Abstract
Steroid-resistant nephrotic syndrome (SRNS) causes 15% of chronic kidney disease cases. A mutation in 1 of over 40 monogenic genes can be detected in approximately 30% of individuals with SRNS whose symptoms manifest before 25 years of age. However, in many patients, the genetic etiology remains unknown. Here, we have performed whole exome sequencing to identify recessive causes of SRNS. In 7 families with SRNS and facultative ichthyosis, adrenal insufficiency, immunodeficiency, and neurological defects, we identified 9 different recessive mutations in SGPL1, which encodes sphingosine-1-phosphate (S1P) lyase. All mutations resulted in reduced or absent SGPL1 protein and/or enzyme activity. Overexpression of cDNA representing SGPL1 mutations resulted in subcellular mislocalization of SGPL1. Furthermore, expression of WT human SGPL1 rescued growth of SGPL1-deficient dpl1Δ yeast strains, whereas expression of disease-associated variants did not. Immunofluorescence revealed SGPL1 expression in mouse podocytes and mesangial cells. Knockdown of Sgpl1 in rat mesangial cells inhibited cell migration, which was partially rescued by VPC23109, an S1P receptor antagonist. In Drosophila, Sply mutants, which lack SGPL1, displayed a phenotype reminiscent of nephrotic syndrome in nephrocytes. WT Sply, but not the disease-associated variants, rescued this phenotype. Together, these results indicate that SGPL1 mutations cause a syndromic form of SRNS.
Introduction
Steroid-resistant nephrotic syndrome (SRNS), when also resistant to other immunosuppressive agents, leads to chronic kidney disease (CKD) within a few years of onset, requiring renal replacement therapy for survival. It causes 15% of all end-stage kidney disease that manifests by 25 years of age (1). Histologically, SRNS manifests mostly as focal segmental glomerulosclerosis (FSGS) (2).
The first insights into the pathogenesis of SRNS were gained by the discovery of monogenic causes of SRNS, revealing that the encoded proteins are essential for the function of the renal glomerular cells called podocytes (3, 4). We recently demonstrated in a worldwide cohort of 1,783 families that a monogenic cause of SRNS can be detected in 1 of 27 genes in approximately 30% of SRNS cases manifesting before age 25 years (5). Currently, more than 40 monogenic forms of SRNS have been identified (6).
Interestingly, syndromic forms of SRNS have been increasingly characterized and display variable involvement of other organs besides the kidney, in most of the cases the central nervous system, but also the genital tract, eye, muscle, bone, and the immune system. To date, mutations have been identified in transcription factors and nuclear proteins (WT1, LMX1B, SMARCAL1, NXF5, NUP93, NUP205, and XPO5), lysosomal proteins (SCARB2), basement membrane proteins (LAMB2), and proteins involved in COQ10 biosynthesis (COQ2, COQ6, PDSS2, and ADCK4) (6). Because genetic mapping data indicated a multitude of potential additional loci for SRNS, we here performed whole exome sequencing (WES) to identify additional monogenic SRNS genes and identified 9 different mutations in sphingosine-1-phosphate lyase (SGPL1) in 7 families as causing a previously unrecognized syndromic SRNS with a combination of ichthyosis/acanthosis, adrenal insufficiency, immunodeficiency, and/or neuronal dysfunction. We characterized mechanisms of molecular loss of function for the mutations detected and implicated sphingosine-1-phosphate (S1P) metabolism in the pathogenesis of SRNS. The syndromic features resulting from SGPL1 mutations indicate the pivotal role of S1P metabolism in multiple tissues including kidney.
Results
SGPL1 mutations cause nephrotic syndrome, ichthyosis, facultative adrenal insufficiency, immunodeficiency, and neurologic defects in humans.
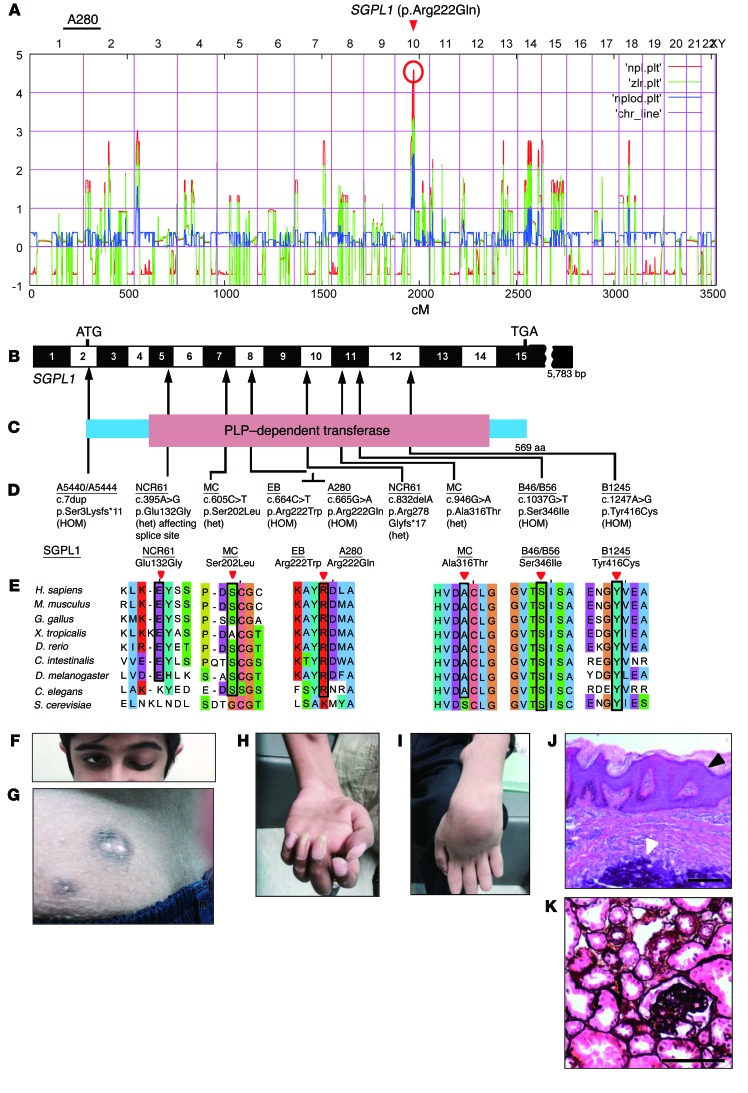
Using WES in 7 families with a disease phenotype of SRNS, with adrenal insufficiency, ichthyosis-like acanthosis, immunodeficiency, or neurologic abnormalities (Table 1, Table 2, and Supplemental Figures 1–3; supplemental material available online with this article; https://doi.org/10.1172/JCI89626DS1), we identified recessive mutations in SGPL1 (Figure 1, A–E). Homozygosity mapping (HM) in a Pakistani family (A280) (Supplemental Figure 1) with 3 siblings yielded a nonparametric lod score peak on chromosome 10, which did not coincide with any of the known recessive SRNS loci (Figure 1A) (7). Using WES, we detected a homozygous missense mutation (p.Arg222Gln) in a highly conserved (Caenorhabditis elegans) amino acid residue encoded by SGPL1 (Table 1 and Figure 1, B–E). Notably, affected individuals from another consanguineous family (EB) showed a homozygous missense mutation involving the same amino acid residue (p. Arg222Trp), but presented with a more severe phenotype, with neonatal onset and profound immunodeficiency (Table 1 and Figure 1, B–E). A homozygous frameshift mutation of SGPL1 (p.Ser3Lysfs*11) (Table 2 and Figure 1, B–E) was identified in affected individuals from family A5444, showing a phenotype of SRNS and cortisol deficiency. In family B46/B56, in whom 3 siblings had congenital nephrotic syndrome (NS) and severe extrarenal involvement combined with lymphopenia (Table 2 and Supplemental Figure 1), WES detected another homozygous missense mutation of SGPL1 (p.Ser346Ile) (Table 2 and Figure 1, B–E) and in another individual with a related disease phenotype, B1245, a homozygous missense mutation, p.Tyr416Cys (Table 2 and Figure 1, B–E), with both mutations evolutionarily conserved to S. cerevisiae. In an individual MC with mononeuritis multiplex, SRNS, lymphopenia, and progressive contractures of extremities, WES identified compound heterozygous mutations in SGPL1 (Table 2 and Figure 1, B–E). Both mutations are missense and affect highly conserved amino acid residues.
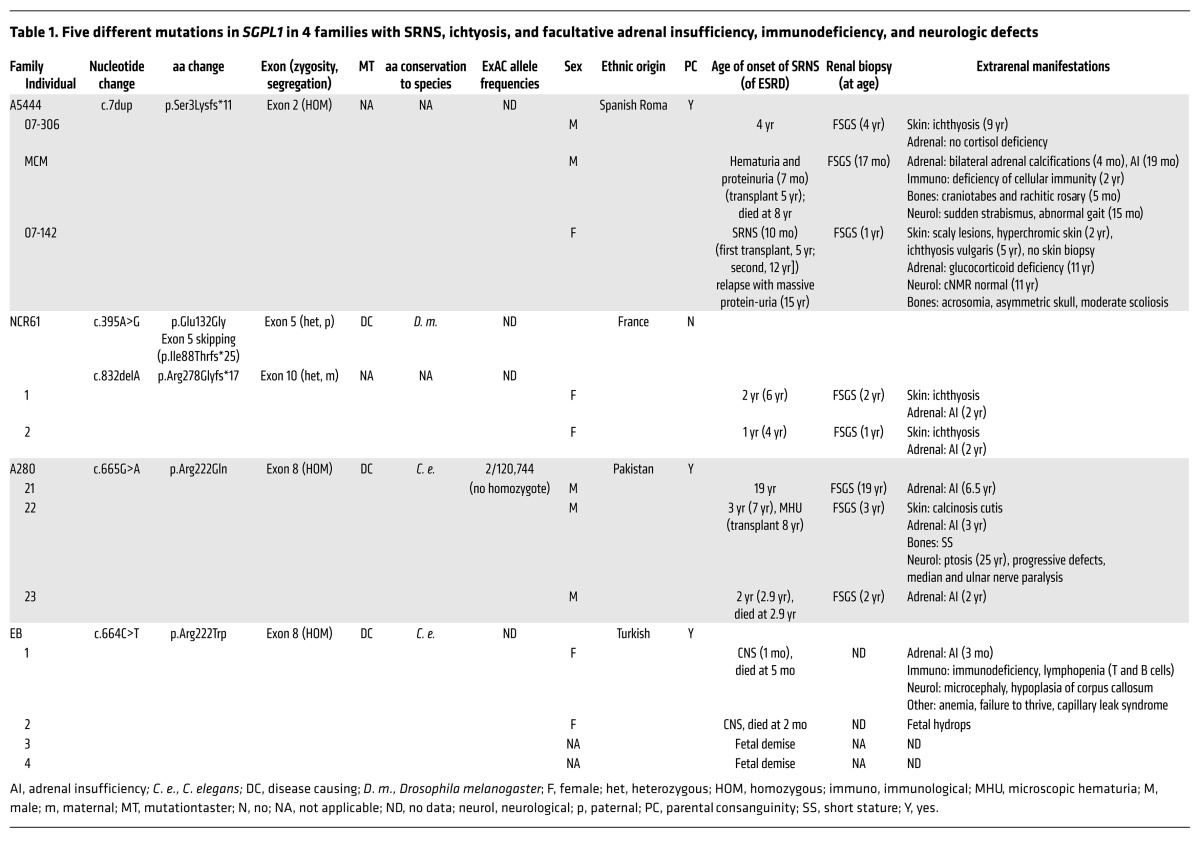
Table 1. Five different mutations in SGPL1 in 4 families with SRNS, ichtyosis, and facultative adrenal insufficiency, immunodeficiency, and neurologic defects.
