Abstract
The most frequent focal alterations in human retinoblastoma are mutations in the tumor-suppressor gene retinoblastoma (RB) and amplification of the oncogene MYCN. Whether MYCN overexpression drives retinoblastoma has not been assessed in model systems. Here, we have shown that Rb inactivation collaborates strongly with MYCN overexpression and leads to retinoblastoma in mice. Overexpression of human MYCN in the context of Rb inactivation increased the expression of MYC-, E2F-, and ribosome-related gene sets, promoted excessive proliferation, and led to retinoblastoma with anaplastic changes. We then modeled responses to MYCN-directed therapy by suppressing MYCN expression in MYCN-driven retinoblastomas. Initially, MYCN suppression led to proliferation arrest and partial tumor regression with loss of anaplasia. However, over time, retinoblastomas reemerged, typically without reactivation of human MYCN or amplification of murine Mycn. A subset of returning retinoblastomas showed genomic amplification of a Mycn target gene encoding the miR cluster miR-17~92, while most retinoblastomas reemerged without clear genetic alterations in either Mycn or known Mycn targets. This Rb/MYCN model recapitulates key genetic driver alterations seen in human retinoblastoma and reveals the emergence of MYCN independence in an initially MYCN-driven tumor.
Introduction
MYC family members (MYC, MYCN, MYCL) are among the most frequently amplified oncogenes in human cancers. MYCN amplifications have long been known to contribute to human retinoblastoma, often together with retinoblastoma (RB) loss (1, 2). MYCN is also amplified in a small subset of human retinoblastomas harboring an intact RB gene (3). Despite frequent amplifications of MYCN in human retinoblastomas, the mechanisms underlying MYCN oncogenic activity in retinoblastoma have been largely unexplored. MYCN and RB also cooperate in tumorigenesis beyond retinoblastoma. For example, MYCN is frequently amplified in neuroblastoma, and Rb deletion in mice accelerated MYCN-driven neuroblastoma (4). Also, in human small-cell lung cancer, RB deletions and MYCN amplifications co-occur (5). A better understanding of how RB loss cooperates with MYCN overexpression in tumorigenesis is needed.
MYCN is a basic helix-loop-helix transcription factor that dimerizes with MYC-associated factor X (MAX) and can activate transcription via binding to E-box elements and noncanonical sites. MYCN and MYC can activate target genes such as Mir-17-92, which encodes the oncogenic miR cluster miR-17~92 (6–8). MYC family members regulate RNA Pol I, II, and III activities and broadly increase mRNA translation (9). MYC family members also globally increase the transcriptional output of actively transcribed genes (10, 11), in part by recruiting the pause-release factor pTEFb to promote transcriptional elongation (12). Inactivation of Mycn in mice led to nuclear condensation, reduced histone acetylation, and global changes in histone methylation (13). MYCN regulates proliferation, differentiation, and global chromatin organization; with these pleiotropic activities, the key oncogenic functions of MYCN remain unclear.
Recent strategies have emerged to therapeutically target MYCN-dependent tumors by reducing MYCN expression. MYCN stability is regulated by aurora kinase A, whereby binding to aurora kinase A can protect MYCN protein from degradation (14). Aurora kinase A inhibitors destabilized MYCN, suggesting a therapeutic strategy for combating MYCN-amplified tumors (15, 16). Other promising approaches for targeting MYCN-amplified tumors include inhibition of BET domain proteins (17) or of CDK7 (18), both of which lead to reduced MYCN transcription. Despite excitement about the potential of targeted therapies to decrease MYCN expression in MYCN-amplified tumors, whether resistance to MYCN-directed therapies will occur has not been determined in any model system.
Mouse models provide ideal tools to understand the mechanisms underlying oncogene function and to model the development of therapy resistance. Inducible models can be used to test the continued requirement for an initiating oncogenic alteration. Unlike in humans, germline Rb heterozygosity in mice does not lead to retinoblastoma (19–21). Inactivation of both copies of Rb in the earliest stages of retinal development led to extension of the proliferative period of retinal development, but eventually, cells exited the cell cycle or underwent apoptosis (22, 23). For mice to develop retinoblastoma in past models, Rb needed to be inactivated together with another Rb family member, Rb1 (also known as p107) (22, 24–26) or Rbl2 (also known as p130) (23, 27), or the CDK inhibitor Cdkn1b (also known as p27) (28). MYCN is the most frequently focally altered gene in human retinoblastoma beyond RB (2), and so we sought to control MYCN expression in the mouse retina harboring Rb family member deletion. We report a model of retinoblastoma that precisely mimics key genetic alterations in human retinoblastoma tumors and obviates any need to genetically inactivate Rb family members beyond Rb. This model was an ideal system for determining whether MYCN is required for retinoblastoma maintenance and for identifying features of tumors that become MYCN independent.
Results
To explore roles for MYCN in retinoblastoma, we used a TET-ON–inducible system, in which human MYCN can be controllably overexpressed (29) (Figure 1A). Here, a retina-specific Cre driver (Pax6 α enhancer Cre) (30) is coupled with a lox-stop-lox reverse tetracycline transactivator (rtTA) allele to lead to rtTA expression specifically in early retinal progenitor cells (beginning near E10) and their daughter cells. In the presence of a tetracycline-responsive element–MYCN/firefly luciferase (TRE-MYCN/LUC) allele (29) containing a tetracycline response element, human MYCN, and a luciferase reporter, doxycycline (DOX) addition to the food leads to overexpression of both MYCN and luciferase that can be monitored in vivo. Mice with the combination of LoxP-Stop-LoxP–reverse Tet transactivator (LSL-rtTA) TRE-MYCN/LUC Pax6 α enhancer Cre alleles are referred to herein as TET-MYCN mice.
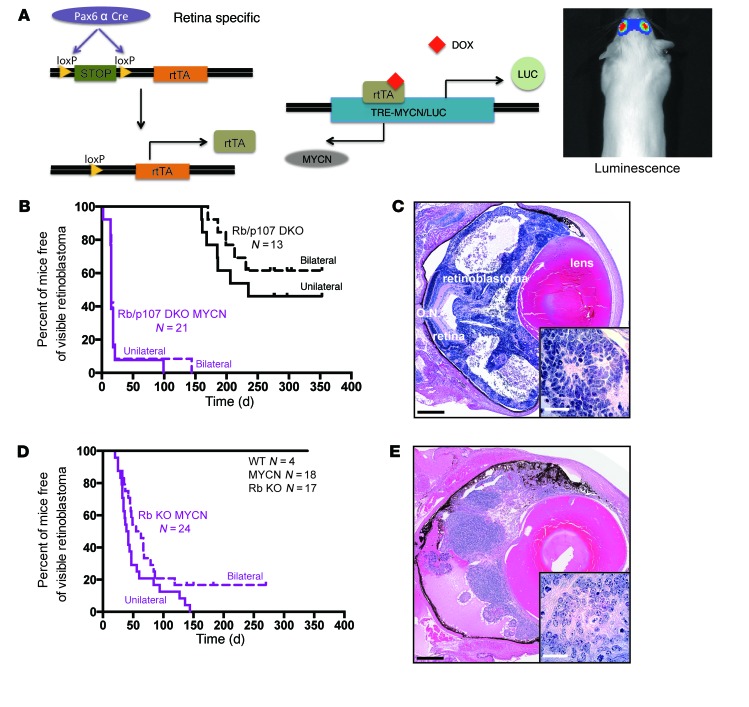
Figure 1. MYCN overexpression cooperates with Rb loss to drive retinoblastoma.
