Abstract
Broadly neutralizing antibodies have been associated with spontaneous clearance of the hepatitis C infection as well as viral persistence by immune escape. Further study of neutralizing antibody epitopes is needed to unravel pathways of resistance to virus neutralization, and to identify conserved regions for vaccine design. All reported broadly neutralizing antibody (BNAb) epitopes in the HCV Envelope (E2) glycoprotein were identified. The critical contact residues of these epitopes were mapped onto the linear E2 sequence. All publicly available E2 sequences were then downloaded and the contact residues within the BNAb epitopes were assessed for the level of conservation, as well as the frequency of occurrence of experimentally-proven resistance mutations. Epitopes were also compared between two sequence datasets obtained from samples collected at well-defined time points from acute (<180 days) and chronic (>180 days) infections, to identify any significant differences in residue usage.
The contact residues for all BNAbs were contained within 3 linear regions of the E2 protein sequence. An analysis of 1749 full length E2 sequences from public databases showed that only 10 out of 29 experimentally-proven resistance mutations were present at a frequency greater than 5%. Comparison of subtype 1a viral sequences obtained from samples collected during acute or chronic infection revealed significant differences at positions 610 and 655 with changes in residue (p<0.05), and at position 422 (p<0.001) with a significant difference in variability (entropy).
The majority of experimentally-described escape variants do not occur frequently in nature. The observed differences between acute and chronically isolated sequences suggest constraints on residue usage early in infection.
Keywords: Broadly neutralizing antibodies, hepatitis C, epitopes, glycoprotein E2, InC3 collaboration
1. Introduction
Hepatitis C virus (HCV) infection is a global health problem with an estimated 80 million infections and 700,000 deaths annually (Gower et al., 2014; Hajarizadeh et al., 2013). The hepatitis C virion consists of a positive sense, single strand RNA genome enclosed by a capsid and an envelope (E) with two immunogenic glycoproteins, E1 and E2 (Ashfaq et al., 2011). The enormous genetic diversity of HCV, which currently spreads across seven genotypes and 67 subtypes is a major barrier to the successful development of a prophylactic vaccine (Smith et al., 2014). This diversity is a result of the viral RNA-dependent RNA polymerase that has no proof-reading capacity, resulting in a high mutation rate and the development of genetically diverse intra-host viral variants termed a quasi-species (Liang, 2013; Wang et al., 2011). This diversity underpins the considerable antigenic complexity observed in HCV infection.
Despite this diversity, several observations offer hope for a HCV vaccine. Firstly, analogous to Human Immunodeficiency Virus (HIV), only one, or very few, of the large number of circulating quasi-species are capable of being transmitted and establishing infection in a new host (so-called transmitted founder variants, T/F) (Bull et al., 2011), thereby potentially offering a restricted range of targets for neutralisation. In HIV, this genetic bottleneck has been shown to be driven by the viral fitness of the founder and the affinity of the T/F virus for the entry receptors (Boeras et al., 2011). Secondly, unlike HIV infection, 25% of primary HCV infections are naturally cleared by the host (Grebely et al., 2014). Thirdly, some individuals repeatedly clear infections from different genotypes (May et al., 2015; Osburn et al., 2010; Pham et al., 2010; Sacks-Davis et al., 2015). One factor that has been implicated in successful clearance is the development of broadly neutralizing antibodies (BNAb) (Osburn et al., 2014), which are defined as antibodies with cross neutralization capacity against genotypes and subtypes other than that to which the host has been exposed. Existence of such antibodies has been demonstrated against other RNA viruses such as HIV and influenza, as well as HCV (Corti and Lanzavecchia, 2013). BNAbs against HCV have been shown to confer passive immunity to immunodeficient mice transplanted with human hepatocytes (Law et al., 2008).
BNAbs bind to viruses directly, generally targeting epitopes across the viral envelope, thereby interfering with virus-cell interactions and subsequent entry (Wang et al., 2011). The viral entry to host hepatocytes is mainly modulated by E1 and E2 glycoproteins of the HCV Envelope (Zeisel et al., 2011) and several key molecules on the host hepatocyte cell surface, namely: CD81, occludin, scavenger receptor B1, and claudin (Wang et al., 2011). The CD81 – E2 interaction has been of particular interest to researchers seeking to identify strategies to prevent virus entry into hepatocytes.
