing all significantly changed ATAC-Seq peaks ± 500 bp between control (CTRL) and cko (KO) GCps (n = 3). Note reduced DNA accessibility in KO samples for the majority of peaks (CTRL/KO peaks), with a few showing increased DNA accessibility in the KO (KO/CTRL peaks). (B) DNase-Seq (purple) (45) and ATAC-Seq reads mapped over the Reln gene in control (CTRL ATAC-Seq, green) and cko (KO ATAC-Seq, orange) P7 GCps. Peaks are indicative of regions of “open” chromatin with high DNA accessibility. Blue stars indicate ATAC-Seq peaks that are significantly different in CTRL and KO cells. (C) Visualization of normalized CTRL (green) and KO ATAC-Seq (orange) reads, and H3K4me1 ChIP-Seq reads from WT GCps (red) that mapped to intron 8, the Reln transcriptional start site (TSS), and a region 127 kb upstream of the Reln TSS. Note significantly reduced DNA accessibility at the intronic and upstream regions in KO cells compared with CTRL cells.
Next, we determined whether any of these differentially regulated regions were in close vicinity of differentially expressed genes. Four hundred seventy-two (9.6%) of the peaks with reduced accessibility mapped to a location within 1 kb of a differentially expressed gene identified by RNA-Seq. Of these regions, 333 (70.6%) and 139 (29.4%) were located within 1 kb of a downregulated and upregulated gene, respectively. Thus, although there appears to be some association of downregulated gene expression with reduced DNA accessibility, this is by no means absolute. A similar observation was made for regions that showed increased accessibility in cko cells. Of the 210 peaks, 20 (9.5%) mapped to a location within 1 kb of a differentially expressed gene. Of these peaks, 11 (55%) and 9 (45%) were within 1 kb of a downregulated and upregulated gene, respectively.
Finally, we asked whether the accessibility of any potential regulatory elements at the Reln locus was altered in Chd7-deficient GCps. A direct comparison of DNase-Seq and ATAC-Seq profiles over the Reln locus (Figure 7B) clearly showed the superiority of the ATAC-Seq approach in being able to unambiguously identify regions of DNA accessibility. A statistically significant reduction in DNA accessibility was evident at 6 putative regulatory elements at the Reln locus (blue stars, Figure 7B). As examples of regions with reduced DNA accessibility, zoomed-in views of a region in intron 8 and 127 kb upstream of the transcriptional start site are shown (Figure 7C). Although accessibility at the Reln transcriptional start site also appears to be reduced, these changes did not reach statistical significance. Finally, we confirmed that these regions were positive for the H3K4me1 histone modification, a marker of regulatory regions to which CHD7 can be recruited (8). Thus, our findings are consistent with a model whereby CHD7 is recruited to regulatory elements at the Reln locus, where it directly modulates chromatin structure. Together, these observations suggest a crucial role for CHD7 in maintaining an accessible, “open” chromatin structure throughout the genome and at the Reln locus.
Discussion
In this article we define an important role for CHD7 in the expansion of the GCp pool in the perinatal cerebellum. Together with our previous work (11), we conclude that CHD7 controls cerebellar growth at 2 distinct stages of cerebellar development. CHD7 is essential for the maintenance of rhombomere 1 identity and Fgf8 expression in the embryonic isthmus organizer, with disruption of the latter predisposing the embryo to cerebellar vermis hypoplasia. In this article, we show that CHD7 expression becomes localized to cerebellar GCps where it regulates GCp proliferation and survival. CHD7 deletion from GCps as well as PC progenitors in the VZ from E12.5 with nestin-Cre resulted in a more severe phenotype, potentially implicating CHD7 expression in the VZ in cerebellar development. Chd7 gene deletion specifically in the cerebellar VZ using Ptf1a-Cre will be necessary to address this possibility directly. We show that reduced RELN signaling in CHD7-deficient GCps can partially account for the cerebellar hypoplasia. The RelnTG line we used in these experiments does not express the Reln transgene past P5, likely explaining the limited phenotypic rescue achieved by P21. Nevertheless, the possibility that other genes deregulated in CHD7-deficient GCps also contribute to reduced GCp expansion remains likely.
The exact mechanism whereby reduced RELN signaling results in GCp proliferation defects is not known. In homozygous reeler (Relnrl/rl) mutants, PC migration from the VZ is disrupted, presumably leading to a deficit in PC-derived mitogenic signals (23). PC positioning is abnormal in our mutants, especially in the cerebellar hemispheres, suggesting that PC disruption may contribute to the hypoplastic phenotype. Expression analysis of several genes transcriptionally regulated by SHH signaling (Gli1, Ptch1, and Mycn) by in situ hybridization and quantitative RT-PCR revealed no significant downregulation in their expression, suggesting that PC disruption in our mutants may affect GCp proliferation in an SHH-independent manner. Alternatively, PC defects may not be responsible for GCp proliferation defects, and RELN downregulation may affect GCp proliferation in a cell-autonomous manner. Consistent with this latter possibility, transcripts encoding the RELN receptors ApoER2, VLDLR, and ephrin B1, B2, and B3 are present in GCps (RNA-Seq data, Supplemental Table 2). GCp-specific deletion of Reln and combinations of genes encoding the different RELN receptor genes will be necessary to examine this possibility.