(A) Strategy to overexpress MYCN in Rb or Rb/p107-deficient retina. Retina-specific Pax6 α enhancer Cre (Pax6 α Cre) drives the expression of rtTA in the developing retina. The TRE-MYCN allele allowed for DOX-dependent expression of MYCN and luciferase in the retina. Image shows an in vivo assessment of bioluminescence in a tumor-bearing animal. (B) Kaplan-Meier curve shows the time to externally evident retinoblastoma in Rb/p107 (Rb/p107 DKO) compared with Rb/p107/TET-MYCN (Rb/p107 DKO MYCN) mice. The time to tumor presence in the first eye (unilateral curve) and second eye (bilateral curve) is shown. (C) H&E-stained eye from an Rb/p107/TET-MYCN mouse at P12, showing extensive retinoblastoma. The inset image is of tumor cells (original magnification, ×100) and shows Homer Wright rosettes typical of human and mouse retinoblastomas. O.N., optic nerve. (D) Kaplan-Meier curve showing the time to unilateral and then bilateral retinoblastoma formation in Rb/TET-MYCN (Rb KO MYCN) mice. (E) H&E staining of retinoblastoma-containing eye from an Rb/TET-MYCN model (inset: original magnification, ×100). Black scale bar: 500 μm; white scale bar: 25 μm.
MYCN overexpression accelerates Rb/p107-deleted retinoblastoma.
We examined whether TET-MYCN would synergize with Rb family member loss to promote retinoblastoma. In a previous mouse model harboring conditional Rb and p107 deletion in the retina (genotype Pax6 α enhancer Cre Rbfl/fl p107fl/fl, referred to hereafter as Rb/p107 mice), retinoblastomas arose with long latency. MYCN was spontaneously amplified in a subset of mice from this model (24), just as MYCN is amplified in human retinoblastomas. We used the Rb/p107 model to now overexpress MYCN (referred to hereafter as Rb/p107/TET-MYCN mice). To induce MYCN during retinal development, we fed DOX-containing chow to breeders throughout their pregnancy and before weaning, such that MYCN was overexpressed throughout retinal development and into adulthood. Rb/p107/TET-MYCN animals were followed until they developed advanced retinoblastoma. We found that overexpression of MYCN dramatically accelerated the time to retinoblastoma (Figure 1B). Rb/p107 mice developed retinoblastoma with only partial penetrance, and starting at about 5 months of age, Rb/p107/TET-MYCN mice exhibited bilateral retinoblastomas, typically from the time that their eyes opened (Figure 1, B and C). Thus, MYCN potently synergizes with Rb/p107 loss to promote retinoblastoma.
MYCN overexpression with Rb deletion alone drives retinoblastoma.
We next examined the effects of MYCN overexpression on retinae lacking Rb but with p107 intact (Rb/TET-MYCN mice). Remarkably, MYCN overexpression promoted retinoblastoma in the Rb-mutant context, bypassing any need to inactivate p107 or p130 (Figure 1, D and E). Rb/TET-MYCN mice developed retinoblastoma with very rapid kinetics (average of 54 days) and complete penetrance. While a subset of retinoblastoma patients who harbor MYCN amplification but not RB mutation has been described (3), we found no effect of MYCN overexpression on retinoblastoma when Rb was WT (Figure 1D). MYCN overexpression cooperated with Rb loss to lead to rapid retinoblastoma when DOX was present during retinal development (Figure 1, D and E). Interesting morphological changes were also noted when comparing retinoblastomas in the Rb/TET-MYCN mouse model with those in the Rb/p107 and Rb/p107/TET-MYCN mouse models. Tumors arising in Rb/p107 mice were composed of small, round blue cells with scattered apoptotic bodies and mitotic figures (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI88508DS1). In some animals, neuroblastic Homer Wright rosettes were present. In Rb/TET-MYCN mice, tumor cells were somewhat larger and more often elongated, angular, or molded together (Supplemental Figure 1B) — anaplastic features that have been associated with MYCN amplification in human retinoblastoma and worse clinical outcomes (3, 31). We found that anaplastic changes were even more prominent in Rb/p107/TET-MYCN tumors (Supplemental Figure 1C).
We hypothesized that retinoblastoma, a pediatric cancer, would require MYCN overexpression to occur during retinal development to lead to retinoblastoma. Indeed, when we administered DOX to adult 3-week-old mice, retinoblastoma emerged in only 1 of 20 Rb/TET-MYCN mice (Supplemental Figure 2). This result indicates that there is a critical window during retinal development, prior to P21, for MYCN overexpression to efficiently cooperate with Rb loss in promoting retinoblastoma.
MYCN overexpression synergizes with Rb loss to bypass a proliferative block.
