Abstract
Influenza A viruses (IAV) infect many host species, including humans and pigs. Severe combined immunodeficiency (SCID) is a condition characterized by a deficiency of T, B, and/or natural killer (NK) cells. Animal models of SCID have great value for biomedical research. Here, we evaluated the pathogenesis and the innate immune response to the 2009 H1N1 pandemic IAV (H1N1pdm09) using a recently identified line of naturally occurring SCID pigs deficient in T and B lymphocytes that still have functional NK cells. SCID pigs challenged with H1N1pdm09 showed milder lung pathology compared to the non-SCID heterozygous carrier pigs. Viral titers in the lungs and nasal swabs of challenged SCID pigs were significantly higher than in carrier pigs 7 days postinfection, despite higher levels of IL-1β and IFN-α in the lungs of SCID pigs. The lower levels of pulmonary pathology were associated with the T and B cell absence in response to infection. The higher viral titers, prolonged shedding, and delayed viral clearance indicated that innate immunity was insufficient for controlling IAV in pigs. This recently identified line of SCID pigs provides a valuable model to understand the immune mechanisms associated with influenza protection and recovery in a natural host.
Keywords: Severe combined immunodeficiency, Influenza, Pig, Innate, Immunity
Introduction
Severe combined immunodeficiency (SCID) is characterized by a life-threatening deficiency of T, B, and/or natural killer (NK) cells. Spontaneous SCID has been discovered in humans, horses, and dogs, and has been artificially created by transgenics in mice and rats [1, 2]. An X-linked SCID phenotype has been artificially induced in pigs through the disruption of the interleukin-2 receptor gamma chain gene (IL2RG) [3, 4], as well as the RAG1 and/or RAG2 genes required for the development of B and T lymphocytes [5, 6]. Pigs from a line of purebred Yorkshires under selection for increased feed efficiency at Iowa State University [7] were affected by naturally occurring SCID [8, 9, 10]. These SCID pigs were shown to have decreased numbers of B and T cells [11] and were unable to reject xenografted human cancer cells [9]. Genetic analyses revealed 2 mutations in the Artemis (DCLRE1C) gene that were responsible for the naturally occurring SCID in this line of pigs [10]. Mutations in DCLRE1C were first linked to human SCID in 2001 [[12], reviewed in [13]]. Some Artemis-deficient patients were reported to have recurring respiratory infections [14], but none were specifically associated with influenza virus infection. The discovery of SCID in pigs gives rise to new avenues of biomedical research, such as studies to further examine the role of Artemis in immunity as well as in cancer and clinical therapy of immunodeficient patients, or leveraged for improvements in xenotransplantation. Development of gene tests for the loci that confer the naturally occurring SCID phenotype may also be valuable to commercial swine producers.
Influenza A virus (IAV) is a segmented, negative-sense RNA virus that is pathogenic in many host species, including humans and pigs [15]. Swine are a natural host of influenza infection and can serve as intermediary hosts in the evolution of influenza viruses of risk to humans [16], representing an ideal model for the study of influenza infection. Clinical signs of influenza in pigs are similar to those observed in humans and are manifested as acute respiratory disease characterized by fever, inactivity, decreased food intake, respiratory distress, coughing, sneezing, conjunctivitis, and nasal discharge [17]. The clinical disease in pigs and humans ranges from clinically unaffected to severe respiratory signs; the variation is due to prior immunity, properties of the virus, and many other contributing health and environmental factors [17]. The disease incubation period is between 1 and 3 days, with rapid recovery beginning 4–7 days after onset in healthy individuals without complicating infections of multiple etiology [17]. A primary virulence mechanism of IAV is disruption of the innate immune response by interfering with the type I interferon response early in infection [18]. However, as early as 5–7 days after infection, compensating innate mechanisms and the adaptive arm of the immune system begin to clear the virus from the respiratory tract. Protective antibody responses can typically be measured within 2 weeks of viral exposure [19].
Although IAV infection has been characterized in SCID mice as an absence of cell-mediated and humoral immune responses postvaccination [20] and prolonged weight loss with failure to clear virus [21], mice are not a natural host for IAV. Furthermore, laboratory-adapted viral strains used in most of these studies may not be relevant to fully virulent circulating strains. Mice susceptible to IAV have a large mutation in the Mx gene and fail to synthesize Mx protein, a type I interferon-induced protein that can selectively inhibit IAV replication [22]. Immunocompromised human patients affected with SCID often develop complications after influenza infection [23, 24]; however, the course of influenza infection in humans affected by the genetic defect causing the SCID in this pig model has not been described. Here, we evaluated IAV pathogenesis and the immune response in SCID pigs after inoculation with the 2009 H1N1 pandemic IAV strain (H1N1pdm09) that now circulates globally as a seasonal human strain. H1N1pdm09 is also endemically or sporadically detected in pigs globally [25]. Our objective was to utilize this unique SCID pig model to evaluate the ability of the innate immune response in swine, a natural host for IAV, to defend against acute influenza infection in the absence of the adaptive immune response. As H1N1pdm09 is a pathogen for humans and immunodeficiency is known to be a risk for severe influenza disease in humans [26], this swine SCID model provides an excellent opportunity for evaluating potential therapies for human patients.
Material and Methods
Study Design
Three SCID-carrier sows were used as the source of SCID pigs in this study. These sows were seropositive for IAV antibodies, indicating natural exposure to IAV prior to start of the study, but were shown to be free of IAV by virus isolation. The SCID carrier sows were artificially inseminated using semen collected from a boar that was a compound heterozygote for the 2 mutations identified in Artemis and rescued by bone marrow transfer. All inseminations were within a window of 1 week and all 3 sows farrowed without surgical intervention. In order to obtain seronegative SCID offspring from the 3 carrier sows, piglets were removed from their dams prior to suckling colostrum and fostered onto 4 healthy sows from an IAV-negative source herd within 6 h of life. Genomic DNA isolated from tail tissue was used for individual DNA testing to check for the SCID-associated genotype described below.
Twenty-six piglets were weaned at approximately 3 weeks of age and divided into 4 groups according to SCID genotype screening and challenge (Table 1). All pigs were treated with ceftiofur crystalline-free acid (Zoetis, Parsippany, NJ, USA) and enrofloxacin (Bayer Animal Health, Shawnee Mission, KS, USA) at weaning to reduce respiratory bacterial contaminants. Pigs were demonstrated to be free of IAV antibodies and IAV based on nasal swab samples prior to the start of the study. Pigs were housed in biosafety level 2 containment and cared for in compliance with the Institutional Animal Care and Use Committee of the National Animal Disease Center.