The epitopes targeted by BNAbs can be linear or conformational (Wang et al., 2011). Linear epitopes are regions of the viral envelope that interact with the antibody as a whole. Conformational epitopes consist of sites that sit in close proximity on the three dimensional folded protein structure. Elucidation of the crystal structure of E2 has given much insight into interactions of linear and conformational epitopes, helping to reveal potential targets for vaccine design (Kong et al., 2013). Currently, the sites/regions these epitopes encompass have been variably defined as domains A-E, antigenic regions (AR) and epitopes I-III (Giang et al., 2012; Sautto et al., 2013). However, many of these epitopes overlap with each other when plotted on the linear protein sequence of E2 (Supplementary figure 1).
Non-synonymous mutations within BNAb epitopes have been shown to facilitate escape from humoral immune responses (i.e. escape variants) (Dhillon et al., 2010; Gal-Tanamy et al., 2008). However, the prevalence of escape variants in the HCV-infected population is understudied, and in vitro studies have limitations as escape mutations induced in the laboratory may not occur in nature, especially if such mutations are associated with a fitness cost. In addition, it is of particular interest to resolve whether the genetic bottleneck encountered when the T/F variant establishes infection in a new host can be strategically targeted to facilitate immune protection (Bull et al., 2011). In this regard, it is crucial to study the diversity within BNAb epitopes in early acute infections, as well as in chronic infections. This has not been done previously as the asymptomatic nature of acute HCV has made it difficult to obtain samples for sequencing in acute infection.
This study aimed to: i) review binding epitopes of all BNAbs identified against HCV to recognize patterns of epitope localisation on linear regions of the E2 protein; ii) identify conserved and variable residues within these epitopes across all genotypes from publicly available sequence data; iii) identify experimentally-proven resistance variations within BNAb epitopes and their frequency of occurrence in the population; and iv) compare similarities and differences in BNAb epitopes in samples from acute and chronic HCV infections.
1. Methods
2.1 Review of BNAbs, contact residues and resistance mutations
A systematic literature search was carried out to identify all neutralizing and broadly neutralizing antibodies directed against HCV envelope proteins described to date. MEDLINE, PUBMED, EMBASE and Web of Science were searched for articles with the keywords ‘hepatitis C’ in the abstract and ‘neutralizing’ or ‘neutralizing antibodies’ in any field. There were no time or language restrictions to the search (last date of search: 6 January 2016). There were 1145 abstracts in the original search after duplicates were removed. Endnote X7 software (Thomson Reuters, Carlsbad, CA 92011, USA) was used to filter the articles. Bibliographies of cited literature were also searched. All abstracts were read by the first author, and relevant articles were selected. Fifty-eight full text articles were selected for the final synthesis, which included in vitro experimental studies, animal studies, observational human studies and clinical trials. After reading the final selection of articles, the data on BNAb epitopes, as well as the breadth of neutralization and potency of antibodies as expressed in EC50 values (minimum concentration of antibody that reduces infectivity by 50%) were summarized. Experimentally-proven resistance mutations within BNAb binding epitopes were also recorded.
2.2 Characterization of the variation between epitope regions in the host population
All available full-length HCV E2 sequences derived from natural infection (coding for glycoprotein E2) were downloaded from the Los Almos HCV sequence database (http://hcv.lanl.gov/content/index). Only the genotypes 1a, 1b, 2a, 2b, 3a, 4a, 5a and 6a were considered for further analysis to ensure the availability of at least 10 sequences per subtype. An additional 190 recently generated sequences from subjects with acute HCV were added (Rodrigo C, 2015). Short partial E2 sequences were excluded by setting a limit on the minimum sequence length at 1000 nucleotides. All sequences were processed in Geneious software (version 8.0.5, Biomatters Inc.) for manual curation and removal of ambiguous sequences with inappropriate stop codons. Clonal sequences were manually removed after tracing individual papers (to avoid duplication of counts when calculating frequencies of mutations), and using meta-data available on GenBank. Sequences were realigned with MUSCLE algorithm with all genotypes combined. All nucleotide sequences were translated to amino acid sequences for further analysis.
The individual epitope regions identified from the literature were mapped onto the final alignment. When there were multiple overlapping epitopes described by different research groups, a linear region of the genome inclusive of all overlapping epitopes was selected for further analysis. These composite epitope areas are henceforth referred to as “epitope regions”. Within the selected epitope regions, the frequency of occurrence of individual amino acids was plotted to identify conserved residues. For each residue, Shannon entropy was calculated as a measure of complexity using an online tool available from the Los Alamos HCV database (Kuiken et al., 2008). The experimentally-proven escape mutants were specifically sought to determine their prevalence in the HCV infected host population.