The finding that increased Reln expression had a significant effect on the size of the central lobules in female cko mice, but not males (Figure 6K), suggested that perhaps female mice were more sensitive to changes in Reln expression. Interestingly, sex-genotype interactions appear to be important in the expression of psychiatric diseases associated with RELN. Specifically, genome-wide association studies have identified common variants in the RELN gene that increase the risk of schizophrenia and bipolar disorder only in women (46, 47). Sex-specific differences in the anatomical and behavioral phenotype of heterozygous reeler mice (Relnrl/+) have also been reported (48). PC degeneration has been identified from postnatal day 15 in male Relnrl/+ mice, and motor dysfunction has been reported in adult males (49, 50). Reduced Reln expression and neuroactive steroid levels appear to be critical in the pathophysiology of the Relnrl/+ phenotype, since administration of 17β-estradiol protects against PC degeneration in males (49). Taken together, these studies are indicative of a complex interplay between RELN signaling and sex hormones during cerebellar development. In the present study, we ruled out a simple model whereby Reln gene expression in GCps differs in the 2 sexes (Figure 6M).
We report here that adult GCp-specific Chd7 cko mutant mice with cerebellar hypoplasia exhibit a range of behavioral deficits. The identification of developmental motor delay and mild motor deficits in these animals implies that cerebellar hypoplasia may underlie some of these features in CHARGE syndrome patients. We propose that clinical studies to assess and compare the severity of motor delay and coordination deficits in CHARGE syndrome patients with or without cerebellar hypoplasia will be important to delineate the contribution of cerebellar defects to these deficits. Our findings may also have implications for understanding other clinical features of CHARGE syndrome. We note that several CHD7-regulated genes implicated in other cell types and tissues affected in CHARGE syndrome (e.g., Sema3a, neural crest; ref. 51) are also dysregulated in GCps, perhaps indicative of common gene regulatory mechanisms in different developmental contexts.
Interestingly, we detected no deficits in social behavior in these mutants. This finding is important to consider in the context of cerebellar anomalies described in patients diagnosed with autism. For example, hypoplasia of the central vermis lobules VI–VII has been reported in autism spectrum disorder (ASD) cases (52, 53). Our data suggest that marked hypoplasia of the central cerebellar lobules, associated with reduced PC numbers in these lobules, is not sufficient to cause social deficits in mice. This conclusion has to be tempered by consideration of several caveats. First, mouse models might not be ideal for addressing this question, as the nature of cerebellar–prefrontal cortex connections may be qualitatively or quantitatively different from that in primates and humans (12). Second, the behavioral tests used to evaluate social investigations in mice may not be sensitive enough to detect appropriate changes in behavior. Another neuroanatomical anomaly frequently reported in the context of ASD is the reduction in PC number in postmortem brain samples from ASD patients (reviewed in ref. 54). It was reported recently that conditional deletion of the Tsc1 gene from cerebellar PCs alone was sufficient to cause autism-like behaviors in a mouse model (55). Our data indicate that a reduction in PC number as a consequence of disruption of the normal developmental process is, by itself, not sufficient to cause autism-like social behaviors in mice.
A number of nucleosome remodeling factors appear to be critical for normal brain development, and mutations that disrupt their normal function are associated with neurodevelopmental disorders (56). Identifying the changes in chromatin structure that result from disrupted function of these factors is an important step toward understanding the mechanisms that underlie these conditions. The prevailing model for CHD7 function is that CHD7 remodels chromatin by sliding nucleosomes along chromatin to modulate DNA accessibility at key developmental regulatory elements (57). We identified a striking reduction in DNA accessibility throughout the genome in CHD7-deficient cells, consistent with a role for CHD7 in the maintenance of regions of open, accessible chromatin. This finding is in keeping with the activity of the CHD7 homolog, kismet, that was operationally identified as a Trithorax family member that functions as an antagonist of Polycomb factors (58), which promote chromatin compaction and gene repression (59).
In conclusion, we have defined a critical role for CHD7 in cerebellar GCp expansion and identified CHD7 as a regulator of Reln gene expression, thereby linking CHD7 to a pathway associated with cerebellar hypoplasia and neuropsychiatric disease in humans. Behavioral analyses of GCp-specific mouse mutants revealed important impacts of cerebellar hypoplasia on motor development and function. These findings have implications for understanding the neurodevelopmental basis of motor delay in CHARGE syndrome. Intriguingly, our data do not support a view where cerebellar hypoplasia per se is sufficient to cause behavioral deficits in the social domain, at least not in mouse models. Finally, we identify a crucial molecular function of CHD7 in the maintenance of open, accessible chromatin in primary neuronal progenitors.
Methods
Mice.