Table 1.
Experimental design and group assignment according to immune deficiency genotype
| Treatment group | Genotype | Challenge strain | n |
|---|---|---|---|
| SCID/NC | SCID | None | 5 |
| SCID/CH | SCID | H1N1pdm09 | 6 |
| Carrier/NC | Carrier | None | 7 |
| Carrier/CH | Carrier | H1N1pdm09 | 8 |
NC, nonchallenged controls; CH, challenged; SCID, severe combined immunodeficiency; H1N1pdm09, pandemic H1N1 A/ California/04/2009.
At weaning, pigs in the challenge groups were inoculated intranasally with 2 mL of 1 × 106 TCID50/mL of A/California/04/2009 (H1N1pdm09). The challenge virus was propagated in Madin-Darby canine kidney (MDCK) cells following standard procedures. Clinical signs of respiratory disease and rectal temperatures were observed daily. At 7 days postinfection (dpi), the pigs were humanely euthanized with a lethal dose of pentobarbital (Fatal Plus, Vortech Pharmaceuticals, Dearborn, MI, USA) and necropsied. Necropsy samples included bronchoalveolar lavage fluid (BALF), trachea, and right cardiac or affected lung lobe in 10% buffered formalin.
Molecular Determination of SCID Genotype
Allele-specific primer sets were designed for 7 single-nucleotide polymorphisms (SNPs) in the region surrounding the Artemis gene using web-based allele-specific PCR (WASP) [27]. PCR cycling conditions included an initial hot start at 95°C for 2 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at temperatures indicated for each primer pair in Table 2 for 30 s, and elongation at 72°C for the time indicated in Table 2, ending with a final elongation step at 72°C for 5 min. Primer sequences, location, and genotypes for the 7 SNPs for which allele-specific primers were designed are given in Table 2. These SNPs were chosen as they mark specific haplotypes that segregated with SCID phenotype in prior litters [10]. The causative mutations and the 2 haplotypes (h12 and h16) that carry the mutations are described by Waide et al. [10].
Table 2.
Primers and PCR conditions used to identify immune-deficient piglets by molecular genotype
| SNP name | Allele | Product size | Forward primer sequence | Reverse primer sequence | T, °C | Extension time, s |
|---|---|---|---|---|---|---|
| ASGA- | A | 177 | TCCTCTGACCAAGCCTCTGT | TCGTCCATGTACCAGAGCCT | 56 | 30 |
| 0048074 | C | 177 | TCCTCTGACCAAGCCTCTGT | CGTCCATGTACCAGAGCCG | 56 | 30 |
| ASGA- | A | 659 | AACCAGTCCCTGACCAACTG | TCCATATTTGTTAAGGGCAGTAATCT | 54 | 30 |
| 0048114 | G | 131 | TGCTCAGAGCTTTACATGGATTTAG | GGCCCATGTTGACATAAAGC | 54 | 30 |
| ALGA- | A | 643 | TCCTCTGCAGGGTTTCAAAG | CAGGGTGTGGGACTTTGTT | 54 | 30 |
| 0059043 | G | 127 | TCAGCTTGGGCAGCTAGG | CCACAGGCACATTGATCTTG | 54 | 30 |
| H3GA- | A | 576 | AGTTGAAATCAAAGTATCCCAA | AACTGTAACAAGCGTCCCTTTCT | 55 | 30 |
| 0030245 | C | 576 | AGTTGAAATCAAAGTATCCCAA | AACTGTAACAAGCGTCCCTTTCG | 55 | 30 |
| ALGA- | A | 593 | GGTATTCTCCTCCTCTACCTCT | CTGGATTGGCAGAGGCTCTTTAT | 55 | 30 |
| 059061 | G | 593 | GGTATTCTCCTCCTCTACCTCT | CTGGATTGGCAGAGGCTCTTTAC | 55 | 30 |
| ALGA- | A | 475 | GAATGGGAGGTGAGTAAGTAAA | CCAGCTGCAAGGGAGACT | 55 | 30 |
| 0059066 | C | 475 | GAATGGGAGGTGAGTAAGTAAA | CCAGCTGCAAGGGAGACG | 55 | 30 |
| ALGA- | A | 425 | AGCATTAAGACTGTGTGTGTGT | GGTCAAAGTCGTGGGTGTGTTT | 55 | 30 |
| 0059080 | G | 425 | AGCATTAAGACTGTGTGTGTGT | GGTCAAAGTCGTGGGTGTGTTC | 55 | 30 |
| ASGA- | ASGA- | ALGA- | H3GA- | ALGA- | ALGA- | ALGA- | |
|---|---|---|---|---|---|---|---|
| 0048074 | 0048114 | 0059043 | 0030245 | 059061 | 0059066 | 0059080 | |
| Location | 51153137 | 51812252 | 51975024 | 52066694 | 52086867 | 52109172 | 52174549 |
| hl2 | C | G | G | C | A | C | A |
| h16 | C | A | G | C | A | C | A |
| Normal | A | G | A | A | G | A | G |
The bottom section of the table shows the locations and genotypes of SNPs for chromosomes that carry normal, h12, and h16 haplotypes, as described previously [10]. T, annealing temperature.
Peripheral Lymphocyte Enumeration by Flow Cytometry
Blood was collected by venipuncture into an EDTA-treated vacutainer tube before (0 dpi) and after challenge (7 dpi) to determine the number of lymphocytes in circulation using a single-tube whole blood staining technique as previously described [28], with minor modifications. Briefly, 50 μL of whole blood was stained with a pan-lymphocyte antibody (PG106A; Washington State University, Pullman, WA, USA) and a fluorochrome-conjugated secondary antibody specific to mouse IgM. For enumeration, 50 μL of beads (106 beads/mL, Spherotech, Lake Forest, IL, USA) was added and 1 mL of FACSLyse (BD Biosciences, Franklin Lakes, NJ, USA) to lyse red blood cells and fix leukocytes. Data were acquired using an LSRII with FACSDiva software (BD Biosciences, Franklin Lakes, NJ, USA) and analyzed using FlowJo software.