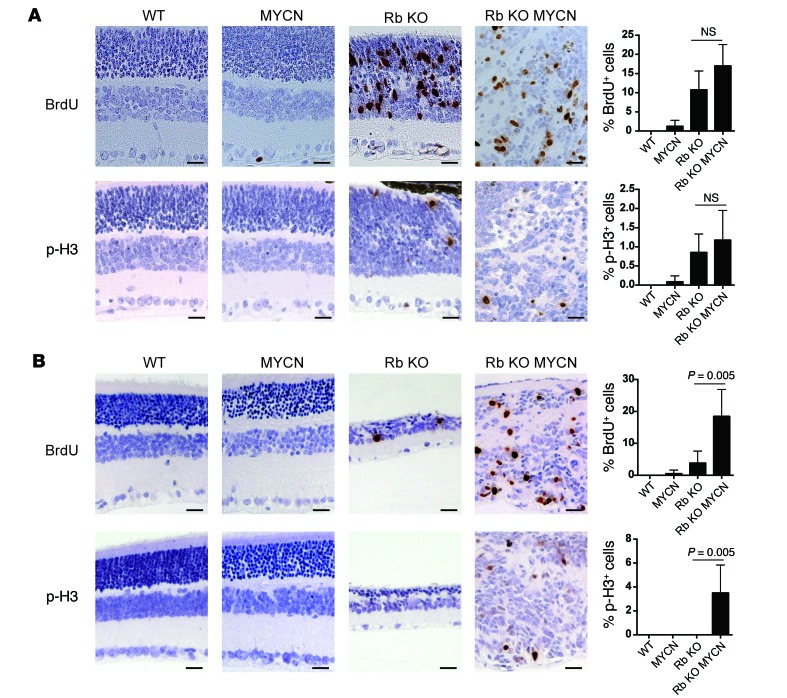
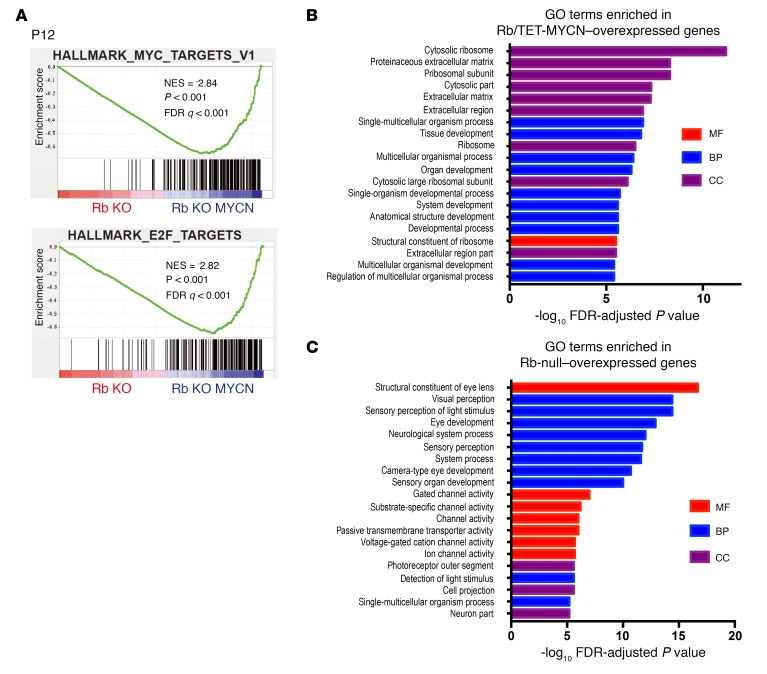
To examine mechanisms underlying the observed synergy between MYCN and Rb loss, we analyzed mutant retinae at 2 time points, P12 and P22. We examined proliferation using BrdU and anti-Ser10 phosphorylated histone H3 (p-H3) immunohistochemical analysis and apoptosis using a TUNEL assay. At P12, when cells in control retinae had stopped proliferating, Rb and Rb/TET-MYCN retinae exhibited extensive proliferation and apoptosis (Figure 2A and Supplemental Figure 3). We found no significant differences in proliferation or apoptosis levels between these genotypes. By P22, however, proliferation in the Rb-mutant retina had nearly ceased, and retinae in the Rb KO model were hypocellular owing to prior apoptosis. In contrast, proliferation continued unabated in the Rb/TET-MYCN retinae (Figure 2B). Thus, MYCN overexpression bypasses a proliferation block that otherwise occurs in the Rb-deficient retina. To identify gene expression changes driven by MYCN in the Rb-deficient retina, we focused on the earlier P12 stage, when there was no significant difference in overall proliferation levels between Rb and Rb/TET-MYCN–mutant retinae. Gene set enrichment analysis (GSEA) using RNA-sequencing (RNA-seq) data revealed both MYC-associated and E2F-associated gene sets as the top 2 most highly enriched gene sets in the Rb/MYCN condition compared with Rb alone (Figure 3A and Supplemental Table 1). Enrichment of E2F-associated gene sets compared with Rb-mutant retinae suggests that gene expression programs promoted by Rb loss may be strengthened by MYCN overexpression. We also used the edgeR package (32) to identify significant differentially expressed genes (Supplemental Table 2). Analyses of enriched Gene Ontology (GO) terms also revealed ribosome-, extracellular matrix–, and tissue development–associated genes to be enriched in the Rb/MYCN-overexpressed transcripts when compared with Rb-null retinae (Figure 3B). The most significant GO term was “cytosolic ribosome” (FDR-adjusted P value = 6.6 × 10–12), consistent with MYCN activation of ribosomal biogenesis–associated gene expression programs (33). Genes overexpressed in the Rb-null relative to Rb/TET-MYCN retinae at P12 included genes involved in eye/retina development (Figure 3C). The phenotypic data, along with gene expression changes, including enrichment of E2F target gene sets with MYCN overexpression, support the notion that MYCN drives proliferation programs in Rb-null cells to promote retinoblastoma. This is consistent with our finding that MYCN overexpression obviates the need for inactivation of other negative cell-cycle regulators to induce retinoblastoma (such as the other Rb family members p107, p130, and p27).
Figure 2. MYCN cooperates with Rb inactivation to increase proliferation in the retina.
(A) Brdu and p-H3 analysis of retinae at P12 showing increased proliferation in Rb-null (Rb KO) and Rb/TET-MYCN (Rb KO MYCN) retinae relative to control or MYCN-only retinae. N = 4–7 independent retinae, with 1 section examined per retina. (B) BRDU and p-H3 analysis at P22 shows increased proliferation in Rb/TET-MYCN retinae relative to Rb-deficient retinae that had largely exited the cell cycle. N = 4–6 independent retinae, with 1 section per retina. Scale bar: 25 μm. Error bars represent the SD. P values shown in B were determined by 2-tailed Student’s t test.
Figure 3. Gene expression changes with MYCN overexpression in Rb-deleted retina.
(A) GSEA comparing Rb-null (n = 4) with Rb/MYCN (n = 4) retinae at P12. The 50-gene “Hallmark signatures” set from MSigDB was queried and showed MYC- and E2F-related gene sets as the top 2 most significant gene sets enriched upon MYCN overexpression in the Rb-null retina. (B) Top 10 gene sets enriched in Rb/TET-MYCN and top 3 gene sets in Rb-null P12 retinae, with the normalized enrichment score (NES) shown. (C) Enriched GO terms in genes upregulated in Rb/TET-MYCN relative to Rb-null P12 retinae. (D) Enriched GO terms in genes upregulated in Rb-null relative to Rb/TET-MYCN P12 retinae. MF, molecular function; BP, biological process; CC, cellular component.
Suppression of MYCN expression in retinoblastoma cells.