The Chd7flox (28), nestin-Reln transgenic (RelnTG) (43), Math1-Cre (30), and nestin-Cre (29) mouse lines, provided by Pete Scambler (Institute of Child Health, London, United Kingdom), Tom Curran (Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA), David Rowitch (UCSF, San Francisco, California, USA), and Gill Bates (University College London, London, United Kingdom), respectively, were genotyped by PCR using tail or ear DNA. Mouse lines were backcrossed with C57BL/6J mice for at least 3 generations. Conditional neural-specific mutants were obtained from nestin-Cre Chd7fl/+ × Chd7fl/fl matings. Conditional GCp-specific mutants were obtained from Math1-Cre Chd7fl/fl × Chd7fl/fl matings. For behavioral tests, sex-balanced matings were used to produce animals for testing to control for any sex-specific parental genotype effects. Juvenile (3-week-old) C57BL/6J mice used as conspecifics were purchased from Charles River Laboratories and group-housed by sex.
Mice were bred in the Biological Services Unit at Guy’s Campus or the Institute of Psychiatry, Psychology & Neuroscience, King’s College London.
Histology.
Samples were dissected in PBS, fixed overnight in 4% paraformaldehyde (PFA) at 4°C, dehydrated, and embedded in paraffin wax. Serial, sagittal sections were cut at 10 μm and left to dry overnight at 42°C.
Volumetric analysis and PC counts.
The surface area of serial, Cresyl violet (0.1%)–stained sections was measured in ImageJ (NIH) and the vermis volume calculated by multiplication of the total surface area by the section thickness (10 μm). PCs were counted in 5 separate, nonadjacent 10-μm vermis sections (3 cerebella per genotype) in H&E-stained sections.
Immunohistochemistry.
Sections were deparaffinized in xylene and rehydrated, heated in a sodium citrate buffer (pH 6.0), permeabilized in 0.2% Triton X in PBS (PBSTx), and blocked in 10% heat-inactivated goat serum (GS) in PBSTx for 1 hour before incubation in primary antibody in 5% GS-PBSTx overnight at 4°C. Antibodies were: rabbit anti–DAB-1 (1:150; Sigma-Aldrich, D1569), rabbit anti–Purkinje cell protein 2 (anti-PCP2) (1:200; gift from Brad Denker, Harvard University, Boston, Massachusetts, USA), mouse anti-BrdU (1:100; BD Biosciences 347580), and cleaved caspase-3 (Asp 175) (1:150; Cell Signaling Technology 9661). For DAB-1 and PCP2 detection, sections were incubated with Alexa Fluor–labeled secondary antibodies (1:200; Life Technologies) and counterstained with DAPI. Slides were mounted with Citifluor (Citifluor Ltd.), and fluorescent images were captured using a Nikon Eclipse 80i microscope with a Nikon Y-QT Hamamatsu C4742-95 camera. For BrdU and cleaved caspase antibodies, sections were incubated with polyclonal goat anti-mouse or goat anti-rabbit biotinylated secondary antibody (1:200; Dako) for 1 hour. Signal was amplified using the VECTASTAIN Avidin/Biotin Complex (ABC) kit (1:200; Vector Laboratories Ltd.), and visualized with 3,3′-diaminobenzidine substrate (0.03%; Sigma-Aldrich). Sections were then counterstained with hematoxylin (30 seconds), dehydrated, mounted in DPX, and imaged with a Nikon Eclipse 80i.
BrdU.
Pregnant females or pups were given 1 i.p. injection of 50 mg/kg or 20 mg/kg 5′-bromo-2′-deoxyuridine (BrdU), respectively. Mice were sacrificed 1 hour later, and brains were fixed with 4% PFA overnight at 4°C, washed in PBS, and processed as described above. The BrdU labeling index was calculated by counting of the number of BrdU-labeled cells and the total number of cells in the EGL in 100-by-100-μm areas (2–3 fields per lobule) using a Nikon Eclipse 80i microscope. The BrdU-positive GCps were then plotted as a fraction of the total number of GCps per lobule.
Western blot.
P7 cerebella were dissected in PBS and lobules IX and X separated from lobules I–VIII. Tissue was lysed in 8 M urea, 1% CHAPS, 50 mM Tris (pH 7.9) followed by the removal of DNA by centrifugation. Proteins (10 μg per lane) were resolved on a Mini-PROTEAN precast gel (Bio-Rad, United Kingdom) and transferred to a nitrocellulose membrane (Bio-Rad, United Kingdom). After blocking with 0.5% nonfat milk powder in TBS with 0.5% Tween-20 (TBST), the membrane was incubated with primary antibodies (rabbit anti–DAB-1 [C-terminal], Sigma-Aldrich, D1569, 1:5,000; rabbit anti-GAPDH, Abcam, ab181602, 1:10,000) in 5% BSA, TBST overnight at 4°C. After washing and incubation with HRP-conjugated secondary antibody (Millipore) for 1 hour at room temperature, HRP conjugates were detected using Clarity Western ECL reagent (Bio-Rad). Gels were visualized on a Bio-Rad gel doc system and images analyzed with Image Lab (Bio-Rad) and prepared using Adobe Photoshop.
In situ hybridization.