2.3 Comparison of BNAb epitopes between acute and chronic infection
As there were inadequate numbers of sequences from other genotypes, comparison of acute (within 180 days since the estimated date of infection) and chronic sequences was restricted to genotype 1a. The late infection time points were selected from the sequencing project described by Kuntzen et al. (Kuntzen et al., 2008), which reported treatment-naïve patients who had been infected for at least one year prior to the sequencing time point. This cohort included patients from United States (Boston, Memphis, New York and Denver), Germany, and Switzerland. This dataset was rich in subtype 1a and 1b sequences. Additional chronic sequences representative of Australian isolates were obtained from an Australian cohort, the Hepatitis Incidence and Transmission Study in prisons study (HITS-p)(Bull et al., 2011; Luciani et al., 2014). The acute infection sequences were obtained from the International Collaboration of Incident HIV and Hepatitis C in Injecting Cohorts Study (InC3)(Grebely et al., 2013). Our recent report regarding the InC3 acute infection sequencing project is described elsewhere (Rodrigo C, 2015). Briefly, the InC3 is a collaboration of nine international prospective cohorts (Australia-4, USA–3, Canada-1, The Netherlands–1) studying acute HCV infection in people who inject drugs. The earliest viraemic time points of all incident infections have been sequenced using next generation sequencing (Rodrigo C, 2015). All individuals included in this dataset were HCV antibody negative on enrolment and were observed to be RNA or antibody positive during follow up. There were no reinfections or super-infections with a second genotype identified within the first 180 days of the infection. The consensus sequences of the E2 protein generated from these samples (enriched in subtype 1a and 3a) were used as the acute infection dataset in this analysis.
The pairwise genetic distances between each of the BNAb epitope regions was firstly compared between sequences obtained from acute and chronic infection samples. For this purpose, the amino acid sequences of recognized epitope regions, and the hypervariable region 1 (HVR1), were separated from the remainder of E2. The pairwise distances (p-distances) between sequences within epitope regions were calculated using MEGA 6.0 software separately for acute and chronic infection samples. The summary statistics from this analysis were standardized by adjusting for sequence length (p-distance per amino acid residue), and compared across the epitope regions and between acute and chronic samples.
In order to identify differences in individual residues between acute and chronic infection samples, consensus sequences of each alignment and entropies per residue were compared. To calculate significant entropy differences between a query sequence (acute infection dataset) and the comparator sequence (chronic infection dataset), a tool available at the Los Almos HCV sequence database was used (http://hcv.lanl.gov/content/sequence/ENTROPY/entropy.html). This analysis was repeated for each position within each epitope region separately, with a moderately conservative level of significance set at p<0.001. The amino acid residues in this paper are represented according to their standard IUPAC single letter abbreviation (IUPAC, 1984).
2. Results
3.1 BNAbs and their binding epitopes
Comparison of the neutralization capacity of BNAbs reported in the literature revealed some antibodies to have a greater cross-genotype neutralization capacity than others. A summary of BNAbs with neutralization capacity against at least four of the most common HCV subtypes worldwide (1a, 1b, 2a, 2b, 3a) is given in Table 1. The binding residues of all BNAbs are shown in supplementary table 1. When the residues targeted by these BNAbs were plotted on the linear HCV polypeptide, the majority of the contact residues lay within three distinct regions on the linear E2 polypeptide. These “epitope regions” extend between amino acid residues 412 – 447 (often otherwise referred to as epitopes I and II), 523–540 (often referred to as epitope III) and 610 – 659 (Supplementary figure 1).
Table 1.
