Abstract
STAT3 is a master transcriptional regulator that plays an important role in the induction of both immune activation and immune tolerance in dendritic cells (DCs). The transcriptional targets of STAT3 in promoting DC activation are becoming increasingly understood; however, the mechanisms underpinning its role in causing DC suppression remain largely unknown. To determine the functional gene targets of STAT3, we compared the genome-wide binding of STAT3 using ChIP sequencing coupled with gene expression microarrays to determine STAT3-dependent gene regulation in DCs after histone deacetylase (HDAC) inhibition. HDAC inhibition boosted the ability of STAT3 to bind to distinct DNA targets and regulate gene expression. Among the top 500 STAT3 binding sites, the frequency of canonical motifs was significantly higher than that of noncanonical motifs. Functional analysis revealed that after treatment with an HDAC inhibitor, the upregulated STAT3 target genes were those that were primarily the negative regulators of proinflammatory cytokines and those in the IL-10 signaling pathway. The downregulated STAT3-dependent targets were those involved in immune effector processes and antigen processing/presentation. The expression and functional relevance of these genes were validated. Specifically, functional studies confirmed that the upregulation of IL-10Ra by STAT3 contributed to the suppressive function of DCs following HDAC inhibition.
Keywords: Acetylation, Dendritic cell, ChIP sequencing, Histone deacetylase inhibitor, STAT3
Introduction
Dendritic cells (DCs) play an important role not only in the activation of proinflammatory responses and activation of antigen-specific T cell responses, but also in the suppression of inflammation and the induction of T cell tolerance [1]. STAT3 is critical for the ability of DCs to promote immune activation, but emerging data demonstrate that STAT3 also plays an important role as a negative regulator of inflammatory responses [2, 3, 4] and in the induction of tolerance by DCs and macrophages [5]. Genetic data suggest that targeted disruption of the STAT3 gene induces an exaggerated inflammatory response, further demonstrating the role of STAT3 in suppressing inflammatory responses by DCs and macrophages [5, 6]. However, the molecular mechanism of STAT3 in causing mutually distinct responses, activation or suppression of DCs remains unknown.
Exposure of DCs to stimuli that cause immune activation, such as LPS or inflammatory cytokines, causes tyrosine phosphorylation, dimerization and activation of STAT3, which lead to the expression of STAT3-responsive immune activation genes. However, studies have also reported phosphorylation-independent regulation of STAT3 dimerization [7, 8] and target gene expression [9, 10, 11]. Acetylation (the addition of a functional acetyl group) of proteins is an important posttranslational modification (PTM). HDACs remove the acetyl groups from lysine residues. HDAC inhibitors function as antitumor agents by inhibiting the activity of HDAC, and they exhibit anti-inflammatory effects at low doses. Emerging data have identified reversible acetylation by the inhibition of HDACs as another modulator of STAT3 dimerization [12, 13]. CBP/p300 has been reported to be a transcriptional coactivator that regulates STAT3 activity in vivo [14]. Upon cytokine treatment, CBP/p300 adds acetyl groups to amino acids to acetylate STAT3 on a single amino acid residue, lysine 685, thereby enhancing STAT3 DNA binding, transactivation activity and nuclear localization [12, 13]. However, it is not clear whether acetylation of STAT3 modulates the expression of STAT3-responsive genes.
Studies have also demonstrated that HDAC inhibitors exert potent anti-inflammatory or immunomodulatory effects at noncytotoxic doses [15, 16, 17, 18, 19, 20]. Using immunomodulatory approaches, allogeneic bone marrow (BM) transplantation in several experimental models of graft-versus-host-disease has shown potential therapeutic benefits [17, 21, 22, 23]. These effects result, at least in part, from the ability of HDAC inhibitors to suppress the function of host antigen-presenting cells, which include DCs, by enhancing the STAT3-dependent expression of the indoleamine 2,3-dioxygenase 1 gene (Ido1) [21, 23, 24]. In addition to regulating Ido1 expression, whether STAT3 acetylation by HDAC inhibition negatively regulates DC function by orchestrating a broader transcriptional cascade via a unique genome-wide functional network remains unknown.
Here, we describe a systematic experimental/computational approach to explore the STAT3-regulated genes that are responsible for the negative regulation of DC function following HDAC inhibition. We determined STAT3-dependent gene targets by analyzing STAT3 binding to the entire genome following HDAC inhibition of DCs using ChIP (chromatin immunoprecipitation) coupled with next-generation sequencing (ChIP-seq) [25]. Specific STAT3 binding profiles at associated gene loci across the genome were obtained using ChIP-seq, and gene expression profiles were obtained using Affymetrix microarrays following HDAC inhibition. Genome-wide STAT3 binding coupled with gene expression profiling revealed that, following HDAC inhibition, STAT3 either positively or negatively regulates the transcription of target genes that underlie important pathways that are responsible for the negative regulation of DC function. Specifically, we demonstrate that the induction of STAT3 acetylation by HDAC inhibition regulates target genes by upregulating genes that are primarily involved in the negative regulation of cytokine production and IL-10 signaling or by downregulating genes that are primarily involved in immune effector processes and antigen processing/presentation. Furthermore, the upregulation of IL-10Ra by STAT3 contributed to the negative regulation of DC function following HDAC inhibition.
Materials and Methods
BMDC Differentiation, Purification and Treatment
To obtain BM-derived DCs (BMDCs), BM cells isolated from the tibia and fibula of C57BL/6 (B6) or BALB/c female mice, aged 8–12 weeks, were cultured with murine recombinant GM-CSF (10 ng/ml; BD Biosciences Pharmingen) and IL-4 (10 ng/ml; PeproTech) for 7 days, harvested and positively selected for CD11c cells with 95% purity using an autoMACS Pro Separator (Miltenyi Biotec) as previously described [21, 24]. Purified BMDCs were treated with SAHA (500 nm, 12 h), IL-10 (100 ng/ml, 12 h), IL-6 (100 ng/ml, 6 h), IL-10Ra antibody (10 μg/ml, 12 h), rat IgG (10 μg/ml, 12 h), 1-MT (1-Methyl-D-tryptophan; 200 μm, 12 h) or LPS (500 ng/ml, 6 h).
ChIP and High-Throughput ChIP-Seq Analysis
Chromatin for ChIP was prepared from BMDCs (107 cells each) treated with SAHA (500 nm/ml) or diluent for 12 h by fixing the BMDCs in 1% formaldehyde for 10 min, followed by quenching with 125 mM of glycine for 5 min. ChIP was performed using antibodies against STAT3 (catalog No. 9132; Cell Signaling), H3K4me3 (catalog No. 9751; Cell Signaling) and a matched IgG control (catalog No. 2729; Cell Signaling). Chromatin (approx. 200 ng of DNA) was sonicated into 200- to 500-bp fragments. DNA ends were repaired with polynucleotide kinase and Klenow enzyme and treated with Taq polymerase for the generation of a protruding 3′ ‘A’ nucleotide for adaptor ligation. Libraries were prepared with the Illumina ChIP-seq Sample Prep Kit and sequenced using a genome analyzer (Illumina) following the manufacturer's protocols [25, 26, 27]. The sequencing raw data were aligned using BWA (Burrows-Wheeler Aligner; http://bio-bwa.sourceforge.net/) mapping software version 0.5.8a [28]. The enriched ‘peaks’ were called using the Model-Based Analysis of ChIP-Seq (MACS) algorithm (http://liulab.dfci.harvard.edu/MACS) [29]. Called peaks were edited in the BED format and input into GREAT (Genomic Regions Enrichment of Annotations Tool, version 1.8; http://bejerano.stanford.edu/great/public/html/) [30] for defining the binding sites in noncoding genomic regions by analyzing the annotations of the nearby genes and functional prediction. ChIP-seq reads were mapped to the most recent mouse genome (mm9) using IGV (Integrative Genomics Viewer; https://www.broadinstitute.org/igv/v1.3.1) [31]. The ChIP-seq data reported here can be found in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) with the series accession ID GSE74262.
ChIP Assay
Chromatin for ChIP was prepared using antibodies against acetyl (685)-STAT3 (catalog No. 2523; Cell Signaling) and matched IgG control. A tenth of the sheared chromatin was saved and used as a positive control as previously described [21, 24]. The fold enrichment at each site was determined based on normalized values in the acetyl (685)-STAT3-IP samples versus IgG-IP samples for both SAHA- and diluent-treated samples. qPCR was conducted using an Eppendorf thermocycler (Reallex2). The primer sequences are listed in online supplementary table S1-A (for all online suppl. material, see www.karger.com/doi/10.1159/000450681).
Motif Analysis
We utilized the STAT3 motif from the TRANSFAC database (http://transfac.gbf.de) [32] to study the difference in binding signal strength between the SAHA- and diluent-treated samples. We matched the canonical STAT3 motif (TTCnnnGAA) at 200-bp flanking regions of the peak summits of the top 500 peaks ranked by p value in SAHA-treated BMDCs (GSE74262) in comparison to the relevant STAT3 binding sites in diluent-treated samples using a binomial test. We further analyzed the distributions of the canonical and noncanonical motif consensuses of approximately 1,000-bp flanking regions of the peak summits. The canonical motifs and single-nucleotide variant noncanonical motifs were analyzed around the peak summit regions of SAHA-induced STAT3 binding sites in comparison to STAT3 binding sites in diluent-treated samples. We extracted the peak sequence from the reference genome and matched for the consensus motif. The position of each motif sequence in relation to the summit position of SAHA-induced STAT3 binding sites was collected, and the distribution across all binding sites was plotted against those for STAT3 binding in untreated samples.