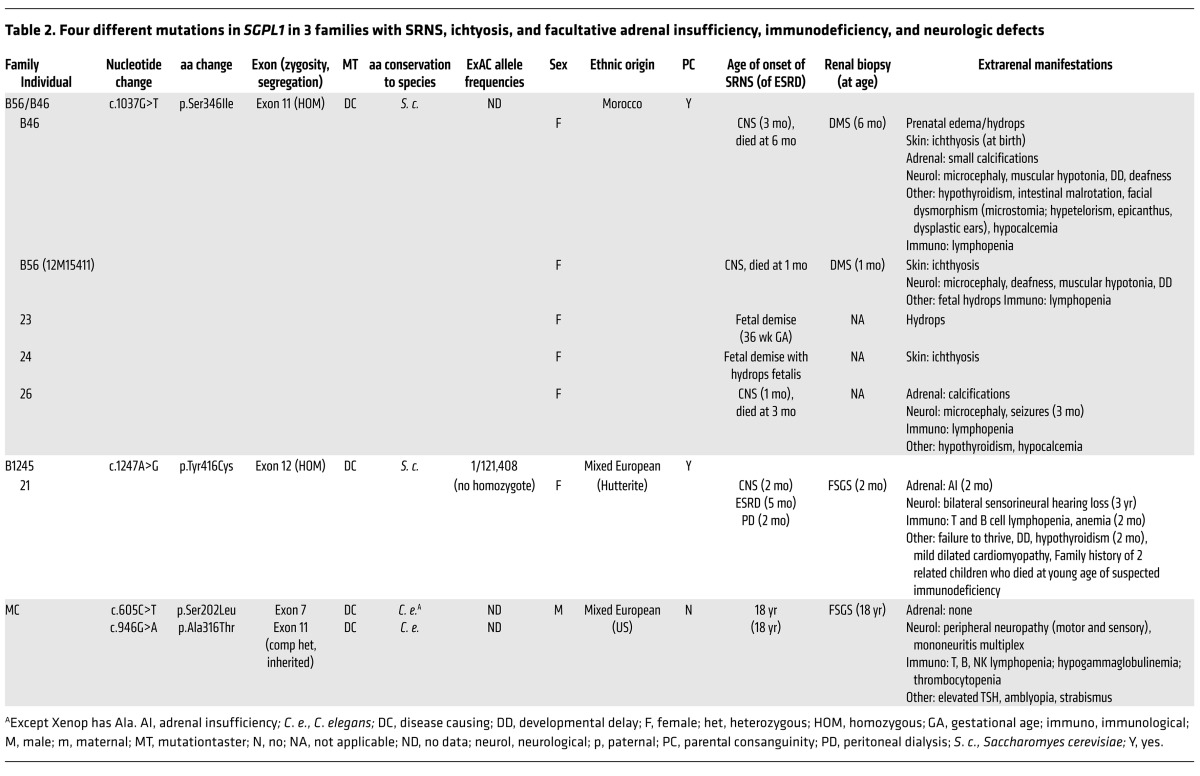
Table 2. Four different mutations in SGPL1 in 3 families with SRNS, ichtyosis, and facultative adrenal insufficiency, immunodeficiency, and neurologic defects.
Figure 1. HM and WES reveal SGPL1 mutations as causing SRNS with ichthyosis and facultative adrenal insufficiency or neurologic defects (NPHS type 14).
(A) Nonparametric lod scores across the human genome in 3 siblings of consanguineous family A280 with SRNS, acanthosis, and ichtyosis with facultative adrenal insufficiency. The x axis shows single-nucleotide polymorphism positions on human chromosomes concatenated from p-ter (left) to q-ter (right). Genetic distance is given in cM. The SGPL1 locus (arrowhead) is positioned within the maximum nonparametric lod peak on chromosome 10. (B) Exon structure of human SGPL1 cDNA. SGPL1 contains 15 exons. Positions of start codon (ATG) and of stop codon (TGA) are indicated. (C) Domain structure of SGPL1. The extent of the PLP-dependent transferase domain is shown. (D) Five homozygous (HOM) and 4 compound-heterozygous SGPL1 mutations (het) detected in 7 families with NPHS type 14. Family numbers (underlined), mutations, and predicted translational changes are indicated (see also Tables 1 and 2). (E) Evolutionary conservation of altered amino acid residues of SGPL1. Note that c.395A>G also resulted in p.Ile88Thrfs*25 through exon 5 skipping. (F) Ptosis in individual A280-22. (G) Skin image from individual A280-22 showing brownish black desquamation on sebostatic skin with multiple radial papules with a blueish/black erythema and central calcinosis. (H and I) Median (H) and ulnar nerve (I) paralysis in individual A280-22. (J) H&E-stained epidermal section from individual A280-22 showing acanthosis/orthokeratotic hyperkeratosis (black arrowhead) and calcinosis (white arrowhead). (K) Renal histology (silver staining) of individual A280-22, showing FSGS. Scale bars: 100 μm.
Patients from the nonconsanguineous family NCR61 (Table 1 and Figure 1, B–E) were found to be compound heterozygous for a frameshift mutation (p.Arg278Glyfs*17) and a missense mutation (p.Glu132Gly) not predicted to be deleterious by the PolyPhen (http://genetics.bwh.harvard.edu/pph2/) and SIFT (http://sift.jcvi.org/) prediction tools, but predicted to induce skipping of exon 5 due to the introduction of additional exon splicing silencer elements (EX-SKIP, http://ex-skip.img.cas.cz) (8). Fibroblasts of patient NCR61-1 had almost no SGPL1 protein detectable by Western blot at the predicted size (Supplemental Figure 4A). Therefore, we hypothesized that the deleterious effect of c.395A>G was indeed related to the predicted RNA splicing defect and not to the p.Glu132Gly amino acid substitution. Amplification of the patient cDNA between exons 4 and 8 by PCR revealed a light band at the expected size and an intense smaller band with a size corresponding to loss of exon 5 (Supplemental Figure 4B). Sequencing of the PCR products confirmed the absence of exon 5 for the abundant smaller fragment and showed that the normal size fragment corresponded to the frameshift allele p.Arg278Glyfs*17. Skipping of exon 5 leads to a frameshift at amino acid position 88 and a stop codon 24 amino acids downstream (p. Ile88Thrfs*25).
By high-throughput exon sequencing (5, 9) in a worldwide cohort of approximately 800 additional families with NS, we did not detect any additional families with biallelic mutations of SGPL1. In all 7 families with SGPL1 mutations (Tables 1 and 2 and Figure 1, B–K), direct inspection of sequence alignments did not yield a mutation in any of the 40 known SRNS genes. Most mutations identified in this study were absent from more than 60,000 control individuals in the ExAC server (http://exac.broadinstitute.org/) (Tables 1 and 2). Two missense mutations (c.665G>A; p.Arg222Gln and c.1247A>G; p.Tyr416Cys) are reported in the ExAC server, but their allele frequencies are extremely rare and they never occur in the homozygous state (Tables 1 and 2). All mutations segregated with the disease phenotype (Supplemental Figures 1 and 2). We thereby identified recessive SGPL1 mutations as a cause of syndromic SRNS with ichthyosis/acanthocystosis, adrenal insufficiency, immunodeficiency, or neurologic involvement. We introduced the term NPHS14 for this syndromic SRNS caused by mutations of SGPL1.
In the majority of cases, NS, manifested as congenital NS or in the first year of life, showed no response to steroid therapy and rapidly progressed to end-stage renal disease (ESRD). Histologically, FSGS was the main finding, but diffuse mesangial sclerosis (DMS) was found in cases with congenital NS (Tables 1 and 2 and Supplemental Figure 3). Extrarenal manifestations included ichthyosis and primary adrenal insufficiency, present in almost all cases except in those patients deceased at very early ages. About half of the affected individuals had a severe immunodeficiency. Patients presented with lymphopenia and multiple bacterial infections. A more detailed immunological work-up performed in patient EB-1 revealed severe lymphopenia with markedly reduced CD4 and CD8 T lymphocytes as well as B lymphocytes. In vitro cytokine generation upon stimulation was also deficient (Supplemental Table 3). Neurological deficits were also present in half of the patients and included sensorineural deafness, microcephaly (Supplemental Figure 3), corpus callosum hypoplasia in 1 case, peripheral nerve paralysis of the median and ulnar nerves, and ptosis of the left eye (Tables 1 and 2 and Figure 1, F–I). In a few cases, dysmorphic features and bone defects were also present. Two families had a history of fetal demise and hydrops fetalis.
SGPL1 encodes S1P lyase, an intracellular enzyme responsible for the final step in sphingolipid breakdown, converting its main substrate S1P into ethanolamine phosphate and hexadecenal. S1P is a bioactive sphingolipid that acts extracellularly by binding to protein-coupled receptors of the lysophospholipid receptor family and intracellularly through S1P receptor–independent (S1PR-independent) mechanisms. Through receptor activation, S1P mediates autocrine and paracrine signals, controlling cell migration and proliferation as well as lamellipodia dynamics (10). An Sgpl1-deficient mouse model exhibits glomerular proteinuria, a skin phenotype of acanthosis with orthokeratotic hyperkeratosis and platelet activation (11).
Sgpl1 localizes to the endoplasmic reticulum of renal glomerular cells.