Viral Titers in Lungs and Nasal Swabs
Nasal swabs (Nylon Minitip Flocked Dry Swabs; Copan Diagnostics, Murrieta, CA, USA) taken at 0, 1, 3, 5, and 7 dpi were filtered (0.45-mm syringe tip filters; Millipore, Billerica, MA, USA) and plated onto 24-well plates with confluent phosphate-buffered saline-washed MDCK cells for virus isolation, as previously described [29]. For viral titration, 10-fold serial dilutions of virus isolation-positive nasal swabs and BALF samples in serum-free MEM supplemented with 1 μg/mL tolylsulfonyl phenylalanyl chloromethyl ketone-trypsin and antibiotics were plated in triplicate onto phosphate-buffered saline-washed confluent MDCK cells in 96-well plates. The plates were fixed with 4% phosphate-buffered formalin after 48 h of incubation and stained using an anti-IAV nucleoprotein monoclonal antibody and TCID50/mL titers were calculated for each sample as previously described [29].
Pathological Examination of the Trachea and Lungs
The percentage of the lung affected with macroscopic lesions typical of IAV infection was evaluated at necropsy, with the percentage of the affected surface calculated based on weighted proportions for each lobe as previously described [30]. Fixed trachea and lung tissue samples were routinely processed and stained with hematoxylin and eosin for histopathologic examination. A veterinary pathologist blinded to the treatment groups evaluated and scored microscopic lesions according to previously described parameters [30]. Individual categorical scores were totaled into a composite score for lung and trachea microscopic lesions. Individual scores were also obtained for IAV-specific antigen immunohistochemical (IHC) staining in lung and trachea tissues, as previously described [30].
IAV Antibody Detection
Serum samples were collected by venipuncture before (0 dpi) and after challenge (7 dpi) and used to detect IAV-specific systemic antibodies. An enzyme-linked immunosorbent assay (ELISA; AI MultiS-Screen Ab Test, IDEXX, Westbrook, ME, USA) was used to detect anti-IAV nucleoprotein antibodies in serum according to the manufacturer's recommendation with modifications to include a suspect threshold. The presence or absence of antibodies was calculated based on the sample to negative (S/N) ratio, and the means of each treatment group were compared. For use in hemagglutination inhibition (HI) assays, sera were heat inactivated at 56°C for 30 min, treated with a 20% suspension of kaolin (Sigma-Aldrich, St. Louis, MO, USA), and adsorbed with 0.5% turkey red blood cells to remove nonspecific hemagglutinin inhibitors and natural serum agglutinins. HI assays were performed with H1N1pdm09 as the antigen and 0.5% turkey red blood cells using standard techniques [31].
Cytokine Assays
Upon collection, an aliquot of BALF was centrifuged at 400 g for 10 min to pellet cells and debris. The cell-free BALF was used to determine the concentration of IFN-α, IFN-γ, IL-2, IL-1β, IL-6, IL-8, TNF-α, IL-10, and IL-4 by multiplex ELISA (Aushon Biosystems, Billerica, MA, USA). Samples were analyzed in duplicate and averaged concentrations were used for statistical analysis.
Statistical Analysis
Statistically significant differences were identified by 1-way fixed-effects analysis of variance for each individual parameter: macroscopic lesions, composite microscopic lung and trachea scores, log10-transformed virus titers, mean OD for ELISA, and cytokine levels, with p ≤ 0.05 considered significant using Prism 6.0 (GraphPad, La Jolla, CA, USA). Variables shown to have significant effects were subjected to pairwise mean comparisons using the Bonferroni test and p values were corrected for multiple comparisons.
Results
SCID Phenotype Confirmed by Enumerating Circulating Lymphocytes
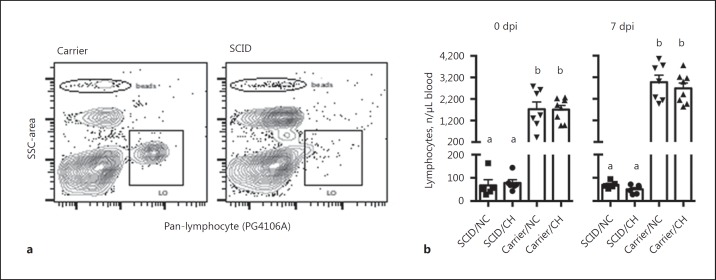
Eleven pigs were shown by DNA testing to have the SNP genotype that predicts a SCID phenotype, and 15 were classified as carriers. The number of lymphocytes in circulation on the day of challenge and at necropsy (dpi 7) was enumerated using flow cytometry to confirm the SCID phenotype in the pigs with a predicted SCID genotype, and the results are shown in Figure 1. Pigs with the SCID genotype displayed typical flow cytometric plots, with means of less than 100 stained lymphocytes per microliter compared to means over 1,000 lymphocytes per microliter in the carriers. These remaining lymphocytes are likely to be primarily NK cells, which have been shown to circulate in these SCID pigs [10, 11].
Fig. 1.
Enumeration of peripheral lymphocytes by flow cytometry confirmed the SCID phenotype. Peripheral blood was collected and the number of circulating lymphocytes labeled with a pan-lymphocyte antibody (clone PG106A) as described in the Materials and Methods. a Representative flow plots of lymphocyte populations (PG106A+) in carrier and SCID pigs. Beads were used to enumerate the number of lymphocytes (LO). b The number of lymphocytes in the indicated groups on the day of challenge (0 dpi) and necropsy (7 dpi). Each individual symbol indicates the value for a pig in that respective treatment group, with mean and SEM also indicated. Bars with different letters indicate statistical differences (p ≤ 0.05). CH, challenged; NC, nonchallenged control.