Affymetrix Microarrays and GSEA Analyses
Mouse BMDCs were treated with SAHA or diluent for 12 h; tcRNA was extracted with TRIzol reagent and cleaned over Qiagen RNeasy columns. After the quality of the total RNA was verified by an Agilent 2100 Bioanalyzer, the samples were processed using the WT-Ovation™ Pico System (Affymetrix), and a single round of amplification was performed on samples with even stricter concentration restraints. This system incorporates oligo(dT) and random primers for amplification at the 3′ end and throughout the entire transcriptome. Affymetrix mouse genome 430 2.0 arrays (Affymetrix, Santa Clara, Calif., USA), which contain 45,000 transcripts of annotated genes and expressed sequence tags, were used. The stained arrays were scanned on an Agilent gene array scanner (Affymetrix) with a 560-nm filter. The data were published and analyzed using the R statistical environment (http://cran.r-project.org/) provided by Bioconductor (http://www.bioconductor.org/), and the data were examined for quality control by showing the same distribution of the PM probes for each chip and no degradation. The expression values were then calculated for each gene using a robust multiarray average. This modeling strategy converts the PM probe values into an expression value (log2-transformed) for each gene. The probe sets with a fold change of 2 or greater were selected. These sets were subjected to an additional constraint in which 1 of the 2 samples had an enrichment value of 26 or greater to prevent the selection of genes with large fold changes based on 2 small numbers. The correlation of the ChIP-seq data with the microarray analysis of gene expression was performed using Gene Set Enrichment Analysis (GSEA) software version 2.2 (http://www.broadinstitute.org/gsea/index.jsp) [26]. The microarray data were deposited in the GEO database (GSE74306).
PCR
Total RNA was isolated from BMDCs treated with SAHA, IL-10, IL-10Ra antibody or diluent using TRIzol reagent. Briefly, 2 μg of total RNA was reverse-transcribed into cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) in the presence of random hexamers. All of the reactions were performed in triplicate with SYBR Green Master Mix (Applied Biosystems) and 25 ng of both the forward and reverse primers according to the manufacturer's recommended thermocycling conditions, and the reactions were subjected to a melt curve analysis. The threshold levels for each experiment were set during the exponential phase of the reaction. The DNA in each sample was quantified by interpolation of its threshold cycle (Ct) value from a standard curve of Ct values. The calculated quantity of the target gene for each sample was divided by the average sample quantity of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to obtain the relative gene expression. All of the oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, Iowa, USA). The primer sequences are listed in online suppl. table S1-A.
Western Blot Analysis
BMDCs were treated with SAHA, IL-10, IL-6 or LPS. Cell lysates were prepared with lysis buffer containing proteinase inhibitors (Pierce) and PhosSTOP (Roche). One hundred micrograms of protein was resolved on 10% SDS-PAGE gels, transferred to polyvinylidene fluoride membranes, detected with specific antibodies against acetyl (685)-STAT3 (catalog No. 2523; Cell Signaling), phospho-STAT3 (Tyr705) (catalog No. 9131; Cell Signaling), STAT3 (catalog No. 9139; Cell Signaling), IL-10Ra (catalog No. 06-1067; Millipore) and β-actin (catalog No. 8226; Amcom), probed with secondary antibodies (either HRP-conjugated goat anti-mouse IgG or goat anti-rabbit IgG antibodies), and detected by chemiluminescence followed by autoradiography.
Mixed Lymphocyte Cultures
Splenocytes from BALB/c mice were processed for T cell isolation by negative selection (Pan T cell Isolation Kit II; Miltenyi Biotec). BMDCs were obtained as described above and treated with or without SAHA, followed by treatment with or without IL-10, IL-10Ra antibody or IgG control. The BMDCs were then washed with culture medium. BALB/c T cells were cultured with BALB/c BMDCs or treated B6 BMDCs at a ratio of 40:1 (T cells vs. DCs) for 96 h using 96-well flat-bottomed plates (Falcon Labware, Lincoln Park, N.J., USA). Supernatants were collected for the measurement of cytokine concentrations by ELISA. Proliferation was determined by incubating cells with 3H-thymidine (1 μ Ci/well [0.037 MBq]) for the last 16 h, and proliferation was calculated using a 1205 Betaplate reader (Wallac, Turku, Finland).
ELISA
The concentrations of IFN-γ and IL-6 were measured by ELISA with specific anti-mouse mAbs for capture and detection, and the appropriate standards were purchased from BD Biosciences Pharmingen. The assays were performed according to the manufacturer's protocol and read at 450 nm with subtraction of the absorbance at 570 nm using a microplate reader (Bio-Rad).
Statistical Analyses
Data were analyzed using Prism GraphPad version 6. Comparisons between 2 groups were calculated using a t test. A p value <0.05 was considered statistically significant.
Results
Genome-Wide STAT3 Binding Regions following HDAC Inhibition
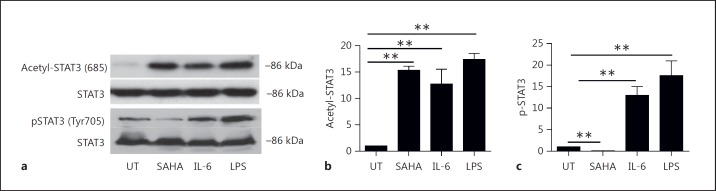
STAT3 activation can be triggered by phosphorylation following cytokine stimulation or upon its acetylation induced by p300 following HDAC inhibition [11, 12, 13, 24]. Previous observations have demonstrated that proinflammatory cytokines induce STAT3 phosphorylation and activate DCs [2, 11, 12, 13], whereas HDAC inhibition-induced STAT3 acetylation suppresses DCs [23, 24]. To first confirm these observations, we treated BMDCs with SAHA, a clinically used HDAC inhibitor [15, 16, 17, 18, 19, 20, 21, 33] that is known to suppress DC responses, or with IL-6 or LPS, which are known to activate DCs. We observed that HDAC inhibition induced STAT3 acetylation at lysine 685 (fig. 1a, b) but inhibited its basal phosphorylation (compared to diluent-treated samples; fig. 1a, c). These results demonstrated that HDAC inhibition posttranslationally modifies STAT3 primarily by promoting acetylation. By contrast, we also observed that both IL-6 and LPS induced STAT3 phosphorylation at tyrosine 705 and enhanced acetylation at lysine 685 (fig. 1). Our results confirmed previous observations and further suggest that the primary PTM of STAT3 following DC suppression by HDAC inhibition is acetylation, whereas both phosphorylation and acetylation PTMs of STAT3 are induced in DCs by the activating stimuli IL-6 and LPS. These results also suggest that STAT3 might bind to different gene targets following its activation by HDAC inhibition, and that characterization of the unique binding regions by acetyl-STAT3 in the DC genome following HDAC inhibition is a potentially feasible approach.
Fig. 1.
HDAC inhibition induces STAT3 acetylation in DCs. a A representative Western blot image showing acetylation and phosphorylation of STAT3 in BMDCs which were treated with SAHA, IL-6 or LPS, and processed for immunoblotting with antibodies against STAT3, pSTAT3 (T705) and acetyl (685)-STAT3. b Densities of acetyl-STAT3 from 3 similar experiments were measured based on comparison with untreated samples (mean ± SEM); ** p < 0.001. p values were obtained using an unpaired t test. c Densities of phosphorylated STAT3 from 3 similar experiments were measured based on comparison with untreated samples (mean ± SEM); ** p < 0.001. p values were obtained using an unpaired t test.
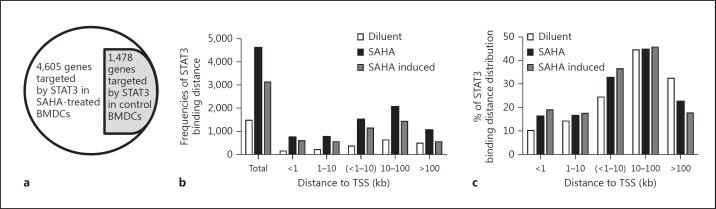
Therefore, we next sought to determine STAT3 genomic binding sites following its acetylation upon HDAC inhibition. To this end, we performed ChIP using a STAT3 antibody because there is no suitable acetyl-STAT3 antibody that carries multiple signals for all possible acetyl-lysines in STAT3 [12, 13, 34] and because the STAT3 antibody binds to all the genome sites, which are likely to be enriched for STAT3 but not specific for acetyl-685 STAT3 following HDAC inhibition (fig. 1). We utilized a matched IgG as a control for both SAHA- or diluent-treated BMDCs. We coupled ChIP with high-throughput sequencing (ChIP-seq). To additionally monitor the possible impact of HDAC inhibition on protein-chromatin association, we performed a similar ChIP-seq using an H3K4me3 antibody with both SAHA- and diluent-treated BMDCs. The sequences were aligned using BWA mapping software version 0.5.8a [28]. The enriched ‘peaks’ were called using the MACS algorithm [29]. To rigorously define high-stringency ChIP-seq binding sites, STAT3 binding peaks were called at a threshold of p < 0.0001 with a fragment size of approximately 200 bp. Using this standard, 3,598 binding peaks were obtained with SAHA-treated BMDCs, and 976 binding peaks were obtained with control BMDCs (GSE74262). Because gene expression can be regulated by transcription factors binding at a broad range of distances, even extending farther than 100 kb upstream or downstream of the transcription start site (TSS) [26, 27], we next utilized the GREAT tool for analysis [30], which considers the nonrandom distribution of the genome and is specifically suited for the genome-wide analysis of ChIP-seq data. Additionally, it can interpret the functional properties of genes that are associated with genome-wide binding sites. The gene search range was set in the genome from 5 kb upstream of the TSS to 1 kb downstream of the TSS, with a 1,000-kb max extension using GREAT. These parameters identified 4,605 STAT3-targeted genes (online suppl. table S1-B; fig. 2a) from 3,598 binding peaks (online suppl. table S1-C) in SAHA-treated BMDCs, whereas 1,478 STAT3-targeted genes (online suppl. table S1-D; fig. 2a) were identified from 976 binding peaks (online suppl. table S1-E) in control BMDCs (referred as basal STAT3 binding). Interestingly, all 1,478 STAT3-targeted genes in the control BMDCs were targeted in SAHA-treated BMDCs, thus showing a 100% overlap of the 1,478 basal STAT3 target genes in diluent- and SAHA-treated BMDCs (fig. 2a). Importantly, enhanced STAT3 binding following HDAC inhibition in SAHA-treated BMDCs identified 3,127 additional STAT3-targeted genes from 2,620 SAHA-induced STAT3 binding regions (online suppl. table S1-B-E; fig. 2a; online suppl. fig. S1). These data demonstrated that SAHA treatment-induced STAT3 acetylation increased the binding of this protein to target DNA sites in the genome, suggesting that STAT3 acetylation might play an important role in the regulation of BMDC function via newly targeted genes.