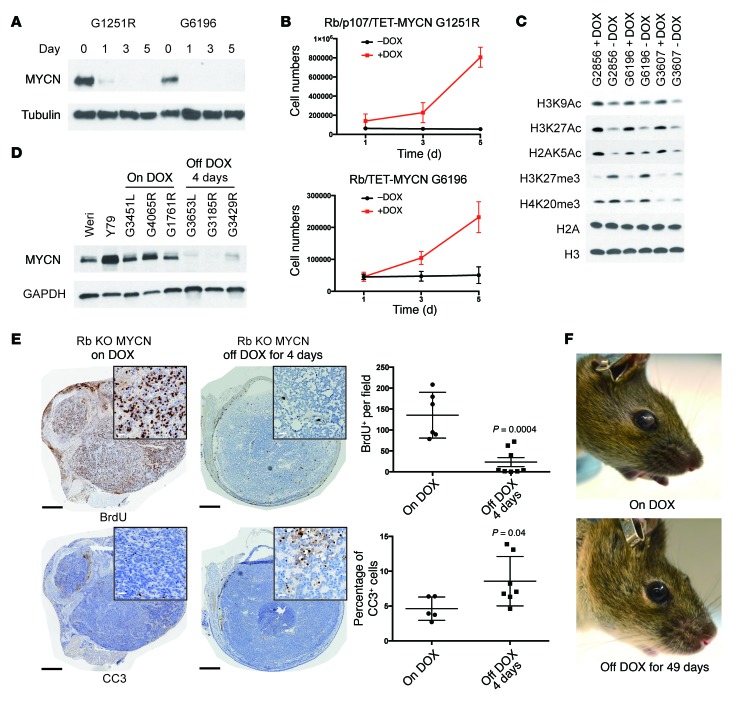
To determine whether maintenance of MYCN expression is important for retinoblastoma cells in culture, we generated cell lines from Rb/TET-MYCN and Rb/p107/TET-MYCN retinoblastomas in the presence of DOX. Removal of DOX led to a rapid loss of MYCN protein (Figure 4A) and suppression of proliferation (Figure 4B) that was associated with G1 block (Supplemental Figure 4A). The cells exhibited an enlarged and flattened morphology (Supplemental Figure 4B) and stained positively for senescence-associated β-gal (SA-β-gal) (Supplemental Figure 4C). MYCN has been found to control global chromatin structure, with MYCN-deficient neural progenitor cells exhibiting global hypoacetylation (13). To examine the effects of MYCN suppression on global histone marks, we performed Western blotting of extracted histones. Removal of DOX led to global reduction in acetylation at a number of sites associated with active chromatin, including H3K9, H3K27, and H2AK5, and led to increases in repressive marks such as H3K27 and H4K20 trimethylation (Figure 4C and Supplemental Figure 4D). Acute MYCN suppression in retinoblastoma cells led to the suppression of proliferation and changes in histone modifications that are generally associated with repressive chromatin.
Figure 4. Sustained MYCN expression is initially required for retinoblastoma cell proliferation.
DOX removal in cell lines derived from Rb/TET-MYCN tumors led to (A) reduced MYCN protein expression on Western blotting and (B) proliferation impairment, as assessed by counting cells 1, 3, and 5 days after plating. Data were pooled from 3 independent experiments. (C) Western blot of histone extracts from 3 Rb/TET-MYCN cell lines showing reduced histone H3K9, H3K27, and H2A K5 acetylation (Ac) and increased H3K27 and H4K20 trimethylation (me3) upon removal of DOX from the media. Individual blots indicating equal loading are shown Supplemental Figure 4D. (D) DOX removal in vivo initially resulted in suppression of MYCN protein expression by day 4. (E) Immunohistochemical analysis shows decreased BrdU-positive cells and increased cleaved caspase 3 (CC3) levels 4 days after DOX removal. Black scale bars: 500 μm; white scale bars (insets): 25 μm. Plot shows quantification from 5 to 8 independent retinae per genotype. The P values were determined by Student's t test. (F) Photographs showing a mouse with retinoblastoma in the anterior chamber, compared with 49 days later when the tumor had regressed.
Suppression of MYCN expression in MYCN-driven tumors leads to cell-cycle arrest and partial tumor regression.
Several studies have reported on potential MYCN-directed therapeutic approaches to reduce MYCN expression levels (14–18). One approach involves inhibition of aurora kinase A, which is required for MYCN protein stability (14–16). We found that MYCN overexpression in retinoblastoma cell lines conferred sensitivity to aurora kinase A inhibition using MLN8237 (alisertib) (Supplemental Figure 5, A–C). MLN8237 treatment led to reduced MYCN protein levels and activation of p53 in an Rb/TET-MYCN retinoblastoma cell line (Supplemental Figure 5B). Two Rb/TET-MYCN and two Rb/p107/TET-MYCN cell lines were highly sensitive to MLN8237 in the DOX-ON condition (Supplemental Figure 5C). Removal of DOX from the media of these cell lines suppressed MYCN expression and strongly reduced the effectiveness of MLN8237, suggesting that the effectiveness of aurora kinase A inhibition was MYCN dependent. DOX removal led to decreased proliferation upon MYCN suppression, which could have affected the MLN8237 response; thus, we also compared MLN8237 activity in retinoblastoma cells that differed in MYCN expression levels but that had similar proliferation rates. In cell lines derived from Rb/p107 retinoblastomas, in which MYCN protein levels were very low, there was a strikingly reduced effect of MLN8237 compared with cell lines from Rb/p107/TET-MYCN retinoblastomas (Supplemental Figure 5, D and E). Thus, the inhibitory effect of MLN8237 was specific to MYCN-overexpressing retinoblastoma cells. These data indicate that MYCN-directed therapy acts in a MYCN-specific manner in retinoblastoma cells.
We hypothesized that retinoblastomas initiated by Rb inactivation and MYCN overexpression would maintain a continued requirement for MYCN expression, given the complete absence of retinoblastoma in mice lacking Rb alone (22, 23, 34). To test this hypothesis, we removed DOX from the feed of retinoblastoma-bearing Rb/TET-MYCN animals. Four days after DOX removal, MYCN protein levels were suppressed (Figure 4D), and we found a strong decrease in BrdU-positive cells along with an increase in apoptosis, as shown by cleaved caspase 3 immunohistochemical analysis (Figure 4E). Histological changes were also evident 4 days after DOX removal, as the reduction in MYCN expression was associated with a round, regular tumor cell morphology, suggesting increased differentiation as well as a dramatic reduction in mitotic figures (Supplemental Figure 1D). In contrast to the phenotype observed in cell culture, where DOX removal led to features of senescence, we found no evidence of senescence in vivo 4 days or 27 days after DOX removal. Tumors did not stain for SA-β-gal 4 or 27 days after DOX removal (Supplemental Figure 6A), and we did not observe an upregulation of a panel of senescence-associated transcripts such as p21 or p16 or senescence-associated cytokines (Supplemental Figure 6B). In 66 of 71 tumor-bearing eyes, complete elimination of retinoblastoma in the anterior chamber of the eye was eventually observed with DOX removal (average time of 21 ± 13 days) (Figure 4F and Figure 5B). The early response to MYCN suppression was suppression of tumor proliferation, increased cell death, and tumor elimination from the anterior chamber of the eye.
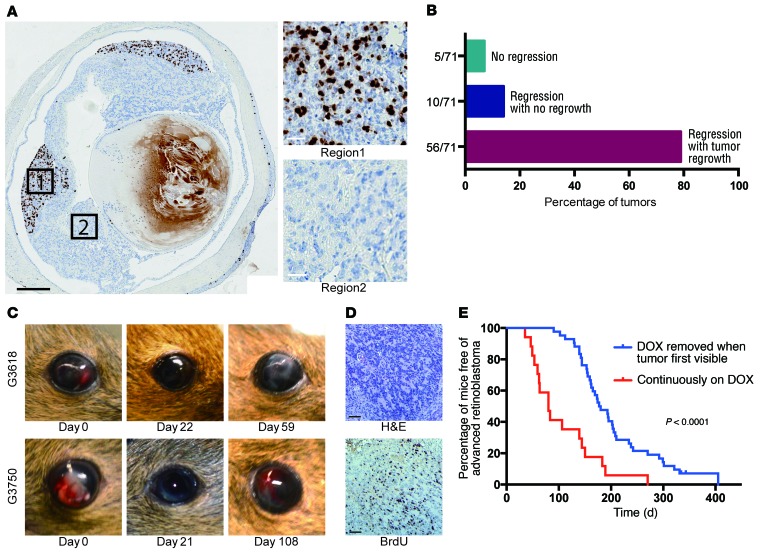
Figure 5. MYCN-independent retinoblastomas emerge in the absence of DOX.