Virus Replication Was Not Controlled in SCID Pigs
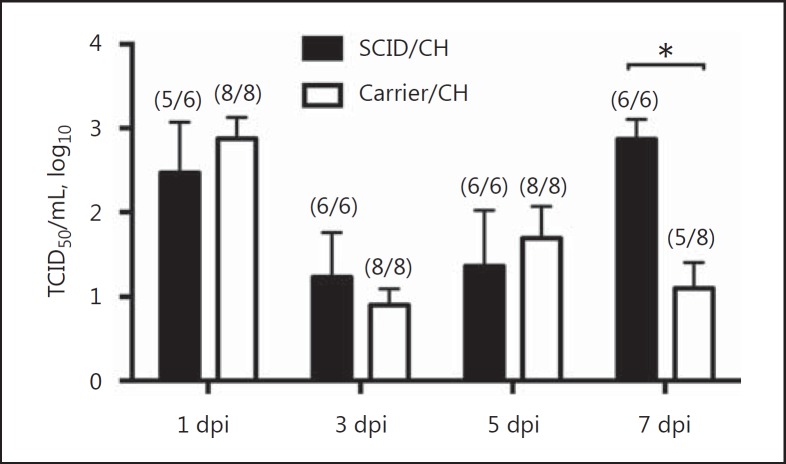
Nasal swabs were collected at 0, 3, 5, and 7 dpi to evaluate viral shedding, and the nonchallenged pigs were negative throughout the study. Virus shedding was similar between SCID and carrier challenged groups until 5 dpi, however SCID pigs continued to shed with significantly higher titers at 7 dpi when compared to carrier pigs (p < 0.0001; Fig. 2).
Fig. 2.
Virus titers in nasal swabs at 1, 3, 5, and 7 dpi of pigs with SCID or carrier control pigs challenged (CH) with A/California/04/2009. The numbers of infected pigs/total number of pigs are indicated in parentheses. Results are shown as means and SEM of log10 titers. Significant differences (* p ≤ 0.05) within each dpi are indicated.
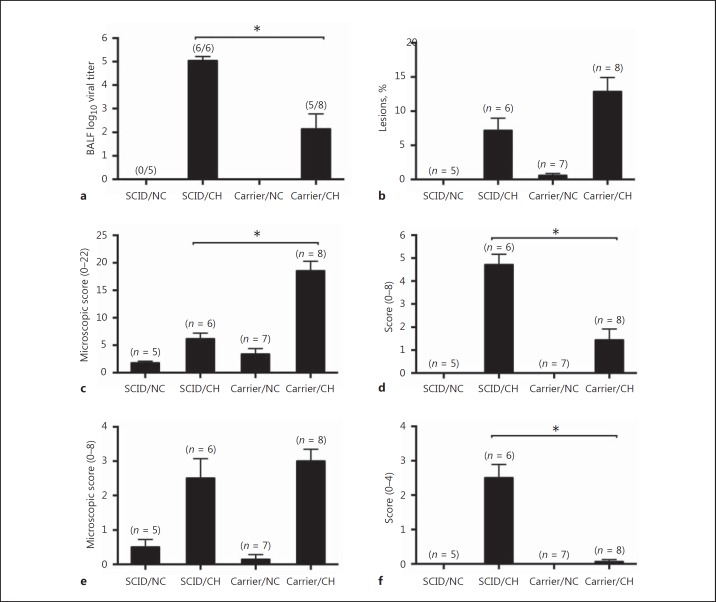
At 7 dpi, virus titers were determined in BALF. IAV was not isolated from the BALF of any nonchallenged pigs. Virus was detected in BALF of 6/6 and 5/8 challenged SCID and carrier pigs, respectively; however, the viral titers in BALF from challenged SCID pigs were significantly higher than those from carrier pigs (p = 0.0001; Fig. 3a), similar to the 7 dpi nasal swab virus titers.
Fig. 3.
Influenza effects on the lower respiratory tract differ in SCID pigs. Virus titers in BALF at 7 dpi (a), macroscopic pneumonia (b), lung microscopic pathology (c), IAV antigen detection scores in lungs (d), trachea microscopic pathology (e), and IAV antigen detection scores in trachea (f) in pigs with SCID or carrier control pigs challenged (CH) with A/California/04/2009, and nonchallenged controls (NC). The numbers of pigs per group or numbers of infected pigs/total number of pigs are indicated in parentheses. The results are shown as means and SEM. Significant differences (* p ≤ 0.05) between challenged groups are indicated.
Clinical Disease and Lung Pathology
Challenge with H1N1pdm09 resulted in mild apparent clinical signs in both SCID and carrier pigs during the 7-day challenge period. Body temperatures were not significantly different between challenged and nonchallenged groups (data not shown), with an exception of lower temperatures observed for carrier/CH pigs compared to nonchallenged and SCID/CH pigs at 5 dpi, perhaps related to a difference in inflammatory response.
Macroscopic lung lesions and microscopic lung and trachea lesions were evaluated at 7 dpi. SCID and carrier pigs challenged with H1N1pdm09 displayed moderate pathologic changes characterized by cranioventral purple-colored consolidation typical of IAV infection. Of note, SCID pigs challenged with H1N1pdm09 displayed a significantly lower percentage of macroscopic lung lesions than carrier/CH pigs (p = 0.04; Fig. 3b). Following the same trend, microscopic pneumonia scores were significantly lower for SCID/CH pigs compared to challenged carrier pigs (p < 0.0001; Fig. 3c), with SCID/CH pigs having only mild necrotizing bronchiolitis and minimal peribronchiolar lymphocytic cuffing. However, trachea microscopic scores were similar for the 2 challenged groups (Fig. 3e).
In contrast to the lung pathology findings, detection of IAV-specific antigen in lungs and trachea by IHC was significantly higher in challenged SCID pigs compared to carrier pigs (p < 0.0001; Fig. 3d, f). The IHC scores were consistent with virus titers in BALF and nasal shedding at 7 dpi, as more virus was detected in challenged SCID pigs compared to challenged carrier pigs.