Fig. 2.
Genome-wide binding and distribution of STAT3 following HDAC inhibition in DCs. a Diagram of the gene numbers targeted by STAT3 in BMDCs treated with SAHA or diluent control. 4,605 genes were targeted by STAT3 in SAHA-treated BMDCs compared to 1,478 genes targeted in BMDCs treated with diluent. b Distribution frequencies of STAT3 binding sites classified by distance from the target gene TSS. c Percent of STAT3 binding sites at different distances from the target gene TSS.
Distribution of STAT3 Binding Site Distances from the TSS in Targeted Gene Loci
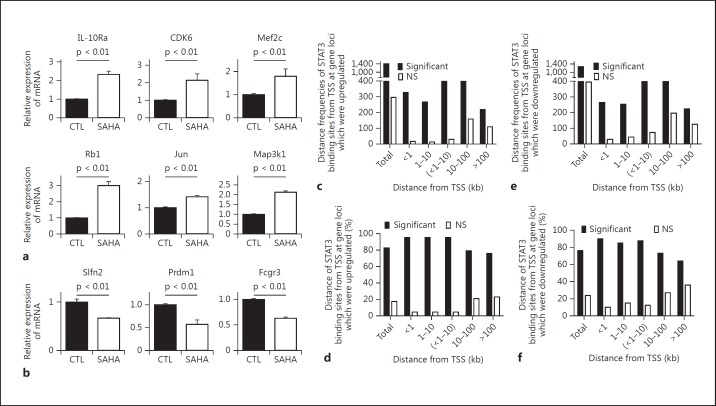
Next, to examine the distribution of STAT3 binding sites at STAT3-regulated gene loci, we extracted STAT3 binding distances from the TSSs of target genes using GREAT analyses (online suppl. table S1-B, S1-C) and classified them as <1, 1–10 (or together <1–10), 10–100 or >100 kb. The relative proportions of STAT3 binding sites within <1, 1–10 or <1–10 kb of the TSS were 16.2, 16.5 or 32.7%, respectively, in SAHA-treated BMDCs and 10.2, 14.2 or 24.4%, respectively, in control BMDCs. The corresponding proportions of SAHA-induced newly targeted gene loci were 19, 17.5 or 36.5%, respectively, indicating that more proximal STAT3 binding events were induced by HDAC inhibition. For STAT3 binding sites within 10–100 kb, similar distribution proportions were observed among SAHA-treated BMDCs (44.7%), control BMDCs (44.5%), and SAHA-induced newly targeted gene loci (45.7%; online suppl. fig. S1; fig. 2b, c). The proportions of STAT3 binding sites greater than 100 kb from TSSs were 22.6% in SAHA-treated BMDC, 32.4% in control BMDCs and 17.7% in SAHA-induced newly targeted gene loci, indicating that fewer STAT3 binding sites at distances greater than 100 kb were induced by HDAC inhibition (online suppl. fig. S1; fig. 2b, c). These observations demonstrated that SAHA-induced STAT3 binding is generally more proximal than distal to the TSS of target genes.
STAT3 Binding Induced by HDAC Inhibition Is Transcriptionally Functional
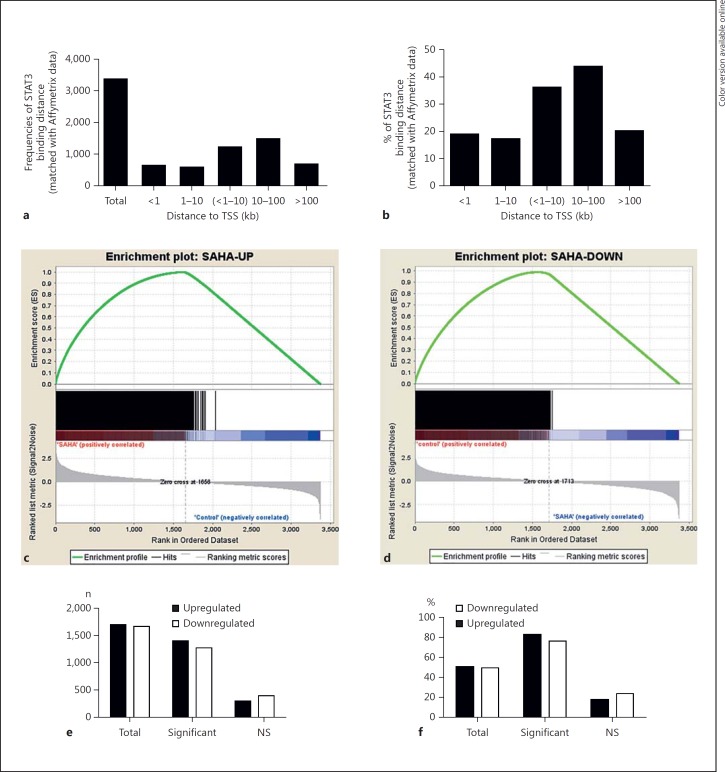
To assess the genome-wide functional correlation between STAT3 binding and the associated gene expression, we used gene expression microarrays to determine the gene expression profiles of acetyl-STAT3-targeted genes that were identified by GREAT analysis. A total of 3,369 genes matched to Affymetrix microarray chips among the 4,605 genes identified by GREAT, and a distance analysis reflected a similar distribution pattern of STAT3 binding distances from the TSSs of matched genes, thus facilitating an additional functional analysis using GSEA [26] (table 1; fig. 3a, b). Among the 3,369 matched genes, 1,701 genes were upregulated (50.5%, 1,701/3,369), and 1,668 genes were downregulated (49.5%, 1,668/3,369; fig. 3c, d). Notably, 1,405 genes (82.6%, 1,405/1,701) were significantly upregulated (fig. 3c; table 1; online suppl. table S1-F), and 1,273 genes (76.3%, 1,273/1,668) were significantly downregulated (fig. 3d; table 1; online suppl. table S1-G). These observations suggested that, following HDAC inhibition, STAT3 plays an important role in both the positive and negative transcriptional regulation of gene expression.
Table 1.
GSEA analyses of the impact of STAT3 binding distances on the target gene expression
| Total target genes (n = 3,369) | <1 kb | 1 – 10 kb | <1 – 10 kb | 10 – 100 kb | Over 100 kb | |
|---|---|---|---|---|---|---|
| Upregulated | 1,701 (50.5) | 343 (20.2) | 280 (16.5) | 623 (36.7) | 751 (44.1) | 327 (19.2) |
| Significant | 1,405 (82.6) | 327 (95.3) | 267 (95.3) | 594 (95.3) | 593 (79) | 218 (76) |
| Not significant | 296 (17.4) | 16 (4.7) | 13 (4.7) | 29 (4.7) | 158 (21) | 109 (23) |
| Downregulated | 1,668 (49.5) | 294 (17.6) | 298 (17.9) | 592 (35.5) | 727 (43.7) | 349 (21) |
| Significant | 1,273 (76.3) | 265 (90) | 254 (85) | 519 (87.7) | 531 (73) | 223 (64) |
| Not significant | 395 (23.7) | 29 (10) | 44 (15) | 73 (12.3) | 196 (27) | 126 (36) |
Values in parentheses are percentages.
Fig. 3.
STAT3 bindings induced by HDAC inhibition are transcriptionally functional. a Distribution frequencies (numbers) of genes targeted by STAT3, matched with Affymetrix expression microarrays and classified by the distance to the target gene TSS. b Percent of genes targeted by STAT3, matched with Affymetrix expression microarrays and calculated based on distances from the target gene TSS. c The expression of STAT3 target genes, which were upregulated, was analyzed with GSEA. Genes were ranked by enrichment score (ES). d The expression of STAT3 target genes, which were downregulated, was analyzed with GSEA. Genes were ranked by ES. e Numbers of the genes which were upregulated or downregulated, significant or not significant (NS). f Percentages of genes which were upregulated or downregulated, significant or not.