(A) BrdU analysis showing regions with extensive proliferation in Rb/TET-MYCN retinae in the DOX-OFF condition 27 days after DOX removal, while other regions were nonproliferative. Black scale bar: 500 μm; white scale bars: 25 μm. Five of twelve eyes examined showed pockets of proliferation, with one section examined per eye. (B) Histogram showing the proportion of eyes with no tumor regression upon DOX removal, with regression but no regrowth, and with regression followed by DOX-independent tumor regrowth. (C) Photographs showing tumor regression upon DOX removal and then reappearance in the anterior chamber of the eye in 2 animals. (D) BrdU analysis showing a high proliferation rate in DOX-independent returned tumor. Scale bars: 50 μm. (E) Kaplan-Meier curves showing the time to development of late-stage retinoblastoma filling the eye. Rb/TET-MYCN mice were either maintained continuously on DOX (n = 17), or had DOX removed from their diet when the retinoblastoma was first externally visible in the eye (n = 42), usually with tumor evident in the anterior chamber. The P value shown was determined by log-rank test.
MYCN-independent proliferation and tumor reemergence.
We next determined whether the proliferation arrest associated with MYCN suppression in vivo would be sustained over time. Twenty-seven days after DOX removal, we performed BRDU analysis. In 5 of 12 eyes, we found pockets of extensive proliferation, while other regions of the retinoblastoma were nonproliferative (Figure 5A). We followed mice for a return of tumor to the anterior chamber of the eye. In 56 of the 66 eyes that showed tumor regression from the anterior chamber, we found that tumors eventually returned (Figure 5, B and C). The average time to relapse was 87 ± 51 days, and when tumors returned, they were highly proliferative (Figure 5D). We followed mice beyond the time of the tumor’s first appearance in the eye to when morbidity from advanced retinoblastoma filling the eye occurred and euthanasia was required. We compared cohorts with or without DOX removal at the first appearance of tumor cells in the eye and found that the mice with DOX removal had significantly increased survival, free of advanced retinoblastoma filling the eye (Figure 5E). One mechanism associated with DOX-independent growth of tumors driven by a DOX-inducible oncogene could be reactivation of the initiating oncogene. For example, rtTA mutations could occur that render the allele DOX independent (35). Examination of 27 tumors that had returned using quantitative real-time PCR to assess transgenic human MYCN expression showed that only 1 tumor had high MYCN expression levels (Supplemental Figure 7A). This tumor also had high levels of luciferase bioluminescence (not shown), suggesting a rare occurrence of DOX-independent activation of rtTA in this sample. We did not observe widespread upregulation of endogenous murine Mycn, Myc, or Mycl in the DOX-independent tumors (Supplemental Figure 7B). Thus, reactivating the initiating oncogene MYCN was not a frequent cause of DOX-independent retinoblastoma regrowth in this model. Previous mouse models relied on mutation of other Rb family members or p27, together with Rb inactivation, to induce retinoblastoma. We found no loss of p107, p130, or p27 in a group of tumors that returned in the absence of DOX (Supplemental Figure 8).
Amplification of Mir-17-92 in a subset of DOX-independent tumors.
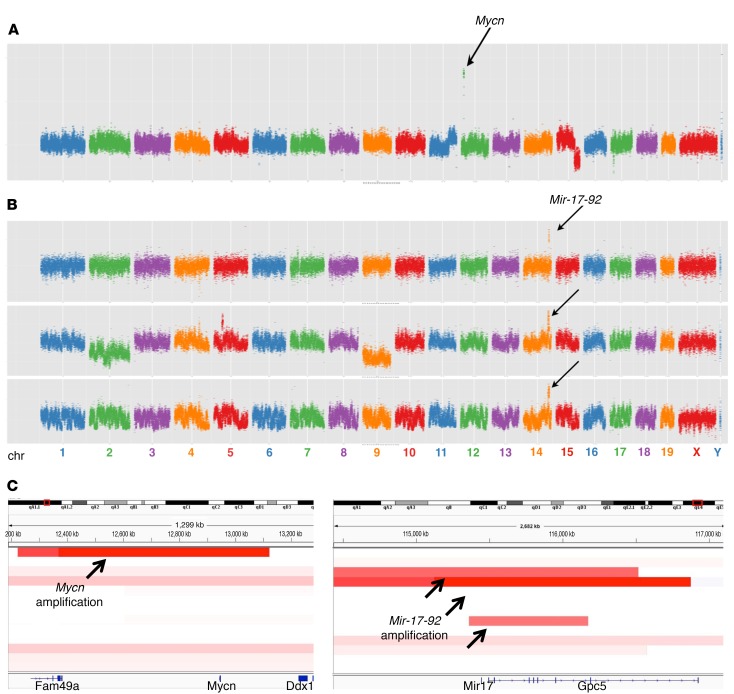
To identify genetic changes that occur in MYCN-independent retinoblastomas, we performed copy number variation (CNV) analyses, comparing DNA copy numbers in the tumor relative to normal tail DNA. We used low-coverage whole-genome sequencing to identify regions of deletion and amplification in Rb/TET-MYCN (DOX-ON) versus Rb/TET-MYCN (DOX-OFF) retinoblastomas and found whole chromosome gains and losses in the MYCN-ON condition but did not see focal amplifications. In contrast, we now observed focal amplifications in the MYCN-OFF condition. One of thirteen DOX-independent samples showed Mycn genomic amplification (Figure 6, A and C). More frequently, recurrent focal amplifications were found not in Mycn but rather in a key MYCN target gene, Mir-17-92, which encodes the miR-17~92 cluster (Figure 6, B and C). To expand upon this result, we performed quantitative real-time PCR for Mir-17-92 copy numbers across a larger sample set and found that 4 of 26 tumors harbored Mir-17-92 amplifications in the MYCN-OFF group, while 0 of 25 harbored Mir-17-92 amplifications in the MYCN-ON group (Supplemental Figure 9A). We did not expect to see Mir-17-92 amplifications in the MYCN-ON group, as Mir-17-92 is a well-characterized direct MYCN target gene (7, 36–38). We next determined whether enforced expression of Mir-17-92 would allow Rb/TET-MYCN cells to continue to cycle upon DOX removal and the consequent MYCN suppression. We used a murine stem cell virus–Mir-17-92 (MSCV–Mir-17-92) retroviral vector (39) to overexpress Mir-17-92 in 2 Rb/TET-MYCN cell lines. We achieved expression of miR-17 at levels similar to those found in the Rb/TET-MYCN DOX-ON condition (Supplemental Figure 9B). However, Mir-17-92 overexpression did not rescue the proliferation arrest caused by DOX removal and MYCN suppression in either cell line (Supplemental Figure 9C). Thus, a subset of retinoblastomas that had evolved DOX independence exhibited amplification of a key MYCN target gene, while the majority of these tumors did not show focal amplifications or deletions of MYCN or its targets.