In situ hybridization was performed using standard methods (60). DNA templates were produced by PCR amplification from mouse genomic DNA. The PCR primers used were: Chd7 (exon3) forward 5′-TTGGTAAAGATGACTTCCCTGGTG-3′, reverse 5′-GTTTTGGCGTGACAGTTTTTGC-3′; Reln (exon 65) forward 5′-CAACAGAAGACGAAGGTCGCTTAG-3′, reverse 5′-ACAAGTAGGTCAAAGTCCAGCAGC-3′. Digoxigenin-labeled antisense probes for Chd7 and Reln were produced by in vitro transcription using a DIG RNA labeling kit with T7 RNA polymerase (Roche).
GCp purification and quantitative PCR.
P7 cerebella were dissected in DPBS (Gibco), and lobules IX and X were removed. The cerebella of the individuals from each genotype were pooled. Cells were dissociated using Papain I (100 U; Worthington) followed by trituration. Granule cell precursors were isolated by Percoll fractionation with a 35% and 65% Percoll gradient (Sigma-Aldrich) as previously described (14).
Total RNA was extracted from the purified granule cells of 3 samples of each genotype using Trizol (Invitrogen). cDNA was synthesized from 200 ng of RNA using the nanoScript 2 Reverse Transcription kit (Primerdesign Ltd.) with random hexamer primers. Quantitative RT-PCR was performed on a Stratagene Mx3000P (Agilent Technologies) using the Precision qPCR master mix with SYBR Green (Primerdesign Ltd.). All reactions were run in triplicate, and first-strand DNA synthesis reactions without reverse transcriptase were used as controls. The quantification cycle (Cq) threshold values were normalized to the housekeeping gene Gapdh to calculate the ΔCq value and the ΔΔCq calculated relative to the control sample. The primer sequences were: Chd7 forward 5′-TCACCAGCCTTGGGCACAACTC-3′, reverse 5′-TAGCTGAGCGTTCTGTGCGCTG-3′; Reln forward 5′-TTACTCGCACCTTGCTGAAAT-3′, reverse 5′-CAGTTGCTGGTAGGAGTCAAAG-3′; Gapdh forward 5′-AGGTCGGTGTGAACGGATTTG-3′, reverse 5′-TGTAGACCATGTAGTTGAGGTCA-3′.
RNA-Seq.
GCps were purified from P7 control (Chd7fl/+) and cko (Math1-Cre Chd7fl/fl) mice as described above. Total RNA was extracted from freshly isolated cells (5 × 106 per cerebellum) with an Agilent Absolutely RNA miniprep kit (Agilent Technologies). RNA quality and integrity were assessed on a 2100 Bioanalyzer and 2200 Tapestation using an R6K screenTape (Agilent Technologies). After quantification on Qubit using Quant-iT RNA assay kit (Life Technologies), 2 μg total RNA was used from each sample for library preparation using an Illumina TruSeq kit. Libraries were resolved by agarose gel electrophoresis, and 250- to 550-bp DNA fragments were isolated and gel-purified with a Qiagen DNA gel extraction kit (Qiagen). Two libraries with unique barcodes were combined and sequenced per lane on a HiSeq2000 (Illumina) at the Biomedical Research Centre, King’s Health Partners (London, United Kingdom). RNA-Seq reads were aligned to the mouse reference genome (mm10) using TopHat (version 2.0.14) (61) with an expected inner distance between mate pairs of 150 bp and SD for the distribution on inner distances between mate pairs of 50 bp. The transcriptome index was created using Ensembl build 75. HTseq (version 0.6.1p1) (62) was used to estimate the number of reads (ftp://ftp.ensembl.org/pub/release-75/gtf/mus_musculus/). An intersection-strict mode was used and all reads with an alignment quality lower than 3 skipped. Differential expression was analyzed with DESeq2 1.8.1. (https://bioconductor.org/packages/release/bioc/html/DESeq2.html). Genes with an FDR less than 0.05 were considered differentially expressed. RNA-Seq, ChIP-Seq, and ATAC-Seq data have been deposited in the NCBI’s Gene Expression Omnibus (GEO 18162240).
ChIP-Seq.
GCps were purified from P7 control (Chd7fl/+) mice as described above. Freshly isolated cells (2 × 107 to 3 × 107) of each genotype were fixed, chromatin isolated, and precipitated with an antibody against H3K4me1 (Abcam ab8895), and ChIP-Seq libraries were produced and sequenced as previously described (63). The ChIP DNA libraries were sequenced (50 bp, single-end) on an Illumina HiSeq 2000 sequencer at the New York University Genome Technology Center (New York, New York, USA). The reads were aligned to the mouse reference genome (mm10) using Bowtie (version 1.1.0), allowing only uniquely mapping reads, with an insert size of 1,000 bases. Default values were used for all other parameters. Following mapping, PCR duplicates were removed using the SAMtools “rmdup” command (64). Identification of enriched regions was performed by comparison of the ChIP to the input reads using MACS2 (2.1.0.20140616) (65), with a P value cutoff of 0.0001 and 3,90 as the model building parameters. The bedGraph output generated by MACS2 was used to visualize the ChIP and input signals.
ATAC-Seq.