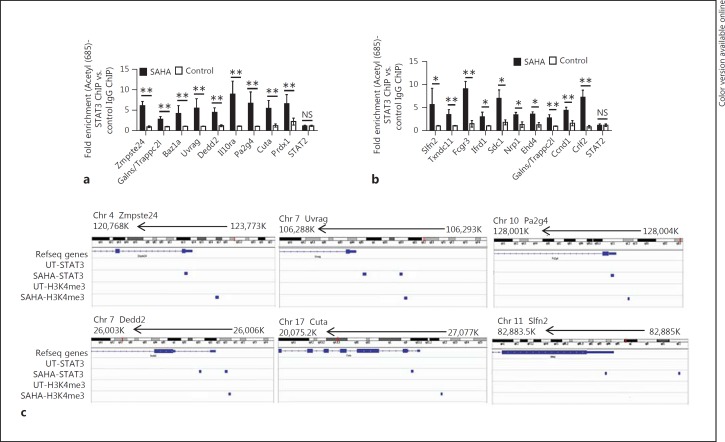
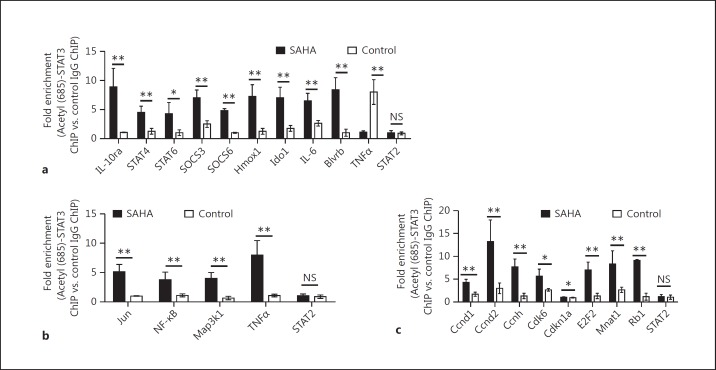
It has been suggested that the acetylation of lysine 685 in STAT3 is required for STAT3-dependent [12, 13, 34] and STAT3 phosphorylation-independent gene expression [9, 10]. To independently validate whether acetyl (685)-STAT3 binds to target gene loci, we used either an anti-acetyl (685)-STAT3 antibody or matched IgG control for ChIP assays following the treatment of BMDCs with SAHA or diluent. We assessed acetyl-STAT3 binding to a random selection of 18 STAT3 binding peaks that were associated with upregulated genes, including Zmpste24, Galns/Trappc2l, Baz1a, Uvrag, Dedd2, Il10Ra, Pa2g4, Cuta, and Prdx1 (fig. 4a), downregulated genes, including Slfn2, Txndc11, Fcgr3, Ifrd1, Sdc1, Nrp1, Ehd4, Ccnd1, and Crlf2 (fig. 4b), and a non-STAT3-occupied region, a STAT2 promoter fragment, to verify STAT3 binding specificity. Our results demonstrated the veracity and specificity of acetyl-STAT3 binding following HDAC inhibition to the gene loci that are associated with either upregulated (fig. 4a) or downregulated genes (fig. 4b), but no gene enrichment was detected in the non-STAT3 occupied regions (fig. 4a, b).
Fig. 4.
Validation of enrichments of acetyl (685)-STAT3 following HDAC inhibition at the loci of genes with either up or downregulated expression. a, b Chromatin for ChIP was prepared from SAHA or diluent-treated BMDSCs using antibodies against acetyl (685)-STAT3 and matched IgG control. Fold enrichment at each site was determined based on normalized values in the manner of acetyl (685)-STAT3-IP samples versus IgG-IP samples. Acetyl-STAT3 bindings to a random selection of 18 STAT3 binding peaks that were associated with upregulated (a) or with downregulated genes (b) were assessed by ChIP-qPCR. Data were from 3 independent experiments (mean ± SEM). p values were obtained using an unpaired t test. * p < 0.05, ** p < 0.01. NS = Not significant. c The enrichments of STAT3 and H3K4me3 at gene loci, including Zmpste24, Uvrag and Pa2g4, were shown by IGV.
H3K4me3 is a chromatin modification that highlights the start sites of active genes [35, 36, 37]. Therefore, to monitor whether HDAC inhibition is likely to impact protein-chromatin association in BMDCs, we investigated H3K4m3 binding in the genome of SAHA- or diluent-treated BMDCs using IGV [31]. IGV is a high-performance visualization tool for the interactive exploration of large, integrated genomic datasets including next-generation sequencing data that allows for the examination of whether and how STAT3 and H3K4m3 are enriched in these targeted gene loci. We observed that STAT3 was enriched in targeted gene loci including Zmpste24, Uvrag, Pa2g4, Dedd2, Cuta, and Slfn2 (fig. 4c), and that H3K4m3 was enriched at different distances from the TSS in transcriptionally upregulated gene loci, such as Zmpste24, Uvrag, Pa2g4, Dedd2, and Cuta, but not in downregulated gene loci, such as Slfn2 (fig. 4c). These data indicate that upon HDAC inhibitor treatment of DCs, the enrichment of H3K4me3 is concomitant with STAT3-mediated transcription activation but not with STAT3-mediated transcription inactivation. These data are consistent with and further expand previous reports [35, 36, 37] and suggest that SAHA treatment does not overtly disturb protein-chromatin association.
Functional Correlation between STAT3 Binding Distances from the TSS and Transcriptional Activity after HDAC Inhibition
Next, to validate the functional relevance of STAT3 acetylation, we verified the expression levels of genes that were either upregulated or downregulated following SAHA treatment in BMDCs. Utilizing qPCR, we observed that SAHA treatment not only increased the expression of genes, such as Il10Ra, Cdk6, Mef2c, Rb1, Map3k1, and Jun (fig. 5a), but also decreased the expression of other genes, such as Slfn2, Prdm1, and Fcgr3 (fig. 5b), in accord with the data obtained from the Affymetrix gene expression arrays (GSE74306).
Fig. 5.
Correlation of binding distance of STAT3 with gene expression. Confirmation with qPCR of a random selection of genes associated with STAT3 binding following HDAC inhibition which were upregulated (a) or downregulated (b). Data were combined from 3 independent experiments (mean ± SEM). p values were obtained using an unpaired t test. CTL = Control. c Frequencies (numbers) of distance distribution of STAT3 binding sites from TSS at the gene loci which were significantly or not significantly upregulated. d Percentages of STAT3 binding sites at different distances from TSS at gene loci which were significantly or not significantly upregulated by STAT3. e Frequencies of distances of STAT3 binding sites from TSS at the gene loci which were significantly or not significantly downregulated. f Percentages of STAT3 binding sites at different distances from TSS at gene loci which were significantly or not significantly downregulated by STAT3. NS = Not significant.
Next, because STAT3, similar to other transcription factors [26, 27, 30], bound at distances that ranged from proximal to distal with distances that could exceed 100 kb from the TSS, we examined whether the distance between a STAT3 binding site and its corresponding TSS affect the transcription capability. Using a GSEA analysis, we observed that the transcriptional activation of target genes driven by proximal STAT3 binding sites (<1 kb or 1–10 kb from the TSS) was stronger (with similar 95% significant upregulation in both distance groups) than that driven by distal STAT3 binding sites (the 10–100 and >100 kb groups showed 79 and 76% significant upregulation, respectively) (table 1; fig. 5c, d; online suppl. table S2). Similarly, we observed that the proximal STAT3 binding sites (<1 kb or 1–10 kb from the TSS) functioned more effectively on the negative regulation of target gene expression (90 and 85% significant downregulation, respectively) compared with distal STAT3 binding sites (the 10–100 kb and >100 kb groups showed 73 and 64% significant downregulation, respectively; table 1; fig. 5e, f; online suppl. table S3). These results suggested that the distance between a STAT3 binding site and its TSS can affect both the upregulation and downregulation of transcription, and regulation by proximal STAT3 binding sites is more effective than that by distal STAT3 binding sites.
STAT3 Binds to Both Its Canonical and Noncanonical Motifs after HDAC Inhibition
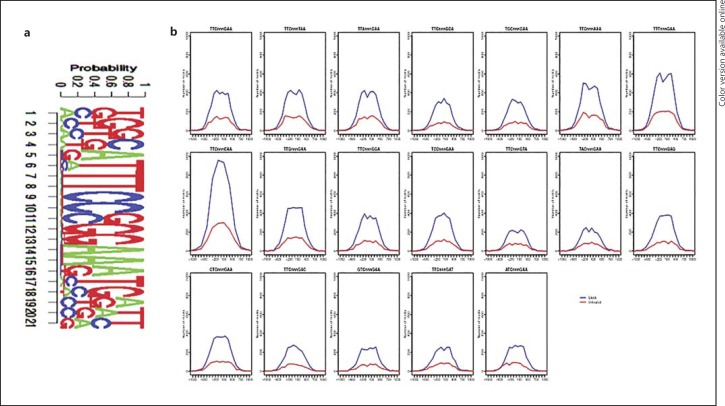
Following acetylation, STAT3 binds to palindromic sequences, such as TTCnnnGAA, termed GASs (IFN-γ activation sites), and these canonical motifs are broadly present throughout the genome [11]. Therefore, we next examined whether STAT3 bound to its canonical TTCnnnGAA motif upon treatment of BMDCs with SAHA. The STAT3 motif was defined using TRANSFAC [32]. To determine the strength of STAT3 binding signals in SAHA-treated and control BMDCs, we ranked the binding strength according to the p value determined by MACS analysis (GSE74262) and matched the canonical GAS motif to 200 bp flanking regions of the peak summits of the top 500 peaks. As a result, 349 canonical GAS binding motifs were identified in SAHA-treated BMDCs, compared with 87 binding sites identified in control BMDCs (p value 3.312e-60, binomial test; fig. 6a). These results indicated that the signals for STAT3 binding to canonical motifs in SAHA-treated BMDCs are significantly stronger than those in control BMDCs.
Fig. 6.
Characterization of STAT3 binding motifs following HDAC inhibition. a The top 500 STAT3 binding peaks were examined for the canonical GAS motif TTCnnnGAA using TRANSFAC. b STAT3 binding peaks were examined for canonical and noncanonical GAS motifs. Binding frequencies in SAHA-treated (blue line) and control BMDCS (red line) were shown.
