Abstract
Long-acting anti-HIV products can substantively change the standard of care for patients with HIV/AIDS. To this end, hydrophobic antiretroviral drugs (ARVs) were recently developed for parenteral administration at monthly or longer intervals. While shorter-acting hydrophilic drugs can be made into nanocarrier-encased prodrugs, the nanocarrier encasement must be boosted to establish long-acting ARV depots. The mixed-lineage kinase 3 (MLK-3) inhibitor URMC-099 provides this function by affecting autophagy. Here, we have shown that URMC-099 facilitates ARV sequestration and its antiretroviral responses by promoting the nuclear translocation of the transcription factor EB (TFEB). In monocyte-derived macrophages, URMC-099 induction of autophagy led to retention of nanoparticles containing the antiretroviral protease inhibitor atazanavir. These nanoparticles were localized within macrophage autophagosomes, leading to a 4-fold enhancement of mitochondrial and cell vitality. In rodents, URMC-099 activation of autophagy led to 50-fold increases in the plasma drug concentration of the viral integrase inhibitor dolutegravir. These data paralleled URMC-099–mediated induction of autophagy and the previously reported antiretroviral responses in HIV-1–infected humanized mice. We conclude that pharmacologic induction of autophagy provides a means to extend the action of a long-acting, slow, effective release of antiretroviral therapy.
Introduction
The next frontier for antiretroviral therapy (ART) rests in easing drug use, targeting viral reservoirs, extending drug half-life, reducing viral mutations, and overcoming systemic toxicity (1–3). We previously developed, characterized, and tested therapeutic responses of long-acting nanoformulated ART (nanoART) (4–11). Recent successes have come from encasing hydrophobic crystalline antiretroviral drugs (ARVs) into excipients, decorating the formed particles with ligands that target cells and tissue reservoirs or by pharmacologically boosting intracellular depot formations (4, 11–13). Such approaches extended the drugs’ half-life by controlling the rate of drug-particle dissociation and its release hydrolysis to the systemic circulation. However, the production of more hydrophobic and protein-bound ARVs provides only a partial solution. Controlling the dynamics of particle trafficking inside the cell and within early, recycling, and late endosomes is necessary to sustain intracellular drug particle depots and limit hepatic and renal metabolism and drug excretion (4, 6, 11, 14, 15).
We previously showed that nanoparticles of hydrophobic ARVs encased in polymer excipients and coadministered with the mixed-lineage kinase 3 (MLK-3) inhibitor URMC-099 facilitated particle endocytic trafficking and enhanced intracellular drug retention (11). However, the mechanisms for how the MLK-3 inhibitor restricts viral growth were not known. Here, we demonstrate that URMC-099 enhancement of antiretroviral responses is due to the stimulation of autophagy and leads to the subsequent sequestration of drug particles into autophagosomes. An extension of the ARV half-life by URMC-099 was observed for atazanavir (ATV) and dolutegravir (DTG), viral protease and integrase inhibitors, respectively. An increased understanding of how nanoformulated ARV depots are maintained will facilitate the development of a spectrum of anti-HIV slow-release products and in turn enhance the effectiveness of clinical drug regimens. We believe that our work, in toto, represents a notable step forward in the development of sustained-release ARVs for human use.
Results
URMC-099 facilitates nanoART activity.
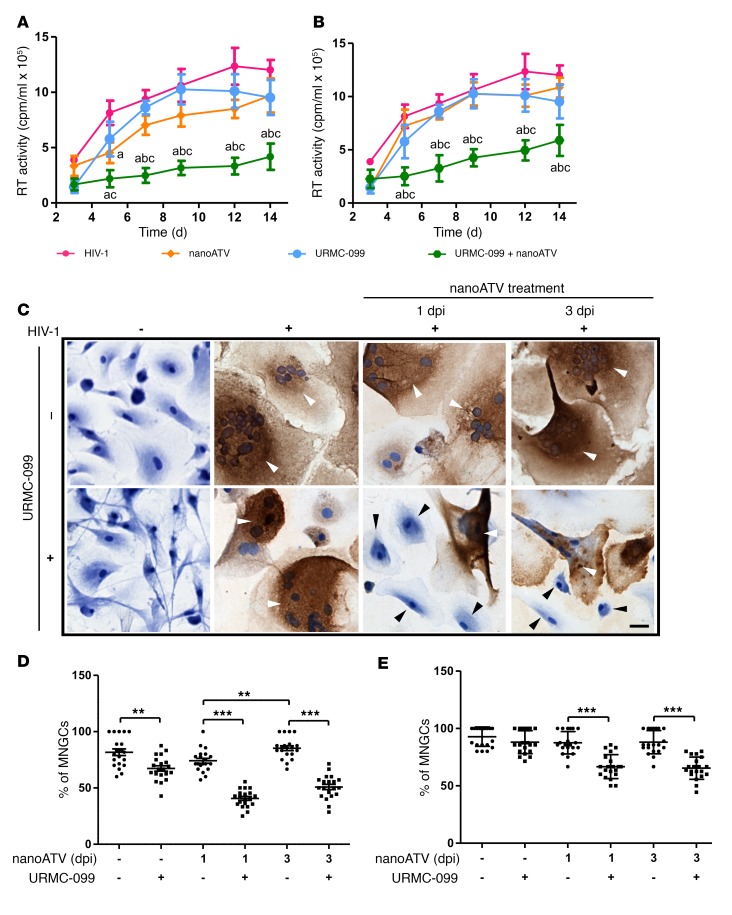
We evaluated the ability of URMC-099 to affect the anti-HIV activity of nanoformulated ARVs in infected human monocyte-derived macrophages (MDMs). We treated HIV-1 ADA strain–infected (HIV-1ADA–infected) MDMs with 400 ng/ml URMC-099, followed by subtherapeutic concentrations (1 μM) of nanoformulated ATV (nanoATV) at 1, 3, and 5 days after infection at a multiplicity of 0.1 infectious virions per cell. The concentration of URMC-099 used elicited a maximal therapeutic efficacy that included drug particle retentions, amyloid β clearance, cellular vitality, and robust neuroprotective and antiinflammatory responses (11, 16–20). As concentrations of nanoATV of 100 μM or greater suppress viral replication, we performed URMC-099 dose-response determinations using subtherapeutic nanoATV concentrations for the measurement of viral growth. Progeny virion formation in culture supernatant fluids was determined by reverse transcriptase (RT) activity. Notably, we found that URMC-099 administered with 1 μM nanoATV reduced virion production by 4-fold compared with virion production when nanoATV was given alone (Figure 1, A and B and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI90025DS1). In addition, coadministration of nanoATV and URMC-099 resulted in reductions of up to 4-fold in the numbers of HIV-1p24+ cells (Figure 1C). We found that the percentages of multinucleated giant cells (MNGCs) were also reduced with URMC-099 and nanoATV cotreatment on both days 1 and 3 after infection (Figure 1, D and E). In the presence of URMC-099, we observed mononucleated MDMs, whereas, without URMC-099, most of the cells were MNGCs by 14 days after infection (Figure 1E and Supplemental Figure 1B). On day 7 after infection, URMC-099 treatment alone showed a reduction in MNGCs compared with that observed with HIV-1 infection (Figure 1D); however, the effect was lost by day 14 (Figure 1E). On day 7 after infection, nanoART treatment initiated on day 3 after HIV-1 infection resulted in a greater number of MNGCs compared with those detected on day 1 of treatment initiation, and even this effect was lost by day 14 after infection.
Figure 1. URMC-099 potentiates antiretroviral activity of ARV nanoformulations.
HIV-1ADA–infected human MDMs were treated with 1 μM nanoATV on (A) day 1 or (B) day 3 after infection in the presence or absence of 400 ng/ml URMC-099. Supernatants were collected on different days after infection to measure HIV-1 RT activity. Values represent the mean ± SD (n = 5). The same HIV control plot is presented in A and B. For a better comparison, see the HIV control plot in Figure 5, A and B, and in Supplemental Figure 5, A, B, E, and F. The mean was compared by 2-way ANOVA, which showed a time-dependent effect (P < 0.0001). Pairwise comparisons were performed with Bonferroni’s post-hoc test for P < 0.05 compared with acontrol, bNanoATV, and cURMC-099. (C) On day 7 after infection, the cells were fixed and stained for HIV-1 p24 antigen and counterstained with hematoxylin. Multi- and mononucleated cells are marked with white and black arrowheads, respectively. Scale bar: 20 μm. On (D) day 7 and (E) day 14 after infection, the percentage of MNGCs was quantified from the total number of cells in 20 different fields. Values represent the mean ± SD. **P ≤ 0.01 and ***P ≤ 0.001, by Student’s t test. Multiple comparisons were corrected for the FDR by the Benjamini-Hochberg method, and data are representative of 3 independent experiments. dpi, days post infection.
URMC-099 and mTORC1-dependent transcription factor EB nuclear translocation.