GCps were purified from P7 mice as described above. Intact nuclei were isolated (EZ Prep Nuclear Isolation Kit, Sigma-Aldrich) from 50,000 freshly isolated GCps of each genotype, and ATAC-Seq libraries were produced as previously described (44). Library quality was assessed on a 2100 Bioanalyzer and quantified using the KAPA library quantification kit (Kapa Biosystems). Six libraries were multiplexed (3 per genotype) and 75 bp paired-end reads were sequenced on a HiSeq4000 (Wellcome Trust Centre for Human Genetics, Oxford, United Kingdom). Reads were uniquely aligned to the mouse reference genome (mm10) using Bowtie (version 1.1.1) (66) with a maximum insert size for valid paired-end alignments of 2,000 and a maximum number of attempts of 200 to match an alignment for 1 mate with an alignment for the opposite mate. Peaks were called in 3 control replicates by comparing them to 3 cko samples and vice versa using MultiGPS (version 0.5) (67) with the minimum q value for reported peaks set at 0.05 and the mitochondrial genome excluded. The bedtools window function from BEDTools (version 2.17.0) (68) was used to identify ATAC-Seq peaks within 1,000 bp of differentially expressed genes (DEGs).
DNase-Seq.
Raw reads were obtained from Frank et al. (45), and analyzed in the same way as the ATAC-Seq reads, except in the mapping step, where reads mapping with up to 4 locations were retained for further analysis.
Heatmaps.
Reads that fell within ±500 bp of all changed ATAC-Seq peaks were plotted with ngs.plot.r (69), and regions were clustered according to the peak value of the first profile, using a global color scale across all plots.
Behavioral tests.
Three batches of mice were used for testing of developmental milestones, recording of ultrasonic vocalizations, and testing of juvenile-adult behavior. For the juvenile-adult behavior, tests were carried out in the following order: juvenile social investigation, rotarod, open field, light/dark test, adult social investigation, 3-chamber social approach, marble burying, olfactory habituation/dishabituation, Morris water maze, and grip strength.
Using a battery of tests (70, 71), key behavioral milestones observed in mice prior to weaning (P2–P20) were used to assess core trajectories in the development of motor and sensory abilities (70) (see Supplemental Methods for detailed information). Ultrasonic vocalizations were recorded in pups across 3-minute sessions in response to social separation from the mother and siblings at P2, P4, P6, P8, and P12, in a dimly lit (<10 lux) soundproof chamber. The mean number of calls was used as a quantitative measure of communicative ability, as described previously (72). Motor coordination and learning were assessed on a rotating rod (Ugo Basile) as described previously (73) when the mice reached 42–45 days of age. The latency to fall for any particular day was calculated as the mean of 2 trials. Repetitive digging behavior to bury marbles was measured (74) in a dimly lit test room (<10 lux). Twelve blue glass marbles were arranged in a symmetrical 4-by-3-cm grid on top of 5-cm-deep sawdust (Litaspen premium, Datesand Ltd.) in a clean, standard housing cage (32 × 16 × 14 cm). Each mouse was given 30 minutes to freely explore the cage, and the number of marbles buried was counted. Social investigation was assessed at different ages to evaluate trajectories across developmental stages from juvenile to adulthood (33, 75, 76). Social investigation of age-matched C57BL/6J sex-matched conspecifics was assessed in juvenile test mice (P21) as described previously (75). Social investigation of juvenile C57BL/6J sex-matched conspecifics was assessed in adult test mice as described previously (76).
Statistics.
Statistical tests used are reported in the figure legends, where all data presented indicate mean ± SEM, unless otherwise specified. A P value less than 0.05 was considered significant. All representative experiments were conducted on a minimum of 3 separate occasions with a minimum of 3 individual samples. All statistical analysis was conducted using SPSS (Statistics 22, IBM).
Study approval.
All animal housing and experimental procedures were performed in compliance with the local ethical review panel of King’s College London, and the UK Home Office Animals Scientific Procedures Act 1986. The work was carried out under licenses (PPL70/6694 and PPL70/7184), and all efforts were made to minimize animal suffering and to reduce the number of animals used.
Author contributions
DEW designed and performed most experiments, analyzed results, and prepared figures. KLHR and CF designed, performed, and analyzed behavioral experiments. BPS, KLHR, and MAB designed and performed ATAC-Seq experiments, and analyzed next-generation sequencing (NGS) data with C. Mohan, HH, and FCW. SK performed the analysis of the nestin-Cre cko mice. TY, AS, and MESS performed initial analysis of mutant phenotypes and contributed data to the manuscript. AC performed and analyzed ultrasonic vocalizations and contributed to developmental milestone experiments. C. Michetti, MS, and OK analyzed behavioral data. ACM analyzed and interpreted RNA-Seq data. WWT and DR assisted with ChIP-Seq experiments. MLS supervised ultrasonic vocalization analysis and assisted with behavioral data analysis. HV and IM contributed to data analysis. DEW, KLHR, C. Mohan, BPS, HH, HV, IM, and CF contributed to the writing of the manuscript. MAB designed the study, performed the RNA-Seq and ChIP-Seq experiments, supervised data analysis and interpretation, and wrote the manuscript. All authors read and approved the final version.