However, many transcription factors, including STAT3, may also bind to variant motifs called noncanonical motifs [26, 27]. For example, IL-21-activated STAT3 binds to not only the canonical TTCnnnGAA sequence but also the variant GAS motif variant TTCnnnTAA [27]. Therefore, we examined whether acetyl-STAT3 binds to noncanonical GAS motifs with single nucleotide variants. We hypothesized that the positions of motifs bound by STAT3 should coincide with the maximum peak density. We investigated the distributions of the consensus sequences of canonical and all 18 noncanonical GAS motifs among 3,598 STAT3 binding peaks in SAHA-treated BMDCs and the 976 STAT3 binding peaks in control BMDCs. We extracted the peak sequences from the reference genome and matched for the motif consensus sequences, setting the search to a 1-kb range for each motif sequence associated with the summit position of a STAT3 binding site and ignoring all motifs further than 1 kb from the peak summit. All hits were collected, and the distribution across all binding sites in SAHA-treated BMDCs was plotted (in blue) against that of STAT3 binding sites in control BMDCs (in red; fig. 6b). We observed a higher overall binding strength in SAHA-treated BMDCs compared with control BMDCs based on the frequency of the motif sequences (fig. 6b). Although we observed similar distribution patterns for STAT3 binding to the canonical GAS motif and to the 18 noncanonical motifs in SAHA-induced STAT3 binding sites (blue lines; fig. 6b), the top 5 motifs, which were most frequent and centered, were TTTnnnGAA, TTCnnnCAA, TTGnnnGAA, TTCnnnAAA, and the canonical motif TTCnnnGAA (fig. 6b). However, only 7 motifs (the canonical TTCnnnGAA and the noncanonical TTCnnnTAA, TTAnnnGAA, TTCnnnAAA, TTTnnnGAA, TTCnnnCAA, and TTGnnnGAA) were adequately frequent and centered in the basal nonacetyl-STAT3 binding plot in control BMDCs (red lines; fig. 6b). These observations demonstrated that following HDAC inhibition STAT3 bound to both its canonical GAS motif and its noncanonical motifs.
HDAC-Induced Gene Expression Associated with STAT3 Binding Sites Is Critical for DC Function
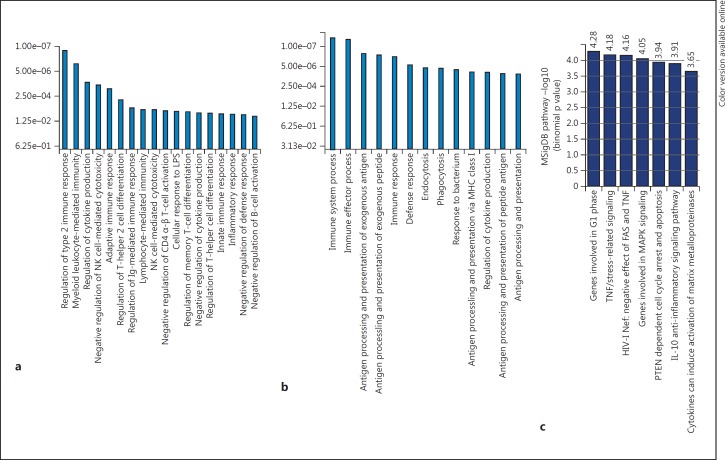
We next examined whether the genes regulated by SAHA-induced STAT3 binding are involved in the immune function of BMDCs using 2 different bioinformatics tools, namely Ingenuity Pathway Analysis (IPA; http://www.ingenuity.com/) [38] and the Molecular Signatures Database (MSigDB; www.broadinstitute.org/gsea/msigdb) [26, 30]. IPA, a broadly adopted functional analysis tool for accurately interpreting the biological meaning of genomic data, was used to analyze the functional pathways that depend on genes that were upregulated or downregulated by SAHA-induced acetyl-STAT3 binding. A GSEA analysis of 1,405 significantly upregulated genes (table 1; online suppl. table S1-F) and 1,273 significantly downregulated genes (table 1; online suppl. table S1-G) revealed a clear enrichment of key immune functions. The top functions identified in the biological process category based on the upregulated genes included the regulation of type 2 immune response/T helper type 2 cell differentiation and the negative regulation of cytokine production (fig. 7a). However, the top functions based on the downregulated genes were immune effector processes and antigen processing/presentation (fig. 7b). MSigDB, a collection of annotated gene sets used with GSEA and coupled with GREAT [30], was used to interpret the functional properties of the genes associated with the 3,598 STAT3 binding sites in SAHA-treated BMDCs and the 976 STAT3 binding sites in control-treated BMDCs. The analysis revealed a clear enrichment in 7 pathways, including IL-10 anti-inflammatory signaling, PTEN-dependent cell cycle arrest and apoptosis, TNF/stress-related signaling, MAPK signaling and the G1 phase of the cell cycle (fig. 7c). These results suggested that genome-wide binding of STAT3 following HDAC inhibition generates an important gene/function network that is critical for negative regulation of BMDC function.
Fig. 7.
STAT3 significantly binds to the loci of genes that are associated with pathways that negatively regulate immune response after HDAC inhibition. a Overrepresented GO terms resulting from 1,405 significantly upregulated genes with IPA show the regulation of type 2 immune response/Th2 cell differentiation and the negative regulation of cytokine production. b Overrepresented GO terms resulting from 1,273 significantly downregulated genes with IPA show immune effector processes and antigen processing/presentation. c Overrepresented PANTHER pathways (MSigDB) based on 3,598 STAT3 bindings in SAHA-treated BMDCs and 976 STAT3 bindings in control BMDCs, which were imported into GREAT, display IL-10 anti-inflammatory signaling, PTEN-dependent cell cycle arrest and apoptosis, TNF/stress-related signaling, genes involved in MAPK signaling and genes involved in the G1 phase of the cell cycle.
Direct Upregulation of IL-10Ra by STAT3 following HDAC Inhibition Promotes IL-10-Dependent Responses
Next, to confirm the preceding insights from bioinformatics analyses and to directly assess and validate whether HDAC inhibition-induced acetylation of STAT3 is relevant to the activation of gene loci involved in IL-10 responses, TNF/stress-related signaling and the G1 phase of the cell cycle, we performed ChIP assays. The anti-acetyl (685)-STAT3 antibody, matched IgG control, specific primers for binding peaks, and control primers for the non-STAT3 occupied region (STAT2 promoter) were utilized to verify acetyly-STAT3 binding specificity. As shown in figure 8, the results demonstrated specific binding of acetyl-STAT3 at identified gene loci, including those involved in IL-10 signaling (fig. 8a), TNF/stress-related signaling (fig. 8b) and the G1 phase of the cell cycle (fig. 8c).
Fig. 8.
Validation of enrichments for acetyl (685)-STAT3 binding at gene loci that are related to immune regulation pathways as predicted by MSigDB. a–c Chromatin for ChIP was prepared from SAHA or diluent-treated BMDSCs using antibodies against acetyl (685)-STAT3 and matched IgG control. Fold enrichment at each site was determined based on normalized values in the manner of acetyl (685)-STAT3-IP samples versus IgG-IP samples. a Confirmation of genes targeted by acetyl-STAT3 and involved in the IL-10 signaling pathway. b Confirmation of genes targeted by acetyl (685)-STAT3 and involved in the cell cycle G1 phase. c Confirmation of genes targeted by acetyl (685)-STAT3 and involved in TNF/stress-related signaling pathway. Combined results (mean ± SEM) were from at least 3 independent experiments. p values were obtained using an unpaired t test. ** p < 0.01, * p < 0.05. NS = Not significant.
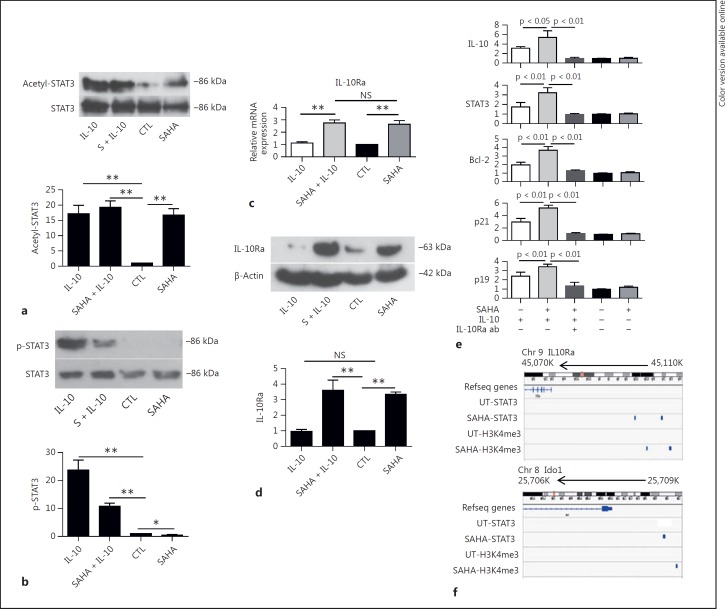
We next focused on defining the biological relevance of the IL-10 signaling pathway, which has potent immunoregulatory effects on both the maturation of DCs and their antigen-presenting function [39]. We observed that SAHA treatment induced STAT3 acetylation (fig. 9a), consistent with a previous report [24], whereas IL-10 treatment induced STAT3 phosphorylation and acetylation (fig. 9a, b), similar to the observations when BMDCs were treated with IL-6 and LPS (fig. 1). We observed and confirmed significant upregulation of IL-10Ra mRNA and protein in BMDCs following SAHA treatment (fig. 5a). By contrast, treatment with IL-10 did not alter the expression of IL-10Ra (fig. 9c, d). These observations suggest that IL-10Ra expression may be competently transcribed by STAT3 acetylation alone but not by phospho-STAT3 or a mixture of phospho-STAT3 and acetyl-STAT3. This finding prompted further examination of the role of IL-10 when IL-10Ra expression was increased by HDAC inhibition-induced STAT3 acetylation in the regulation of its transcriptional target genes, such as IL-10 [40], STAT3 [41], Bcl-2 [42], and p19 and p21 [43]. To this end, BMDCs were treated with or without SAHA, followed by incubation with IL-10, IL-10Ra blocking antibody or matched IgG control, and then harvested and processed for qPCR analysis. We observed that IL-10 induced the expression of its target genes, including IL-10, STAT3, Bcl-2, p19 and p21, in accord with previous reports. Importantly, pretreatment with SAHA increased the transcriptional expression of IL-10 target genes. However, blocking IL-10 signaling with IL-10Ra blocking antibodies abolished HDAC inhibition-induced increased expression of these genes (fig. 9e). These observations indicated that the increased expression of IL-10Ra by HDAC inhibition enhanced the IL-10 responses in BMDCs. These data also imply a potential synergy between STAT3 acetylation and IL-10 responses in BMDCs treated with the HDAC inhibitor SAHA.