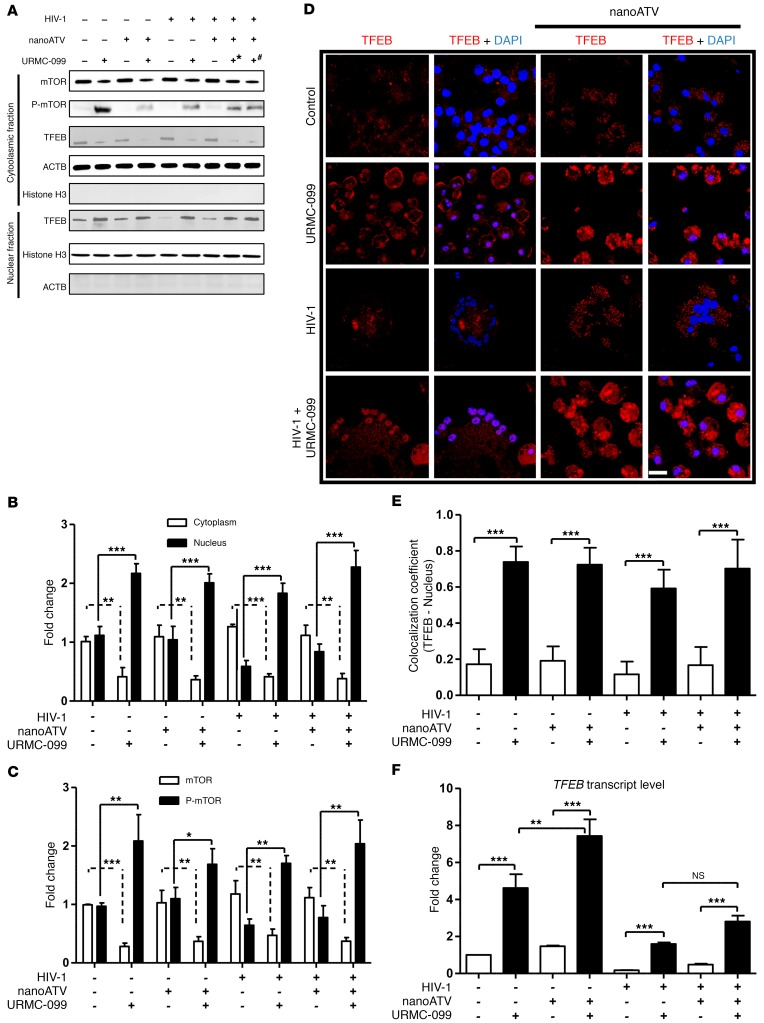
Since URMC-099 regulates JNK phosphorylation (18), we reasoned that URMC-099 would affect the mechanistic target of mTOR complex 1 (mTORC1) through JNK (21), which in turn would modulate vesicular trafficking through transcription factor EB (TFEB) (22). We found that URMC-099 could affect endosomal trafficking through any one or all of these factors (Supplemental Figure 2A). To understand the URMC-099 mechanism, we pretreated MDMs with 100 μM nanoATV, followed by HIV-1 infection and treatment with 400 ng/ml URMC-099. Here, we used the therapeutic level of 100 μM nanoATV to eliminate the virus-induced effect on signaling proteins and to differentiate the added outcome of URMC-099 from that of nanoATV alone. Moreover, a higher dose of nanoATV ensures the effect of URMC-099 on depot formation rather than on drug synergy (between ATV and URMC-099). URMC-099 was administered throughout the experimental period, while nanoATV was administered only once at the start of the experiment. After 14 days, TFEB levels were quantified in the cytoplasmic and nuclear fractions by Western blot analysis. With URMC-099 treatment, TFEB levels were increased by 2- to 3-fold in the nuclear cell fraction and reduced by 2-fold in the cytoplasmic fraction when compared with the levels detected with nanoATV treatment alone (Figure 2, A and B). Since the inhibition of JNK phosphorylation negatively regulates mTORC1, we tested whether URMC-099 could inhibit mTORC1 function. Ser2448 phosphorylation of the kinase mTORC1 regulates nuclear translocation of TFEB (23, 24). We observed that URMC-099 treatment increased Ser2448 phosphorylation by 2.2 ± 0.51-fold (Figure 2, A and C). HIV-1 sequesters TFEB in the cytoplasm through HIV-1 Nef (25), and we observed sequestration of TFEB in the cytoplasmic fraction following viral infection. This was reversed by URMC-099 within HIV-1–infected MDMs and MNGCs (Figure 2, A, B and D). URMC-099 treatment led to 80% localization of TFEB in the nuclei of MDMs compared with only 20% localization without URMC-099 treatment (Figure 2, D and E). URMC-099 also triggered increases in the transcription of TFEB mRNA by approximately 4- to 8-fold under different treatment conditions (Figure 2F). With HIV-1 infection, TFEB mRNA levels were reduced by 2- to 3-fold compared with levels in uninfected controls, and URMC-099 could recover the phenotype. Increased TFEB translocation was delayed and did not reach significant levels until day 7 after URMC-099 treatment (Supplemental Figure 2B). These data confirm that URMC-099 regulates TFEB nuclear translocation in an mTORC1-dependent manner.
Figure 2. URMC-099 regulates TFEB nuclear localization.
Human MDMs were treated for 14 days with 100 (+*) or 400 ng/ml (+#) URMC-099 in the presence or absence of 100 μM nanoATV, with or without HIV-1ADA infection. (A) MDMs were fractionated and analyzed by Western blotting. (B) TFEB and (C) mTOR quantification of Western blots. Protein bands were quantified and normalized to actin β (ACTB) or histone H3 using ImageJ2 software. (D) Human MDMs were stained for TFEB (red) and DAPI (blue) and analyzed using a confocal microscope to visualize nuclear localization of TFEB. Scale bar: 20 μm. (E) Quantification of colocalization coefficient for TFEB and DAPI. (F) Total RNA was isolated on day 14, and real-time qPCR was performed to determine TFEB expression. Values represent the mean ± SD (n = 3). *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, by Student’s t test. (B, C, E, and F) Data were corrected for multiple comparisons using the Benjamini-Hochberg method. Data are representative of 3 independent experiments. All URMC-099 treatments not specified used 400 ng/ml.
URMC-099 stimulates autophagy in human MDMs.
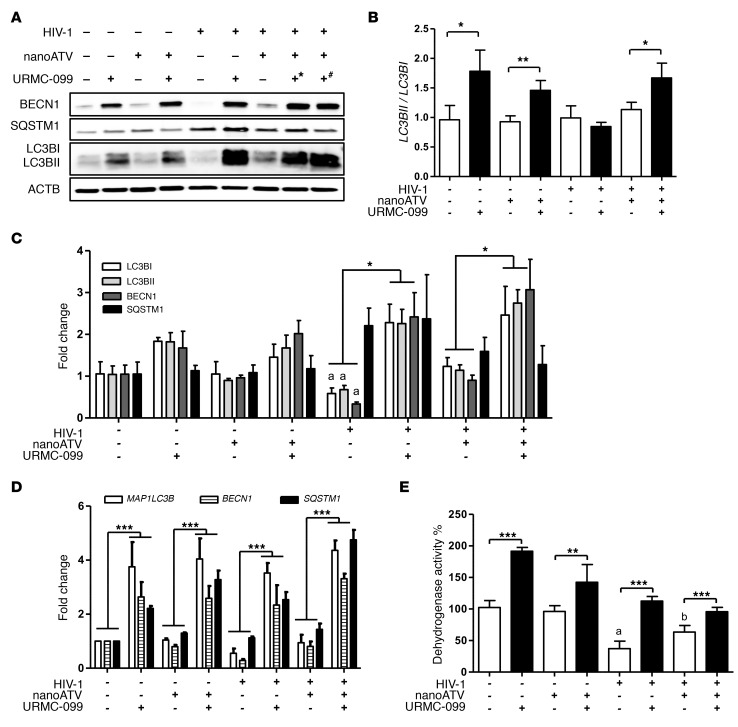
On the basis of the preceding data sets, we theorized that nuclear translocation of TFEB induces autophagy and lysosomal biogenesis. TFEB serves as the master regulator of both autophagy and lysosomal biogenesis (26), and such regulation could have a profound role in nanoART intracellular sequestration in MDMs. Thus, we examined the autophagosome markers microtubule-associated protein 1 light chain 3 β (LC3B, also known as MAP1LC3B) and beclin 1 (BECN1). Using Western blot assays, we found that each marker was significantly upregulated by URMC-099 (Figure 3, A–C and Supplemental Figure 3A). URMC-099 increased the expression of both LC3BI and LC3BII from day 7 onward (Supplemental Figure 3A). Significant increases in LC3BII/LC3BI ratios were induced by URMC-099 (Figure 3B) and support autophagosome formation. These data support the finding of TFEB nuclear localization and confirm that autophagy is induced through TFEB. Degradation of sequestosome 1 (SQSTM1), a marker of autophagosome sequestration, was subsequently investigated for its role in autophagosome-lysosome fusion, which is known to occur during the completion of autophagy (27). Interestingly, URMC-099 did not induce significant increases in SQSTM1 levels compared with those in controls (Figure 3, A and C and Supplemental Figure 3A). Furthermore, while increases in LC3B (MAP1LC3B), BECN1, and SQSTM1 gene expression were detected with URMC-099 treatment (Figure 3D and Supplemental Figure 3B), these increases affected only LC3B and BECN1, but not SQSTM1, protein expression (Figure 3C and Supplemental Figure 3A). The results imply that autophagosomes are increased as a consequence of URMC-099 treatment and fuse with lysosomes, but that a delayed response is operational (Supplemental Figure 3A). TFEB nuclear translocation leads to increases in TFEB mRNA levels and the induction of autophagy (28, 29). This likely represents the lack of any distinctive differences in autophagy-specific proteins among URMC-099–treated groups, irrespective of HIV-1 infection or nanoATV. HIV-1 differentially regulates autophagy at early and later stages of viral infection (25). Fourteen days after infection of MDMs, we found that HIV-1 inhibited autophagy, as indicated by decreased levels of LC3B and BECN1, whereas SQSTM1 levels were increased following HIV-1 infection. These findings were independent of URMC-099 treatments and are linked, in measure, to the known inhibition of autophagy by HIV-1 and transcriptional activation of SQSTM1 by URMC-099. Importantly, URMC-099 reversed HIV-1–induced downregulation of autophagy during the later stages of viral infection of MDMs (Figure 3, A, C and D).
Figure 3. URMC-099 induces autophagy in macrophages.
Human MDMs were treated for 14 days with 100 (+*) or 400 ng/ml (+#) URMC 099, in the presence or absence of 100 μM nanoATV, with or without HIV-1ADA infection. (A) Total cell lysates were analyzed for different autophagy markers by Western blotting. (B and C) Quantification of Western blots. (B) LC3BII/LC3B1 ratio and (C) quantification of protein expression of LC3BI (white bars), LC3BII (light gray bars), BECN1 (dark gray bars), and SQSTM1 (black bars). Protein bands were quantified and normalized to ACTB using ImageJ2 software (n = 3). (D) Total RNA was isolated on day 14, and real-time qPCR was performed to determine MAP1LC3B, BECN1, and SQSTM1 expression (n = 3). (E) On day 14, an MTT assay was performed to assess mitochondrial activity (n = 5). Values represent the mean ± SD. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and P ≤ 0.05, acompared with uninfected control MDMs and bcompared with HIV-1–infected MDMs; Student’s t test. (C–E) Data were corrected for multiple comparisons using the Benjamini-Hochberg method. Data are representative of 3 independent experiments. All URMC-099 treatments not specified used 400 ng/ml.
URMC-099 improves mitochondrial activity and cell vitality.
Autophagy is a known mechanism for mitochondrial turnover (30), and increased autophagy leads to increased cell viability (31). In order to better understand the relationships between autophagy and cellular recycling processes, we measured cellular metabolic activity by the MTT assay, which is an index of tetrazolium dye reduction due to NAD(P)H-dependent oxidoreductase activity and thus can be used to assess mitochondrial health. MTT activity was determined in infected and uninfected MDMs with or without URMC-099 and/or nanoATV treatment. Our findings showed significant and specific increases in MTT activity as the sole result of URMC-099 treatment (Figure 3E and Supplemental Figure 3C). With similar treatments, a trypan blue exclusion assay showed no difference in cell viability with URMC-099 alone (Supplemental Figure 3, D and E). Though there was no change in cell viability with URMC-099 treatment, we observed increased mitochondrial activity. However, HIV-1 infection led to cytotoxicity in macrophages and yielded reduced cell viability compared with what we observed in uninfected controls (Supplemental Figure 3D).
URMC-099 retains ARV nanoparticles in autophagosomes.