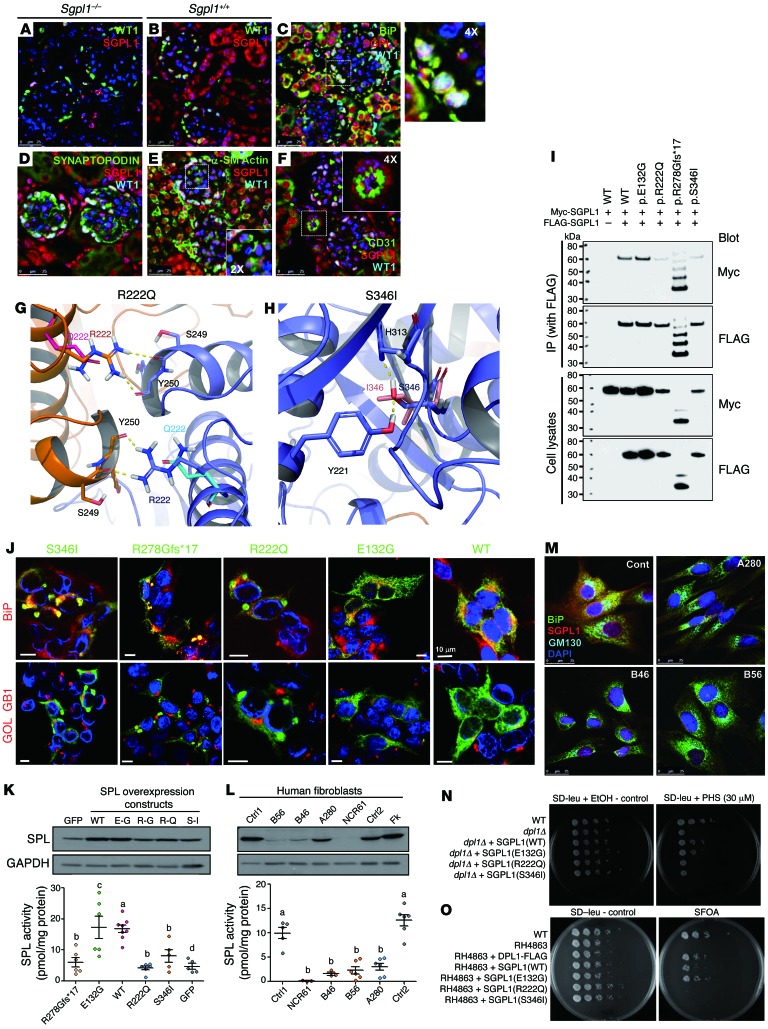
All of the 40 monogenic genes that, when mutated, cause SRNS are highly expressed in podocytes. We therefore examined cell-type–specific and subcellular localization of SGPL1 in mouse kidney. To assess the specificity of an anti-SGPL1 antibody used in immunofluorescence, we first stained kidney sections of Sgpl1–/– and Sgpl1+/+ mice (Figure 2, A and B). Most of the SGPL1 signal observed in kidneys of Sgpl1+/+ mice was absent from kidneys of Sgpl1–/– mice (Figure 2A), demonstrating specificity of the signal. To determine cellular localization of SGPL1, immunofluorescence was performed using various cell- and organelle-specific markers (Figure 2, C–F). SGPL1 localized to podocytes, whose nuclei were marked with WT1 (Figure 2C). As shown by the colocalization of SGPL1 with the ER marker BiP, it localized to ER in podocytes, although this colocalization was more obvious in proximal tubules (Figure 2C). In addition, SGPL1 appeared to be present in other renal glomerular cell types, for example, in mesangial cells stained with α–smooth muscle actin (Figure 2E) and endothelial cells stained with CD31 (Figure 2F).
Figure 2. Biological and biochemical consequences of recessive SGPL1 mutations.
(A–F) Kidney sections of Sgpl1–/– mice (A) and Sgpl1+/+ mice (B) were stained with anti-SGPL1 (red) and WT1 antibodies (green). (B–F) Coimmunofluorescence of SGPL1 with marker proteins (green) in Sgpl1+/+ kidney: podocyte (B, WT1), ER (C, BiP), podocyte foot processes (D, synaptopodin), mesangial cells (E, α–smooth muscle actin), and endothelial cells (F, CD31). Scale bars: 25 μm. (G and H) Structural modeling of SGPL1 mutations. The 2 monomers of the SGPL1 homodimer are shown in the drawing in blue and orange, respectively. (G) p.Arg222Gln; (H) p.Ser346Ile. (I) Coimmunoprecipitation to assess dimerization of WT vs. mutant SGPL1 proteins (see also Supplemental Figure 5). Coimmunoprecipitation is representative of 3 experiments. (J) Mislocalization of variant SGPL1 proteins upon overexpression in HEK293T cells. BiP (red), or Golgi marker GOLGB1 (red), and anti-Myc antibody (green). Scale bars: 10 μm. (K) SGPL1 enzyme activity levels in transformed HEK293T cells. HEK293T cells expressing a GFP indicate endogenous SGPL1 activity levels (a vs. b and a vs. d, P < 0.0025; c vs. d, P = 0.013; a vs. c, no significant difference). (L) SGPL1 protein expression and enzyme activity levels in fibroblasts from 2 control individuals (Ctrl 1 and 2), normal human foreskin fibroblasts (Fk), and 4 individuals with SGPL1 mutations. (a vs. b, P < 0.0001.) Results are from the averages of triplicates in K and L. (M) Immunofluorescence of SGPL1 in fibroblasts. BiP (green), Golgi marker GM130 (blue), and anti-SGPL1 antibody (red). Scale bars: 25 μm. (N) PHS toxicity test. Ability to complement dpl1Δ deletion on medium containing PHS was tested for human SGPL1 WT and mutants. (O) Synthetic lethality test. Human WT and p.Glu132Gly SGPL1 expressing RH4863 survived on 5-FOA plates. However, p.Arg222Gln and p.Ser346Ile mutants did not allow for survival of DPL1 (SGPL1) deficient strains.
In silico modeling of SGPL1 missense mutations.
To predict potential structural changes in SGPL1 protein that might arise from the mutations (p.Arg222Gln and p.Ser346Ile) detected in patients with NPHS type 14 Tables 1 and 2), we performed in silico analyses (Figure 2, G and H). SGPL1 forms a symmetric homodimer. Two subunits form a tightly intertwined dimer with both chains contributing to the catalytic cavity defined by the covalently bound cofactor pyridoxal phosphate (PLP) (Figure 2, G and H). Arg222 is located at the symmetric dimer interface and forms 2 important hydrogen bonds with the carbonyl backbone moieties of Tyr250 and Ser249, contributing to the binding affinity of the homodimer. The p.Arg222Gln mutation leads to a loss of these 2 hydrogen bonds in both chains (Figure 2G), which is reflected in the delta affinity being predicted as unfavorable by about 10 kcal/mol. In addition, the mutation seems to be destabilizing the protein, since Arg222 also forms hydrogen bonds with adjacent residues in its own symmetric dimer chain. Ser346 is not at the dimer interface, but buried within each chain (Figure 2H). Its hydroxyl group is involved in 2 hydrogen bonds, accepting one from Tyr221 and donating one onto His313 (Figure 2H). The p.Ser346Ile mutation is predicted to be considerably disfavored because it not only breaks this hydrogen-bond network, but because isoleucine is also much bulkier than serine, leading to steric clashes with its surrounding residues (Figure 2H). Hence the delta stability is predicted to be +130 kcal/mol.
Mutations alter subcellular localization of SGPL1.
Because in silico analysis predicted that the p.Arg222Gln mutation may affect the interface of the SGPL1 homodimer, we examined this hypothesis by coimmunoprecipitation. We transfected HEK293T cells with plasmids with 2 alternative tags (Myc and FLAG). Compared with WT SGPL1, p.Arg222Gln and p.Ser346Ile mutant proteins exhibited reduced expression levels, whereas p.Glu132Gly had a similar expression level upon overexpression (Figure 2I and Supplemental Figure 5). Neither missense (p.Glu132Gly, p.Arg222Gln, and p.Ser346Ile) nor truncating (p.Arg278Glyfs*17) mutations abrogated dimer formation (Figure 2I and Supplemental Figure 5). However, when compared with overexpression of the SGPL1 WT construct, overexpression of p.Arg222Gln, p.Ser346Ile, and p.Arg278Glyfs*17 SGPL1 proteins in HEK293T cells led to abnormal cytoplasmic aggregates (Figure 2J). Aggregation of SGPL1 mutant proteins was confirmed upon overexpression in human podocytes (Supplemental Figure 6).
Mutations in SGPL1 result in decreased expression and reduced lyase activity.
To assess whether mutations detected in individuals with NPHS type 14 altered SGPL1 enzyme activity, we measured SGPL1 and enzyme activity levels in HEK293T cells transiently expressing either WT or mutant SGPL1 (Figure 2K). We found that overexpression of the p.Arg222Gln, p.Ser346Ile, and p.Arg278Glyfs*17 mutant alleles resulted in strongly reduced enzyme activity levels, whereas the p.Glu132Gly mutant (c.395A>G) did not, consistent with our finding that c.395A>G leads to skipping of exon 5 and protein truncation (p.Ile88Thrfs*25) (Supplemental Figure 4). We also found that protein expression levels and corresponding SGPL1 enzyme activity levels in patient dermal fibroblasts (Figure 2L) exhibited reduced SGPL1 enzyme activity for individuals B46 (p.Ser346Ile), B56 (p.Ser346Ile), NCR 61-1 (p.R278fs*17 and p.Ile88Thrfs*25), and A280-21 (p.Arg222Gln) compared with controls (Figure 2L). In addition, there was reduced expression of SGPL1 protein when examined by immunofluorescence microscopy in patient fibroblasts (Figure 2M). Taken together, these results suggest that the primary impact of the patient’s mutations is on SGPL1 expression level and protein stability.
SGPL1 mutations fail to rescue growth in DPL1-deficient yeast.
Functionality of human WT and mutant SGPL1 proteins were tested in an in vivo yeast complementation assay by measuring their ability to complement the deletion of the SGPL1 yeast ortholog DPL1 (dpl1Δ) on medium containing phytosphingosine (PHS) (Figure 2N and Supplemental Figure 7A). Inability to degrade toxic long-chain bases led to decreased viability and slowed growth. Human WT and p.Glu132Gly mutant SGPL1 were found to be able to partially restore dpl1Δ, while p.Arg222Gln, p.Ser346Ile, and p.Tyr416Cys as well as frameshift mutants p.Arg278Glyfs*17 and p.Ser3Lysfs*11 showed no improved growth compared with dpl1Δ (Figure 2N and Supplemental Figure 7A), consistent with our finding that the c.395A>G allele most likely conveys loss of function through its splice defect (Supplemental Figure 4B). The results were also reproduced on plates containing C17 and C18 dihydrosphingosine (DHS).
To further investigate functional consequences of SGPL1 mutants, we carried out a synthetic lethality assay in yeast (Figure 2O and Supplemental Figure 7B) using an RH4863 strain in which DPL1 and LCB3 were deleted, rendering the strain synthetically lethal. Viability of the strain was maintained by expressing LCB3 from a URA3 plasmid. We tested functionality of SGPL1 and corresponding mutants expressed in RH4863 by forcing them to lose LCB3-coding URA3 plasmid on plates containing 5-fluoroorotic acid (5-FOA), which selects against URA3-expressing cells. Survival on 5-FOA plates indicates that cells have regained the ability to degrade long-chain bases and thus compensate for the loss of LCB3. In agreement with the PHS toxicity test, synthetic lethality assay demonstrated functional integrity of WT and p.Glu132Gly mutant SGPL1 proteins, but not p.Arg222Gln, p.Ser346Ile, p.Tyr416Cys, p.Arg278Glyfs*17, and p.Ser3Lysfs*11 (Figure 2O and Supplemental Figure 7B).
SGPL1 mutations alter ceramide composition of patient fibroblast-conditioned medium.
S1P participates in regulating multiple cellular processes, and its intracellular levels are tightly regulated. Since SGPL1 controls the only exit point for sphingolipid catabolism, inactivation of the enzyme can result in accumulation of various bioactive sphingolipid intermediates, including phosphorylated and nonphosphorylated sphingoid bases and ceramides.
Conditioned medium from patient fibroblast cultures showed significantly elevated C22:0, C24:0, and C24:1 ceramides compared with control fibroblast-conditioned medium, whereas S1P levels were below the limits of detection (Supplemental Figure 8). These very long chain ceramides are produced by ceramide synthase 2 (12).
Sgpl1–/– mice exhibit podocyte foot process effacement.