Properties of selected broadly neutralizing antibodies described against HCV
| Antibody | Epitopes with critical interacting residues on E1 or E2 | Neutralization capacity | Comments |
|---|---|---|---|
| IGH505 and IGH526 (Meunier et al., 2008) | Residues 313–327 | Genotypes 1a, 1b, 4a, 5a, 6a were neutralized, EC50 : <2–25 μg/ml Genotypes 3a was not neutralized and 2a gave mixed results (2a HCVpp were not neutralized but JFH-1 clone was neutralized) |
These are one of the few anti E1 antibodies with broadly neutralizing capacity. Both had overlapping but non-identical epitopes within residues 313–327. |
| AP 33(Dhillon et al., 2010; Gal-Tanamy et al., 2008; Owsianka et al., 2005; Tarr et al., 2006) | Linear epitope, residues 412–423 (L413, N415, G418, W420) | Against genotypes 1–6 (tested with HCVpp) ** EC50 : 0.6–32 μg/ml |
First monoclonal antibody from animal sera to show significant broadly neutralizing capacity across all genotypes. Inhibits CD81–E2 interaction. |
| MRCT 10.v362 (Pantua et al., 2013) | Linear epitope Residues 412–423 |
Neutralized HCVpp of genotypes 1a, 1b, 2a and HCVcc of 2a EC90: 0.07–1.76 μg/ml |
Humanised version of murine antibody, AP33 |
| Hu5B3.v3 (Pantua et al., 2013) | Linear epitope Residues 412–423 (N415, N417, W420) |
Neutralized HCVpp of genotypes 1a, 1b, 2a and HCVcc of 2a EC90: 0.7–17.4 μg/ml |
Anti E2 antibodies Interferes with CD81 binding |
| Mab 24(Alhammad et al., 2015) | Conformational residues; L413, N415, G418, W420, and H421 | Inhibited infectivity of all genotypes 1–7 to varying extents. Over 90% inhibition for subtypes 1a, 2a, 4a, 5a. EC50: 1–6 μg/ml (against subtype 1a) |
Blocks CD81-E2 interaction |
| HC33.1, HC33.4(Keck et al., 2013; Keck et al., 2014) | Linear epitope, residues 412–423 | EC50 for chimeric JFH1 HCVcc of genotypes 1a, 2a, 3a, 4a, 5a, 6a ranged from 0.1 – >50 μg/ml | Interferes with CD81 – E2 interaction. |
| 95-2 and HCV 1 (Broering et al., 2009) | Linear epitope, residues 412–423 (L413, W420) | Neutralized HCVpp of genotypes 1a, 1b, 2b, 3a,4a EC50 : 1–100 nM (approximations based on a graph in the paper) |
Probably interferes with CD81-E2 binding |
| HC-84.1, HC84.26(Fofana et al., 2012) | Conformational epitopes, residues 418–446, 611–616 | EC50 for chimeric JFH1 HCVcc of genotypes 1a, 2a, 3a, 4a, 5a, 6a ranged from 0.043 – >50 μg/ml for HC 84.1 and 0.005–12.91 μg/ml for HC 84.26 | Interferes with CD81 – E2 interaction. |
| 3/11(Tarr et al., 2006) | Linear epitope, residues 412–423 (N415, W420, H421) | Neutralized HCVpp of genotypes 1–6 except genotype 5 when tested at a fixed concentration of 50μg/ml. Percentage of neutralization varied from 10–80% for different genotypes. | Less potent than AP33. Interferes with CD81 – E2 interaction. |
| 1:7 and A8(Johansson et al., 2007) | Conformational epitope, residues 523–535 (G523, W529, G530, D535) | Neutralization across genotypes 1–6 (tested with HCVpp and JFH - 1) EC50 : 0.06–0.56 μg/ml for JFH1. EC50 for HCVpp not mentioned |
Inhibits CD81 – E2 interaction. |
| HC-1, 2, 11,12,13(Keck et al., 2008) | Mapped for HC 1 and 12 only (as other antibodies were similar to one of these two) Conformational epitope, residues 523–540 (W529, G530, D535) |