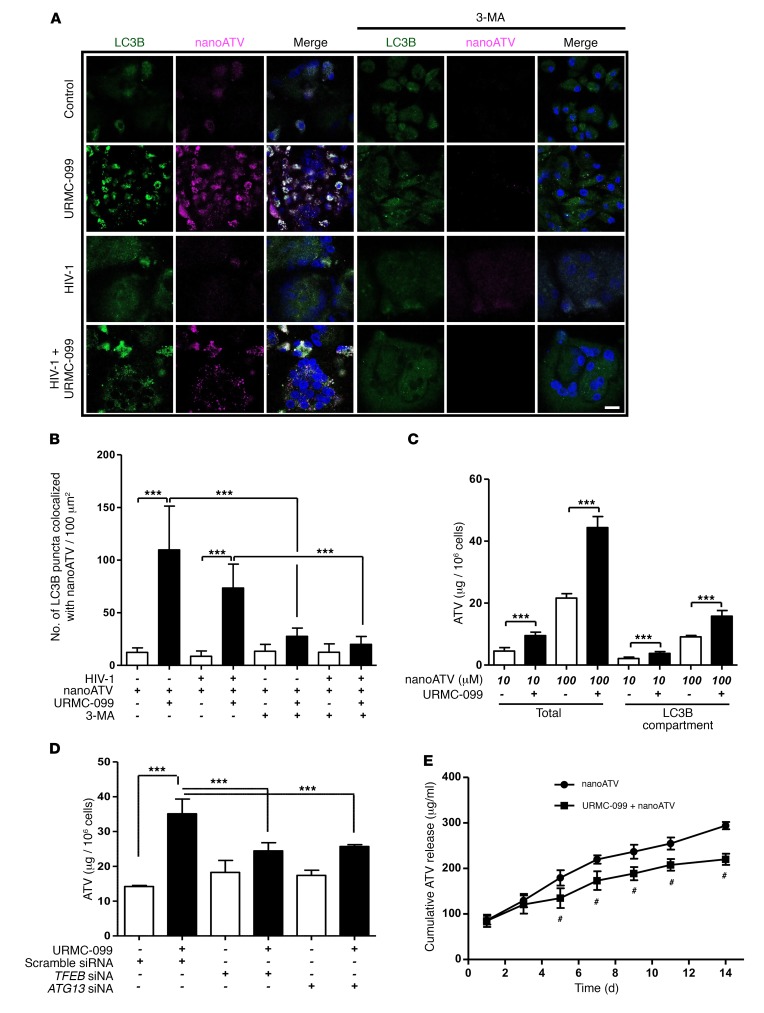
We next reasoned that autophagy facilitated by URMC-099 might affect the retention of nanoATV that we observed previously (11). This was studied by treating uninfected or HIV-1–infected MDMs with 10 μM CF633-labeled nanoATV. Cells were also cultured in the presence or absence of URMC-099 for 12 days and then transfected with a GFP-tagged LC3B construct (LC3B-GFP). We observed increased numbers of LC3B puncta and localization of nanoATV in LC3B+ autophagosomes in the presence of URMC-099 (Figure 4, A and B). In addition, we detected increased nanoATV fluorescent signals from URMC-099–treated cells compared with untreated control cells. Inhibition of autophagy by 3-methyladenine (3-MA) decreased autophagosome numbers and nanoATV fluorescent signals (Figure 4, A and B). To confirm these observations, ATV levels were quantified in total cell lysates and autophagosomes. For the latter, LC3B+ autophagosomes were isolated by magnetic bead pull-down (32), and ATV levels were measured by HPLC. Drug levels increased by 1.5- to 2.5-fold in both cells and autophagosomes as a consequence of URMC-099 treatment when compared with levels detected in the untreated cells (Figure 4C). The use of TFEB or autophagy-related 13 (ATG13) siRNA demonstrated a reversal of URMC-099–induced cellular drug retention (Figure 4D). These findings confirmed that URMC-099 induces macrophage drug depots and that these effects are mediated through TFEB and the formation of autophagosomes. The process, notably, requires ATG13 (33). Although nanoATV was localized in compartments of Rab-related proteins 7 and 11 (Rab7 and Rab11), regardless of URMC-099 treatment (11), the subcellular fate of nanoATV in Rab compartments, fused with autophagosomes, or rather in phagosomes at different stages of maturation, remained unclear. URMC-099 treatment increased the colocalization of LC3B and endosomal Rab proteins (Rab7 and Rab11) (Supplemental Figure 4, A and B), suggesting that fusion occurred between endosomes and autophagosomes (34–38). Although there were no differences in the uptake of nanoATV in URMC-099–treated MDMs (11), the increased retention of nanoATV by URMC-099 reduced the release kinetics of nanoATV, resulting in a sustained, slower release of the drug (Figure 4E). Together, these data suggest that induction of autophagy leads to increased retention of nanoATV and a sustained slow release of drug.
Figure 4. URMC-099 increases nanoATV retention in macrophage autophagosomes.
(A) Human MDMs were treated for 14 days with 400 ng/ml URMC-099 in the presence of fluorescently tagged nanoATV (10 μM) (purple), with or without HIV-1ADA infection. MDMs were transfected with an LC3B-tagged GFP construct (green), stained with DAPI (blue), and analyzed by confocal microscopy. Scale bar: 20 μm. (B) Quantification of LC3B puncta that colocalized with nanoATV. (C) Human MDMs were treated with nanoATV (10 μM or 100 μM) in the presence or absence of 400 ng/ml URMC-099. After 14 days, the ATV concentration was determined in total cells, and LC3B autophagosomal compartments were isolated using magnetic bead separation (n = 3). (D) Human MDMs were treated with 100 μM nanoART and 400 ng/ml URMC-099. Cells were transfected with siRNA for TFEB or ATG13, and on day 14, the ATV concentration was determined in total cells (n = 3). (E) URMC-099 regulated the release of nanoATV. Human MDMs were treated with 100 μM nanoATV for 16 hours, washed with PBS, and incubated with or without 400 ng/ml URMC-099. On different days, supernatant was collected, ATV concentration was quantified using HPLC, and the cumulative ATV release was plotted according to treatment duration (n = 3). Comparison of the means by 2-way factorial ANOVA showed a time-dependent treatment effect (P = 0.0002), with pairwise comparisons made using Bonferroni’s post-hoc test (#P < 0.05). Data are representative of 3 independent experiments. (B–D) Values represent the mean ± SD. *P ≤ 0.05 and ***P ≤ 0.001, by Student’s t test. Multiple comparisons were corrected for the FDR using the Benjamini-Hochberg method.
Autophagy facilitates nanoformulated ARV activity.
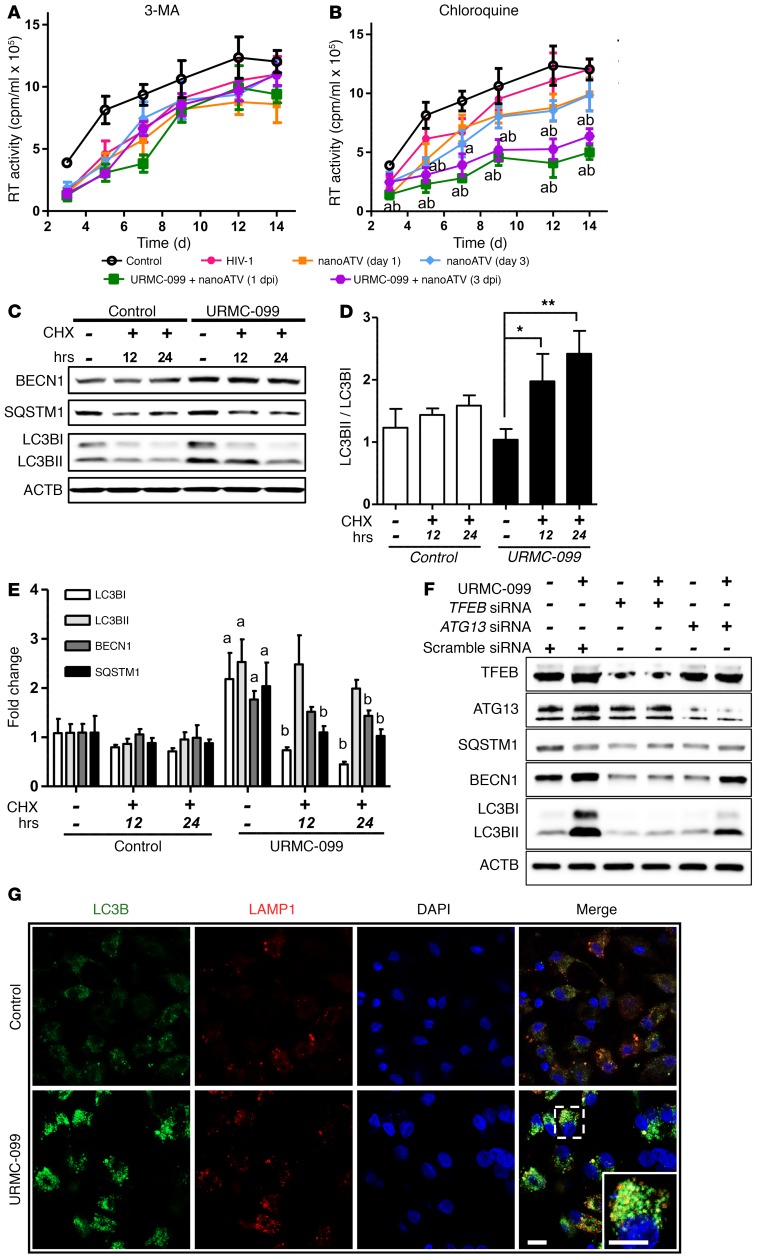
It is well accepted that autophagosomes are used for HIV-1 assembly and maturation (25, 39, 40). Thus, we investigated whether autophagy could affect HIV-1 replication in MDMs in the setting of nanoATV treatment. In these experiments, MDMs were treated with a subtherapeutic dose of 1 μM nanoATV at 1 or 3 days after HIV-1 infection, with or without URMC-099 cotreatment. When autophagy was inhibited in the initial stage of regulation with the PI3K inhibitors 3-MA or wortmannin, the viral-suppressive effect of URMC-099 shown in Figure 1, A and B, was reversed (Figure 5A and Supplemental Figure 5A). We used these PI3K inhibitors, since PI3K is upstream of mTOR signaling and is required for autophagy. JNK pathway crosstalk with PI3K-mediated mTOR signaling is not directly involved in mTOR regulation (41). Thus, inhibiting PI3K signaling affects autophagy independently of URMC-099, and these PI3K inhibitors revealed the involvement of autophagy in URMC-099–enhanced ARV activity. Inhibition with chloroquine or bafilomycin A1 in the later stages of autophagy that include autophagosome-lysosome fusion did not alter the URMC-099 effect (Figure 5B and Supplemental Figure 5B). However, the URMC-099–induced increase in MTT activity in MDMs was reduced with either 3-MA or chloroquine, independently of HIV-1 infection (Supplemental Figure 5C). These data indicate that autophagosome accumulation is required to facilitate maximal nanoATV effectiveness but that autophagosome-lysosme fusion is required for autophagy-induced mitochondrial activity. We observed a similar reduction in HIV-1 infection when rapamycin, an mTOR inhibitor and a known autophagy inducer, was used with 1 μM nanoATV (Supplemental Figure 5D). The rapamycin effect was lost with the autophagy inhibitor 3-MA, but not with chloroquine (Supplemental Figure 5, E and F). Taking into account the action of ATV as an HIV-1 protease inhibitor and the autophagosome involvement in HIV-1 biogenesis, we posit that drug and virus interactions enhanced by URMC-099 could be the reasons for further attenuation of HIV-1 infection.