Sgpl1–/– mice exhibit glomerular proteinuria, a skin phenotype of acanthosis with orthokeratotic hyperkeratosis, platelet activation, and immunodeficiency (11). In order to determine the renal histology of Sgpl1–/– mice, we performed transmission electron microscopy (TEM) of kidneys harvested from Sgpl1+/+ and Sgpl1–/– mice to directly examine glomerular structures upon loss of Sgpl1. Sgpl1–/– kidneys exhibited complete foot process effacement and absence of slit diaphragms (Supplemental Figure 9). In addition, Sgpl1–/– mice exhibited hypoalbuminemia (serum albumin KO 2.2 ± 0.6 g/dl vs. WT or heterozygote [HET] 3.7 ± 0.4 g/dl, n = 4 KO and n = 6 WT or HET) and an elevated urinary albumin/creatinine ratio (ACR) (KO 1176.4 ± 932.8 vs. WT or HET 103.5 ± 94.2, n = 3 KO, n = 5 WT or HET, P value, no significant difference).
However, when we examined apoptosis in response to S1P in cultured podocytes, we did not observe an effect in a dose range expected to be present in SGPL1-induced pathology (Supplemental Figure 10). Likewise, we did not observe any effect of SGPL1 knockdown on podocyte migration rate, a pathogenic effect that has previously been demonstrated in many monogenic forms of SRNS (Supplemental Figure 11) (13).
Decreased mesangial cell migration rate upon SGPL1 knockdown is reversed by an S1PR inhibitor.
Because we did not observe any cellular pathologic effect in cultured podocytes regarding apoptosis or cell migration, because SGPL1 was present in mesangial cells (Figure 2E), and because Sgpl1–/– mice exhibited reduced glomerular mesangial cell numbers (11), we examined pathogenic effects of siRNA knockdown of Sgpl1 in mesangial cells (Figure 3).
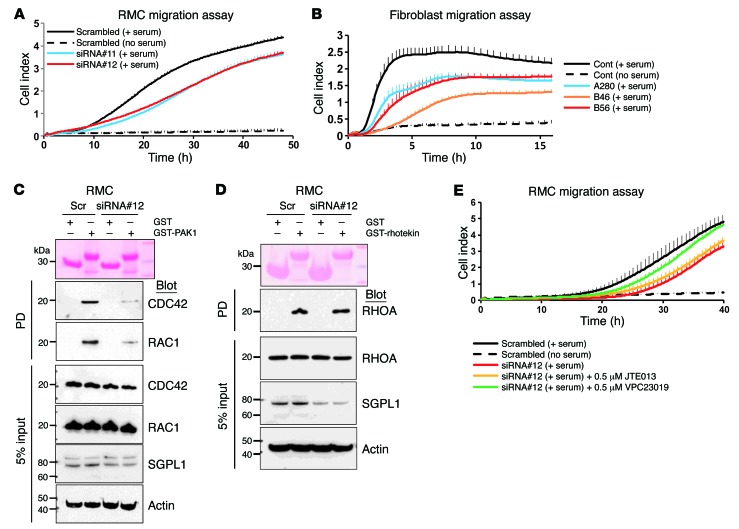
Figure 3. Effect of Sgpl1 knockdown on RHO GTPase activity in RMCs and patient fibroblasts.
(A) Effect of SGPL1 knockdown on rat RMCs using the xCELLigence system. RMC transfected with 2 different SGPL1 siRNAs exhibited decreased serum-induced migration rate (blue and red lines) compared with scrambled siRNA control (black line). (B) Diminished cell migration rate in patients with SGPL1 mutations. Using the xCELLigence system, fibroblasts from individuals with SGPL1 mutations (blue, red, and orange lines) showed decreased migration rate compared with control (black solid line). (C) Active GTP-bound RAC1 and CDC42 precipitated from RMCs transfected with scrambled (Scr) or SGPL1 siRNA using a GST-PAK1 (CRIB) pulldown assay. Compared with control cells, RMCs transfected with SGPL1 siRNA exhibited a significant decrease in relative CDC42 and RAC1 activity. The efficiency of knockdown by siRNA was confirmed by immunoblotting with an anti-SGPL1 antibody (second to lowest panel). (D) Active GTP-bound RHOA precipitated from RMCs using a GST-rhotekin (RBD) pulldown assay. RMCs transfected with scrambled control siRNA versus SGPL1 siRNA exhibited no significant differences in relative RHOA activity. C and D represent 3 experiments each. (E) Effect of S1PR antagonists on RMC migration rate. SGPL1 knockdown caused decreased migration rate (red line) (Supplemental Figure 3B), which was partially rescued by VPC23109 (green line), but not by JTE013 (orange line). VPC23109 is an antagonist that selectively inhibits S1PR1 and S1PR3, whereas JTE013 is an antagonist for S1PR2. Each cell index value corresponds to the average of more than triplicates and SD is in only 1 direction for clarity in A, B, and E.
To test whether SGPL1 or S1P is involved in cell survival or proliferation, we investigated the effect of silencing SGPL1 on cell apoptosis and proliferation in rat mesangial cells (RMCs). Knockdown of Sgpl1 did not affect apoptosis or proliferation in RMCs (Supplemental Figure 12). We then examined the effect of Sgpl1 knockdown on migration of RMCs and found that migration of RMCs was reduced (Figure 3A) upon knockdown of Sgpl1 by transfection with either of 2 siRNAs (Supplemental Figure 12). In addition, we confirmed this effect by showing that cell migration was significantly reduced in fibroblasts from individuals with SGPL1 mutations (Figure 3B).
S1P has previously been implicated in the regulation of RHO-like small GTPases (RHOA/RAC1/CDC42), and disruption of RAC1 signaling was previously implicated in the pathogenesis of SRNS (13–15). We therefore tested whether knockdown of SGPL1 would affect activation of the RHO-like small GTPases. When Sgpl1 was silenced by siRNA in RMCs, we observed a decrease in active GTP forms of CDC42 and RAC1 (Figure 3C), whereas active RHOA (Figure 3D) was unchanged.
To test for specificity of the migration defect observed upon Sgpl1 knockdown, we employed S1PR antagonists. Among 5 S1PR (S1PR1-5), S1PR1, S1PR2, and S1PR3 are expressed in kidney (16). Therefore, we chose VPC23109, which preferentially acts on S1PR1 and S1PR3, and JTE013, which is an antagonist for S1PR2. Interestingly, VPC23109 partially rescued the decreased migration conferred by Sgpl1 knockdown in RMCs, whereas JTE013 failed to rescue the migration defect, suggesting that the S1P effect on RMC migration rate is mediated through S1PR1 and/or S1PR3 (Figure 3E).
SGPL1 is required for Drosophila nephrocyte function.
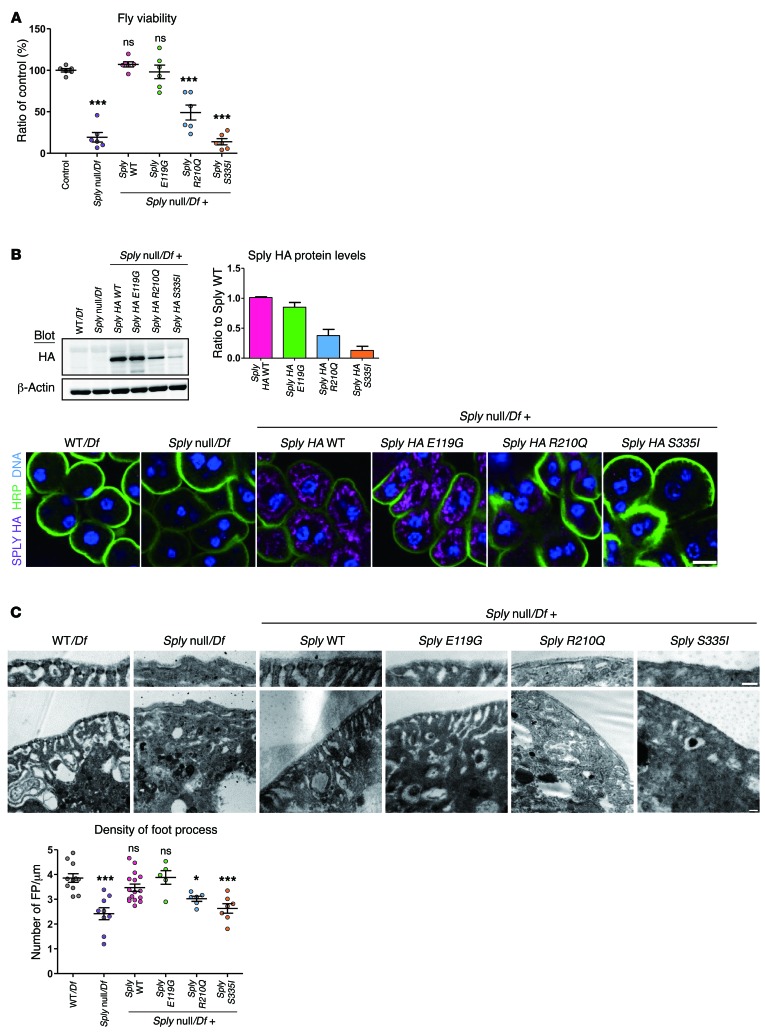
Sply is the SGPL1 ortholog of Drosophila, and a Sply null allele has previously been characterized and shown to decrease fly viability (17). Accordingly, Sply null hemizygous flies (Sply null/Df) had decreased viability (Figure 4A).
Figure 4. SGPL1 missense mutations fail to rescue the phenotype of the Drosophila SGPL1 ortholog (Sply) KO.
Human SGPL1 mutations p.Glu132Gly, p.Arg222Gln, and p.Ser346Ile are equivalent to Drosophila Sply mutations p.Glu119Gly, p.Arg210Gln, and p.Ser335Ile. (A) Viability defects of Sply null hemizygous and Sply mutant flies. Viability was calculated as the percentage of Sply null hemizygous offspring of heterozygous parents. Values are normalized to the viable control Df(2R)BSC433/Df(2R)247. More than 650 flies per genotype; 6 independent experiments. (B) Western blot of HA-tagged Sply in third instar larvae (top panel) and immunofluorescence of third instar garland nephrocytes stained for HA (purple) (bottom panel). Membrane and nuclei were labeled with HRP (green) and Hoechst (blue), respectively. Five or more larvae/genotype; 3 independent experiments. Scale bar: 10 μm. (C) Foot process density in Sply null hemizygous and Sply mutant third instar garland nephrocytes. TEM images and quantification. Six or more nephrocytes/genotype; 2 independent experiments. Scale bars: 200 nm. Statistical analysis performed by Bonferroni’s test following ANOVA. ***P < 0.0005; *P < 0.05. All graphs show mean ± SEM. ns, not significant.