Fig. 9.
Direct upregulation of IL-10Ra by STAT3 following HDAC inhibition promotes IL-10-dependent responses. a Acetylation of STAT3 was induced by SAHA and IL-10 treatment. BMDCs were treated with SAHA, IL-10 and SAHA plus IL-10, and processed for immunoblotting with antibodies against acetyl (685)-STAT3 and STAT3. Top panel: a representative Western blot image showing acetylation of STAT3. Lower panel: densities of acetyl-STAT3 from 3 similar experiments were measured based on comparison with untreated samples (mean ± SEM). ** p < 0.001; p values were obtained using an unpaired t test. b Phosphorylation of STAT3 was induced by IL-10 and inhibited by SAHA. BMDCs were treated as in a, and processed for immunoblotting with antibodies against pY705-STAT3 and SΤΑΤ3. Top panel: a representative Western blot image showing phosphorylation of STAT3. Lower panel: densities of phosphorylated STAT3 from 3 similar experiments were measured based on comparison with untreated samples (mean ± SEM). ** p < 0.001, * p < 0.05; p values were obtained using an unpaired t test. c Quantification of IL-10Ra mRNA expression by qPCR. Data were from 3 independent experiments (mean ± SEM). ** p < 0.01; p values were obtained using an unpaired t test. NS = Not significant. d Western blot with anti-IL-10Ra antibody showing protein expression of IL-10Ra. BMDCs were treated with SAHA, IL-10 or SAHA plus IL-10, and processed for immunoblotting with antibodies against IL-10Ra and β-actin. Top panel: a representative image of Western blot showing expression levels of IL-10Ra protein. Lower panel: densities of expression levels of IL-10Ra protein from 3 similar experiments were measured based on comparison with untreated samples (mean ± SEM). ** p < 0.001; p values were obtained using an unpaired t test. NS = Not significant; CTL = control. e Enhanced IL-10 signaling by SAHA treatment facilitated the expression of IL-10 transcriptional target genes. Quantification of mRNA expression of IL-10 transcriptional target genes was determined by qPCR. Combined results (mean ± SEM) were from 3 independent experiments. p values were obtained using an unpaired t test. f The enrichments of STAT3 and H3K4me3 following HDAC inhibition in IL-10Ra and Ido1 gene loci were shown by IGV.
Furthermore, IGV analysis revealed STAT3 binding peaks, along with H3K4me3 enrichment, at the IL-10Ra and Ido1 gene loci following SAHA treatment (fig. 9f). These data, when taken collectively with our previous observations that HDAC inhibition induces the expression of IDO in BMDCs or PBMCs [23, 24], demonstrate that STAT3 transcriptionally targets the IL-10Ra and Ido1 genes.
Enhanced IL-10 Responses following HDAC Inhibition Contribute to the Negative Regulation of DC Functions
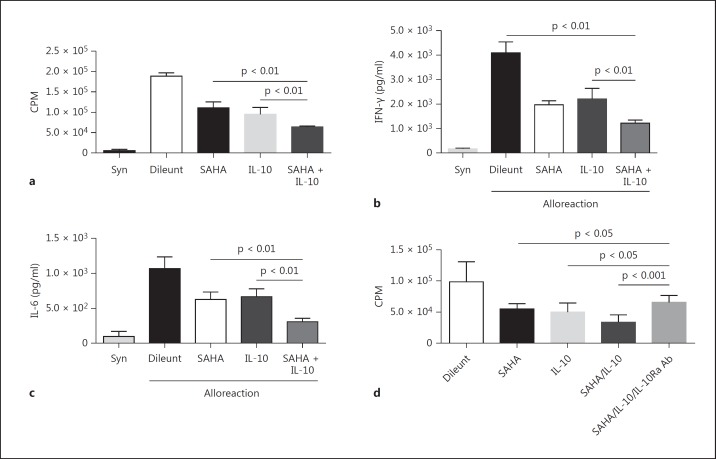
IL-10 acts directly on antigen-presenting cells, such as DCs, to negatively regulate T cell activation [39, 44, 45, 46, 47]. The above data demonstrate increased expression of STAT3-dependent IL-10Ra and the resultant responses to IL-10 upon treatment of DCs with the HDAC inhibitor SAHA. Therefore, we hypothesized that the increased responsiveness to IL-10 induced by HDAC inhibition plays a function role in the ability of DCs to affect T cell responses. To examine this hypothesis, we utilized well-established allogeneic mixed lymphocyte reactions. B6 BMDCs were treated with or without SAHA before incubation with or without IL-10. The DCs were then harvested and utilized as stimulators in a mixed lymphocyte reaction with allogeneic BALB/c T cells. Syngeneic BALB/c DCs were utilized as controls. As shown in figure 10, allogeneic T cell expansion and the expression of the cytokines IFN-γ and IL-6 were attenuated by treatment with either SAHA or IL-10, and these processes were further suppressed by combinatorial treatment with SAHA and IL-10 (fig. 10a-c). To further confirm a critical role for STAT3-mediated IL-10Ra expression in the negative regulation of BMDC function upon HDAC inhibitor treatment, IL-10Ra-blocking antibodies were utilized. Blocking IL-10 responses with the IL-10Ra antibodies significantly improved allogeneic T cell expansion (fig. 10d). These data demonstrate that the upregulation of IL-10Ra by STAT3 upon HDAC inhibition contributes to enhanced IL-10 responses and the negative regulation of DC function.
Fig. 10.
HDAC inhibition facilitates IL-10-dependent negative regulation of BMDC function. a Allogeneic T cell expansion was attenuated by treatment with either SAHA or IL-10, and further suppressed by combinatorial treatment with SAHA and IL-10. Expression of IFN-γ (b) or IL-6 (c) was attenuated by treatment with either SAHA or IL-10, and further suppressed by combinatorial treatment with SAHA and IL-10. d Blocking IL-10 signaling significantly improved allogeneic T cell expansion which was mitigated by SAHA plus IL-10 treatment. Data were from 3 independent experiments (mean ± SEM). p values were obtained using an unpaired t test. Syn = Syngeneic cells.
Discussion
Acetylation acts as an important PTM of proteins by introducing an acetyl functional group, and HDACs remove the acetyl groups from lysine residues. HDAC inhibitors are antitumor agents that inhibit the activity of HDAC, and at low doses they exhibit anti-inflammatory effects in part by acetylating transcription factors such as STAT3.
STAT3 is a master transcriptional regulator of immune responses. It is known to play an important role in the activation of DCs. However, emerging studies have shown that HDAC inhibition suppresses macrophage and DC functions in a STAT3-dependent manner [24], and mitigates experimental graft-versus-host-disease [17, 21, 22, 23] and other immune diseases [15, 18, 19, 20]. To explore the molecular mechanisms by which STAT3 modulates the immune responses of DCs following HDAC inhibition, we performed STAT3-ChIP-seq to define the genome-wide binding of STAT3 following HDAC inhibition in BMDCs and coupled those data with gene expression profiling. We observed that HDAC inhibition contributes to both the upregulation and downregulation of STAT3 target genes. We validated the expression of these genes and further explored the relevance of STAT3 in their expression and function in modulating DC responses. HDAC inhibition promoted the acetylation of STAT3, facilitated its binding to both canonical and noncanonical motifs, and increased the expression of negative regulators of DC function. Specifically, the HDAC inhibition-enhanced STAT3-dependent increase in IL-10Ra expression is one of the mechanisms for the suppressive function of DCs.
To drive target gene expression, STAT3, in addition to undergoing PTMs, is also largely dependent on the recruitment of and interaction with nuclear coactivators [14, 34] or corepressors [48, 49, 50, 51], which collectively determine the final direction of its target gene expression. For example, the protein inhibitor of activated STAT3 (PIAS3) blocks the DNA binding activity of STAT3 and inhibits STAT3-mediated gene activation [48, 49, 50]. Upon cytokine treatment, CBP/p300 acetylates STAT3, thereby enhancing the DNA binding, transactivation activity and nuclear localization of STAT3 [12, 13, 23, 24, 34]. However, deacetylation by HDACs removes acetyl groups from amino acids, thus inhibiting the transcription of STAT3 target genes [12, 13, 34]. Our data show that HDAC inhibition induces or increases STAT3 acetylation [12, 13, 23, 24] and upregulates or downregulates several genes. However, our data do not address the role of coactivators or corepressors in mediating the STAT3-dependent transcriptional regulation after HDAC inhibition. Future studies will determine the precise cooperating partners that are required to mediate STAT3-dependent gene regulation after HDAC inhibition.