EC50 for 1a and 2a HCVcc varied from 0.03–4.6 μg/ml. Inhibition of infectivity of HCVpp of genotypes 1–6 (by a fixed dose of 20μg of each antibody) ranged between 0–90%. | The inhibition of 1a and 1b HCVpp were high for all antibodies (>78%). HC-1 had the best neutralization capacity across all HCVpp genotypes (29–86% inhibition of infectivity). |
| AR3A, AR3B, AR3C, AR3D (Law et al., 2008) | Conformational epitopes, residues, 396–424), 436–447 and 523–540 (S424, G523, P525, G530, D535, N540) | Neutralization against genotypes 1–6 (tested with HCVpp and JFH - 1) EC50 : 1–50 μg/ml |
Interferes with CD81–E2 interaction. |
| CBH-5(Keck et al., 2007; Owsianka et al., 2008) | Conformational epitope, residues 523–540 (G523, P525, G530, D535, N540) | CBH-5 had neutralization capacity against all genotypes (tested with HCVpp and JFH - 1) EC50 : 0.04–13μg/ml |
Interfered with CD81-E2 binding. CBH-2, another monoclonal antibody also had broadly neutralizing capacity but not against all genotypes |
| e20, e137(Mancini et al., 2009; Perotti et al., 2008; Sautto et al., 2012) | Conformational epitope, residues 412–423, 523–540 (T416, W420, W529, G530, D535) | e137 neutralized infectivity of HCVpp of genotypes 1a, 1b, 2b and 4 by 20–75% when tested at a fixed dose of 15μg/ml A second paper describes that e20 and e137 both additionally neutralized HCVpp of genotypes 2a and 5 along with HCVcc of JFH-1 EC50 of e20: 0.02–1.2 μg/ml EC50 of e137: 0.03–1.4μg/ml |
Interfered with CD81-E2 binding |
| AR4A (Giang et al., 2012) | Conformational, residues, 201–206 (E1), 657–659, 698 (E2). | Neutralized HCVcc and HCVpp of all six genotypes EC50: 0.03–38.5 μg/ml |
Anti E1 and E2 antibody, does not interfere with CD81 binding |
| Human monoclonal antibody 55 (Shimizu et al., 2013) | Conformational, residues 508–607 | Inhibited infectivity of all genotypes 1–6 to varying extents EC50: 1.3 – 230μg/ml (Genotype 7 excluded as it was not neutralized effectively) |
Probably blocks CD81-E2 interaction |
| D03(Tarr et al., 2013) | Probably conformational, residues 415, 523 and 526 appear critical for binding | Neutralized infectivity of genotypes 1–6 EC50: 1 – 10μg/ml |
Alpaca nanobody. Mechanism of action is probably not exclusive to inhibition of E2-CD81 binding. First BNAb to inhibit cell to cell transmission |
EC50 : Minimum concentration of antibody that reduces infectivity by 50%
3.2 Understanding conservation of amino acid residues at the population level
The full length E2 sequences from the Los Alamos HCV database included genotype 1a sequences (n= 2803), 1b (n=922), 3a (n= 891) and 2b (n=197). All other genotypes (2a, 4a, 5a and 6a) had less than 60 sequences available. After removal of clonal sequences, a total of 1749 sequences remained for alignment using the MUSCLE algorithm. The epitope regions highlighted in figure 1 (412 – 447, 523 – 540 and 610 – 659+698) were extracted from this alignment.
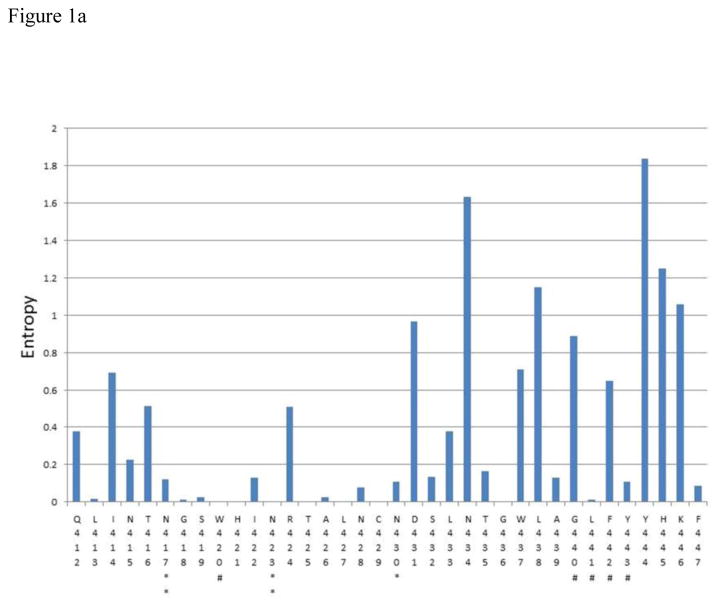
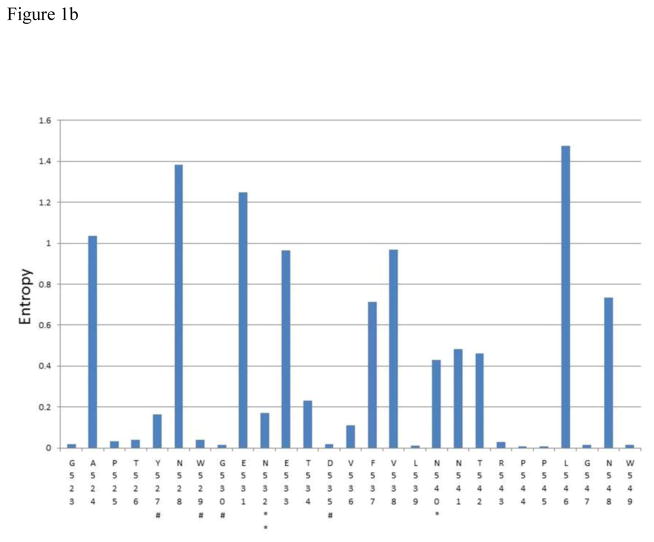
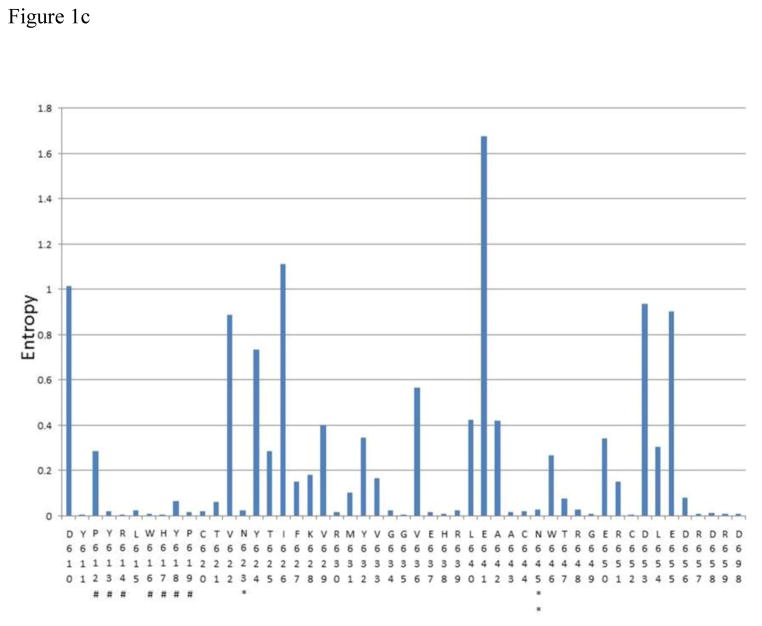
Figure 1.