Supplementary Material
Acknowledgments
We thank Kei Miyamoto and Angela Pisco for helpful discussions on ATAC-Seq, and Andrea Streit, Chris Ponting, and our laboratory colleagues for critical comments on the manuscript. Sequencing was conducted at the Biomedical Research Centre Genomics Core at Guy’s Hospital, which is supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the National Health Service (NHS), the NIHR, or the Department of Health. Sequencing of ATAC-Seq samples was performed at the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z). This work was supported by a project grant from the Medical Research Council (MR/K022377/1) to MAB and CF, a Clinical Training Fellowship from the Wellcome Trust (WT096385MA) to DEW, and a Travelling Fellowship from the Company of Biologists (Development) (CB24 9LF) to C. Michetti.
Footnotes
ACM’s present address is: Department of Physiology, University of Lausanne, Lausanne, Switzerland.
WWT’s present address is: Institute of Molecular and Cell Biology, Agency for Science, Technology and Research, Singapore.
Conflict of interest: The authors have declared that no conflict of interest exists.
License: This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reference information:J Clin Invest. 2017;127(3):874–887.https://doi.org/10.1172/JCI83408.
Contributor Information
Sahrunizam Kasah, Email: sahrunizam.kasah@kcl.ac.uk.
Conor Mohan, Email: conor.mohan@kcl.ac.uk.
Tian Yu, Email: tian.yu@ucl.ac.uk.
Blanca Pijuan Sala, Email: blanca.pijuan@gmail.com.
Husam Hebaishi, Email: hebaishi.husam@gmail.com.
Angela Caruso, Email: angyca@live.it.
Ana Claudia Marques, Email: anaclaudia.marques@unil.ch.
Caterina Michetti, Email: caterinamichetti@hotmail.com.
Apar Shah, Email: aparshah_18@yahoo.co.uk.
Mara Sabbioni, Email: mara.sabbioni@hotmail.it.
Omer Kulhanci, Email: omerkulhanci@outlook.com.
Wee-Wei Tee, Email: wwtee@imcb.a-star.edu.sg.
Danny Reinberg, Email: danny.reinberg@nyumc.org.
Maria Luisa Scattoni, Email: marialuisa.scattoni@iss.it.
Holger Volk, Email: hvolk@rvc.ac.uk.
Imelda McGonnell, Email: Imcgonnell@RVC.AC.UK.
Cathy Fernandes, Email: cathy.fernandes@kcl.ac.uk.
M. Albert Basson, Email: albert.basson@kcl.ac.uk.
References
- 1.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14(5):347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake KD, et al. CHARGE association: an update and review for the primary pediatrician. Clin Pediatr (Phila) 1998;37(3):159–173. doi: 10.1177/000992289803700302. [DOI] [PubMed] [Google Scholar]
- 4.Verloes A. Updated diagnostic criteria for CHARGE syndrome: a proposal. Am J Med Genet A. 2005;133A(3):306–308. doi: 10.1002/ajmg.a.30559. [DOI] [PubMed] [Google Scholar]
- 5.Vissers LE, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36(9):955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 6.Bergman JE, Janssen N, Hoefsloot LH, Jongmans MC, Hofstra RM, van Ravenswaaij-Arts CM. CHD7 mutations and CHARGE syndrome: the clinical implications of an expanding phenotype. J Med Genet. 2011;48(5):334–342. doi: 10.1136/jmg.2010.087106. [DOI] [PubMed] [Google Scholar]
- 7.Engelen E, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43(6):607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 8.Schnetz MP, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19(4):590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnetz MP, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6(7):e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouazoune K, Kingston RE. Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc Natl Acad Sci U S A. 2012;109(47):19238–19243. doi: 10.1073/pnas.1213825109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu T, et al. Deregulated FGF and homeotic gene expression underlies cerebellar vermis hypoplasia in CHARGE syndrome. Elife. 2013;2:e01305. doi: 10.7554/eLife.01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basson MA, Wingate RJ. Congenital hypoplasia of the cerebellum: developmental causes and behavioral consequences. Front Neuroanat. 2013;7:29. doi: 10.3389/fnana.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basson MA, et al. Specific regions within the embryonic midbrain and cerebellum require different levels of FGF signaling during development. Development. 2008;135(5):889–898. doi: 10.1242/dev.011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu T, Yaguchi Y, Echevarria D, Martinez S, Basson MA. Sprouty genes prevent excessive FGF signalling in multiple cell types throughout development of the cerebellum. Development. 2011;138(14):2957–2968. doi: 10.1242/dev.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126(14):3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 16.Lewis PM, Gritli-Linde A, Smeyne R, Kottmann A, McMahon AP. Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev Biol. 2004;270(2):393–410. doi: 10.1016/j.ydbio.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22(1):103–114. doi: 10.1016/S0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 18.Barkovich AJ, Millen KJ, Dobyns WB. A developmental and genetic classification for midbrain-hindbrain malformations. Brain. 2009;132(pt 12):3199–3230. doi: 10.1093/brain/awp247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong SE, et al. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat Genet. 2000;26(1):93–96. doi: 10.1038/79246. [DOI] [PubMed] [Google Scholar]