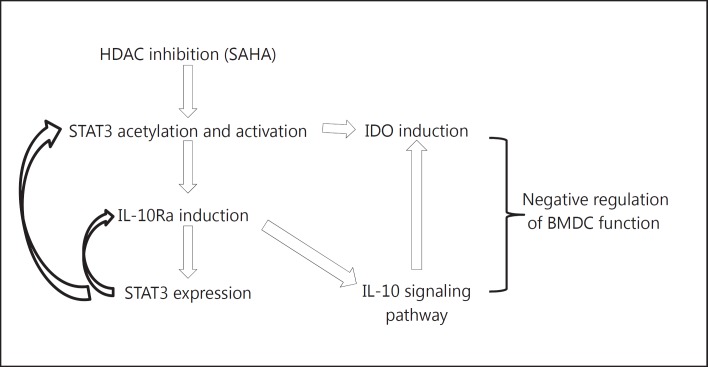
As approximately 10–15% of genes are arranged in a head-to-head orientation in eukaryotic genomes, the distance between the TSSs of adjacent genes can be less than 1 kb [52]. GREAT predicts target genes based on bidirectional promoters, and in the present study, 4,605 genes were predicted from 3,598 STAT3 binding peaks following HDAC inhibition. We validated STAT3 binding sites by independent ChIP assays using an acetyl-(685) STAT3 antibody, thus confirming the veracity and specificity of either proximal or distal binding up to 100 kb from the TSS. Consistent with previous ChIP-seq studies [26, 27, 53], we observed that following HDAC inhibition STAT3 binds to not only the canonical motif but also the noncanonical motifs. Among the top 500 STAT3 binding peaks, the canonical GAS motif demonstrated a significantly higher binding strength in HDAC inhibitor-treated BMDCs when compared with control BMDCs, indicating that following HDAC inhibition STAT3 binding prefers the canonical GAS motif. The net effect of HDAC inhibition on DCs is likely a consequence of STAT3-dependent and independent effects of global histone acetylation; our data nonetheless point to a novel impact on the genome-wide binding of STAT3. Furthermore, STAT3 appears to bind to novel sites, resulting in the upregulation of genes involved in the negative regulation of DC responses. Mechanistic studies demonstrated that following HDAC inhibition STAT3 is acetylated and induces IL-10Ra expression, thus increasing IL-10-dependent inhibition of DC functions. Furthermore, an IGV analysis revealed an enrichment of both STAT3 and H3K4me3 at the Ido1 gene locus following HDAC inhibition, in accordance with a previous study showing that HDAC inhibition acetylates and activates STAT3, which then regulates BMDCs by promoting Ido1 transcription [21, 23, 24]. However, 1-MT, an inhibitor of IDO, did not affect SAHA-induced IL-10Ra expression (online suppl. fig. S2A). Additionally, blocking IL-10Ra did not affect HDAC inhibitor-induced IDO expression (online suppl. fig. S2B). These data suggest that HDAC inhibitor-induced IDO and IL-10Ra are mutually exclusive. Future studies will determine whether the independent effects on IDO and IL-10Ra expression have additive or synergistic effects. HDAC inhibition enhanced the response of IL-10 signaling by upregulating IL-10Ra, which may accentuate and sustain Ido1 expression in the presence of IL-10 in DCs [54, 55] (fig. 11). These findings demonstrate that following HDAC inhibition STAT3 orchestrates a network, at least in part, by directly upregulating both Ido1 and IL-10Ra to negatively regulate DC function, thereby mimicking a specific feature of regulatory DCs [54, 55, 56].
Fig. 11.
Model of STAT3-dependent negative regulation of DCs induced by HDAC inhibition. In BMDCs, SAHA treatment induced STAT3 acetylation, which directly induced expression of Ido and IL-10Ra. The upregulation of IL-10Ra enhanced the IL-10 signaling pathway which supports accentuation of IDO expression, forming a positive feedforward mechanism for sustaining the tolerogenic function of BMDCs following SAHA treatment.
To monitor the possible disturbance of protein-chromatin association by the effect of HDAC inhibition on histone acetyltransferase and HDAC, we performed a similar ChIP-seq using an H3K4me3 antibody with both SAHA- and diluent-treated BMDCs and expected normal enrichment on activated gene loci. We did not observe an obvious change in the protein-chromatin association affected by HDAC inhibition in our experiments as shown with the proper enrichment of H3K4me3 in the target loci as previously reported [35, 36, 37]. In fact, cytokine stimulation induced not only STAT3 phosphorylation but also acetylation [2, 11, 12, 13, 27] (fig. 1, 9a, b), indicating possible changes in the balance between histone acetyltransferase and HDAC, which leads to STAT3 acetylation. STAT3 undergoes a PTM, such as phosphorylation or acetylation [11]. These data suggest that STAT3 acetylation, but not STAT3 phosphorylation, likely contributes to the negative regulation of BMDC function, at least in part, via enhancing the IL-10 signaling pathway. In fact, these results were obtained by GREAT based on MSigDB, which determines the changes in genome-wide STAT3 binding based on a comparison between SAHA-treated BMDCs (which showed STAT3 acetylation, but inhibited STAT3 phosphorylation) and diluent-treated BMDCs (which showed basal level STAT3 phosphorylation) to exclude the possible disturbance from the basal level of phosphorylated STAT3 in diluent-treated BMDC controls. Phosphorylated STAT3 is known to activate target genes in DCs [56] or macrophages [2, 40], but the target genes of acetyl-STAT3 in DCs are not well understood. The phosphorylation of STAT3 upon cytokine stimulation has been emphasized in previous STAT3-ChIP-seq studies [2, 27, 57]. Importantly, in the present study, the effect of HDAC inhibition on the PTM of STAT3 is likely to be primarily dependent on acetylated STAT3.
However, STAT3 is acetylated at multiple lysine residues following HDAC inhibition [34]. Because of the unavailability of a suitable acetyl-STAT3 antibody that has complete signals for all acetyl-lysines in STAT3 following HDAC inhibition, we used a STAT3 antibody in our ChIP-seq experiments, and we utilized an acetyl (685)-STAT3 antibody in our confirmation ChIP assay, which is critical for STAT3 dimerization [10, 11, 12, 13]. Thus, our study is limited in its ability to definitively analyze the specific and exclusive impact of acetyl-STAT3 binding and transcriptional regulation. Nonetheless, based on genome-wide binding analyses of STAT3 using ChIP-seq coupled with gene expression microarrays, we have provided the first characterization of the genomic targets of STAT3 following HDAC inhibition. Our data collectively demonstrate that following HDAC inhibition STAT3 binds to multiple targets and activates a transcriptional program that negatively regulates DC functions, at least in part, through the increased expression of IL-10Ra.
Author Contributions
Y.S. designed and performed experiments, analyzed data, and wrote the paper; M.I., M.Z. and R.M. analyzed data; Y.W., X.C., K.O., C.Z., N.M., J.W., C.R. and T.T. performed experiments; Z.S.Q. and M.C. designed experiments and analyzed data; P.R. designed experiments, analyzed data, and wrote the paper.
Disclosure Statement
The authors have declared that no conflicts of interest exist in relation to this work.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
This work was supported by NIH grants CA-0173878 and CA-143379 (National Cancer Institute) and HL-090775 (National Heart, Lung and Blood Institute) to P. Reddy.