Local Cytokine Response following IAV Challenge
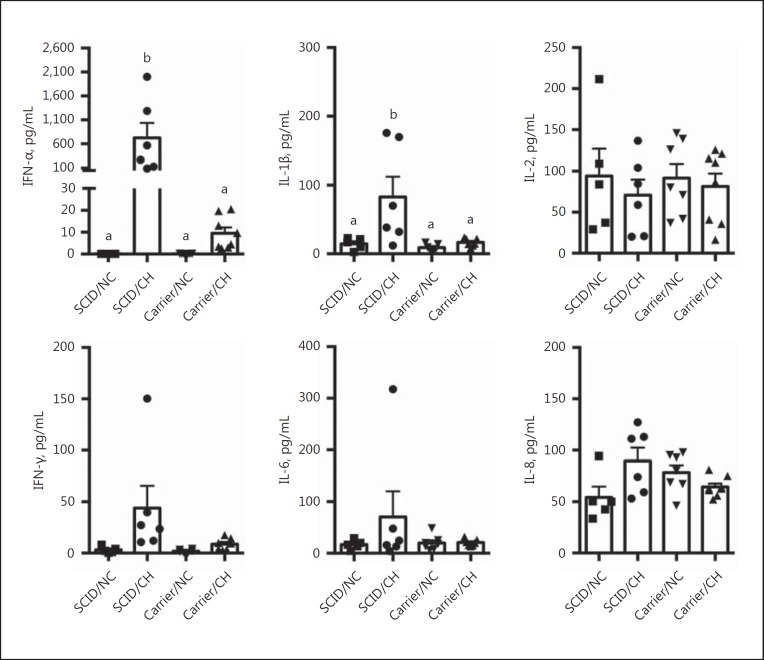
To assess differences in the local immune response following IAV infection between SCID and carrier pigs, the levels of 9 cytokines in cell-free lung lavage collected at necropsy (dpi 7) were measured (Fig. 4). Cytokines IL-4, TNF-α, and IL-10 were below the limit of detection (data not shown). There was a significant increase in the amount of IFN-α and IL-1β detected in the lungs of SCID pigs challenged with IAV when compared to levels in the lungs of challenged carrier pigs (p < 0.05). The average amount of IFN-α in the lavage collected from the SCID/CH group was 724.9 pg/mL, compared to 9.6 pg/mL in the carrier/CH group, and average levels of IL-1β were 83 pg/mL compared to 16.6 pg/mL for the same respective groups. While there was a trend for increased levels of IFN-γ and IL-6 in the lung lavage collected from challenged SCID pigs, these levels were not statistically different from those of challenged carrier pigs. The levels of IL-2 and IL-8 in the lungs were also not statistically different between the 2 challenged groups.
Fig. 4.
Levels of IFN-α, IL-1β, IL-2, IFN-γ, IL-6, and IL-8 in the lungs of pigs with SCID or carrier control pigs challenged (CH) with A/California/04/2009, and nonchallenged controls (NC). Lung lavage was collected on day 7 following challenge, and the amount of each indicated cytokine was measured by multiplex ELISA. Each individual symbol indicates the value for a pig in that respective treatment group, with the mean and SEM also indicated. Bars with different letters indicate statistical differences (p ≤ 0.05).
Induction of IAV-Specific Antibody Response in Carrier but Not SCID Pigs
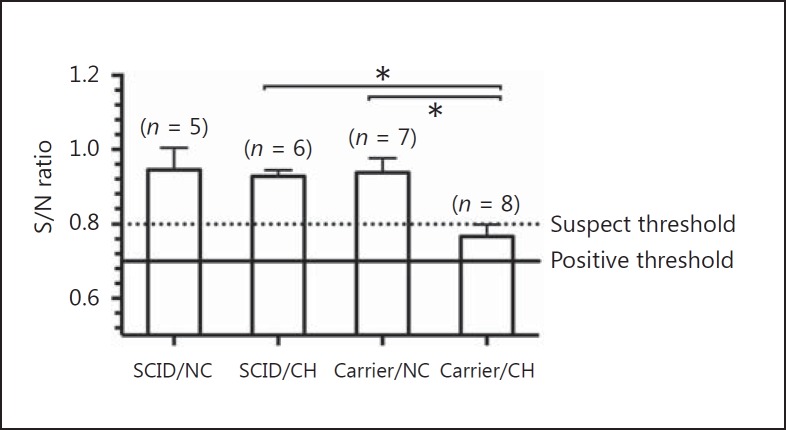
Serum anti-IAV nucleoprotein antibodies were not detected in challenged SCID pigs. Challenged carrier pigs showed significantly greater levels of detectable nucleoprotein antibodies at 7 dpi compared to the nonchallenged controls and challenged SCID pigs (p < 0.0001; Fig. 5). Although the group average was within the suspect range by test standards, 5/8 animals were considered positive or suspects, indicating early specific antibody responses. HI antibody titers were below the detection limit for all pigs (data not shown).
Fig. 5.
Serum antibody detection at 7 dpi in pigs with SCID or carrier control pigs challenged (CH) with A/California/04/2009, and nonchallenged controls (NC) by blocking ELISA. Values are shown as mean S/N ratios and SEM, with lower S/N ratios indicative of the presence of IAV-specific antibodies. The cut-off value for a negative sample is indicated by the dotted line, and the maximum value for a sample to be considered positive is indicated by the solid line (positive threshold) with a suspect range between the two. The number of pigs per group is indicated in parentheses. Significant differences (* p ≤ 0.05) between groups are indicated.
Discussion
Swine influenza is typically an acute, self-limiting respiratory disease that affects animals of all ages and has a short course of infection [17], resembling the clinical onset of IAV infection in humans. Both innate and adaptive immunity are important for the effective protection against influenza virus infection, resolution of clinical signs, and viral clearance [18]. The same immune response factors necessary for elimination of IAV and recovery can also initiate or exacerbate tissue damage as a result of dysregulated immune responses [32]. Here, we assessed the course of IAV infection and the immune response in SCID pigs in which B and T cells were absent or in very low numbers, but NK cell numbers and their in vitro function were unaffected [11, 33]. Our results show that the lack of adaptive immune response in the SCID pigs resulted in delayed viral clearance from the lungs and prolonged viral shedding, despite clinical signs being milder than in carrier controls.
The SCID pigs used here were deficient in B and T cells but retained NK cells and, therefore, did not have the common cytokine receptor γ-chain (γc) deficiency that results in X-linked SCID, the most common form of SCID in humans (T-B+NK-) [34]. Genetic causes of SCID are heterogeneous, and X-linked SCID is estimated to constitute 17% of SCID occurrences, while T-B-NK+ cases (primarily caused by Artemis and RAG-1/2 mutations) make up approximately 11% of SCID patients worldwide [35]. Immunocompromised patients, such as individuals affected with SCID or who received bone marrow or organ transplants, are at increased risk of developing severe complications after influenza infection and often show greater mortality and prolonged viral shedding [23, 24]. Mouse models of SCID are primarily based on RAG1/2 knockouts, which result in a similar immune system disfunction as the SCID pigs used here. Thus, these pigs provide an additional model to study the immune response mechanisms involved in the protection and/or resolution of IAV infection, particularly in regards to the adaptive compartment of the immune system. Because swine are natural hosts for IAV, are genetically and anatomically similar to humans [36], and also have an immune genome component that is very similar to humans [37], these SCID pigs represent an improved model to the use of SCID mice for evaluating potential therapies and preventive strategies for human influenza [38].