- 20.Ozcelik T, et al. Mutations in the very low-density lipoprotein receptor VLDLR cause cerebellar hypoplasia and quadrupedal locomotion in humans. Proc Natl Acad Sci U S A. 2008;105(11):4232–4236. doi: 10.1073/pnas.0710010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rice DS, Sheldon M, D’Arcangelo G, Nakajima K, Goldowitz D, Curran T. Disabled-1 acts downstream of Reelin in a signaling pathway that controls laminar organization in the mammalian brain. Development. 1998;125(18):3719–3729. doi: 10.1242/dev.125.18.3719. [DOI] [PubMed] [Google Scholar]
- 22.Castagna C, Aimar P, Alasia S, Lossi L. Post-natal development of the Reeler mouse cerebellum: An ultrastructural study. Ann Anat. 2014;196(4):224–235. doi: 10.1016/j.aanat.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Miyata T, et al. Migration, early axonogenesis, and Reelin-dependent layer-forming behavior of early/posterior-born Purkinje cells in the developing mouse lateral cerebellum. Neural Dev. 2010;5:23. doi: 10.1186/1749-8104-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fatemi SH, Stary JM, Halt AR, Realmuto GR. Dysregulation of Reelin and Bcl-2 proteins in autistic cerebellum. J Autism Dev Disord. 2001;31(6):529–535. doi: 10.1023/A:1013234708757. [DOI] [PubMed] [Google Scholar]
- 25.Fatemi SH, et al. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. 2012;11(3):777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Micucci JA, et al. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum Mol Genet. 2014;23(2):434–448. doi: 10.1093/hmg/ddt435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng W, et al. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell. 2013;13(1):62–72. doi: 10.1016/j.stem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Jones KM, Sarić N, Russell JP, Andoniadou CL, Scambler PJ, Basson MA. CHD7 maintains neural stem cell quiescence and prevents premature stem cell depletion in the adult hippocampus. Stem Cells. 2015;33(1):196–210. doi: 10.1002/stem.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23(1):99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 30.Matei V, et al. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234(3):633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orvis GD, et al. The engrailed homeobox genes are required in multiple cell lineages to coordinate sequential formation of fissures and growth of the cerebellum. Dev Biol. 2012;367(1):25–39. doi: 10.1016/j.ydbio.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fatemi SH, et al. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. 2012;11(3):777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11(7):490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Layman WS, et al. Defects in neural stem cell proliferation and olfaction in Chd7 deficient mice indicate a mechanism for hyposmia in human CHARGE syndrome. Hum Mol Genet. 2009;18(11):1909–1923. doi: 10.1093/hmg/ddp112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergman JE, Bosman EA, van Ravenswaaij-Arts CM, Steel KP. Study of smell and reproductive organs in a mouse model for CHARGE syndrome. Eur J Hum Genet. 2010;18(2):171–177. doi: 10.1038/ejhg.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strick PL, Dum RP, Fiez JA. Cerebellum and nonmotor function. Annu Rev Neurosci. 2009;32:413–434. doi: 10.1146/annurev.neuro.31.060407.125606. [DOI] [PubMed] [Google Scholar]
- 37.Stoodley CJ. The cerebellum and cognition: evidence from functional imaging studies. Cerebellum. 2012;11(2):352–365. doi: 10.1007/s12311-011-0260-7. [DOI] [PubMed] [Google Scholar]
- 38.Stoodley CJ, Schmahmann JD. Evidence for topographic organization in the cerebellum of motor control versus cognitive and affective processing. Cortex. 2010;46(7):831–844. doi: 10.1016/j.cortex.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mariani J, Crepel F, Mikoshiba K, Changeux JP, Sotelo C. Anatomical, physiological and biochemical studies of the cerebellum from Reeler mutant mouse. Philos Trans R Soc Lond B Biol Sci. 1977;281(978):1–28. doi: 10.1098/rstb.1977.0121. [DOI] [PubMed] [Google Scholar]
- 40.Trommsdorff M, et al. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97(6):689–701. doi: 10.1016/S0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 41.Arnaud L, Ballif BA, Cooper JA. Regulation of protein tyrosine kinase signaling by substrate degradation during brain development. Mol Cell Biol. 2003;23(24):9293–9302. doi: 10.1128/MCB.23.24.9293-9302.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bock HH, Jossin Y, May P, Bergner O, Herz J. Apolipoprotein E receptors are required for reelin-induced proteasomal degradation of the neuronal adaptor protein Disabled-1. J Biol Chem. 2004;279(32):33471–33479. doi: 10.1074/jbc.M401770200. [DOI] [PubMed] [Google Scholar]
- 43.Magdaleno S, Keshvara L, Curran T. Rescue of ataxia and preplate splitting by ectopic expression of Reelin in reeler mice. Neuron. 2002;33(4):573–586. doi: 10.1016/S0896-6273(02)00582-2. [DOI] [PubMed] [Google Scholar]