References
- 1.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 2.Laouar Y, Welte T, Fu XY, Flavell RA. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity. 2003;19:903–912. doi: 10.1016/s1074-7613(03)00332-7. [DOI] [PubMed] [Google Scholar]
- 3.Hutchins AP, Poulain S, Miranda-Saavedra D. Genome-wide analysis of STAT3 binding in vivo predicts effectors of the anti-inflammatory response in macrophages. Blood. 2012;119:e110–e119. doi: 10.1182/blood-2011-09-381483. [DOI] [PubMed] [Google Scholar]
- 4.El Kasmi KC, Smith AM, Williams L, Neale G, Panopoulos AD, Watowich SS, Hacker H, Foxwell BM, Murray PJ. Cutting edge: a transcriptional repressor and corepressor induced by the STAT3-regulated anti-inflammatory signaling pathway. J Immunol. 2007;179:7215–7219. doi: 10.4049/jimmunol.179.11.7215. [DOI] [PubMed] [Google Scholar]
- 5.Cheng F, Wang HW, Cuenca A, Huang M, Ghansah T, Brayer J, Kerr WG, Takeda K, Akira S, Schoenberger SP, Yu H, Jove R, Sotomayor EM. A critical role for Stat3 signaling in immune tolerance. Immunity. 2003;19:425–436. doi: 10.1016/s1074-7613(03)00232-2. [DOI] [PubMed] [Google Scholar]
- 6.Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, Akira S. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 7.Braunstein J, Brutsaert S, Olson R, Schindler C. Stats dimerize in the absence of phosphorylation. J Biol Chem. 2003;278:34133–34140. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- 8.Kretzschmar AK, Dinger MC, Henze C, Brocke-Heidrich K, Horn F. Analysis of Stat3 (signal transducer and activator of transcription 3) dimerization by fluorescence resonance energy transfer in living cells. Biochem J. 2004;377:289–297. doi: 10.1042/BJ20030708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFκB. Genes Dev. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasgupta M, Unal H, Willard B, Yang J, Karnik SS, Stark GR. Critical role for lysine 685 in gene expression mediated by transcription factor unphosphorylated STAT3. J Biol Chem. 2014;289:30763–30771. doi: 10.1074/jbc.M114.603894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brierley MM, Fish EN. Stats: Multifaceted regulators of transcription. J Interferon Cytokine Res. 2005;25:733–744. doi: 10.1089/jir.2005.25.733. [DOI] [PubMed] [Google Scholar]
- 12.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 13.Wang R, Cherukuri P, Luo J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J Biol Chem. 2005;280:11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- 14.Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B, Chin YE. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 15.Leoni F, Zaliani A, Bertolini G, Porro G, Pagani P, Pozzi P, Dona G, Fossati G, Sozzani S, Azam T, Bufler P, Fantuzzi G, Goncharov I, Kim SH, Pomerantz BJ, Reznikov LL, Siegmund B, Dinarello CA, Mascagni P. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc Natl Acad Sci USA. 2002;99:2995–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skov S, Rieneck K, Bovin LF, Skak K, Tomra S, Michelsen BK, Odum N. Histone deacetylase inhibitors: a new class of immunosuppressors targeting a novel signal pathway essential for CD154 expression. Blood. 2003;101:1430–1438. doi: 10.1182/blood-2002-07-2073. [DOI] [PubMed] [Google Scholar]
- 17.Reddy P, Maeda Y, Hotary K, Liu C, Reznikov LL, Dinarello CA, Ferrara JL. Histone deacetylase inhibitor suberoylanilide hydroxamic acid reduces acute graft-versus-host disease and preserves graft-versus-leukemia effect. Proc Natl Acad Sci USA. 2004;101:3921–3926. doi: 10.1073/pnas.0400380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra N, Reilly CM, Brown DR, Ruiz P, Gilkeson GS. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J Clin Invest. 2003;111:539–552. doi: 10.1172/JCI16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reilly CM, Mishra N, Miller JM, Joshi D, Ruiz P, Richon VM, Marks PA, Gilkeson GS. Modulation of renal disease in MRL/lpr mice by suberoylanilide hydroxamic acid. J Immunol. 2004;173:4171–4178. doi: 10.4049/jimmunol.173.6.4171. [DOI] [PubMed] [Google Scholar]
- 20.Leoni F, Fossati G, Lewis EC, Lee JK, Porro G, Pagani P, Modena D, Moras ML, Pozzi P, Reznikov LL, Siegmund B, Fantuzzi G, Dinarello CA, Mascagni P. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Mol Med. 2005;11:1–15. doi: 10.2119/2006-00005.Dinarello. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy P, Sun Y, Toubai T, Duran-Struuck R, Clouthier SG, Weisiger E, Maeda Y, Tawara I, Krijanovski O, Gatza E, Liu C, Malter C, Mascagni P, Dinarello CA, Ferrara JL. Histone deacetylase inhibition modulates indoleamine 2,3-dioxygenase-dependent DC functions and regulates experimental graft-versus-host disease in mice. J Clin Invest. 2008;118:2562–2573. doi: 10.1172/JCI34712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi SW, Braun T, Chang L, Ferrara JL, Pawarode A, Magenau JM, Hou G, Beumer JH, Levine JE, Goldstein S, Couriel DR, Stockerl-Goldstein K, Krijanovski OI, Kitko C, Yanik GA, Lehmann MH, Tawara I, Sun Y, Paczesny S, Mapara MY, Dinarello CA, DiPersio JF, Reddy P. Vorinostat plus tacrolimus and mycophenolate to prevent graft- versus-host disease after related-donor reduced-intensity conditioning allogeneic haemopoietic stem-cell transplantation: a phase 1/2 trial. Lancet Oncol. 2014;15:87–95. doi: 10.1016/S1470-2045(13)70512-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi SW, Gatza E, Hou G, Sun Y, Whitfield J, Song Y, Oravecz-Wilson K, Tawara I, Dinarello CA, Reddy P. Histone deacetylase inhibition regulates inflammation and enhances Tregs after allogeneic hematopoietic cell transplantation in humans. Blood. 2015;125:815–819. doi: 10.1182/blood-2014-10-605238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Chin YE, Weisiger E, Malter C, Tawara I, Toubai T, Gatza E, Mascagni P, Dinarello CA, Reddy P. Cutting edge: negative regulation of dendritic cells through acetylation of the nonhistone protein STAT-3. J Immunol. 2009;182:5899–5903. doi: 10.4049/jimmunol.0804388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Northrup DL, Zhao K. Application of ChIP-Seq and related techniques to the study of immune function. Immunity. 2011;34:830–842. doi: 10.1016/j.immuni.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, Ozato K, Levy DE, Nutt SL, Calame K, Leonard WJ. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31:941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 34.Zhuang S. Regulation of STAT signaling by acetylation. Cell Signal. 2013;25:1924–1931. doi: 10.1016/j.cellsig.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, Hitz BC, Gupta R, Rando TA, Baker JC, Snyder MP, Cherry JM, Brunet A. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandstrom RS, Foret MR, Grow DA, Haugen E, Rhodes CT, Cardona AE, Phelix CF, Wang Y, Berger MS, Lin CH. Epigenetic regulation by chromatin activation mark H3K4me3 in primate progenitor cells within adult neurogenic niche. Sci Rep. 2014;4:5371. doi: 10.1038/srep05371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharyya S, Sen P, Wallet M, Long B, Baldwin AS, Jr, Tisch R. Immunoregulation of dendritic cells by IL-10 is mediated through suppression of the PI3K/Akt pathway and of IκB kinase activity. Blood. 2004;104:1100–1109. doi: 10.1182/blood-2003-12-4302. [DOI] [PubMed] [Google Scholar]
- 40.Staples KJ, Smallie T, Williams LM, Foey A, Burke B, Foxwell BM, Ziegler-Heitbrock L. IL-10 induces IL-10 in primary human monocyte-derived macrophages via the transcription factor Stat3. J Immunol. 2007;178:4779–4785. doi: 10.4049/jimmunol.178.8.4779. [DOI] [PubMed] [Google Scholar]
- 41.Liu WH, Liu JJ, Wu J, Zhang LL, Liu F, Yin L, Zhang MM, Yu B. Novel mechanism of inhibition of dendritic cells maturation by mesenchymal stem cells via interleukin-10 and the JAK1/STAT3 signaling pathway. PLoS One. 2013;8:e55487. doi: 10.1371/journal.pone.0055487. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Weber-Nordt RM, Henschler R, Schott E, Wehinger J, Behringer D, Mertelsmann R, Finke J. Interleukin-10 increases Bcl-2 expression and survival in primary human CD34+ hematopoietic progenitor cells. Blood. 1996;88:2549–2558. [PubMed] [Google Scholar]
- 43.O'Farrell AM, Parry DA, Zindy F, Roussel MF, Lees E, Moore KW, Mui AL. Stat3-dependent induction of p19INK4D by IL-10 contributes to inhibition of macrophage proliferation. J Immunol. 2000;164:4607–4615. doi: 10.4049/jimmunol.164.9.4607. [DOI] [PubMed] [Google Scholar]
- 44.Williams L, Bradley L, Smith A, Foxwell B. Signal transducer and activator of transcription 3 is the dominant mediator of the anti-inflammatory effects of IL-10 in human macrophages. J Immunol. 2004;172:567–576. doi: 10.4049/jimmunol.172.1.567. [DOI] [PubMed] [Google Scholar]
- 45.Antoniv TT, Ivashkiv LB. Interleukin-10-induced gene expression and suppressive function are selectively modulated by the PI3K-Akt-GSK3 pathway. Immunology. 2011;132:567–577. doi: 10.1111/j.1365-2567.2010.03402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Waal Malefyt R, Haanen J, Spits H, Roncarolo MG, te Velde A, Figdor C, Johnson K, Kastelein R, Yssel H, de Vries JE. Interleukin 10 (IL-10) and viral IL-10 strongly reduce antigen-specific human T cell proliferation by diminishing the antigen-presenting capacity of monocytes via downregulation of class II major histocompatibility complex expression. J Exp Med. 1991;174:915–924. doi: 10.1084/jem.174.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barton BE. STAT3: a potential therapeutic target in dendritic cells for the induction of transplant tolerance. Expert Opin Ther Targets. 2006;10:459–470. doi: 10.1517/14728222.10.3.459. [DOI] [PubMed] [Google Scholar]
- 48.Shapiro MJ, Shapiro VS. Transcriptional repressors, corepressors and chromatin modifying enzymes in T cell development. Cytokine. 2011;53:271–281. doi: 10.1016/j.cyto.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 50.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt D, Muller S. PIAS/SUMO: new partners in transcriptional regulation. Cell Mol Life Sci. 2003;60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Li Y, Wei J, Li Y. Transcriptional regulation and spatial interactions of head-to-head genes. BMC Genomics. 2014;15:519. doi: 10.1186/1471-2164-15-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wan CK, Oh J, Li P, West EE, Wong EA, Andraski AB, Spolski R, Yu ZX, He J, Kelsall BL, Leonard WJ. The cytokines IL-21 and GM-CSF have opposing regulatory roles in the apoptosis of conventional dendritic cells. Immunity. 2013;38:514–527. doi: 10.1016/j.immuni.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han Y, Chen Z, Yang Y, Jiang Z, Gu Y, Liu Y, Lin C, Pan Z, Yu Y, Jiang M, Zhou W, Cao X. Human CD14+ CTLA-4+ regulatory dendritic cells suppress T-cell response by cytotoxic T-lymphocyte antigen-4-dependent IL-10 and indoleamine-2,3-dioxygenase production in hepatocellular carcinoma. Hepatology. 2014;59:567–579. doi: 10.1002/hep.26694. [DOI] [PubMed] [Google Scholar]
- 55.Finger EB, Bluestone JA. When ligand becomes receptor - tolerance via B7 signaling on DCs. Nat Immunol. 2002;3:1056–1057. doi: 10.1038/ni1102-1056. [DOI] [PubMed] [Google Scholar]
- 56.Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, Nguyen D, Wright KL, Atadja PW, Bhalla K, Pinilla-Ibarz J, Seto E, Sotomayor EM. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009;10:92–100. doi: 10.1038/ni.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun HW, Kanno Y, Powrie F, O'Shea JJ. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data