A – C Shannon entropy of each amino acid residue within epitope clusters of E2. The amino acid residue number and IUPAC abbreviation is indicated in X-axis. * indicates a glycosylation site and ** indicates a glycosylation site known to affect antibody neutralization according to Wang et al.(Wang et al., 2011). # Indicates potential CD81 binding sites (Rothwangl et al., 2008; Wang et al., 2011). All E2 sequences available from Los Almos HCV database for genotypes 1a,1b,2a,2b,3a,4a,5a and 6a (after removing clones) plus InC3 sequencing project data (n=1749) were used for this analysis. Amino acid numbering follows that of the translated HCV polyprotein of the reference subtype 1a genome (H77)
The consensus sequence for each epitope cluster is given below
- Epitope cluster 1: 412-QLINTNGSWHINRTALNCNDSLNTGWLAGLFYYHKF – 447
- Epitope cluster 2: 523-GAPTYNWGENETDVFVLNNTRPPLGNW – 549
- Epitope cluster 3: 610-DYPYRLWHYPCTVNYTIFKVRMYVGGVEHRLEAACNWTRGERCDLEDRDR-659 + D698
The Shannon entropy for each amino acid residue was calculated as a measure of probability of variation (Figure 1). The analysis showed that at 70 sites within epitope regions there was either no observable variation, or very limited variation (Shannon entropy was close to zero and in most occasions less than 0.2). Thirteen of these sites included most of the hypothesized CD81 binding residues, including W420, H421, L441, Y443, Y527, W529, G530, D535, Y613, R614, W616, H617, Y618 and P619. Similarly, glycosylation sites known to be involved in BNAb binding (N417, N423, N532 and N645) also showed very limited variation.
In addition, there were many other sites within the epitope regions to which no function has yet been assigned (with regard to BNAb binding), but were more than 99% conserved across genotypes at the population level (e.g. T425). The proportion of sites which had an entropy below 0.2 were 58.3% for the first epitope region, 55.5% for the second epitope region, and 68% for the third epitope region. The consensus sequences for each epitope region across all genotypes are shown in Figure 1. A genotype specific analysis of the entropy values for subtypes 1a, 1b, 2b and 3a (95.3% of all sequences) is shown in Supplementary table 2.
3.3 Frequency of occurrence of resistance mutations
From the literature review, 27 experimentally-proven resistance mutations were identified within BNAb epitopes (and two others outside the epitope regions). The frequency of occurrence of these mutations in the sequence dataset is shown in Table 2. Only 10 mutations (34%) were present at a frequency greater than 5%. Interestingly, 7 (24%) of the experimentally-observed resistant mutations were not observed in any of the E2 sequences analysed.
Table 2.