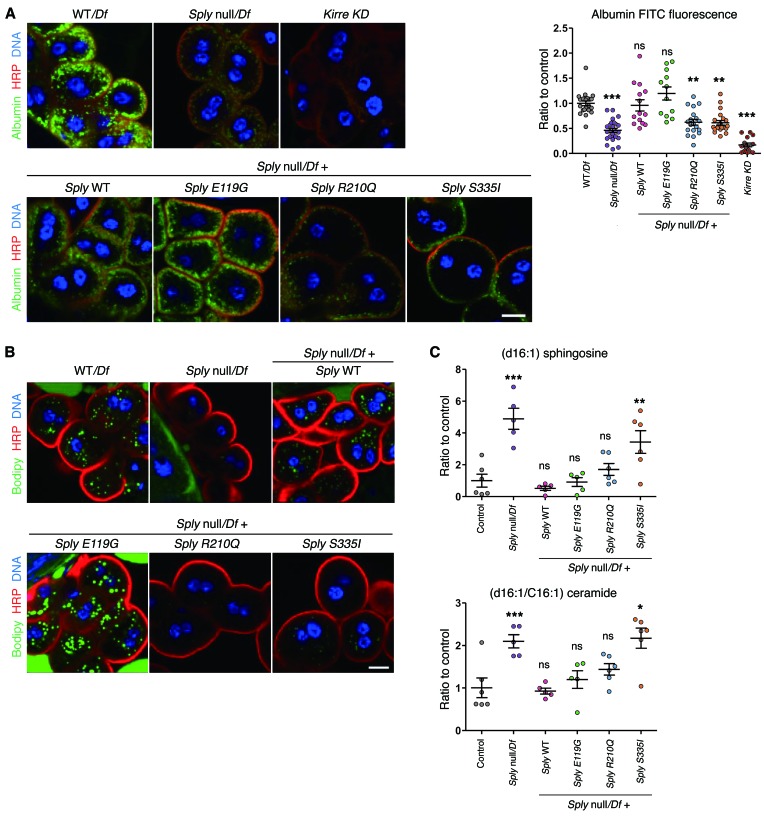
To assess whether Sply deficiency evoked a podocytopathy phenotype in flies, we assessed the structure and function of nephrocytes in Sply null hemizygous flies. Nephrocytes are thought to represent the Drosophila counterparts of podocytes. These cells display invaginations of plasma membrane that form analogous structures to podocyte foot processes and are connected by a slit diaphragm (18). This structure allows filtering of hemolymph and macromolecules that can then undergo endocytosis. We observed no major decrease of the slit diaphragm protein kirre (NEPH1 ortholog) by immunofluorescence (Supplemental Figure 13); however, by TEM, we identified a reduction of foot process density (Figure 4C). To assess the impact of Sply on nephrocyte function, we analyzed the uptake of fluorescent-coupled albumin. Albumin is filtered and endocytosed in nephrocytes under normal conditions, but not when there is disruption of foot processes (18, 19). Consistently, Sply null hemizygous flies had significantly reduced uptake of albumin as compared with control (Figure 5A).
Figure 5. Defects of vesicular transport and sphingosine metabolism in the SGPL1 drosophila ortholog (Sply) KO.
(A) Albumin uptake in Sply null hemizygous and Sply mutant third instar garland nephrocytes. Nephrocytes were incubated for 2.5 minutes with albumin-FITC (green), fixed and stained for the membrane marker HRP (red). Ten or more larvae/genotype; 3 independent experiments. Scale bar: 10 μm. (B) Lipid droplets in Sply null hemizygous and Sply mutant third instar garland nephrocytes, assessed by Bodipy staining. Six or more larvae/genotype; 2 independent experiments. Scale bar: 10 μm. (C) Sphingoid bases accumulation in Sply null hemizygous and Sply mutant third instar larvae assessed by LC/MS. Note that p.Arg210Gln mutant rescues with less efficiency than Sply WT (p.Arg210Gln vs. Sply WT, P = 0.009 for sphingosines and P = 0.02 for ceramides, t test). n = 6 independent experiments. For Sply null, Sply WT, and p.E119G, 1 analysis was removed due to poor quality chromatography. Control corresponds to WT larvae. Statistical analysis performed by Dunnet’s (C) post-hoc tests following ANOVA or Dunn’s post-hoc test following Kruskal-Wallis (A). ***P < 0.0005; **P < 0.005; *P < 0.05. All graphs show mean ± SEM.
Due to the known role of Sply on sphingolipid catabolic pathway and its indirect implication in other lipid pathways, such as the regulation of sterol regulatory element-binding protein processing in Drosophila (20), we also studied the effect of Sply KO on the lipid content of nephrocytes. Sply null hemizygous flies showed an almost complete absence of lipid droplets stained by Bodipy (Figure 5B) or Nile red (data not shown), suggesting a reduction of neutral lipids. Using liquid chromatography–mass spectrometry (LC/MS), we also assessed the accumulation of sphingoid bases and ceramides in Sply null hemizygous flies. Drosophila has been shown to have C14 and C16 sphingosine (21) and C14 and C16 sphingadienes (22), a sphingoid base with an extra double bond compared with sphingosine. Sphingolipid intermediates upstream of S1P, including long chain bases and ceramides, have been shown to accumulate when SGPL1 activity is inhibited, including in Sply mutant flies (17, 22). As expected, we observed a significant increase of C16 sphingosine, C14 sphingadiene, and C16 ceramide in Sply null hemizygous third instar larvae (Figure 5C and Supplemental Figure 14). Together, these data suggest that Sply null hemizygous flies have defects reminiscent of the podocyte disease found in humans with SGPL1 mutations and display altered lipid metabolism due to the disruption of sphingolipid catabolic pathway.
Lack of rescue with mutant alleles in the Drosophila model.
To assess the effect of SGPL1 missense mutations in vivo, we reexpressed the HA-tagged Sply carrying WT or the corresponding human mutations p.Glu119Gly, p.Arg210Gln, and p.Ser335Ile (equivalent to human mutants p.Glu132Gly, p.Arg222Gln, and p.Ser346Ile, respectively). We observed that under its endogenous promoter, HA-tagged Sply is expressed in nephrocytes (Figure 4B). Consistent with our previous results, the expression of mutants p.Arg210Gln and p.Ser335Ile was greatly reduced at the protein level compared with WT (Figure 4B), but without any significant difference at the mRNA level (Supplemental Figure 15).
Reexpression of Sply WT and p.Glu119Gly transgenes rescued Sply mutant viability defect and nephrocyte dysfunction, which was not the case for p.Arg210Gln and p.Ser335Ile mutations (Figure 4 and Figure 5A). Regarding the neutral lipid content of nephrocytes, we observed a rescue of the Sply phenotype with Sply WT and p.Glu119Gly transgenes, but not with p.Arg210Gln or p.Ser335Ile (Figure 5B).
In whole third instar larvae, the phenotype of C16 sphingosine and C16 ceramide accumulation present in Sply null hemizygous flies was rescued with equal efficiency by Sply WT and p.Glu119G transgenes (Figure 5C). However, p.Ser335Ile mutant failed to rescue (Figure 5C). p.Arg210Gln mutant partially rescued the Sply null phenotype, but with significantly less efficiency than Sply WT (Figure 5C). This suggests that p.Arg210Gln and p.Ser335Ile, but not p.Glu119Gly, mutations affect the Sply enzymatic activity and result in the accumulation of upstream sphingolipids, sphingosines, and ceramides. Similar results were observed when analyzing fly sphingadienes, although complementation results for the mutants p.Arg210Gln and p.Ser335Ile did not reach a statistically significant difference from control (Supplemental Figure 14). We conclude that SGPL1 missense mutations p.Arg222Gln and p.Ser346Ile are pathogenic, impair sphingolipid metabolism, and induce nephrocyte defects reminiscent of those found in human podocytopathies. Consistent with our data on the splicing defects (Supplemental Figure 4B) induced by p.Glu132Gly mutation, we found that the corresponding amino acid change did not induce any defect in Drosophila.
Discussion
In this study, we identified recessive SGPL1 mutations as a cause of syndromic SRNS. The disease phenotype entailed SRNS with facultative ichthyosis, adrenal insufficiency, neurologic involvement, and immunodeficiency. We show that SGPL1 mutations are loss-of-function mutations that lead to reduced protein levels and enzyme activity and impaired degradation of long-chain sphingoid bases. In Drosophila, the missense mutations led to decreased viability and nephrocyte defects that are reminiscent of podocyte changes in human NS. WT Sply complemented Sply phenotype and sphingolipid profiles, whereas Sply harboring mutations did not complement.
SGPL1 is ubiquitously expressed in tissues and is an essential enzyme of the sphingolipid catabolic pathway. It is thus considered a main regulator of S1P levels and other sphingoid bases. Loss of Sgpl1 in mouse models phenocopies the human disease. In fact, Sgpl1–/– mice fail to thrive and die soon after weaning, showing defects in the immune system, the urinary system, vasculature, and bone as well as altered lipid metabolism (11, 23–25). However, adrenal insufficiency has not been described in Sgpl1–/– mice, but to our knowledge, adrenal function was not assessed in these animals. In the immune system, changes in local S1P concentration and gradient between tissues (low S1P) and efferent lymph and blood (high S1P) affect T cell egress from lymphoid organs (26, 27), resulting in reduced levels of circulating lymphocytes with overrepresentation of T cells with a memory phenotype over naive T cells (27), as observed in the SGPL1 mutant patient in which a complete immunological work-up has been performed. Interestingly, partial deficiency of Sgpl1 in inducible knockout models led to a less severe phenotype, with decreased lethality and in which prevailing manifestations involved kidney (massive proteinuria with FSGS), the immune system, platelets, and skin (28, 29). The variable organ involvement found in different SGPL1 mutated families may be related to levels of residual functional SGPL1 conveyed by different mutations. However, the intrafamilial variability may be explained by the presence of other genetic factors that may influence the disease manifestations. For instance, variants or polymorphisms in genes involved in S1P metabolism and signaling, such as SGPP1, SGPP2, SPHK1, SPHK2, SPNS2, or S1PR1-5, may contribute to intrafamilial variability.
The phenotypic spectrum of SGPL1 mutations is reminiscent of Schimke immunoosseous dysplasia (MIM 242900), which is caused by recessive mutations in SMARCAL1 encoding SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A-like protein 1. Specifically, mutations in both SMARCAL1 and SGPL1 are associated with FSGS and T cell–related immunodeficiency. Whereas SMARCAL1 is involved in reannealing stably unwound DNA, SGPL1 has been implicated as a mediator of DNA damage response (DDR). Increased SGPL1 expression and activity in DDR promotes apoptosis through a pathway involving p53 and caspase 2 (30). Therefore, defects in DDR may also contribute to the pathogenesis of SGPL1 deficiency. However, it should be noted that the T cell defects in patients with SMARCAL1 mutations are global and involve impairment of T cell proliferation, survival, and function. In contrast, SGPL1 deficiency in mice leads to lymphopenia as a result of trafficking defects, as shown by the sequestration of mature T cells in mutant mouse thymus, whereas T cell proliferation in thymus is unaffected (J.D. Saba and colleagues, unpublished observations).
Patient fibroblasts had significantly elevated C22:0, C24:0, and C24:1 ceramides compared with control fibroblast (Supplemental Figure 8). Interestingly, total serum ceramide levels and C24:0 and C16:0 lactyosylceramides were found to be high in children with CKD, which suggests they may play a causal role in the development of FSGS in our cohort (31). Total ceramide levels and the balance of different ceramide species can influence apoptosis and autophagy, thereby contributing to the pathophysiology of many diseases.