Figure 5. URMC-099–induced autophagy affects HIV-1 clearance.
HIV-1ADA–infected human MDMs were treated with 1 μM nanoATV on day 1 or day 3 after infection and incubated with or without 400 ng/ml URMC-099 and in the presence of the autophagy inhibitors (A) 3-MA (100 μM) or (B) chloroquine (10 μM). Supernatants were collected on different days after infection and analyzed for HIV-1 RT activity (n = 5). The same HIV-1 infection control plot is presented in A and B. (A and B) The mean values of RT activity were assessed by 2-way factorial ANOVA, which showed a significant time-dependent treatment effect (P < 0.02). Pairwise comparisons using Bonferroni’s post-hoc test were assessed for URMC-099–treated cultures, with P < 0.05 compared with HIV-infected controls in the absence (acontrol) or presence (bHIV) of an autophagy inhibitor. (C–E) MDMs were treated in the presence or absence of 400 ng/ml URMC-099 for 14 days. Twenty-four hours or twelve hours before harvesting, cells were treated with 10 μM cycloheximide (CHX) to inhibit translation. Total cell lysates were analyzed by Western blotting. (D) Values represent the mean ± SEM of LC3BII/LC3BI ratios and were compared by Student’s t test and adjusted for multiple comparisons using the Benjamini-Hochberg method. *P ≤ 0.05 and **P ≤ 0.01 (n = 3). (E) Differences in mean fold changes were assessed by 2-way ANOVA and pairwise comparison with the respective proteins was done using Bonferroni’s post-hoc test. P ≤ 0.05 for ano URMC-099/no CHX control and bURMC-099/no CHX control (n = 3). (F) Human MDMs treated with 400 ng/ml URMC-099 were transfected with either TFEB siRNA or ATG13 siRNA on days 3 and 7. On day 14, cell lysates were analyzed for by Western blotting. (G) URMC-099–treated (400 ng/ml) and untreated (control) MDMs were transfected with LC3B-GFP on day 12, and 48 hours later were stained and imaged with a confocal microscope. Scale bars: 20 μm. Data are representative of 3 independent experiments.
To investigate autophagosome-lysosome fusion, we treated MDMs with the protein synthesis inhibitor cycloheximide after the cells were exposed to URMC-099. During this fusion, LC3BI is lapidated to LC3BII and released into the cytoplasm. While increases in both LC3BI and LC3BII were affected by URMC-099, LC3BI was decreased by cycloheximide on day 7 (Supplemental Figure 6, A– C) and day 14 (Figure 5, C–E). Since URMC-099 induces autophagy transcriptionally, new autophagosomes were synthesized continuously, hence the increase we observed in the levels of both LC3BI and LC3BII. When translation was inhibited by cycloheximide, the existing autophagosomes fused with lysosomes, resulting in a reduction of LC3BI levels (Figure 5, C and D). Induction of LC3B with URMC-099 treatment was reversed by treatment with TFEB siRNA, but not with ATG13 siRNA (Figure 5F). Confocal imaging also showed a reduction of LC3B expression with TFEB silencing (Supplemental Figure 6D). This further confirms that TFEB is required for autophagy induction and that URMC-099 acts through TFEB. Since ATG13 is involved in autophagosome formation, ATG13 did not have an impact on the effects of URMC-099 on LC3B or BECN1 expression (Figure 5F), whereas nanoATV retention in the autophagosomes was affected by ATG13 silencing (Figure 4D).
LC3B-positive autophagosomes were also colocalized with lysosomal-associated membrane protein 1 (LAMP1), confirming fusion of autophagosomes with lysosomes (Figure 5G). To further confirm autophagosome-lysosome fusion, we quantified IL-1β levels, as fusion leads to IL-1β degradation and reduced secretion (42). IL-1β levels were quantified by ELISA and a cytokine bead array under different treatment conditions. We confirmed by ELISA that URMC-099 decreased IL-1β secretion by 4.4 ± 0.9-fold (Supplemental Figure 7A) and by 4.2-fold with a cytokine bead array (Supplemental Figure 7B).
Stimulation of autophagy in animals given URMC-099.
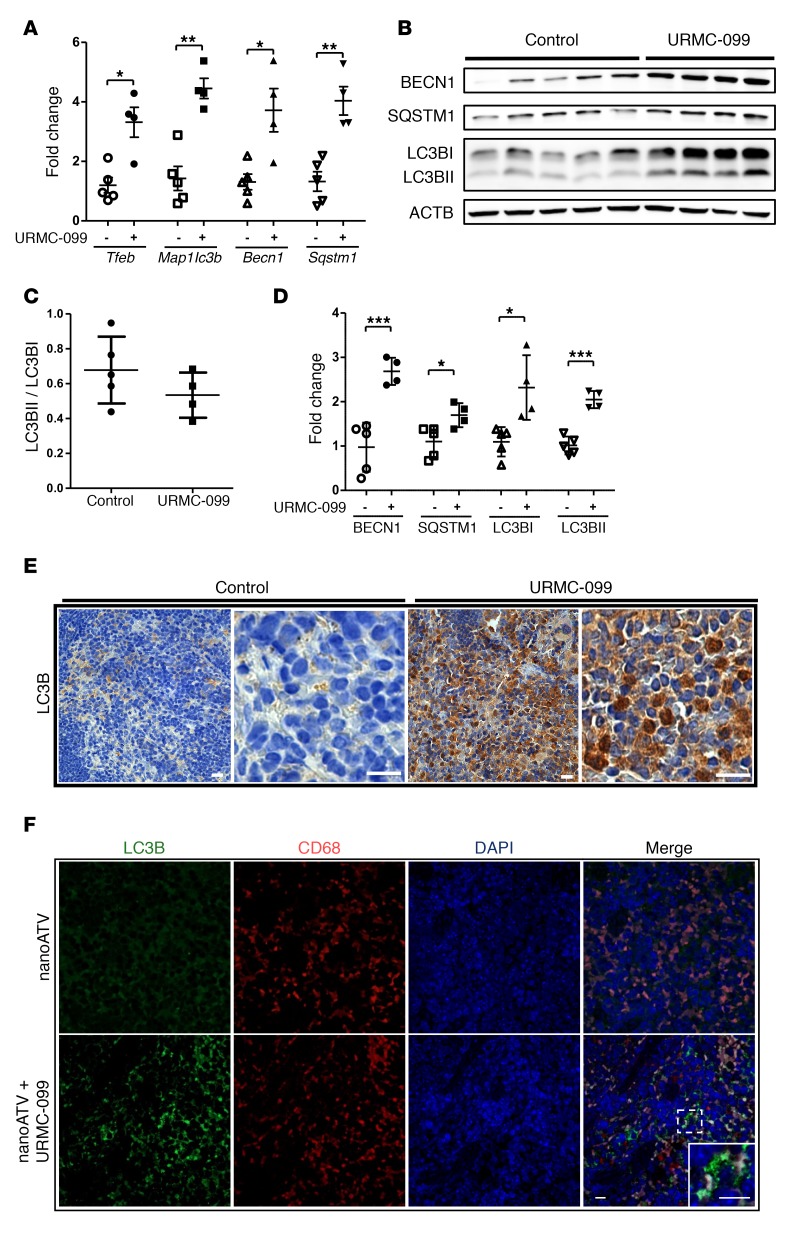
To understand the effect of URMC-099 in vivo, mice were treated with twice-daily i.p. injections of 10 mg/kg URMC-099. A plasma concentration of approximately 1,000 ng/ml (>2 μM) was achieved with this treatment (Supplemental Figure 8A). We found that URMC-099 had no adverse effect on the body weight of the mice (Supplemental Figure 8B). As TFEB induces autophagy in tissue (26, 43, 44), we tested whether URMC-099 treatment leads to activation of autophagy in spleen and liver. Real-time quantitative PCR (qPCR) showed an upregulation of Tfeb, Map1lc3b, Necn1, and Sqstm1 levels in the livers of URMC-099–treated mice (Figure 6A). Similarly, LC3B, BECN1, and SQSTM1 protein expression levels were also increased when mice were treated with URMC-099 (Figure 6, B–D). These data match those from previous studies showing that upregulation of Tfeb triggers a transcriptional activation of autophagy-linked genes (26). Additionally, immunohistological staining of splenic tissue sections showed increased LC3B expression in URMC-099–treated mice (Figure 6E), which further evidenced autophagy induction by URMC-099. In URMC-099–treated mice, we observed increased numbers of LC3B puncta in CD68+ mouse splenic macrophages (Figure 6F).
Figure 6. URMC-099 and autophagy in tissues.
Mice were given twice-daily i.p. injections of buffer (control) or URMC-099 (10 mg/kg), and after 21 days, mice were sacrificed and tissues collected. (A) Total RNA was isolated from liver, and real-time qPCR was performed. (B–D) Liver homogenate was analyzed for different autophagy markers using Western blotting (each lane is from 1 mouse) and is presented as (C) the LC3BII/LC3BI ratio and (D) the quantification normalized to ACTB using ImageJ2 software. Paraffin-embedded, 5-μm-thick splenic sections were stained for (E) LC3B, developed with DAB and counterstained with hematoxylin, and for (F) LC3B (green) and mouse CD68 in macrophages (red). DAPI was used to stain nuclei. Values represent the mean ± SEM. *P ≤ 0.05 and **P ≤ 0.01, and ***P ≤ 0.001, by Mann-Whitney U test. Multiple comparisons were assessed for the FDR using the Benjamini-Hochberg method. Five and four mice were included in the control and URMC-099 groups, respectively. Scale bars: 20 μm.
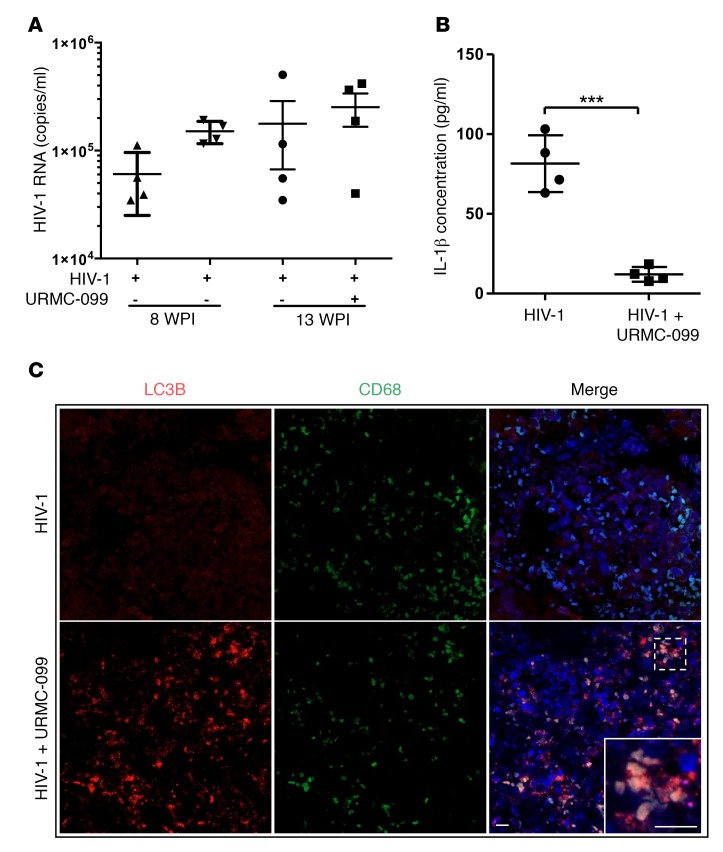
To further elucidate the effect of URMC-099 on HIV-1 infection in vivo, we examined humanized NOD/SCID IL-2Rγc–null (NSG) mice infected with HIV-1 and treated with URMC-099. We found that URMC 099 alone did not affect the plasma HIV levels (Figure 7A). It is known that acute and chronic HIV-1 infection leads to increased IL-1β secretion in vitro and in vivo (45, 46). URMC-099 treatment in our study reduced plasma IL-1β concentrations compared with the plasma concentrations in HIV-1–infected controls (Figure 7B). In URMC-099–treated humanized mice, LC3B puncta in CD68+ macrophages were also readily apparent (Figure 7C). These results suggest that URMC-099 modulates HIV-1–induced cytokine responses in vivo through autophagy (6, 47).