Experimentally recognized resistance mutations against each BNAb and their prevalence of occurrence as estimated by available E2 sequences across all genotypes in Los Almos database after removing clones (n-1749)
| Antibody | Mutation | Sensitive (%) | Resistant (%) | Other residues | Ref. |
|---|---|---|---|---|---|
| AP 33 (Murine) | L413A | 99.8 | 0.0 | 0.2 | (Dhillon et al., 2010; Gal-Tanamy et al., 2008; Keck et al., 2014; Tarr et al., 2006) |
| N415A/Y/D | 96.3 | 2.7 | 1.0 | ||
| N417S | 98.0 | 1.3 | 0.7 | ||
| G418D/A | 99.8 | 0.1 | 0.1 | ||
| W420A | 99.9 | 0.0 | 0.1 | ||
| E655G (?co-occurring mutation with N415Y) | 71.8 | 0.9 | 27.3 | ||
| CBH-2 (Human) | D431G | 65.0 | 0.2 | 37.8 (14.5 is resistant with D431E) | (Keck et al., 2011) |
| A439E | 97.8 | 0.0 | 2.2 | ||
| hu5B3.v3 and MRCT10.v362 (Murine) | N417S/T | 98.0 | 1.3 | 0.7 | (Pantua et al., 2013) |
| HCV-11 (Human) | L438F | 40.7 | 1.4 | 57.9 | (Keck et al., 2011) |
| N434D | 41.6 | 19.1 | 39.3 | ||
| T435A | 96.9 | 2.3 | 0.8 | ||
| CBH-2, HC84.26, CBH-5, HC84.22, AR3A, AR3B, AR3C, and AR3D (Human) | R424S | 71.9 | 27.4 | 0.7 | (Bailey et al., 2015; Wasilewski et al., 2016) |
| D431E | 65.0 | 14.5 | 20.5 | ||
| F442I | 81.9 | 6.0 | 12.1 | ||
| N430D | 98.3 | 0.8 | 0.9 | ||
| L433I | 89.9 | 9.1 | 1.0 | ||
| L438I | 40.8 | 39.2 | 20.1 | ||
| K446E | 66.3 | 0.3 | 33.4 | ||
| A475T | 51.6 | 13.6 | 34.8 | ||
| I538V | 6.8 | 49.7 | 43.5 | ||
| Q546L | 28.1 | 48.9 | 23.0 | ||
| T563V | 46.5 | 49.7 | 3.8 | ||
| MAb24 (Murine) | I414A | 67.8 | 0.0 | 32.2 | (Alhammad et al., 2015) |
| T416A | 85.7 | 1.3 | 13.0 | ||
| HCV-1 and 95-2 (Human) | N415K | 96.3 | 1.0 | 2.7 | (Broering et al., 2009) |
| HC 33.1 (Human) | L413I | 99.8 | 0.0 | 0.2 | (Keck et al., 2014) |
| N417T | 98.0 | 0.0 | 2.0 | ||
| S419N | 99.7 | 0.2 | 0.1 | ||
| N434D | 41.6 | 19.1 | 39.3 | ||
| K610R | 0.0 | 0.0 | 100.0 |
3.4 Comparison of genetic diversity between acute and chronic samples
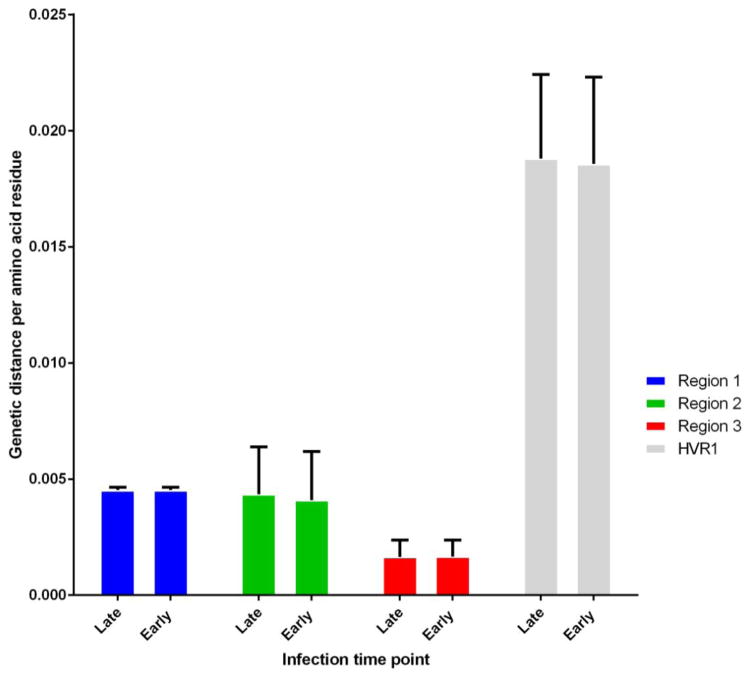
There were 337 genotype 1a sequences available for the chronic infection group, and 89 for the acute infection group. The results of the comparative analysis are summarized in Figure 2. The HVR1 region had a similar mean genetic distance in both datasets (0.019 per amino acid residue) (Figure 2). The pairwise genetic distance per amino acid residue was smallest in the third epitope region (0.002 per residue). There was no significant difference between acute and chronic infections with regard to the variability observed within each epitope region (Figure 2).