Figure 6. Focal amplifications in Rb/TET-MYCN DOX-independent tumors.
CNV profiles of samples harboring focal amplifications in (A) Mycn or (B) Mir-17-92. Low-coverage whole-genome sequencing data were analyzed using CNVSeq. Arrows point to focal amplifications. chr, chromosome. (C) Integrative Genomics Viewer (IGV) (http://software.broadinstitute.org/software/igv/) plot showing the genomic regions of Mycn and Mir-17-92 amplification.
Discussion
We show that MYCN acts as a potent oncogene in Rb-deficient retinoblastoma. RB deletion and MYCN amplification are the two most frequent focal alterations occurring in human retinoblastoma, and these alterations often co-occur in the same tumor (2). MYCN amplifications in the absence of RB mutations or deletions have been described previously (2, 3). In one study, high-level MYCN amplifications were found only in those retinoblastomas in which RB mutations were undetected (3). In a more recent study, however, 8 of 94 retinoblastomas had more than 10 MYCN copies, and 6 of 8 of these MYCN-amplified retinoblastomas had at least 1 hit in the RB gene detected (2). Thus, a subset of retinoblastomas shows MYCN amplification, even in the absence of detected RB mutation, while in other retinoblastomas, MYCN amplification and RB mutations co-occur. Our mouse model did not show that MYCN overexpression alone was sufficient for retinoblastoma but revealed extremely potent cooperation with Rb loss in driving retinoblastoma. Previous mouse retinoblastoma models required deletion of multiple Rb family members (22, 23, 25, 40) or inactivation of Rb and the CDK inhibitor Cdkn1b (28), events that do not occur in human retinoblastomas. To our knowledge, this is the first mouse retinoblastoma model to recapitulate precise genetic alterations, Rb deletion, and the MYCN amplifications found in human retinoblastomas.
The key activities of MYCN that drive cancer are unclear, given the pleiotropic functions of MYCN. In the developing retina, we found that MYCN overexpression conferred the ability to bypass a proliferative block that otherwise limited the extent of inappropriate proliferation of Rb-deficient cells. Recently, it was shown that, upon Rb deletion, MYC can superactivate E2F transcriptional programs, in particular, G1/S phase–related genes (41). This role for MYC in superactivating E2F targets and driving proliferation in Rb-deleted cells may be highly relevant to the retinoblastoma that occurs with Rb deletion and MYCN amplification, as E2F target gene sets were upregulated in the Rb/TET-MYCN retinae compared with what was observed in Rb-mutant retinae. The ability of MYCN overexpression to obviate any need to inactivate Rbl1 or Rbl2 or Cdkn1b in driving retinoblastoma is also consistent with key MYCN oncogenic functions occurring via proliferation control.
The inducible nature of the current mouse model allowed us to test the requirement of MYCN overexpression for retinoblastoma tumor maintenance. Despite the fact that MYCN was required for initial tumor emergence in the context of Rb deficiency, these tumors evolved to not require MYCN. This suggests that therapeutic approaches to reduce MYCN RNA or protein in MYCN-driven retinoblastoma will also likely result in the reemergence of tumors independently of MYCN. In some cases, activation of a MYCN target gene, the Mir-17-92 cluster, occurred. We previously found that Mir-17-92 functions as a potent oncogene, accelerating retinoblastoma in the Rb/p107 model (42). Mir-17-92 is a direct target of MYCN (7, 36–38) and probably explains the absence of amplifications in the Rb/TET-MYCN DOX-ON condition. Interestingly, a developmental syndrome in humans, Feingold syndrome, is caused by hemizygous MYCN loss (43), while a very similar variant syndrome, Feingold syndrome 2, is caused by hemizygous Mir-17-92 inactivation (44, 45). Given the similar genetic syndromes caused by MYCN and Mir-17-92 hemizygous loss, it is intriguing that Mir-17-92 amplifications only occurred in the DOX-OFF tumors. In lymphoma models, Mir-17-92 could partially replace oncogenic functions of the MYCN relative MYC (46). It is unlikely that Mir-17-92 completely replaces oncogenic activity of MYCN in retinoblastoma, as we could not rescue the impaired proliferation caused by MYCN suppression with Mir-17-92 overexpression (Supplemental Figure 9, B and C). Also, in contrast to our observations with MYCN overexpression, we previously found that overexpression of Mir-17-92 did not cooperate with Rb loss alone to lead to retinoblastoma, though Mir-17-92 accelerated retinoblastoma initiated by both Rb and p107 loss (42). In most cases, our CNV analyses of tumors that reemerged in a MYCN-independent fashion did not reveal clear amplifications or deletions that could explain the development of MYCN independence. In many cases, epigenetic changes in the absence of genetic alterations may contribute to MYCN independence. Identification of the mechanisms that retinoblastoma cells in this model undergo to evolve MYCN independence is now critical.
In this study, we developed a mouse model that we believe will be invaluable for retinoblastoma preclinical studies, as this is the first mouse retinoblastoma model to our knowledge to be initiated by manipulating only genes that are also mutated in human retinoblastoma. This model revealed that MYCN-driven retinoblastoma can evolve to become MYCN independent and, we believe, represents an ideal tool for understanding the evolution of MYCN independence in MYCN-driven cancers.
Methods
Mice.
We crossed Rbfl/fl Pax6 α enhancer Cre Rosa26-Lox-STOP-LOX-rtTA/+ mice (or Rbfl/fl, p107–/–, Pax6 α enhancer Cre, Rosa26-Lox-STOP-LOX-rtTA/+ mice) with TRE-MYCN/LUC mice (29) to produce mice with inducible MYCN transgene expression in distal retina. The Rbfl/fl and p107–/– strains were provided by Tyler Jacks (MIT, Cambridge, Massachusetts, USA); TRE-MYCN/LUC mice were provided by William Weiss (UCSF, San Francisco, California, USA); and Pax6 α enhancer Cre mice were provided by Peter Gruss (Max Planck Institute, Gottingen, Germany). DOX was administered in mouse chow at a dose of 625 mg/kg (Harlan Laboratories) without other food sources. For studies of transgene activation, mice were fed DOX chow either during development (DOX food during breeding) or from P21. Mice were monitored for the first visible sign of retinoblastoma, usually involving tumor seeding in the anterior chamber of the eye. We continued to monitor the mice for advanced tumor burden that filled the eye or led to ulceration, at which point the mice were euthanized and tumors collected.
Cell line generation.
Retinoblastoma tumor tissue from mouse models was collected using a sterile needle and syringe to extract tumor cells from retinoblastoma-bearing eyes shortly following euthanasia. Tumor tissue was dissociated using a pipette, placed into standard retinoblastoma cell culture, and efficient generation of cell lines from retinoblastoma-bearing mice was observed.