Figure 7. URMC-099 treatment in humanized mice.
(A and B) Humanized NSG mice were infected with HIV-1ADA and treated with URMC-099 (n = 4 per group). Ten weeks after infection, (A) plasma viral load and (B) plasma IL-1β concentration were determined (n = 4 mice per group). Values represent the mean ± SEM. ***P ≤ 0.001, by Mann–Whitney U test. (C) Paraffin-embedded spleen tissue sections from URMC-099–treated and control HIV-1–infected humanized mice were stained for LC3B (red) in autophagosomes and CD68 (green) in macrophages. DAPI was used to stain nuclei. The merged panel shows colocalization of LC3B and CD68 in spleens from URMC-099–treated mice. Scale bars: 20 μm.
Improved pharmacokinetic profiles of nanoformulated ARVs with URMC-099 treatment.
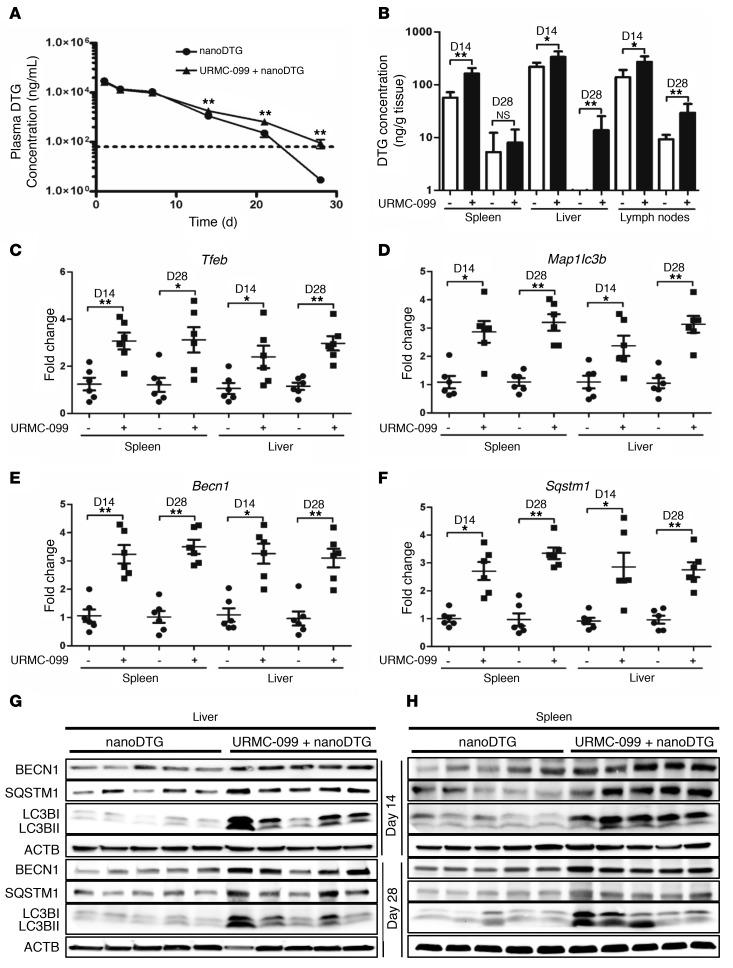
We used the HIV-1 integrase inhibitor DTG to understand the role of autophagy and nanoART in vivo. Mice injected with a single i.m. dose (45 mg/kg) of nanoformulated DTG (nanoDTG), with or without daily i.p. injections of 10 mg/kg URMC-099, showed no adverse effects on their body weight or health (Supplemental Figure 9A). We observed increased plasma DTG levels in URMC-099–treated mice beginning on day 14, and by day 28, there was a 52.3 ± 5.7-fold increase in the plasma drug level compared with levels detected in animals treated with nanoDTG alone (Figure 8A). Similar increases in DTG levels in spleen, liver, and lymph nodes were achieved with URMC-099 treatment (Figure 8B). At early time points, i.e., on days 1 and 3 after nanoDTG administration, we found that the drug levels in both plasma and tissues were not significantly increased with URMC-099, and the autophagy proteins were also not affected with URMC-099 treatment at these time points, suggesting a delayed response (Supplemental Figure 9, B and C).
Figure 8. URMC-099 and nanoART parenteral coadministration sustains plasma DTG levels.
Mice were injected i.m. with a single dose (45 mg/kg) of nanoDTG and treated with or without daily i.p. injections of URMC-099 (10 mg/kg). (A) On different days, blood was collected, and the plasma DTG concentration was determined by UPLC-MS/MS. (B) On day 14 (D14) and day 28 (D28), mice were sacrificed and DTG levels quantified in different tissues by UPLC-MS/MS. (C–F) On days 14 and 28, total RNA was isolated from splenic and liver tissues, and real-time qPCR was performed for different genes. Values represent the mean ± SEM. *P ≤ 0.05 and **P ≤ 0.01, by Mann-Whitney U test (n = 6 mice per group). (G and H) On days 14 and 28, splenic and liver tissues were collected, and total tissue lysate was analyzed by Western blotting for different autophagy markers. Each lane is representative of 5 animals from each of the groups. Multiple comparisons were corrected for the FDR using the Benjamini-Hochberg method. Six mice were assessed per group.
Real-time qPCR of spleen and liver tissues showed a significant increase in Tfeb levels in URMC-099–treated mice, which in turn promoted higher expression of the autophagy genes Map1lc3b, Becn1, and Sqstm1 (Figure 8, C–F). Western blot analyses confirmed a parallel induction of autophagy proteins in spleen and liver by days 14 and 28 of URMC-099 treatment (Figure 8, G and H, and Supplemental Figure 10, A–E). Together, these results show that autophagy induction by URMC-099 enhances nanoARV half-life in vivo.
Discussion
There is an immediate need for long-acting ART in order to meet the challenges of HIV/AIDS regimen adherence. Notably, the secondary gains we achieved in the present study through reductions in drug dosages and improved viral reservoir penetration may prove to be even more significant in the era of viral eradication. The ability to facilitate drug depots and enhance vesicular trafficking of ARV nanoparticles and their sequestration in endosomal compartments is of significance for achieving long-acting ART. We believe that the development of adjunctive drugs such as URMC-099, designed to extend ART action by harnessing a natural cellular process such as autophagy and thereby facilitate drug depots, represents a novel approach to this challenge. The links between TFEB, a master regulator of autophagy, and ARV nanoparticle cellular trafficking by URMC-099 provide a critical new pathway for sustaining antiretroviral activity.
This study is based on our earlier work showing that reductions in viral replication and cytopathicity are due to retention of nanoARVs in Rab compartments by URMC-099 (11). On the basis of the presumptive interactions between endosomal trafficking, cytoskeletal rearrangement, and Rab proteins, we began to explore known pathways that affect cellular trafficking to elucidate the underlying mechanism. Prior associations of URMC-099 with the phosphorylation of p54 and p46 JNK isoforms in HIV-Tat–treated BV-2 microglial cells provided mechanistic clues (48). However, while autophagy is a well-accepted cellular process involved in the recycling of cytoplasmic organelles, macromolecules, and other defective proteins (27), its role in establishing the types of drug depots and sequestration of drug-laden nanoparticles is far from proven. During steady-state conditions, autophagy is maintained at low levels but can be affected by stress, organelle damage, or microbial infections (49, 50). It is also well known that chronic HIV-1 infection inhibits autophagy (51), while its induction inhibits infection (25, 40, 52). Thus, linkages between viral infection, autophagy, and nanoparticle sequestration could be reasoned and were also supported by previous reports involving a range of experimental systems (53, 54).
URMC-099 treatment was shown here, for the first time to our knowledge, to induce autophagy and enhance anti-HIV activities by sustaining depots of nanoformulated ARVs. The puzzle pieces came together, buoyed by a number of observations. First, URMC-099 activated autophagy through nuclear translocation of TFEB. Second, increased autophagy resulted in an increase in both the number of ARV nanoparticles in autophagosomes and ART half-life. Third, by altering HIV-1–regulated autophagy, we observed a suppression of viral replication.
The relationships that we observed in this study among HIV-1, macrophages, URMC-099, TFEB, lysosomal biogenesis, and autophagy are noteworthy. The importance of monocyte-macrophages as viral reservoirs, vehicles for microbial dissemination, secretors of bioactive factors, and clearance vehicles for tissue metabolism and infectious materials cannot be overstated (28, 55). Such seemingly complicated intracellular events can, paradoxically, sustain HIV-1 growth by sequestering cytoplasmic TFEB through HIV-1 Nef and inhibiting autophagy (25). Nevertheless, URMC-099 overcame these HIV-1 effects by transcriptional control of TFEB and, as such, limited the effects of HIV-1 Nef on TFEB control.
We now posit that TFEB-dependent autophagy leads to retained nanoformulated ARVs in autophagosomes. In previous studies, we showed that injection of nanoformulated ARVs leads to macrophage recruitment and particle endocytosis. Storage then ensues in a spectrum of endosomal compartments (56). The particle-laden phagophores mature into autophagosomes and fuse with late endosomes and lysosomes. All of these organelles show the capacity to store ARV nanoparticles (26, 55). Here, we found that the number of autophagosomes was increased by URMC-099, and fusion of endosomes with autophagosomes led to the preferential localization of nanoformulated ARV particles in autophagosomes.
Autophagosomes play a vital role in HIV-1 biogenesis (51, 52). In macrophages, autophagy is stimulated by HIV-1 through the TLR8 signaling that occurs at early stages of infection, while during chronic infection, autophagy is inhibited. The process is regulated in part by the HIV-1 Nef protein (25), whereby HIV-1 inhibits autophagosome maturation to evade its own destruction by lysosomal degradation (52). URMC-099 activation of autophagy can overcome HIV-1–mediated inhibition of autophagy. Thus, a dual antiretroviral response is seen: one through intracellular degradation of virus and the other through sustainment of ARV particles that preclude new infections.