S1P is a bioactive lipid implicated in the regulation of cell survival, apoptosis, proliferation, and migration (32, 33). It can act in a paracrine and autocrine fashion by acting on G protein–coupled S1PRs that account for activation of signaling pathways such as Akt, mTOR, and RHO/RAC/CDC42 (34). However, S1P can also act intracellularly by regulating proapoptotic effector molecules such as BAK and BAX (35), TRAF2 (36), HDAC activity, and thus, epigenetic programs in the nucleus (37). The pathogenesis of disease within the target organs could result from both an excess of intracellular S1P and an imbalance of other sphingoid bases, but also from S1P signaling through the S1PRs. Podocytes are known to exhibit S1PRs, and as components of the glomerular filtration barrier, they could exhibit increased sensitivity to circulating S1P levels, especially under conditions of reduced SGPL1 activity, as has been shown in neuron primary cultures (38). In Drosophila, the phenotype we observed in nephrocytes upon constitutive Sply deletion is most likely to be secondary to intracellular accumulation of S1P, but also to increased interstitial S1P levels, and cannot argue against or in favor of a cell no-autonomous effect of SGPL1 deficiency. However, in our study, cultured podocytes stimulated with S1P did not show increased apoptosis at the S1P expected disease level. Additionally, SGPL1 knockdown in podocytes also did not lead to a phenotype in cell migration or cell proliferation, although these results could be due to insufficient SGPL1 decrease, since in neurons, a 70% reduction of SGPL1 using an RNA strategy is insufficient to induce accumulation of S1P (38). In Drosophila, there has still been no identification of a S1PR orthologue. However, we cannot exclude the possibility of a Drosophila S1PR. Accordingly, the phenotype we observed in nephrocytes upon the constitutive Sply knockout cannot argue against or in favor of a cell-nonautonomous effect of SGPL1 deficiency.
However, in mesangial cells, SGPL1 knockdown induced a decrease of active RAC1 and CDC42 and a phenotype of decreased cell migration, which was partially mitigated by S1PR inhibition. Our findings in RMCs and Drosophila nephrocytes suggest that both cell types can be affected by SGPL1 deficiency, but it is not clear which cell type is primary in the pathogenesis. To address this, generation of transgenic mice with glomerular cell type–specific deletion of Sgpl1 is under way.
Methods
Study participants.
We obtained clinical data, blood samples, and skin biopsies from SRNS patients from worldwide sources. The diagnosis of NS was made by nephrologists based on standardized clinical and histologic criteria (39). Renal biopsies were evaluated by pathologists. Additional, clinical data were obtained using a standardized questionnaire (http://www.renalgenes.org)
WES.
In consanguineous families, we combined WES with HM, as established previously (40, 41). Genetic regions of homozygosity were plotted across the genome (Figure 1A) (42, 43). Exome capture was performed with the Agilent SureSelect V5 Enrichment Capture Kit. The enriched library was sequenced on an Illumina HiSeq 4000 (100 bases paired end). Variant burden analysis was performed as previously described (44). Sequence reads were mapped against the human reference genome (NCBI build 37/hg19) using CLC Genomics Workbench (version 6.5.1) software (CLC bio). High-throughput exon resequencing was performed largely as described previously (9). For the nonconsanguineous family NCR61 and consanguineous family EB, exome sequencing was performed in 2 (NCR61-1 and NCR61-2) and 1 (EB-1) affected children. The enriched library was sequenced on an Illumina HiSeq 2500 (V4 chemistry, 2 × 125 bases). Variants were called as described (45).
RNA extraction, RT-PCR, and cDNA cloning.
Human SGPL1 full-length cDNA was obtained (clone MGC: 60255 IMAGE: 6150776) and subcloned into pRK5-N-Myc, pDEST69-N-FLAG, or BABEpuro-gateway (Addgene plasmid no. 51070). Mutations were introduced using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies).
SGPL1 enzymatic activity assay.
SGPL1 activity in whole cell extracts of patient fibroblasts and HEK293T cells expressing WT and mutant SGPL1 cDNA constructs was measured by quantifying the amount of hexadecenal formed over time as described (46).
3D protein structure.
The crystal structure of human SGPL1 (PDB code 4Q6R) was prepared using the protein preparation wizard in Schrödinger Maestro release 2015.1. (version 10.1). After protein preparation, the effects of mutations were computationally assessed using the Residue Scanning module in BioLuminate 1.8 (Biologics Suite 2015-1: BioLuminate, version 1.8).
Cell culture and transfection.
Human podocytes were provided by M. Saleem (University of Bristol, Bristol, United Kingdom) and maintained at the permissive temperature of 33°C in RPMI + GlutaMAX-I (Gibco, Thermo Fisher Scientific) supplemented with 10% FBS, penicillin (50 IU/ml)/streptomycin (50 μg/ml), and insulin–transferrin–selenium-X. RMCs (ATCC CRL-2573) were maintained in DMEM supplemented with 15% FBS, 0.4 mg/ml G418, and penicillin/streptomycin. SGPL1-specific and control scrambled siRNAs were purchased from Dharmacon. siRNAs were transfected into podocytes or RMCs using Lipofectamine RNAiMAX (Invitrogen). The target sequences of siRNAs are in Supplemental Table 1. Human fibroblasts were grown in DMEM supplemented with 15% FBS, penicillin/streptomycin, and nonessential amino acids (Invitrogen). HEK239T cells were used for lentivirus production and transfected by the calcium phosphate precipitation method.
Immunoblotting, immunofluorescence, and GST pulldown assay.
Experiments were performed as described previously (47). GST-PAK1 and GST-Rhotekin beads were purchased from Cytoskeleton Inc. Anti-SGPL1 (AF5535, R&D Systems), anti-BiP (ab21685), anti–β-actin (ab6276, Abcam), anti-CD31 (MA3100, Thermo), anti-RAC1 (610650), anti-CDC42 (610928), anti-GM130 (610822, BD Transduction Laboratories), anti-Myc (sc-789), anti-RHOA (sc-418), anti-WT1 (sc-7385, Santa Cruz Biotechnology Inc.), anti-GOLGB1(HPA011008), anti–α-smooth muscle actin (A2547), anti-FLAG (F3165, Sigma-Aldrich), anti-HA (11867423001, Roche), and anti-synaptopodin (20-4694, American Research Products) antibodies were purchased from commercial sources. Immunoblotting was quantified by densitometry using ImageJ software (NIH). For immunofluorescence in Drosophila nephrocytes, third instar garland nephrocytes were dissected and fixed for 20 minutes in 4% paraformaldehyde at room temperature. For Kirre stainings, an alternative fixation method was used: nephrocytes were heat fixed during 5 seconds at 90°C in 0.7% NaCl, 0.05% Triton X-100 solution (48). The following primary antibodies were used: anti-Kirre (gift from Karl Fischbach, University of Freiburg) anti-HA, and Alexa Fluor 488 or Cyanine Cy3–conjugated anti–horseradish peroxidase (Jackson Immunoresearch). Nile red and Bodipy 493/503 (Thermo Fisher Scientific) were used to stain lipid droplets. For albumin uptake assay, garland nephrocytes were dissected from third instar larvae in Schneider’s medium (Pan-Biotech), incubated for 2.5 minutes with 0.1 mg/ml Alexa Fluor 488–conjugated albumin (Thermo Fisher Scientific) at room temperature, and fixed in 4% PFA for 20 minutes. Confocal images were obtained with a Leica TCS SP8 SMD, and ImageJ (NIH) was used for image analysis.
Migration and proliferation assay.
Migration assays were performed using the xCELLigence system (ACEA Biosciences). For migration assays, 48 hours after transfection, 4 × 104 cells were plated with serum-free media in the upper chamber of CIM-plate 16. The lower chambers were filled with 10% FBS for chemoattraction or with serum-free media. For proliferation assays, 4 × 104 cells were plated in the E-plate 16. The data obtained were analyzed with RTCA software (ACEA Biosciences Inc.). Results are presented as the time versus cell index curve.
Yeast strains and plasmids.
The following yeast strains were used in this study: BY4741-Euroscarf, MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, dpl1Δ::KanMx, RH4863, MATα lcb3Δ::KanMx dpl1Δ::KanMx ura3 leu2 his4 ade2 bar1, pLCB3::URA3 (PCR amplified genomic LCB3 cloned into YCplac33), RH431, and MATα ura3 leu2 his4 bar1 (49). pRS415-ADH2 was used to overexpress human SGPL1 in yeast. pRS415-ADH-SGPL1 was constructed by cloning SGPL1 in the XmaI-XhoI sites of pRS415-ADH.
PHS toxicity test.
Yeast dpl1Δ cells transformed with PRS415-ADH bearing WT or mutant human SGPL1 coding sequences were cultured overnight in synthetic defined minus leucine medium and spotted onto synthetic defined plates containing ethanol (vehicle control) and PHS (40 μM) as previously described (50).
Synthetic lethality test.
RH4863 yeast cells were transformed with plasmids overexpressing human SGPL1 (WT and mutant variants) or yeast (positive control). Cells grown in synthetic defined minus leucine medium were 10-fold serially diluted and spotted onto synthetic defined–agar without leucine, without uracil, or with 1 mg/ml 5-FOA (50).
Fly strains and generation of transgenic flies.
Stocks were maintained on standard cornmeal-yeast food at 25°C. The following fly stocks were used: Sply05091/CyO,Act-GFP (from BL #11393), Df(2R)BSC433/ CyO,Act-GFP (from BL #24937), and Df(2R)247/CyO,Act-GFP (BL #7155), obtained from the Bloomington Stock Center, and Kirre RNAi (GD #14476), obtained from the Vienna Drosophila RNAi Center. Pros-GAL4 (a gift from Barry Denholm, Center for Integrative Physiology, University of Edinburgh, Edinburgh, United Kingdom) y,w1118 was used as a WT control. Due to inbreeding, homozygous Sply05091 (Sply null allele) were almost impossible to obtain. We therefore generated Sply null hemizygous flies by crossing the Sply05091/CyO,Act-GFP flies with Df(2R)BSC433/ CyO,Act-GFP flies that carry a deletion of the Sply locus and 5 other contiguous genes. In the F1 generation, Sply null hemizygous flies were identified by the absence of the balancer chromosome (CyO,Act-GFP). We confirmed the absence of Sply mRNA in Sply null hemizygous flies by reverse-transcriptase PCR (RT-PCR) (Supplemental Figure 15). For Sply rescue constructs, the HA tag (in-frame before the stop codon) and the corresponding SGPL1 human mutations were individually inserted by site-directed mutagenesis in the cloned 4-kb Sply genomic locus, including regulating sequences (–491 bp) and 3′ UTR (+1066 bp). Subsequently, the WT and mutant Sply rescue constructs were cloned into a pattB vector (Konrad Basler, Institute of Molecular Life Sciences, University of Zurich, Zurich, Switzerland) and injected into flies with an attP landing site at 76A2 by BestGene Inc. Primer sequences and cloning details are available in Supplemental Table 2. For the rescue experiments, 1 copy of the Sply rescue constructs was transferred by mating to the Sply null hemizygous background.
Viability assay.
Fly crosses were allowed to lay eggs for 48 hours on standard cornmeal-yeast food. The percentage of hatching adults of the appropriate genotype was recorded. As a control, we crossed the Df(2R)BSC433/CyO,Act-GFP with Df(2R)247/CyO,Act-GFP (carrying a deletion nonoverlapping with Df[2R]BSC433), and the percentage of the normally viable Df(2R)BSC433/Df(2R)247 flies was recorded. On average, a total of 120 F1 hatching flies were counted per condition, and the experiments were repeated 6 times. Values reported were normalized to the control.