The recruitment of NK cells to the lungs of aerosol-infected mice was shown to have a minimal role in controlling IAV infection in SCID mouse models, while specific T cells were shown to mediate recovery from infection with a lethal dose of IAV [39]. Our results suggest that innate immunity may not be sufficient to control IAV infection in pigs. Despite the presence of some peribronchiolar lymphocytic cuffing in the SCID pigs, likely NK cells, and previous reports of the involvement of these cells in early response to influenza in swine [40], the SCID pigs showed a trend for increasing virus titers at 7 dpi, when carrier pigs were beginning to clear infection, underscoring the critical role of the adaptive immune response in IAV clearance. The innate immune response may work efficiently to control viral replication in the early stages of influenza infection, but the adaptive immune response is required at later stages to clear the virus and resolve the infection, as shown in RAG2 knockout mice that were deficient in both B and T cells but still produced NK cells [21]. We recently demonstrated that NK cells isolated from healthy SCID pigs are intrinsically functional, as they could be activated by IL-2, or a combination of IL-12 and IL-18, to express higher levels of perforin and to phagocytose human cancer cells [33]. However, it is not known if NK cells in the Artemis SCID pig model are functional in vivo, which could explain why implanted human cells are not eliminated in these SCID pigs [9].
Although the SCID pigs consistently showed minimal numbers of circulating B and T lymphocytes [11], levels of IL-2 in their lungs were similar to those of carrier control pigs, which suggests another cell type could be the source of this cytokine. Dendritic cells have been associated with the production of IL-2 in response to stimulation by microbes [41, 42]. Furthermore, epithelial cells of the human airways were also identified as potential sources of IL-2 after exposure to inflammatory stimuli [43].
Interestingly, an inverse relationship between virus load and pathology was observed in the infected SCID pigs at 7 dpi. Infected SCID pigs demonstrated minimal cellular infiltrates through the microscopic evaluation of lungs, suggesting that lymphocyte infiltration and adaptive immunity are involved in the immunopathology of influenza. Lung lesions in SCID pigs were milder than in carrier controls and less pronounced inflammation and peribronchiolar cuffing was observed (data included in composite microscopic scores), which could be ascribed to the lack of T cells producing cytotoxic factors in the SCID pigs. In murine models, reduced numbers of CD8+ T cells were associated with attenuation of immune-mediated influenza pneumonia [44], while memory CD4+ T cell response to secondary exposure was associated with lung immunopathology [45]. Because T lymphocytes are essential for IAV clearance from the infected host [46], the SCID pigs were not expected to efficiently eliminate the virus. While healthy carrier pigs had begun to decrease virus titers by 7 dpi, viral titers in SCID pigs remained significantly higher than carriers and increased over previous time points, with no sign of recovery. Type I interferons, including IFN-α, are important for activating innate antiviral mechanisms involved in responses to IAV infection [47]. In SCID pigs there was more IFN-α in the lungs at 7 dpi compared to carrier pigs, and while IFN-α-mediated responses may have contributed to controlling IAV replication, it was not as effective as having an intact adaptive immune system (carrier pigs).
Using a recently identified line of naturally occurring SCID pigs, we demonstrated prolonged viral replication and milder pathology upon influenza infection in the absence of an adaptive B and T cell immune response. Future studies are important to evaluate whether IAV would be eventually cleared if the SCID pigs were followed for a longer time. This SCID pig model could prove beneficial to identify minimal immune factors essential to control IAV infection, using partial immune cell or antibody replacement treatments and to understand the immune mechanisms associated with influenza protection and recovery. Additionally, they may provide a valuable model to study therapeutics for immunodeficient patients infected with influenza.
Disclosure Statement
USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Acknowledgements
We thank Michelle Harland, Gwen Nordholm, Bruce Pesch, and Zahra Olson for technical assistance, Jason Huegel, Jason Crabtree, Tyler Standley, Jennifer Young, and farm staff for assistance with the animals, and Dr. Susan Brockmeier for assisting with bacterial screening. This study was supported by USDA-ARS, USDA-APHIS, NIH grant 1R24OD019813-01, and the following entities at Iowa State University: the Presidential Initiative for Interdisciplinary Research Project on Vaccines against Antigenically Variable Viruses, the Research Foundation, and the Office of Vice President for Research and Economic Development. E.H.W. is a fellow supported by USDA NIFA National Needs grant No. 2010-38420-20328. D.S.R. was a CNPq-Brazil fellowship recipient.