To elucidate whether autophagy could affect nanoART-mediated restriction of viral replication, we examined autophagosome-lysosome fusion events affected by URMC-099. Reduced and increased levels of LC3BI and LC3BII, respectively, confirmed that fusion of autophagosomes with lysosomes occurred when protein synthesis was inhibited. In addition, HIV-1 induces IL-1β macrophage secretion through TLR8 (57), and autophagy controls IL-1β cytokine levels. This occurs by sequestering the cytokine in autophagosomes and degrading it via lysosome fusion through autophagy-related 16-like 1 (ATG16L1) (42, 58). This process is likely important, as degradation of HIV-1–induced IL-1β is closely linked to autophagosome-lysosome fusion and HIV-1 clearance (42). It is certainly possible that autophagosome-lysosome fusion events recycle as a consequence of URMC-099 treatment and lead to further HIV-1 restriction.
URMC-099 affects cytoskeletal rearrangement and improves macrophage vitality by increasing mitochondrial activity. Indeed, combinations of URMC-099 and ATV nanoparticles resulted in reduced formation of MNGCs. This was readily apparent by days 7 and 14 after viral infection. Indeed, HIV-1–associated MNGCs alter the cellular cytoskeleton (59), and such protein rearrangements are autophagy regulated (14, 60). Thus, both processes are affected by nanoART and URMC-099. Indeed, the combination of nanoATV and URMC-099 attenuates giant cell formation, most likely by affecting endosomal and vesicular trafficking and reducing cytoskeletal rearrangements (61). These data support the notion that URMC-099 induces the changes in endosomal trafficking that affect known intracellular ARV particle accumulation (11). Macrophages play a key role in HIV-1 pathogenesis by forming viral reservoirs and selectively clearing HIV-1–infected CD4+ T lymphocytes (62, 63). As nanoART is also taken up by macrophages, increased retention, sustained release, and improved cellular viability by activation of autophagy can facilitate anti-HIV treatment outcomes. Given that our animal studies demonstrate that autophagy is induced in tissue macrophages and that combinations of nanoDTG and URMC-099 improve pharmacokinetic (PK) profiles, such treatment combinations could serve to further reduce HIV disease morbidities. In summary, we have shown that induction of autophagy leads to the retention and sustained release of nanoformulated ARVs. Our results, for the first time to our knowledge, lend credence to the idea that induction of autophagy by URMC-099 treatment activates a unique pathway that facilitates the biodistribution and half-life of long-acting nanoART. Possible mechanisms leading to a prolonged half-life of nanoART by URMC-099 are illustrated in Figure 9. As autophagosomes intersect with the HIV-1 life cycle, storage of the nanoformulated ARVs in intracellular compartments may further facilitate HIV-1 disposal by harnessing intracellular events that contain viral growth. Moreover, the effect of URMC-099 on TFEB nuclear localization and its outcome on autophagy could have other beneficial effects in combating a range of degenerative disorders that include, but are not limited to, Batten disease, Pompe disease, Gaucher disease, cystinosis, Parkinson’s disease, Huntington’s disease, and Alzheimer’s disease. TFEB is used as a therapeutic target in the treatment of these diseases (64).
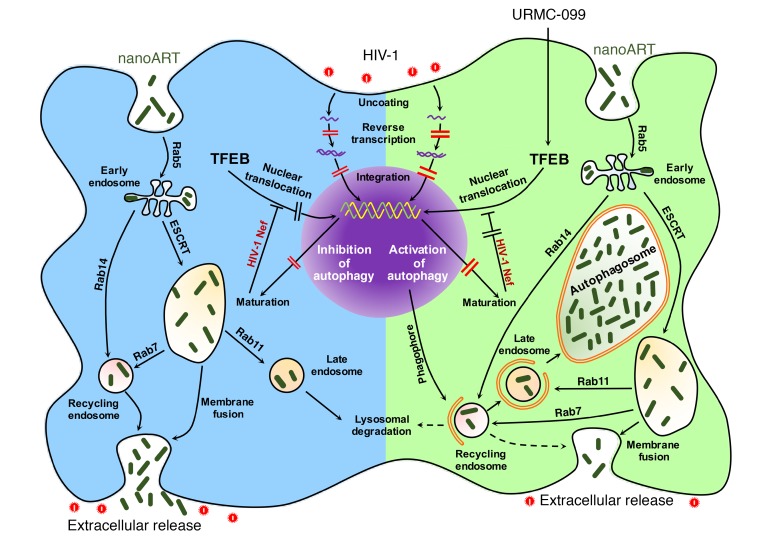
Figure 9. URMC-099 facilitates depots of nanoART in macrophage autophagosomes.
Nanoformulated ARVs enter the MDM via clathrin-coated pits and are transported to the early endosomes. Parts of the nanoARV are recycled by getting into recycling endosomes by Rab14-mediated fast recycling or trafficking regulated by endosomal sorting complexes required for transport machinery (ESCRT) and Rab7. The particles reach late endosomes via ESCRT and Rab11 and eventually fuse with lysosomes. HIV-1 fuses with the cell and releases its contents to the cytoplasm. Partial core shell uncoating and reverse transcription lead to the formation of a preintegration complex that enters the nucleus, and provirus is formed following integration. After proviral transcription and translation, viral assembly and maturation take place. HIV-1 Nef inhibits autophagy by sequestering TFEB in the cytoplasm. A subtherapeutic dose of nanoART limits antiretroviral responses. URMC-099–assisted nuclear translocation of TFEB overcomes HIV-1 Nef–mediated inhibition of autophagy. This affects autophagosome formation and retention of nanoformulated drugs specifically in the autophagosomes. In the presence of URMC-099, nanoformulated ARV retention is increased, and release is decreased even with a subtherapeutic drug dose, ultimately attenuating HIV-1 infection.
In conclusion, we demonstrate in the present study that URMC-099 has the potential to reduce the dosage and frequency of ARV administration and enhance antiretroviral activity and believe that the development of this and similar sustained-release agents would be effective additions to the existing arsenal of anti-HIV drugs.
Methods
Preparation of nanoformulated antiretroviral drugs.
Nanoformulations of ATV and DTG were prepared by high-pressure homogenization (Avestin EmulsiFlex-C3; Avestin Inc.) (10, 65) (See the Supplemental Experimental Procedures for further details). NanoATV loading was determined by HPLC with UV/Vis detection as previously described (10). NanoDTG loading was determined by HPLC using a Phenomenex Kinetix C18 Column (5 μm; 150 × 4.6 mm) with a mobile phase of 65% 50 mM KH2PO4, pH 3.2/35% acetonitrile at a flow rate of 1.0 ml/minute, and 254-nm detection. ATV and DTG were quantitated by comparison of peak areas with those of known standards (0.05–50 μg/ml in methanol).
Human monocyte–derived macrophages.
Human monocytes were isolated by leukapheresis from HIV-1/2 and hepatitis seronegative donors. Cells were cultured in DMEM (Invitrogen, Thermo Fisher Scientific) enriched with 10% heat-inactivated, pooled human serum, 2 mM L-glutamine, 50 μg/ml gentamicin, 10 μg/ml ciprofloxacin, and macrophage colony–stimulating factor (MCSF, collected as supernatants from ATCC 10154) conditioned medium. Cells were maintained at 37°C in a 5% CO2 incubator (66).
HIV-1 infection and drug treatments.
MDMs were infected with the HIV-1 ADA strain at an MOI of 0.01. URMC-099 dissolved in DMSO was used for all in vitro experiments. For pretreatments, MDMs were treated with 1, 10, or 100 μM nanoATV for 16 hours, then washed with PBS and infected with HIV-1ADA for 4 hours. After infection, cells were incubated with or without 100 or 400 ng/ml (237 or 950 nM) URMC-099 for an additional 14 days. For the post-treatment procedure, MDMs were infected and then treated with nanoATV on days 1, 3, and 5 after HIV-1 infection. MDMs were incubated with 400 ng/ml URMC-099 throughout the experimental time period. MDMs were treated with the autophagy inhibitors 3-MA (100 μM), wortmannin (50 nM), chloroquine (10 μM), or bafilomycin A1 (200 nM), or the known autophagy inducer rapamycin (20 nM). Cycloheximide (10 μM) was added to the cells 12 hours and 24 hours before terminating the experiments on day 7 or 14 to inhibit new protein synthesis.
HIV-1p24 antigen staining and RT activity assay.
For HIV-1p24 staining, MDMs were fixed with 4% ice-cold paraformaldehyde (PFA) in PBS for 20 minutes. Fixed cells were washed with PBS and permeabilized with 0.5% Triton X-100 in PBS for 15 minutes at room temperature. Cells were blocked using 10% goat serum in PBS for 30 minutes and incubated with mouse mAb against HIV-1p24. The polymer-based HRP-conjugated anti-mouse Dako EnVision system was used as a secondary detection reagent, and DAB (Dako) was used as chromogen. Images were captured with ×63 and ×20 objectives using a Nuance EX multispectral imaging system fixed to a Nikon Eclipse E800 microscope (Nikon Instruments). HIV-1 RT activity was measured as described previously (67). See the Supplemental Experimental Procedures for further details.
Cytokine and autophagy pathway measurements.
IL-1β from MDM culture supernatants or plasma was quantified using an IL-1β ELISA Kit (eBioscience, Affymetrix). Briefly, samples were incubated on a capture antibody–coated 96-well plate overnight at 4˚C after blocking with an ELISPOT diluent. Detection antibody and avidin-HRP were added sequentially, followed by 5 washes. Next 3,3′,5,5′-tetramethylbenzidine (TMB) solution and stop solution were added to stop the reaction. IL-1β levels were quantified spectrophotometrically by measuring absorbance at 450 nm. A bead-based cytokine array assay (BD Biosciences) was performed on MDM culture supernatants according to the manufacturer’s instructions. Autophagy gene expression was analyzed using real-time qPCR. RNA was isolated from cells or tissues using TRIzol (Applied Biosystems, Thermo Fisher Scientific) as per the manufacturer’s protocol. Residual DNA was removed using the TURBO DNA-Free Kit (Thermo Fisher Scientific). The isolated RNA (2 μg) was reverse transcribed to cDNA using a Verso Reverse Transcription cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. TaqMan probes (Thermo Fisher Scientific) (Supplemental Table 1) and Real-time PCR Master Mix (Applied Biosystems) were used for real-time qPCR (StepOne Plus; Applied Biosystems) according to the manufacturer’s instructions. All samples were amplified in triplicate, and data were normalized to GAPDH or actin cDNAs.
Silencing TFEB and ATG13 expression.