Mice.
Sgpl1–/– mice were previously described (11). Sgpl1–/– mice, littermate controls, and heterozygote breeders were handled in pathogen-free conditions.
TEM.
After dissection, third instar garland nephrocytes and kidneys from Sgpl1+/+ and Sgpl1–/– mice were fixed in 2.5% glutaraldehyde at room temperature and processed for TEM with standard techniques.
Sphingoid bases analysis.
Sphingolipid metabolites from fly larvae and patient fibroblast samples were extracted using a protocol described previously (51). Analytical systems used were composed of Agilent 1290 UPLC coupled with Agilent 6490 triple quadruple mass spectrometer. Analysis was carried out using multiple reaction monitoring (MRM) mode. The general source settings in the positive ionization modes were as follows: gas temperature, 200°C; gas flow, 16 minute–1; nebulizer, 20 psi; sheath gas temperature, 250°C; sheath gas flow, 11 minute–1; and capillary voltage, 3.0 kV. Specific MRM transitions for fly sphingoid bases were programmed based on the transitions described previously (52). Chromatographic separation of sphingolipids was performed with a Zorbax RRHD Eclipse Plus C18 (2.1 × 150 mm, 1.8 μm) equilibrated at 50°C. A binary gradient of mobile phase A (water containing 0.2% formic acid and 2 mM ammonium formate) and B (methanol containing 0.2% formic acid and 1 mM ammonium formate) was delivered at a constant flow rate of 1 ml/min. The total run time was 10 minutes. Initial gradient was 70% B and increased to 100% B at 8 minutes and returned to baseline at 8.5 minutes and maintained until 10 minutes. Data processing of all metabolites was performed using MassHunter software (Agilent).
RT-PCR and real-time PCR.
Total RNA was isolated from fibroblasts using a QIAGEN RNA extraction kit (QIAGEN). cDNA was prepared using reverse transcriptase Superscript II (Invitrogen). PCR was performed using ReadyMix Taq PCR (Sigma-Aldrich) and the following primers: SGPL1 exon 4 forward, 5′-CTCACCAGGAAGATGCCCAT-3′ and SGPL1 exon 8 reverse, 5′-GCTTTGCAGGCCATCAGTAT-3′. The amplified PCR products were then cloned into a pGEM–T Easy Vector (Promega) and analyzed by Sanger sequencing. In Drosophila, relative expression levels were determined using the Absolute SYBR Green ROX Mix (ABgene) and specific primers as follows: Sply forward, 5′-CTGGCTCTGGACTGTGATCTG-3′ and reverse, 5′-GACGCTTGCCACGAATGTAA-3′. Expression levels were normalized to actin.
Statistics.
Results are presented as mean ± SEM or SD for the indicated number of experiments. Statistical analysis of continuous data was performed with 2-tailed Student’s t test or 1-way ANOVA, with Dunnet’s, Bonferroni’s, or Dunn’s post hoc test, as appropriate. P < 0.05 was considered statistically significant.
Study approval.
Approval for human subject research was obtained from the University of Michigan (Ann Arbor, Michigan, USA) and Boston Children’s Hospital institutional review boards and the Comité de Protection des Personnes Ile de France II. All subjects gave informed consent. Approval for mouse research was obtained from the University Committee on Use and Care of Animals (UCUCA) of the UCSF Benioff Children’s Hospital Oakland and Boston Children’s Hospital.
Supplemental appendix.
For additional data, including Supplemental Tables 1–3 and Supplemental Figures 1–15, see the supplemental material, available with the full text of this article.
Author contributions
SL, SG, HYG, SS, CES, KS, RPL, OG, MF, MAC, C Antignac, and FH prepared and evaluated genetic mapping and exome sequences. SL, SG, HYG, KS, MAC, C Antignac, and FH identified mutations in the human SGPL1 gene. SG, HYG, SL, BO, WIC, SH, MZ, VC, C Arrondel, OB, VV, JDS, C Antignac, and FH provided and evaluated critical cell lines and animal resources. JDS, BO, JHS, HYG, and WIC performed all mouse studies and analyzed mutant SGPL1 expression or enzyme activity in fibroblasts and HEK293 cells. JDS, BO, HYG, and WIC analyzed the Sgpl1–/– mouse model, critically evaluated mouse data, and wrote mouse studies. SG, N Lachaussée, MS, JP, and C Antignac performed Drosophila studies. AS performed TEM. JHS performed all LC/MS assays for quantification of cellular SPLY activity and SPL distribution. HR and VG performed yeast complementation experiments. SL, HYG, SG, MZ, VC, C Antignac, D Schapiro, MF, DAB, FS, NK, JG, TG, KS, VC, D Schanze, IF, YS, AST, BA, JD, RB, YP, EA, N Lloberas, AM, CES, FPB, JPM, VRD, AMC, MCW, MAC, NW, and FH recruited patients and gathered detailed clinical information for the study. JG and TG performed the immunophenotyping of patient EB-1. HS, RW, and GC performed structural molecular modeling of the SGPL1 enzyme. JDS directed studies undertaken by BO and JHS. All authors critically reviewed the paper. FH and C Antignac conceived of and directed the entire project and wrote the paper with help from SL, SG, HYG, BO, and JDS.
Supplementary Material
Acknowledgments
The authors thank the families who contributed to this study. We thank the Yale Center for Mendelian Genomics for WES analysis, U. Pannicke for help in analyzing data, and S. Braun for technical assistance. FH was supported by grants from the NIH (DK076683, DK068306). FH is the Warren E. Grupe Professor. HYG was supported by the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Education (2015R1D1A1A01056685), by a Nephcure-ASN Foundation Kidney Research Grant, and by a faculty research grant of Yonsei University College of Medicine (6-2015-0175). C Antignac was supported by grants from the Agence Nationale de la Recherche (GenPod project ANR-12-BSV1-0033.01), the European Union’s Seventh Framework Programme (FP7/2007-2013/no 305608-EURenOmics), the Fondation Recherche Médicale (DEQ20150331682), and the ‘‘Investments for the Future’’ program (ANR-10-IAHU-01). SG was supported by the Program Santé-Science (MD-PhD) of Imagine Institute. FS was supported by the European Union’s Seventh Framework Programme (FP7/2007-2013/n°305608-EURenOmics). Immunophenotyping was supported by the German Centre for Infectious Diseases (Thematical Translation Units: Infections of the immunocompromised host). JDS was supported by the John and Edna Beck Chair in Pediatric Cancer Research, the Swim Across America Foundation, and a grant from the NIH (GM66594, NCI CA129438). KS was supported by the Center for Personalized Immunology (supported by the National Health and Medical Research Council of Australia [NHMRC]), the Australian National University, Canberra, Australia. MZ was supported by the German Ministry of Education and Research (Bundesministerium füur Bildung und Forschung, project: GeNeRARe). EW was supported by the Leopoldina Fellowship Program, German National Academy of Sciences Leopoldina (LPDS 2015-07). TJS was supported by grant Jo 1324/1-1 of Deutsche Forschungsgemeinschaft (DFG). MF was supported by grants from the Spanish Society of Nephrology and the Catalan Society of Nephrology. HR was supported by grants from the Swiss National Science Foundation, SystemsX.CH, and the NCCR Chemical Biology. This work was performed under the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, the Ontario Research Fund, Genome Quebec, and the Children’s Hospital of Eastern Ontario Research Foundation. We acknowledge the contribution of the high-throughput sequencing platform of the McGill University and Génome Québec Innovation Centre, Montréal, Canada. The names of Care4Rare Canada steering committee members appear in the Supplemental Acknowledgments.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(3):912–928.https://doi.org/10.1172/JCI89626.
Contributor Information
Svjetlana Lovric, Email: svjetlanalovric@gmail.com.
Sara Goncalves, Email: sara.goncalves@inserm.fr.
Heon Yung Gee, Email: hygee@yuhs.ac.
Babak Oskouian, Email: boskouia@mills.edu.
Honnappa Srinivas, Email: honnappa.srinivas@novartis.com.
Won-Il Choi, Email: octerpus@nate.com.
Shirlee Shril, Email: Shirlee.Shril@childrens.harvard.edu.
Shazia Ashraf, Email: Shazia.Ashraf@childrens.harvard.edu.
Weizhen Tan, Email: Weizhen.Tan@childrens.harvard.edu.
Jia Rao, Email: amayraojia@gmail.com.
Merlin Airik, Email: merlin.airik@chp.edu.
David Schapiro, Email: David.Schapiro@childrens.harvard.edu.
Daniela A. Braun, Email: daniela.braun@childrens.harvard.edu.
Carolin E. Sadowski, Email: carosad@gmx.de.
Eugen Widmeier, Email: Eugen.Widmeier@childrens.harvard.edu.
Tilman Jobst-Schwan, Email: Tilman.Jobst-Schwan@childrens.harvard.edu.
Johanna Magdalena Schmidt, Email: magdalena-schmidt@uni-bonn.de.
Vladimir Girik, Email: Vladimir.Girik@unige.ch.
Guido Capitani, Email: guido.capitani@psi.ch.
Jung H. Suh, Email: jsuh@chori.org.
Noëlle Lachaussée, Email: noelle.lachaussee@gmail.com.
Christelle Arrondel, Email: christelle.arrondel@inserm.fr.
Julie Patat, Email: jdesir@ulb.ac.be.
Olivier Gribouval, Email: olivier.gribouval@inserm.fr.
Olivia Boyer, Email: olivia.boyer@aphp.fr.
Alain Schmitt, Email: alain.schmitt@inserm.fr.
Vincent Vuiblet, Email: vvuiblet@yahoo.fr.
Seema Hashmi, Email: simasiut@yahoo.com.
Rainer Wilcken, Email: rainer.wilcken@novartis.com.
Julian P. Midgley, Email: Julian.Midgley@albertahealthservices.ca.
Nicola Wright, Email: Nicola.Wright@albertahealthservices.ca.
Jacek Majewski, Email: jacek.majewski@mcgill.ca.
Martin Zenker, Email: martin.zenker@med.ovgu.de.
Franz Schaefer, Email: franz.schaefer@med.uni-heidelberg.de.
Navina Kuss, Email: Navina.Kuss@med.uni-heidelberg.de.
Johann Greil, Email: johann.greil@med.uni-heidelberg.de.
Thomas Giese, Email: Giese@uni-hd.de.
Klaus Schwarz, Email: klaus.schwarz@uni-ulm.de.
Denny Schanze, Email: denny.schanze@med.ovgu.de.
Ingolf Franke, Email: ingolf.franke@med.ovgu.de.
Yves Sznajer, Email: yves.sznajer@uclouvain.be.
Anne S. Truant, Email: anneso.truant@hotmail.com.
Brigitte Adams, Email: badams@ulb.ac.be.
Julie Désir, Email: Julie.Desir@erasme.ulb.ac.be.
Ronald Biemann, Email: ronald.biemann@med.ovgu.de.
York Pei, Email: york.pei@uhn.on.ca.
Elisabet Ars, Email: ears@fundacio-puigvert.es.