Histology and IHC.
Eyes were fixed in Bouin’s solution or formalin for 24 hours before paraffin embedment. Paraffin blocks were sectioned at a thickness of 4 μm and stained with H&E. Analysis of general tumor cell morphology was performed by a board-certified pathologist with expertise in ophthalmic pathology (CGE). Immunohistochemical analysis was performed using the following antibodies: p-H3 (1:300; EMD Millipore; catalog 06-570); BrdU (1:300; BD; catalog 347580); and cleaved caspase 3 (1:100; Cell Signaling Technology; catalog 9661). For p-H3 and cleaved caspase 3 staining, paraffin sections were processed from xylene through a graded ethanol series to TBS Tween-20 (TBST). Unmasking was performed using microwave heating in sodium citrate buffer (0.01 M at pH6.0). Endogenous peroxidases were blocked with 3.5% H2O2, and immunohistochemical analysis was performed with overnight incubation in primary antibody. Biotin-conjugated secondary antibodies (Vector Laboratories) were used at a dilution of 1:200, and detection was done via a biotin-peroxidase complex (Vectastain ABC; Vector Laboratories) with DAB substrate (Vector Laboratories). For BrdU staining, pepsin digestion was performed by incubation in 0.2 μg/μl pepsin in 10 mM HCl at 37°C. After digestion, slides were denatured and neutralized. Immunohistochemical analysis was performed with an overnight incubation in the primary antibodies. For BrdU analysis, an i.p. injection of pups or adult mice was done with BrdU, and eyes were collected and fixed 1 hour later. TUNEL assays were performed using the In Situ Cell Death Detection Kit, POD (Roche) according to the manufacturer’s instructions.
Western blotting.
Lysates were prepared in ice-cold RIPA buffer containing protease inhibitor cocktail (Roche) and 1 mM DTT. Western blot assays were conducted with antibodies against MYCN (Santa Cruz Biotechnology Inc.; catalog sc-791); p130 (BD; catalog 610262); p107 (Sigma-Aldrich; catalog P4360); actin (Sigma-Aldrich; catalog A3854); tubulin (Santa Cruz Biotechnology Inc.; catalog sc398103); GAPDH (Santa Cruz Biotechnology Inc.; catalog sc-32233); p21 (Santa Cruz Biotechnology Inc.; catalog sc-397); and p53 (Santa Cruz Biotechnology Inc.; catalog sc-6243). Histone extractions were performed as described previously (47). We used antibodies against histone H3K9Ac (Cell Signaling Technology; catalog 9649P); H3K27Ac (Abcam; catalog ab4729); H3K27me3 (EMD Millipore; catalog 07-449); H4K20me3 (Abcam; catalog ab9053); H2AK5Ac (Cell Signaling Technology; catalog 2576P); H2A (Cell Signaling Technology; catalog 2578P); and H3 (Cell Signaling Technology; catalog 3638).
Quantitative real-time PCR.
Total RNA from tumors or cell pellets was isolated with TRIzol (Invitrogen, Thermo Fisher Scientific) and used to synthesize cDNA with iScript Reverse Transcription Supermix (Bio-Rad). Quantitative real-time PCR was performed with ALL-in-ONE qPCR Mix (Genecopoeia) with primers designed to detect human MYCN and mouse Myc, Mycn, p21, p16, Igfbp1, Igfbp7, Il1b, Ccr1, Il6, PAI-1, and Gadph (as an endogenous control). The primer sequences are provided in Supplemental Table 3. Quantitative expression data were acquired and analyzed with a 7900 Real-Time PCR System (Applied Biosystems). The relative copy numbers for Mycn and Mir-17-92 were determined by quantitative real-time PCR using All-in-One qPCR Mix and primers designed to detect murine genomic regions of these genes and Ant1 (as an endogenous control). Normal tail DNA was used to normalize the data. All samples were analyzed in triplicate.
miR-17~92 overexpression.
Retroviral MSCV vectors (empty or overexpressing Mir-17-92) were a gift of Andrea Ventura (Memorial Sloan Kettering Institute, New York, New York, USA). MSCV plasmid and plasmids encoding the packaging gene and envelope gene were transfected into 293 cells using Lipofectamine 2000 (Thermo Fisher Scientific). Virus was harvested 3 days after transfection and used for infection of mouse retinoblastoma cell lines. Puromycin (1 μg/ml) was added to cell culture medium for 1 week to select for infected cells. Cells were then expanded for the experiments.
TaqMan small RNA assays.
Total RNA from tumors and cell pellets was isolated using TRIzol (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s instructions. A TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) was used to produce cDNA for TaqMan MicroRNA Assays following the manufacturer’s instructions. TaqMan MicroRNA assays to detect miR17 and U6 small nuclear RNA (snRNA) control were obtained from Life Technologies (Thermo Fisher Scientific). Quantitative real-time PCR was performed using TaqMan Universal Master Mix II (Applied Biosystems) on a 7900 Real-Time PCR System (Applied Biosystems).
SA–β-gal activity.
Fresh-frozen tissue sections or cells were washed with PBS and fixed in 2% formaldehyde and 0.2% glutaraldehyde. Cells were covered with staining solution (40 mM citric acid/Na phosphate buffer, 5 mM K4[Fe(CN)6] 3H2O, 5 mM K3[Fe(CN)6], 150 mM sodium chloride, 2 mM magnesium chloride, and 1 mg/ml X-gal in distilled water) at 37°C overnight for a total of 16 hours. After incubation, tissue and cells were washed with PBS and methanol and air dried prior to imaging. Tissue from aging WT kidneys was used as a positive control.
Next-generation sequencing analysis.
For whole-genome analysis of CNV analysis, we performed low-coverage whole-genome sequencing using DNA from retinoblastomas or matched tails. The NEBNext DNA Library Prep Master Mix Set for Illumina (New England BioLabs; catalog E6040L) was used to generate DNA libraries from 1,000 ng genomic DNA. Single-read data (50 bp) were generated using an Illumina HiSeq 2500. Reads were aligned to the mm9 build of the murine genome using the Burrows Wheeler Aligner (48), and we used CNVseq (49) to examine copy numbers. For RNA-seq analyses, the Ultra RNA Library Prep Kit for Illumina (New England BioLabs; catalog E753L) was used to generate libraries from 500 ng total RNA derived from retinae microdissected at P12. All library preparation was conducted according to the manufacturer’s instructions. Single-end sequencing (50 bp) was performed using an Illumina HiSeq 2500, reads were aligned to the mm9 genome using TopHat version 2.08b (50), and Cuffdiff was used to generate the fragment per kilobase per million (FPKM) expression values used for GSEA (http://www.broadinstitute.org/gsea/) (51). Counts were generated from TopHat alignments using the Python package HTSeq v0.5.4p3 (52), using the “intersection-strict” overlap mode. Genes with low counts across conditions were discarded prior to identification of differentially expressed genes using the Bioconductor package edgeR, version 3.4.0 (32). An FDR method was used to correct for multiple testing (53), where differential expression was defined as |log2 (ratio)| ≥0.585 (±1.5-fold), with the FDR set at 5%. Overrepresented GO terms for genes that were either significantly up or downregulated were identified with the Bioconductor package goseq, version 1.24.0 (54), using an FDR method to correct for multiple testing. Next-generation sequencing data for this study were deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE90961).