Human MDMs were incubated with or without 400 ng/ml URMC-099. On day 3 after URMC-099 treatment, MDMs were treated with either 10 μM TFEB or ATG13 siRNA (Ambion, Thermo Fisher Scientific) lipid complexes. siRNA was diluted in Opti-MEM medium (Life Technologies, Thermo Fisher Scientific), and lipid complexes were formed by incubating diluted siRNA in Lipofectamine RNAiMAX Reagent (Invitrogen, Thermo Fisher Scientific). Cells were treated with siRNA for 24 hours. A similar siRNA treatment procedure was followed on day 7. On day 12, MDMs were transfected with LC3B-GFP for confocal microscopy. Another set of MDMs treated similarly with siRNAs were lyzed for Western blot analyses to detect LC3B and for drug measurements.
Confocal microscopy.
Human MDMs after treatment and infection were fixed in 4% PFA for 15 minutes, permeabilized in 0.5% Triton X-100 for 5 minutes, and blocked for 1 hour in 5% BSA solution. MDMs were incubated with anti-TFEB antibody overnight at 4°C, followed by incubation with Alexa Fluor 594–conjugated secondary antibody (Thermo Fisher Scientific). ProLong Gold anti-fade reagent with 2-(4-amidinophenyl)-1H -indole-6-carboxamidine (Thermo Fisher Scientific) (27) was used to mount the coverslip. Human MDMs were pretreated with 10 μM CF633-labeled nanoATV for 16 hours, infected with HIV-1ADA, washed 3 times with PBS, and incubated with or without 400 ng/ml URMC-099 for 14 days in the presence or absence of the autophagy inhibitor 3-MA. On day 12, MDMs were transfected with a Premo Autophagy Sensor LC3B-GFP (Thermo Fisher Scientific) according to the manufacturer’s instructions and further incubated with URMC-099 and 3-MA for 48 hours (out to day 14). Cells were fixed with 4% PFA on day 14 and mounted onto a coverslip using ProLong Gold Anti-fade Reagent with DAPI. Images were captured using a Zeiss LSM810 confocal microscope and analyzed with Zen 2011 software (Carl Zeiss Microimaging).
Autophagosome isolation and drug quantification.
MDMs were treated with 10 or 100 μM nanoATV for 16 hours, washed thrice with PBS, and then treated with or without 400 ng/ml URMC-099 for 14 days. Subsequently, ATV concentrations in total cells or LC3B-positive autophagosomal compartments were determined. LC3B-positive compartments were immunoisolated using LC3B antibody as described elsewhere (6, 56). See the Supplemental Experimental Procedures for additional details. ATV concentration in total cells and in the isolated compartments was quantitated by HPLC as previously described (6, 56).
URMC-099 treatment in mice.
In parallel experiments designed to assess URMC-099–induced autophagy in vivo, male BALB/cJ mice (The Jackson Laboratory) were maintained on a normal diet with free access to food and water throughout the study. Mice were treated with i.p. injections of 10 mg/ml URMC-099 dissolved in poly(ethylene glycol)400/DMSO (8:1) mixture (vehicle) twice daily for 21 days. Subsequent steps were as described above. Tissues were collected, and total RNA was isolated. Real-time qPCR was performed to determine autophagy marker levels. Liver homogenate was analyzed for autophagy markers by Western blot assays. Tissues were also fixed in 4% PFA for immunohistology staining. Blood was collected 5 hours after URMC-099 injection on days 7 and 14 after initiation of treatment and 16 hours after URMC-099 injection on day 21 to measure plasma URMC-099 concentration. See the Supplemental Experimental Procedures for further details.
HIV-1–infected humanized mice.
Humanized NOD/SCID–IL-2Rγc–null (NSG) mice were used for HIV-1 infection (5, 68). NSG mice were obtained from The Jackson Laboratory, and a breeding colony was maintained at the University of Nebraska Medical Center (UNMC). On the day of birth, mice were irradiated at 1 Gy using a C9 Cobalt-60 Source (Picker Corp.), and then CD34+ cells (105 cells/mouse) were transplanted into newborn mice intrahepatically. Starting from 22 weeks after reconstitution, HIV-1ADA was injected i.p. at 104 50% tissue culture infectious dose (TCID50) per mouse. Mice were treated with URMC-099 from 6 weeks after infection until the endpoint. Plasma was collected 10 weeks after infection, and IL-1β levels were quantified. Viral load in the blood was measured using an automated COBAS Amplicor System V1.5 (Roche Molecular Diagnostics). Mouse plasma (20 μl) was diluted with 480 μl of sterile normal human plasma for the assay. The baseline detection of the assay after dilution was 1,250 viral RNA copies per milliliter.
PK evaluations.
Male BALB/cJ mice were injected i.m. with 45 mg/kg nanoDTG, with or without daily i.p injections of 10 mg/kg URMC-099 or equivalent vehicle dosage [poly(ethylene glycol)400/DMSO (8:1) mixture]. Plasma was collected 1, 3, 7, 14, 21, and 28 days after nanoDTG treatment. Animals were sacrificed, and tissues (liver, spleen, and lymph nodes) were collected on days 14 and 28. DTG levels in plasma and tissues were determined by ultra-performance liquid chromatography–tandem mass spectroscopy (UPLC-MS/MS) (see the Supplemental Experimental Procedures for additional details).
Immunohistology.
Spleens and livers were fixed in 4% PFA overnight. Paraffin-embedded, 5-μm-thick sections were stained with LC3B antibodies. An HRP-conjugated secondary antibody against rabbit IgG was added and developed with 3,3′-diaminobenzidine. Images were captured using ×20 and ×40 objectives. For immunofluorescence staining, sections were stained using antibodies against mouse CD68 and LC3B, and the secondary antibodies were conjugated with Alexa Fluor 488 or 594. Tissues were mounted using ProLong Gold with DAPI (Thermo Fisher Scientific) and imaged using a confocal microscope.
Statistics.
Data were analyzed using GraphPad Prism 6.0 software (GraphPad Software) and Microsoft Excel. Two-way factorial ANOVA and multiple comparisons using Bonferroni’s post-hoc tests were performed for the studies on HIV infection over time in MDM cultures, and the results were also assessed by linear regression. For comparisons of 2 groups, a Mann-Whitney U test or a 1-tailed Student’s t test was used. Multiple pairwise comparisons were corrected for the false discovery rate (FDR) using the Benjamini-Hochberg procedure. Significant differences were determined at a P value of less than 0.05.
Study approval.
All animal studies were performed in compliance with UNMC institutional policies and NIH guidelines for laboratory animal housing and care and were approved by the IACUC of UNMC. Human monocytes were isolated by leukapheresis from HIV-1/2 and hepatitis seronegative donors and were deemed exempt from approval by the IRB of UNMC. Human CD34+ hematopoietic stem cells were isolated from umbilical cord blood, which is also exempt from UNMC IRB approval.
Author contributions
Conceptualization: DPG, HEG, and SG; methodology: DPG, PKD, BJS, ZL, ANB, DLP, NG, YA, JM, RLM, and BE; investigation: DPG, PKD, BJS, ZL, and ANB; writing of the original draft: DPG and HEG; review and editing of the manuscript: DPG, SG, HEG, JM, RLM, and HAG; funding acquisition: HEG, SG, and HAG; and supervision: PKD, HEG, and SG.
Supplementary Material
Acknowledgments
We thank Edward Makarov, Li Wu, Hang Su, Pavan Puligujja, James Hilaire, Amy Conaway, and Jenna M. Puccini for their technical assistance and Larisa Poluektova for lively discussions. We thank Myron Toews for critical comments on the manuscript and Jane Meza for statistical analysis. (All of the above-named individuals are from the University of Nebraska Medical Center.) We also thank the members of the UNMC confocal, elutriation, and animal facilities for their experimental support. This work was supported in part by the University of Nebraska Foundation, which includes donations from the Carol Swarts, MD, Emerging Neuroscience Research Laboratory; the Margaret R. Larson Professorship; the Frances and Louie Blumkin Foundation; the Harriet Singer Endowment and the Vice Chancellor’s Office of UNMC for Core Facility Development. We acknowledge support from NIH grants RO1 MH104147, P01 DA028555, R01 NS36126, P01 NS31492, 2R01 NS034239, P01 MH64570, P01 NS43985, P30 MH062261, P30 AI078498, and R01 AG043540. ViiV Healthcare provided grant support for the development of long-acting antiretroviral drugs. BE is a ViiV Healthcare Scholar.
Footnotes
Conflict of interest: H.A. Gelbard and H.E. Gendelman are members of the scientific advisory board for WavoDyne Therapeutics Inc., which is developing URMC-099 as a first-in-class MLK-3 inhibitor for clinical trials. URMC-099 is owned by URMC (patent nos. US 8,846,909 B2; 8,877,772; and 9,181,247 and associated international patents).
Reference information:J Clin Invest. 2017;127(3):857–873.https://doi.org/10.1172/JCI90025.
Contributor Information
Divya Prakash Gnanadhas, Email: divya.prakash@unmc.edu.
Brady Sillman, Email: brady.sillman@unmc.edu.
Zhiyi Lin, Email: Zhiyi.lin@unmc.edu.
Nagsen Gautam, Email: nagsen.gautam@unmc.edu.
Yazen Alnouti, Email: yalnouti@unmc.edu.
Harris A. Gelbard, Email: harris_gelbard@urmc.rochester.edu.
JoEllyn McMillan, Email: jmmcmillan@unmc.edu.
Benson Edagwa, Email: benson.edagwa@unmc.edu.
Howard E. Gendelman, Email: hegendel@unmc.edu.
Santhi Gorantla, Email: sgorantla@unmc.edu.