Nuria Lloberas, Email: nlloberas@ub.edu.
Alvaro Madrid, Email: amadrid@vhebron.net.
Richard P. Lifton, Email: richard.lifton@yale.edu.
Matias Simons, Email: matias.simons@institutimagine.org.
Howard Riezman, Email: Howard.Riezman@unige.ch.
Corinne Antignac, Email: corinne.antignac@inserm.fr.
Julie D. Saba, Email: jsaba@chori.org.
Friedhelm Hildebrandt, Email: friedhelm.hildebrandt@childrens.harvard.edu.
References
- 1.Smith JM, Stablein DM, Munoz R, Hebert D, McDonald RA. Contributions of the Transplant Registry: The 2006 Annual Report of the North American Pediatric Renal Trials and Collaborative Studies (NAPRTCS) Pediatr Transplant. 2007;11(4):366–373. doi: 10.1111/j.1399-3046.2007.00704.x. [DOI] [PubMed] [Google Scholar]
- 2.Hildebrandt F, Heeringa SF. Specific podocin mutations determine age of onset of nephrotic syndrome all the way into adult life. Kidney Int. 2009;75(7):669–671. doi: 10.1038/ki.2008.693. [DOI] [PubMed] [Google Scholar]
- 3.Somlo S, Mundel P. Getting a foothold in nephrotic syndrome. Nat Genet. 2000;24(4):333–335. doi: 10.1038/74139. [DOI] [PubMed] [Google Scholar]
- 4.Tryggvason K, Patrakka J, Wartiovaara J. Hereditary proteinuria syndromes and mechanisms of proteinuria. N Engl J Med. 2006;354(13):1387–1401. doi: 10.1056/NEJMra052131. [DOI] [PubMed] [Google Scholar]
- 5.Sadowski CE, et al. A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2015;26(6):1279–1289. doi: 10.1681/ASN.2014050489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lovric S, Ashraf S, Tan W, Hildebrandt F. Genetic testing in steroid-resistant nephrotic syndrome: when and how? Nephrol Dial Transplant. 2016;31(11):1802–1813. doi: 10.1093/ndt/gfv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lovric S, et al. Rapid detection of monogenic causes of childhood-onset steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2014;9(6):1109–1116. doi: 10.2215/CJN.09010813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raponi M, et al. Prediction of single-nucleotide substitutions that result in exon skipping: identification of a splicing silencer in BRCA1 exon 6. Hum Mutat. 2011;32(4):436–444. doi: 10.1002/humu.21458. [DOI] [PubMed] [Google Scholar]
- 9.Halbritter J, et al. High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J Med Genet. 2012;49(12):756–767. doi: 10.1136/jmedgenet-2012-100973. [DOI] [PubMed] [Google Scholar]
- 10.Saba JD, Nara F, Bielawska A, Garrett S, Hannun YA. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J Biol Chem. 1997;272(42):26087–26090. doi: 10.1074/jbc.272.42.26087. [DOI] [PubMed] [Google Scholar]
- 11.Schmahl J, Raymond CS, Soriano P. PDGF signaling specificity is mediated through multiple immediate early genes. Nat Genet. 2007;39(1):52–60. doi: 10.1038/ng1922. [DOI] [PubMed] [Google Scholar]
- 12.Pewzner-Jung Y, et al. A critical role for ceramide synthase 2 in liver homeostasis: I. alterations in lipid metabolic pathways. J Biol Chem. 2010;285(14):10902–10910. doi: 10.1074/jbc.M109.077594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gee HY, et al. ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest. 2013;123(8):3243–3253. doi: 10.1172/JCI69134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gee HY, et al. FAT1 mutations cause a glomerulotubular nephropathy. Nat Commun. 2016;7:10822. doi: 10.1038/ncomms10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gee HY, et al. KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest. 2015;125(6):2375–2384. doi: 10.1172/JCI79504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosen H, Gonzalez-Cabrera PJ, Sanna MG, Brown S. Sphingosine 1-phosphate receptor signaling. Annu Rev Biochem. 2009;78:743–768. doi: 10.1146/annurev.biochem.78.072407.103733. [DOI] [PubMed] [Google Scholar]
- 17.Herr DR, et al. Sply regulation of sphingolipid signaling molecules is essential for Drosophila development. Development. 2003;130(11):2443–2453. doi: 10.1242/dev.00456. [DOI] [PubMed] [Google Scholar]
- 18.Weavers H, et al. The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature. 2009;457(7227):322–326. doi: 10.1038/nature07526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuang S, Shao H, Guo F, Trimble R, Pearce E, Abmayr SM. Sns and Kirre, the Drosophila orthologs of Nephrin and Neph1, direct adhesion, fusion and formation of a slit diaphragm-like structure in insect nephrocytes. Development. 2009;136(14):2335–2344. doi: 10.1242/dev.031609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobrosotskaya IY, Seegmiller AC, Brown MS, Goldstein JL, Rawson RB. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science. 2002;296(5569):879–883. doi: 10.1126/science.1071124. [DOI] [PubMed] [Google Scholar]
- 21.Fyrst H, Herr DR, Harris GL, Saba JD. Characterization of free endogenous C14 and C16 sphingoid bases from Drosophila melanogaster. J Lipid Res. 2004;45(1):54–62. doi: 10.1194/jlr.M300005-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Fyrst H, et al. Identification and characterization by electrospray mass spectrometry of endogenous Drosophila sphingadienes. J Lipid Res. 2008;49(3):597–606. doi: 10.1194/jlr.M700414-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Vogel P, et al. Incomplete inhibition of sphingosine 1-phosphate lyase modulates immune system function yet prevents early lethality and non-lymphoid lesions. PLoS ONE. 2009;4(1):e4112. doi: 10.1371/journal.pone.0004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allende ML, et al. Sphingosine-1-phosphate lyase deficiency produces a pro-inflammatory response while impairing neutrophil trafficking. J Biol Chem. 2011;286(9):7348–7358. doi: 10.1074/jbc.M110.171819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bektas M, et al. Sphingosine 1-phosphate lyase deficiency disrupts lipid homeostasis in liver. J Biol Chem. 2010;285(14):10880–10889. doi: 10.1074/jbc.M109.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwab SR, Pereira JP, Matloubian M, Xu Y, Huang Y, Cyster JG. Lymphocyte sequestration through S1P lyase inhibition and disruption of S1P gradients. Science. 2005;309(5741):1735–1739. doi: 10.1126/science.1113640. [DOI] [PubMed] [Google Scholar]
- 27.Yu XQ, et al. Pharmacokinetic/pharmacodynamic modelling of 2-acetyl-4(5)-tetrahydroxybutyl imidazole-induced peripheral lymphocyte sequestration through increasing lymphoid sphingosine 1-phosphate. Xenobiotica. 2010;40(5):350–356. doi: 10.3109/00498251003611376. [DOI] [PubMed] [Google Scholar]
- 28.Billich A, et al. Partial deficiency of sphingosine-1-phosphate lyase confers protection in experimental autoimmune encephalomyelitis. PLoS One. 2013;8(3):e59630. doi: 10.1371/journal.pone.0059630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schümann J, et al. Reduced activity of sphingosine-1-phosphate lyase induces podocyte-related glomerular proteinuria, skin irritation, and platelet Activation. Toxicol Pathol. 2015;43(5):694–703. doi: 10.1177/0192623314565650. [DOI] [PubMed] [Google Scholar]
- 30.Oskouian B, et al. Sphingosine-1-phosphate lyase potentiates apoptosis via p53- and p38-dependent pathways and is down-regulated in colon cancer. Proc Natl Acad Sci USA. 2006;103(46):17384–17389. doi: 10.1073/pnas.0600050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitsnefes M, et al. Ceramides and cardiac function in children with chronic kidney disease. Pediatr Nephrol. 2014;29(3):415–422. doi: 10.1007/s00467-013-2642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosen H, Goetzl EJ. Sphingosine 1-phosphate and its receptors: an autocrine and paracrine network. Nat Rev Immunol. 2005;5(7):560–570. doi: 10.1038/nri1650. [DOI] [PubMed] [Google Scholar]
- 33.Fyrst H, Saba JD. An update on sphingosine-1-phosphate and other sphingolipid mediators. Nat Chem Biol. 2010;6(7):489–497. doi: 10.1038/nchembio.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaho VA, Hla T. An update on the biology of sphingosine 1-phosphate receptors. J Lipid Res. 2014;55(8):1596–1608. doi: 10.1194/jlr.R046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chipuk JE, et al. Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell. 2012;148(5):988–1000. doi: 10.1016/j.cell.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvarez SE, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465(7301):1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hait NC, et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325(5945):1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagen N, Van Veldhoven PP, Proia RL, Park H, Merrill AH, van Echten-Deckert G. Subcellular origin of sphingosine 1-phosphate is essential for its toxic effect in lyase-deficient neurons. J Biol Chem. 2009;284(17):11346–11353. doi: 10.1074/jbc.M807336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.[No authors listed] Primary nephrotic syndrome in children: clinical significance of histopathologic variants of minimal change of diffuse mesangial hypercellularity. A Report of the International Study of Kidney Disease in Children. Kidney Int. 1981;20(6):765–771. doi: 10.1038/ki.1981.209. [DOI] [PubMed] [Google Scholar]
- 40.Hildebrandt F, et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5(1):e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gee HY, et al. Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney Int. 2014;85(4):880–887. doi: 10.1038/ki.2013.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hildebrandt F, et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5(1):e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otto EA, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42(10):840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyden LM, et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482(7383):98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Field MA, Cho V, Andrews TD, Goodnow CC. Reliably detecting clinically important variants requires both combined variant calls and optimized filtering strategies. PLoS ONE. 2015;10(11):e0143199. doi: 10.1371/journal.pone.0143199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suh JH, Eltanawy A, Rangan A, Saba JD. A facile stable-isotope dilution method for determination of sphingosine phosphate lyase activity. Chem Phys Lipids. 2016;194:101–109. doi: 10.1016/j.chemphyslip.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gee HY, Tang BL, Kim KH, Lee MG. Syntaxin 16 binds to cystic fibrosis transmembrane conductance regulator and regulates its membrane trafficking in epithelial cells. J Biol Chem. 2010;285(46):35519–35527. doi: 10.1074/jbc.M110.162438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rothwell WF, Sullivan W. Fluorescent analysis of Drosophila embryos. In: Sullivan W, Ashburner RS, ed. Drosophila Protocols, pp. 141–158. New York, NY: Cold Spring Harbor Laboratory Press; 2000:141–158. [Google Scholar]
- 49.Mukhopadhyay D, Howell KS, Riezman H, Capitani G. Identifying key residues of sphinganine-1-phosphate lyase for function in vivo and in vitro. J Biol Chem. 2008;283(29):20159–20169. doi: 10.1074/jbc.M709753200. [DOI] [PubMed] [Google Scholar]
- 50.Bourquin F, Riezman H, Capitani G, Grütter MG. Structure and function of sphingosine-1-phosphate lyase, a key enzyme of sphingolipid metabolism. Structure. 2010;18(8):1054–1065. doi: 10.1016/j.str.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 51.Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39(2):82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Guan XL, et al. Biochemical membrane lipidomics during Drosophila development. Dev Cell. 2013;24(1):98–111. doi: 10.1016/j.devcel.2012.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.