References
- 1.Perryman LE. Molecular pathology of severe combined immunodeficiency in mice, horses, and dogs. Vet Pathol. 2004;41:95–100. doi: 10.1354/vp.41-2-95. [DOI] [PubMed] [Google Scholar]
- 2.Bauer TR, Jr, Adler RL, Hickstein DD. Potential large animal models for gene therapy of human genetic diseases of immune and blood cell systems. ILAR J. 2009;50:168–186. doi: 10.1093/ilar.50.2.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki S, Iwamoto M, Saito Y, Fuchimoto D, Sembon S, Suzuki M, Mikawa S, Hashimoto M, Aoki Y, Najima Y, Takagi S, Suzuki N, Suzuki E, Kubo M, Mimuro J, Kashiwakura Y, Madoiwa S, Sakata Y, Perry AC, Ishikawa F, Onishi A. Il2rg gene-targeted severe combined immunodeficiency pigs. Cell Stem Cell. 2012;10:753–758. doi: 10.1016/j.stem.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe M, Nakano K, Matsunari H, Matsuda T, Maehara M, Kanai T, Kobayashi M, Matsumura Y, Sakai R, Kuramoto M, Hayashida G, Asano Y, Takayanagi S, Arai Y, Umeyama K, Nagaya M, Hanazono Y, Nagashima H. Generation of interleukin-2 receptor gamma gene knockout pigs from somatic cells genetically modified by zinc finger nuclease-encoding mRNA. PLoS One. 2013;8:e76478. doi: 10.1371/journal.pone.0076478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee K, Kwon DN, Ezashi T, Choi YJ, Park C, Ericsson AC, Brown AN, Samuel MS, Park KW, Walters EM, Kim DY, Kim JH, Franklin CL, Murphy CN, Roberts RM, Prather RS, Kim JH. Engraftment of human iPS cells and allogeneic porcine cells into pigs with inactivated RAG2 and accompanying severe combined immunodeficiency. Proc Natl Acad Sci USA. 2014;111:7260–7265. doi: 10.1073/pnas.1406376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, Guo X, Fan N, Song J, Zhao B, Ouyang Z, Liu Z, Zhao Y, Yan Q, Yi X, Schambach A, Frampton J, Esteban MA, Yang D, Yang H, Lai L. RAG1/2 knockout pigs with severe combined immunodeficiency. J Immunol. 2014;193:1496–1503. doi: 10.4049/jimmunol.1400915. [DOI] [PubMed] [Google Scholar]
- 7.Cai W, Casey DS, Dekkers JC. Selection response and genetic parameters for residual feed intake in Yorkshire swine. J Anim Sci. 2008;86:287–298. doi: 10.2527/jas.2007-0396. [DOI] [PubMed] [Google Scholar]
- 8.Ozuna AG, Rowland RR, Nietfeld JC, Kerrigan MA, Dekkers JC, Wyatt CR. Preliminary findings of a previously unrecognized porcine primary immunodeficiency disorder. Vet Pathol. 2013;50:144–146. doi: 10.1177/0300985812457790. [DOI] [PubMed] [Google Scholar]
- 9.Basel MT, Balivada S, Beck AP, Kerrigan MA, Pyle MM, Dekkers JC, Wyatt CR, Rowland RR, Anderson DE, Bossmann SH, Troyer DL. Human xenografts are not rejected in a naturally occurring immunodeficient porcine line: a human tumor model in pigs. Biores Open Access. 2012;1:63–68. doi: 10.1089/biores.2012.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waide EH, Dekkers JC, Ross JW, Rowland RR, Wyatt CR, Ewen CL, Evans AB, Thekkoot DM, Boddicker NJ, Serao NV, Ellinwood NM, Tuggle CK. Not all SCID pigs are created equally: two independent mutations in the Artemis gene cause SCID in pigs. J Immunol. 2015;195:3171–3179. doi: 10.4049/jimmunol.1501132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewen CL, Cino-Ozuna AG, He H, Kerrigan MA, Dekkers JC, Tuggle CK, Rowland RR, Wyatt CR. Analysis of blood leukocytes in a naturally occurring immunodeficiency of pigs shows the defect is localized to B and T cells. Vet Immunol Immunopathol. 2014;162:174–179. doi: 10.1016/j.vetimm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz K, Ma Y, Pannicke U, Lieber MR. Human severe combined immune deficiency and DNA repair. Bioessays. 2003;25:1061–1070. doi: 10.1002/bies.10344. [DOI] [PubMed] [Google Scholar]
- 14.Volk T, Pannicke U, Reisli I, Bulashevska A, Ritter J, Bjorkman A, Schaffer AA, Fliegauf M, Sayar EH, Salzer U, Fisch P, Pfeifer D, Di Virgilio M, Cao H, Yang F, Zimmermann K, Keles S, Caliskaner Z, Guner SU, Schindler D, Hammarstrom L, Rizzi M, Hummel M, Pan-Hammarstrom Q, Schwarz K, Grimbacher B. DCLRE1C (ARTEMIS) mutations causing phenotypes ranging from atypical severe combined immunodeficiency to mere antibody deficiency. Hum Mol Genet. 2015;24:7361–7372. doi: 10.1093/hmg/ddv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palese P, Shaw M, Orthomyxoviridae: The viruses and their replication . Fields Virology. In: Knipe DM, Howley PM, editors. vol 2. Philadelphia: Lippincott, Williams & Wilkins; 2007. pp. pp 1647–1690. [Google Scholar]
- 16.Yoon S-W, Webby R, Webster R, Evolution and ecology of influenza A viruses . Influenza Pathogenesis and Control. In: Compans RW, Oldstone MBA, editors. vol 1. Cham: Springer International; 2014. pp. pp 359–375. [DOI] [PubMed] [Google Scholar]
- 17.Janke BH. Clinicopathological features of Swine influenza. In: Richt JA, Webby RJ, Swine Influenza, editors. vol 370. Berlin: Springer; 2013. pp. pp 69–83. [Google Scholar]
- 18.Kreijtz JH, Fouchier RA, Rimmelzwaan GF. Immune responses to influenza virus infection. Virus Res. 2011;162:19–30. doi: 10.1016/j.virusres.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 19.Larsen DL, Karasin A, Zuckermann F, Olsen CW. Systemic and mucosal immune responses to H1N1 influenza virus infection in pigs. Vet Microbiol. 2000;74:117–131. doi: 10.1016/s0378-1135(00)00172-3. [DOI] [PubMed] [Google Scholar]
- 20.Bot A, Isobe H, Bona C. Immunodeficient mouse models in the characterization of the protective immunity to influenza virus. Folia Microbiol (Praha) 1998;43:477–478. doi: 10.1007/BF02820795. [DOI] [PubMed] [Google Scholar]
- 21.Wu H, Haist V, Baumgartner W, Schughart K. Sustained viral load and late death in Rag2−/− mice after influenza A virus infection. Virol J. 2010;7:172. doi: 10.1186/1743-422X-7-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staeheli P, Grob R, Meier E, Sutcliffe JG, Haller O. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol Cell Biol. 1988;8:4518–4523. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klimov AI, Rocha E, Hayden FG, Shult PA, Roumillat LF, Cox NJ. Prolonged shedding of amantadine-resistant influenzae A viruses by immunodeficient patients: detection by polymerase chain reaction-restriction analysis. J Infect Dis. 1995;172:1352–1355. doi: 10.1093/infdis/172.5.1352. [DOI] [PubMed] [Google Scholar]