References
- 1.Das MK, Sarma A, Chakraborty T. Nano-ART and NeuroAIDS. Drug Deliv Transl Res. 2016;6(5):452–472. doi: 10.1007/s13346-016-0293-z. [DOI] [PubMed] [Google Scholar]
- 2.Kimata JT, Rice AP, Wang J. Challenges and strategies for the eradication of the HIV reservoir. Curr Opin Immunol. 2016;42:65–70. doi: 10.1016/j.coi.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nowacek A, Gendelman HE. NanoART, neuroAIDS and CNS drug delivery. Nanomedicine (Lond) 2009;4(5):557–574. doi: 10.2217/nnm.09.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araínga M, Guo D, Wiederin J, Ciborowski P, McMillan J, Gendelman HE. Opposing regulation of endolysosomal pathways by long-acting nanoformulated antiretroviral therapy and HIV-1 in human macrophages. Retrovirology. 2015;12:5. doi: 10.1186/s12977-014-0133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dash PK, et al. Long-acting nanoformulated antiretroviral therapy elicits potent antiretroviral and neuroprotective responses in HIV-1-infected humanized mice. AIDS. 2012;26(17):2135–2144. doi: 10.1097/QAD.0b013e328357f5ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo D, et al. Endosomal trafficking of nanoformulated antiretroviral therapy facilitates drug particle carriage and HIV clearance. J Virol. 2014;88(17):9504–9513. doi: 10.1128/JVI.01557-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li T, et al. Magnetic resonance imaging of folic acid-coated magnetite nanoparticles reflects tissue biodistribution of long-acting antiretroviral therapy. Int J Nanomedicine. 2015;10:3779–3790. doi: 10.2147/IJN.S83279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Skinner AL, et al. Cellular Responses and Tissue Depots for Nanoformulated Antiretroviral Therapy. PLoS ONE. 2015;10(12):e0145966. doi: 10.1371/journal.pone.0145966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puligujja P, et al. Pharmacodynamics of folic acid receptor targeted antiretroviral nanotherapy in HIV-1-infected humanized mice. Antiviral Res. 2015;120:85–88. doi: 10.1016/j.antiviral.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puligujja P, et al. Macrophage folate receptor-targeted antiretroviral therapy facilitates drug entry, retention, antiretroviral activities and biodistribution for reduction of human immunodeficiency virus infections. Nanomedicine. 2013;9(8):1263–1273. doi: 10.1016/j.nano.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang G, et al. The mixed lineage kinase-3 inhibitor URMC-099 improves therapeutic outcomes for long-acting antiretroviral therapy. Nanomedicine. 2016;12(1):109–122. doi: 10.1016/j.nano.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautam N, et al. Preclinical pharmacokinetics and tissue distribution of long-acting nanoformulated antiretroviral therapy. Antimicrob Agents Chemother. 2013;57(7):3110–3120. doi: 10.1128/AAC.00267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh D, et al. Development and characterization of a long-acting nanoformulated abacavir prodrug. Nanomedicine (Lond) 2016;11(15):1913–1927. doi: 10.2217/nnm-2016-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amaya C, Fader CM, Colombo MI. Autophagy and proteins involved in vesicular trafficking. FEBS Lett. 2015;589(22):3343–3353. doi: 10.1016/j.febslet.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 15.Edagwa BJ, Zhou T, McMillan JM, Liu XM, Gendelman HE. Development of HIV reservoir targeted long acting nanoformulated antiretroviral therapies. Curr Med Chem. 2014;21(36):4186–4198. doi: 10.2174/0929867321666140826114135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong W, et al. The mixed-lineage kinase 3 inhibitor URMC-099 facilitates microglial amyloid-β degradation. J Neuroinflammation. 2016;13(1):184. doi: 10.1186/s12974-016-0646-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodfellow VS, et al. Discovery, synthesis, and characterization of an orally bioavailable, brain penetrant inhibitor of mixed lineage kinase 3. J Med Chem. 2013;56(20):8032–8048. doi: 10.1021/jm401094t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marker DF, et al. The new small-molecule mixed-lineage kinase 3 inhibitor URMC-099 is neuroprotective and anti-inflammatory in models of human immunodeficiency virus-associated neurocognitive disorders. J Neurosci. 2013;33(24):9998–10010. doi: 10.1523/JNEUROSCI.0598-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polesskaya O, et al. MLK3 regulates fMLP-stimulated neutrophil motility. Mol Immunol. 2014;58(2):214–222. doi: 10.1016/j.molimm.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhoo KH, et al. Pharmacologic inhibition of MLK3 kinase activity blocks the in vitro migratory capacity of breast cancer cells but has no effect on breast cancer brain metastasis in a mouse xenograft model. PLoS One. 2014;9(9):e108487. doi: 10.1371/journal.pone.0108487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su KH, et al. HSF1 critically attunes proteotoxic stress sensing by mTORC1 to combat stress and promote growth. Nat Cell Biol. 2016;18(5):527–539. doi: 10.1038/ncb3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roczniak-Ferguson A, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5(228):ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126(Pt 8):1713–1719. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8(6):903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell GR, Rawat P, Bruckman RS, Spector SA. Human immunodeficiency virus type 1 Nef inhibits autophagy through transcription factor EB sequestration. PLoS Pathog. 2015;11(6):e1005018. doi: 10.1371/journal.ppat.1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Napolitano G, Ballabio A. TFEB at a glance. J Cell Sci. 2016;129(13):2475–2481. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Settembre C, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15(6):647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J. 2012;441(2):523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kapuy O, Vinod PK, Bánhegyi G. mTOR inhibition increases cell viability via autophagy induction during endoplasmic reticulum stress — an experimental and modeling study. FEBS Open Bio. 2014;4:704–713. doi: 10.1016/j.fob.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nowacek A, Kadiu I, McMillan J, Gendelman HE. Immunoisolation of nanoparticles containingendocytic vesicles for drug quantitation. In: Weissig V, Elbayoumi T, Olsen M eds. Cellular and Subcellular Nanotechnology: Methods and Protocols. Totowa, NJ: Humana Press; 2013:41–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alers S, Wesselborg S, Stork B. ATG13: just a companion, or an executor of the autophagic program? Autophagy. 2014;10(6):944–956. doi: 10.4161/auto.28987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21(3):348–358. doi: 10.1038/cdd.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117(Pt 13):2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 36.Hyttinen JM, Niittykoski M, Salminen A, Kaarniranta K. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta. 2013;1833(3):503–510. doi: 10.1016/j.bbamcr.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 37.Jäger S, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117(Pt 20):4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 38.Szatmári Z, et al. Rab11 facilitates cross-talk between autophagy and endosomal pathway through regulation of Hook localization. Mol Biol Cell. 2014;25(4):522–531. doi: 10.1091/mbc.E13-10-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell GR, Bruckman RS, Chu YL, Spector SA. Autophagy induction by histone deacetylase inhibitors inhibits HIV type 1. J Biol Chem. 2015;290(8):5028–5040. doi: 10.1074/jbc.M114.605428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell GR, Spector SA. Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy. PLoS Pathog. 2012;8(5):e1002689. doi: 10.1371/journal.ppat.1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shanware NP, Bray K, Abraham RT. The PI3K, metabolic, and autophagy networks: interactive partners in cellular health and disease. Annu Rev Pharmacol Toxicol. 2013;53:89–106. doi: 10.1146/annurev-pharmtox-010611-134717. [DOI] [PubMed] [Google Scholar]
- 42.Harris J, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286(11):9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho J, et al. Autophagy in sepsis: degradation into exhaustion? Autophagy. 2016;12(7):1073–1082. doi: 10.1080/15548627.2016.1179410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pastore N, Ballabio A, Brunetti-Pierri N. Autophagy master regulator TFEB induces clearance of toxic SERPINA1/α-1-antitrypsin polymers. Autophagy. 2013;9(7):1094–1096. doi: 10.4161/auto.24469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molina JM, et al. Production of cytokines by peripheral blood monocytes/macrophages infected with human immunodeficiency virus type 1 (HIV-1) J Infect Dis. 1990;161(5):888–893. doi: 10.1093/infdis/161.5.888. [DOI] [PubMed] [Google Scholar]
- 46.Yamato K, el-Hajjaoui Z, Simon K, Koeffler HP. Modulation of interleukin-1 beta RNA in monocytic cells infected with human immunodeficiency virus-1. J Clin Invest. 1990;86(4):1109–1114. doi: 10.1172/JCI114815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorenec L, et al. The comparison of Th1, Th2, Th9, Th17 and Th22 cytokine profiles in acute and chronic HIV-1 infection. Microb Pathog. 2016;97:125–130. doi: 10.1016/j.micpath.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Marker DF, et al. The new small-molecule mixed-lineage kinase 3 inhibitor URMC-099 is neuroprotective and anti-inflammatory in models of human immunodeficiency virus-associated neurocognitive disorders. J Neurosci. 2013;33(24):9998–10010. doi: 10.1523/JNEUROSCI.0598-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai Y, Arikkath J, Yang L, Guo ML, Periyasamy P, Buch S. Interplay of endoplasmic reticulum stress and autophagy in neurodegenerative disorders. Autophagy. 2016;12(2):225–244. doi: 10.1080/15548627.2015.1121360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Escoll P, Rolando M, Buchrieser C. Modulation of host autophagy during bacterial infection: sabotaging host munitions for pathogen nutrition. Front Immunol. 2016;7:81. doi: 10.3389/fimmu.2016.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dinkins C, Pilli M, Kehrl JH. Roles of autophagy in HIV infection. Immunol Cell Biol. 2015;93(1):11–17. doi: 10.1038/icb.2014.88. [DOI] [PubMed] [Google Scholar]
- 52.Kyei GB, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186(2):255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Simone FI, et al. HIV-1 Tat and cocaine impair survival of cultured primary neuronal cells via a Mitochondrial Pathway. J Neuroimmune Pharmacol. 2016;11(2):358–368. doi: 10.1007/s11481-016-9669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez M, Kaushik A, Lapierre J, Dever SM, El-Hage N, Nair M. Electro-magnetic nano-particle bound Beclin1 siRNA crosses the blood–brain barrier to attenuate the inflammatory effects of HIV-1 infection in vitro. J Neuroimmune Pharmacol. doi: 10.1007/s11481-016-9688-3. [published online ahead of print June 10, 2016]. https://doi.org/10.1007/s11481-016-9688-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martini-Stoica H, Xu Y, Ballabio A, Zheng H. The autophagy-lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci. 2016;39(4):221–234. doi: 10.1016/j.tins.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kadiu I, Nowacek A, McMillan J, Gendelman HE. Macrophage endocytic trafficking of antiretroviral nanoparticles. Nanomedicine (Lond) 2011;6(6):975–994. doi: 10.2217/nnm.11.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo H, Gao J, Taxman DJ, Ting JP, Su L. HIV-1 infection induces interleukin-1β production via TLR8 protein-dependent and NLRP3 inflammasome mechanisms in human monocytes. J Biol Chem. 2014;289(31):21716–21726. doi: 10.1074/jbc.M114.566620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456(7219):264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 59.Kadiu I, Ricardo-Dukelow M, Ciborowski P, Gendelman HE. Cytoskeletal protein transformation in HIV-1-infected macrophage giant cells. J Immunol. 2007;178(10):6404–6415. doi: 10.4049/jimmunol.178.10.6404. [DOI] [PubMed] [Google Scholar]
- 60.Kruppa AJ, Kendrick-Jones J, Buss F. Myosins, actin and autophagy. Traffic. 2016;17(8):878–890. doi: 10.1111/tra.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Granger E, McNee G, Allan V, Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. 2014;31:20–29. doi: 10.1016/j.semcdb.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baxter AE, et al. Macrophage infection via selective capture of HIV-1-infected CD4+ T cells. Cell Host Microbe. 2014;16(6):711–721. doi: 10.1016/j.chom.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sattentau QJ, Stevenson M. Macrophages and HIV-1: an unhealthy constellation. Cell Host Microbe. 2016;19(3):304–310. doi: 10.1016/j.chom.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Napolitano G, Ballabio A. TFEB at a glance. J Cell Sci. 2016;129(13):2475–2481. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balkundi S, et al. Comparative manufacture and cell-based delivery of antiretroviral nanoformulations. Int J Nanomedicine. 2011;6:3393–3404. doi: 10.2147/IJN.S27830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gendelman HE, et al. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J Exp Med. 1988;167(4):1428–1441. doi: 10.1084/jem.167.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kalter DC, et al. Enhanced HIV replication in macrophage colony-stimulating factor-treated monocytes. J Immunol. 1991;146(1):298–306. [PubMed] [Google Scholar]
- 68.Gorantla S, et al. CD8+ cell depletion accelerates HIV-1 immunopathology in humanized mice. J Immunol. 2010;184(12):7082–7091. doi: 10.4049/jimmunol.1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.