Statistics.
Error bars on all graphs reflect the SD. Comparisons of 2 means were performed using a 2-tailed Student’s t test. Differences were considered significant at a P value of less than 0.05. Kaplan-Meier curves were generated using GraphPad Prism 7 (GraphPad Software), with a log-rank test to determine P values for significance.
Study approval.
All mouse experiments were conducted in accordance with protocols approved by the IACUC of the Fred Hutchinson Cancer Research Center.
Author contributions
NW and DM designed research studies, analyzed data, and wrote the manuscript. NW, DJ, and BB conducted experiments and analyzed data. RB analyzed data. CGE analyzed the histology of mouse retinoblastomas.
Supplementary Material
Acknowledgments
We thank members of the Fred Hutchinson Bioinformatics Core, including Jerry Davison, for help with data analysis. This work was supported by National Cancer Institute (NCI) grant R01CA148867 and American Cancer Society Research Scholar Award 120127-RSG-11-270-01-RMC (to DM). We are also grateful to the Elmer and Sylvia Sramek Foundation for study support.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(3):888–898.https://doi.org/10.1172/JCI88508.
Contributor Information
Nan Wu, Email: nwu2@fredhutch.org.
Deshui Jia, Email: djia@fredhutch.org.
Breanna Bates, Email: bbates@fredhutch.org.
Ryan Basom, Email: rbasom@fhcrc.org.
David MacPherson, Email: dmacpher@fhcrc.org.
References
- 1.Lee WH, Murphree AL, Benedict WF. Expression and amplification of the N-myc gene in primary retinoblastoma. Nature. 1984;309(5967):458–460. doi: 10.1038/309458a0. [DOI] [PubMed] [Google Scholar]
- 2.McEvoy J, et al. RB1 gene inactivation by chromothripsis in human retinoblastoma. Oncotarget. 2014;5(2):438–450. doi: 10.18632/oncotarget.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rushlow DE, et al. Characterisation of retinoblastomas without RB1 mutations: genomic, gene expression, and clinical studies. Lancet Oncol. 2013;14(4):327–334. doi: 10.1016/S1470-2045(13)70045-7. [DOI] [PubMed] [Google Scholar]
- 4.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16(11):2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong AJ, Ruppert JM, Eggleston J, Hamilton SR, Baylin SB, Vogelstein B. Gene amplification of c-myc and N-myc in small cell carcinoma of the lung. Science. 1986;233(4762):461–464. doi: 10.1126/science.3014659. [DOI] [PubMed] [Google Scholar]
- 6.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 7.Northcott PA, et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69(8):3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulte JH, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int J Cancer. 2008;122(3):699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- 9.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10(4):301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 10.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25(12):2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otto T, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15(1):67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Brockmann M, et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell. 2013;24(1):75–89. doi: 10.1016/j.ccr.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustafson WC, et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell. 2014;26(3):414–427. doi: 10.1016/j.ccr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puissant A, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3(3):308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chipumuro E, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159(5):1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke AR, et al. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359(6393):328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 20.Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359(6393):295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 21.Lee EY, et al. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359(6393):288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 22.Chen D, Livne-bar I, Vanderluit JL, Slack RS, Agochiya M, Bremner R. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 2004;5(6):539–551. doi: 10.1016/j.ccr.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 23.MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME, Jacks T. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 2004;18(14):1681–1694. doi: 10.1101/gad.1203304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacPherson D, Conkrite K, Tam M, Mukai S, Mu D, Jacks T. Murine bilateral retinoblastoma exhibiting rapid-onset, metastatic progression and N-myc gene amplification. EMBO J. 2007;26(3):784–794. doi: 10.1038/sj.emboj.7601515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Schweers B, Dyer MA. The first knockout mouse model of retinoblastoma. Cell Cycle. 2004;3(7):952–959. [PubMed] [Google Scholar]
- 26.Robanus-Maandag E, et al. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 1998;12(11):1599–1609. doi: 10.1101/gad.12.11.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dannenberg JH, Schuijff L, Dekker M, van der Valk M, te Riele H. Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev. 2004;18(23):2952–2962. doi: 10.1101/gad.322004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sangwan M, et al. Established and new mouse models reveal E2f1 and Cdk2 dependency of retinoblastoma, and expose effective strategies to block tumor initiation. Oncogene. 2012;31(48):5019–5028. doi: 10.1038/onc.2011.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swartling FJ, et al. Pleiotropic role for MYCN in medulloblastoma. Genes Dev. 2010;24(10):1059–1072. doi: 10.1101/gad.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105(1):43–55. doi: 10.1016/S0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 31.Mendoza PR, et al. Histopathologic grading of anaplasia in retinoblastoma. Am J Ophthalmol. 2015;159(4):764–776. doi: 10.1016/j.ajo.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boon K, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20(6):1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, et al. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat Genet. 2004;36(4):351–360. doi: 10.1038/ng1318. [DOI] [PubMed] [Google Scholar]
- 35.Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci USA. 2008;105(13):5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mestdagh P, et al. MYCN/c-MYC-induced microRNAs repress coding gene networks associated with poor outcome in MYCN/c-MYC-activated tumors. Oncogene. 2010;29(9):1394–1404. doi: 10.1038/onc.2009.429. [DOI] [PubMed] [Google Scholar]
- 37.Fontana L, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3(5):e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lovén J, et al. MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc Natl Acad Sci USA. 2010;107(4):1553–1558. doi: 10.1073/pnas.0913517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mu P, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conkrite K, Sundby M, Mu D, Mukai S, MacPherson D. Cooperation between Rb and Arf in suppressing mouse retinoblastoma. J Clin Invest. 2012;122(5):1726–1733. doi: 10.1172/JCI61403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H, et al. Redeployment of Myc and E2f1-3 drives Rb-deficient cell cycles. Nat Cell Biol. 2015;17(8):1036–1048. doi: 10.1038/ncb3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conkrite K, et al. miR-17~92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 2011;25(16):1734–1745. doi: 10.1101/gad.17027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Bokhoven H, et al. MYCN haploinsufficiency is associated with reduced brain size and intestinal atresias in Feingold syndrome. Nat Genet. 2005;37(5):465–467. doi: 10.1038/ng1546. [DOI] [PubMed] [Google Scholar]
- 44.Grote LE, Repnikova EA, Amudhavalli SM. Expanding the phenotype of feingold syndrome-2. Am J Med Genet A. 2015;167A(12):3219–3225. doi: 10.1002/ajmg.a.37368. [DOI] [PubMed] [Google Scholar]
- 45.de Pontual L, et al. Germline deletion of the miR-17~92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43(10):1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014;26(2):262–272. doi: 10.1016/j.ccr.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2(6):1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19(3):368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 54.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.