- 24.Kunisaki KM, Janoff EN. Influenza in immunosuppressed populations: a review of infection frequency, morbidity, mortality, and vaccine responses. Lancet Infect Dis. 2009;9:493–504. doi: 10.1016/S1473-3099(09)70175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson MI, Stratton J, Killian ML, Janas-Martindale A, Vincent AL. Continual re-introduction of human pandemic H1N1 influenza A viruses into US swine, 2009–2014. J Virol. 2015;89:6218–6226. doi: 10.1128/JVI.00459-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheth AN, Althoff KN, Brooks JT. Influenza susceptibility, severity, and shedding in HIV-infected adults: a review of the literature. Clin Infect Dis. 2011;52:219–227. doi: 10.1093/cid/ciq110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wangkumhang P, Chaichoompu K, Ngamphiw C, Ruangrit U, Chanprasert J, Assawamakin A, Tongsima S. WASP: a web-based allele-specific PCR assay designing tool for detecting SNPs and mutations. BMC Genomics. 2007;8:275. doi: 10.1186/1471-2164-8-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loving CL, Lager KM, Vincent AL, Brockmeier SL, Gauger PC, Anderson TK, Kitikoon P, Perez DR, Kehrli ME., Jr Efficacy of inactivated and live-attenuated influenza virus vaccines in pigs against infection and transmission of emerging H3N2 similar to the 2011–2012 H3N2v. J Virol. 2013;87:9895–9903. doi: 10.1128/JVI.01038-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincent AL, Ma W, Lager KM, Richt JA, Janke BH, Sandbulte MR, Gauger PC, Loving CL, Webby RJ, Garcia-Sastre A. Live attenuated influenza vaccine provides superior protection from heterologous infection in pigs with maternal antibodies without inducing vaccine-associated enhanced respiratory disease. J Virol. 2012;86:10597–10605. doi: 10.1128/JVI.01439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gauger PC, Vincent AL, Loving CL, Henningson JN, Lager KM, Janke BH, Kehrli ME, Jr, Roth JA. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Vet Pathol. 2012;49:900–912. doi: 10.1177/0300985812439724. [DOI] [PubMed] [Google Scholar]
- 31.Kitikoon P, Gauger PC, Vincent AL. Hemagglutinin inhibition assay with swine sera. In: Spackman E, Animal Influenza Virus, editor. New York: Springer; 2014. pp. pp 295–301. [DOI] [PubMed] [Google Scholar]
- 32.Damjanovic D, Small CL, Jeyanathan M, McCormick S, Xing Z. Immunopathology in influenza virus infection: uncoupling the friend from foe. Clin Immunol. 2012;144:57–69. doi: 10.1016/j.clim.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Powell EJ, Cunnick JE, Knetter SM, Loving CL, Waide EH, Dekkers JC, Tuggle CK. NK cells are intrinsically functional in pigs with severe combined immunodeficiency (SCID) caused by spontaneous mutations in the Artemis gene. Vet Immunol Immunopathol. 2016;175:1–6. doi: 10.1016/j.vetimm.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, McBride OW, Leonard WJ. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 35.Modell F, Puente D, Modell V. From genotype to phenotype: further studies measuring the impact of a Physician Education and Public Awareness Campaign on early diagnosis and management of primary immunodeficiencies. Immunol Res. 2009;44:132–149. doi: 10.1007/s12026-008-8092-3. [DOI] [PubMed] [Google Scholar]
- 36.Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol. 2012;20:50–57. doi: 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawson HD, Loveland JE, Pascal G, Gilbert JG, Uenishi H, Mann KM, et al. Structural and functional annotation of the porcine immunome. BMC Genomics. 2013;14:332. doi: 10.1186/1471-2164-14-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hohenadl C, Wodal W, Kerschbaum A, Fritz R, Howard MK, Farcet MR, Portsmouth D, McVey JK, Baker DA, Ehrlich HJ, Barrett PN, Kreil TR. Hyperimmune intravenous immunoglobulin containing high titers of pandemic H1N1 hemagglutinin and neuraminidase antibodies provides dose-dependent protection against lethal virus challenge in SCID mice. Virol J. 2014;11:70. doi: 10.1186/1743-422X-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bot A, Reichlin A, Isobe H, Bot S, Schulman J, Yokoyama WM, Bona CA. Cellular mechanisms involved in protection and recovery from influenza virus infection in immunodeficient mice. J Virol. 1996;70:5668–5672. doi: 10.1128/jvi.70.8.5668-5672.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forberg H, Hauge AG, Valheim M, Garcon F, Nunez A, Gerner W, Mair KH, Graham SP, Brookes SM, Storset AK. Early responses of natural killer cells in pigs experimentally infected with 2009 pandemic H1N1 influenza A virus. PLoS One. 2014;9:e100619. doi: 10.1371/journal.pone.0100619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granucci F, Vizzardelli C, Pavelka N, Feau S, Persico M, Virzi E, Rescigno M, Moro G, Ricciardi-Castagnoli P. Inducible IL-2 production by dendritic cells revealed by global gene expression analysis. Nat Immunol. 2001;2:882–888. doi: 10.1038/ni0901-882. [DOI] [PubMed] [Google Scholar]
- 42.Zelante T, Fric J, Wong AY, Ricciardi-Castagnoli P. Interleukin-2 production by dendritic cells and its immuno-regulatory functions. Front Immunol. 2012;3:161. doi: 10.3389/fimmu.2012.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aoki Y, Qiu D, Uyei A, Kao PN. Human airway epithelial cells express interleukin-2 in vitro. Am J Physiol. 1997;272:L276–L286. doi: 10.1152/ajplung.1997.272.2.L276. [DOI] [PubMed] [Google Scholar]
- 44.Sakthivel P, Gereke M, Breithaupt A, Fuchs D, Gigliotti L, Gruber AD, Dianzani U, Bruder D. Attenuation of immune-mediated influenza pneumonia by targeting the inducible co-stimulator (ICOS) molecule on T cells. PLoS One. 2014;9:e100970. doi: 10.1371/journal.pone.0100970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teijaro JR, Njau MN, Verhoeven D, Chandran S, Nadler SG, Hasday J, Farber DL. Costimulation modulation uncouples protection from immunopathology in memory T cell responses to influenza virus. J Immunol. 2009;182:6834–6843. doi: 10.4049/jimmunol.0803860. [DOI] [PubMed] [Google Scholar]
- 46.Thomas PG, Keating R, Hulse-Post DJ, Doherty PC. Cell-mediated protection in influenza infection. Emerg Infect Dis. 2006;12:48–54. doi: 10.3201/eid1201.051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haller O, Kochs G, Weber F. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 2007;18:425–433. doi: 10.1016/j.cytogfr.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]