- 44.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank CL, et al. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat Neurosci. 2015;18(5):647–656. doi: 10.1038/nn.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goes FS, et al. Sex-specific association of the Reelin gene with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):549–553. doi: 10.1002/ajmg.b.31018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shifman S, et al. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4(2):e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magliaro C, Cocito C, Bagatella S, Merighi A, Ahluwalia A, Lossi L. The number of Purkinje neurons and their topology in the cerebellar vermis of normal and reln haplodeficient mouse. Ann Anat. 2016;207:68–75. doi: 10.1016/j.aanat.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 49.Biamonte F, et al. Interactions between neuroactive steroids and reelin haploinsufficiency in Purkinje cell survival. Neurobiol Dis. 2009;36(1):103–115. doi: 10.1016/j.nbd.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Hadj-Sahraoui N, Frédéric F, Delhaye-Bouchaud N, Mariani J. Gender effect on Purkinje cell loss in the cerebellum of the heterozygous reeler mouse. J Neurogenet. 1996;11(1–2):45–58. doi: 10.3109/01677069609107062. [DOI] [PubMed] [Google Scholar]
- 51.Schulz Y, et al. CHD7, the gene mutated in CHARGE syndrome, regulates genes involved in neural crest cell guidance. Hum Genet. 2014;133(8):997–1009. doi: 10.1007/s00439-014-1444-2. [DOI] [PubMed] [Google Scholar]
- 52.Courchesne E, Yeung-Courchesne R, Press GA, Hesselink JR, Jernigan TL. Hypoplasia of cerebellar vermal lobules VI and VII in autism. N Engl J Med. 1988;318(21):1349–1354. doi: 10.1056/NEJM198805263182102. [DOI] [PubMed] [Google Scholar]
- 53.Kaufmann WE, et al. Specificity of cerebellar vermian abnormalities in autism: a quantitative magnetic resonance imaging study. J Child Neurol. 2003;18(7):463–470. doi: 10.1177/08830738030180070501. [DOI] [PubMed] [Google Scholar]
- 54.Donovan AP, Basson MA. The neuroanatomy of autism — a developmental perspective. J Anat. 2017;230(1):4–15. doi: 10.1111/joa.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai PT, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488(7413):647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.López AJ, Wood MA. Role of nucleosome remodeling in neurodevelopmental and intellectual disability disorders. Front Behav Neurosci. 2015;9:100. doi: 10.3389/fnbeh.2015.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Basson MA, van Ravenswaaij-Arts C. Functional Insights into Chromatin Remodelling from Studies on CHARGE Syndrome. Trends Genet. 2015;31(10):600–611. doi: 10.1016/j.tig.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A. 1988;85(21):8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yaguchi Y, Yu T, Ahmed MU, Berry M, Mason I, Basson MA. Fibroblast growth factor (FGF) gene expression in the developing cerebellum suggests multiple roles for FGF signaling during cerebellar morphogenesis and development. Dev Dyn. 2009;238(8):2058–2072. doi: 10.1002/dvdy.22013. [DOI] [PubMed] [Google Scholar]
- 61.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S, Pyl PT, Huber W. HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tee WW, Shen SS, Oksuz O, Narendra V, Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156(4):678–690. doi: 10.1016/j.cell.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mahony S, et al. An integrated model of multiple-condition ChIP-Seq data reveals predeterminants of Cdx2 binding. PLoS Comput Biol. 2014;10(3):e1003501. doi: 10.1371/journal.pcbi.1003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fox WM. Reflex-ontogeny and behavioural development of the mouse. Anim Behav. 1965;13(2):234–241. doi: 10.1016/0003-3472(65)90041-2. [DOI] [PubMed] [Google Scholar]
- 71.Heyser CJ, Wilson MC, Gold LH. Coloboma hyperactive mutant exhibits delayed neurobehavioral developmental milestones. Brain Res Dev Brain Res. 1995;89(2):264–269. doi: 10.1016/0165-3806(95)00130-6. [DOI] [PubMed] [Google Scholar]
- 72.Scattoni ML, Gandhy SU, Ricceri L, Crawley JN. Unusual repertoire of vocalizations in the BTBR T+tf/J mouse model of autism. PLoS One. 2008;3(8):e3067. doi: 10.1371/journal.pone.0003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wöhr M, et al. Developmental delays and reduced pup ultrasonic vocalizations but normal sociability in mice lacking the postsynaptic cell adhesion protein neuroligin2. Behav Brain Res. 2013;251:50–64. doi: 10.1016/j.bbr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thomas A, Burant A, Bui N, Graham D, Yuva-Paylor LA, Paylor R. Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology (Berl) 2009;204(2):361–373. doi: 10.1007/s00213-009-1466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McFarlane HG, Kusek GK, Yang M, Phoenix JL, Bolivar VJ, Crawley JN. Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes Brain Behav. 2008;7(2):152–163. doi: 10.1111/j.1601-183X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- 76.Grayton HM, Missler M, Collier DA, Fernandes C. Altered social behaviours in neurexin 1α knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS One. 2013;8(6):e67114. doi: 10.1371/journal.pone.0067114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Inouye M, Oda SI. Strain-specific variations in the folial pattern of the mouse cerebellum. J Comp Neurol. 1980;190(2):357–362. doi: 10.1002/cne.901900209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.