Figure 2.
Pairwise genetic distances of individual epitope clusters compared with each other and between acute (n-91, < 180 days since infection) and chronic infection (n-337, > 180 days since infection) time points for subtype 1a. Hypervariable region 1(HVR) is also compared as a control. Error bars show standard deviation.
Two significant amino acid differences were noted within the third epitope region, while none were observed for the first and second epitope regions. These differences were observed at positions 610 [chronic infection; histidine (H) 40.9%: aspartic acid (D) 34.4%, acute infection; H 28.6%: D 52.7%, p =0.003] and 655 [chronic infection; glutamic acid (E) 54.9%: aspartic acid (D) 38.6%, acute infection; E 40.7%: D 47.3%, p =0.046]. The change at position 610 was from an acidic residue (D) to a basic residue (H). Residue 610 is on the exposed surface and lies within the centre of the conformational structure that the third epitope region residues encompass (Figure 3). At position 655, both residues were acidic, but resulted in change from a smaller (D), to a larger, side chain (E). Position 655 lies outside the residues solved in the recent crystal structure (Khan et al., 2014; Kong et al., 2013).
Figure 3.
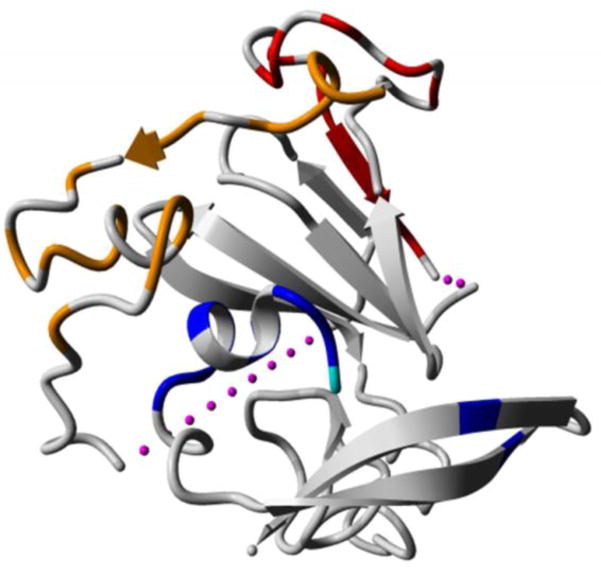
Crystal structure of the HCV E2 core protein (Protein database, accession number 4WMF (Kong et al., 2013)). The residues contained within the three linear epitope regions and identified as critical for interaction with neutralising antibodies are depicted. Orange: indicates critical residues within epitope region I, red: epitope region II and blue: epitope region III. The residue D610, identified to be significantly different between acute and chronically sampled sequences is depicted in cyan and lies within the conformational pocket of region III. The pink dashes depict the regions not solved in the crystal structure
Comparison of the entropy difference calculated for each residue within the epitope regions between acute and chronic infection datasets, revealed a statistically significant difference (p<0.001) at position 422 (epitope region 1) with the higher entropy being observed in the acute infection dataset (Supplementary table 3). The most common replacement for isoleucine seen at position 422 was valine, which was found at a prevalence of 0.6% in chronic infection samples compared to 4.4% in acute infection samples.
3. Discussion
This paper examined the pattern of BNAb binding residues clustering on the linear E2 polypeptide, and identified three distinct regions. Key residues within epitopes, such as proposed glycosylation and CD81 binding sites were highly conserved across genotypes. The additional conserved residues between these putative contact sites may not necessarily be participating in the virus-antibody interactions (i.e. not part of the conformational epitope), or their significance in such interactions is as yet undiscovered. Although speculative, these residues probably have a higher likelihood of being involved in such an interaction compared to any other random residues on E2 that are located outside the epitope regions defined in Figure 1. They may also play a key role in maintaining the correct tertiary protein structure for binding with antibodies.
A significant proportion (24%) of experimentally-proven resistance mutations did not occur in any of the E2 sequences. Also, even though variation was observed at other sites (e.g. N417, G418, A439 and T435) it was the sensitive variant (neutralized by BNAb) that dominated, despite the advantage of immune escape offered by the resistant variants. There are several possible explanations for this finding. Firstly, many BNAbs identified to-date may occur only rarely in vivo and hence exert insufficient immune pressure to drive selection at the population level. Secondly, BNAbs isolated from experimental animal sources, such as mice, commonly do not accurately reflect the in vivo specificity of antibodies in humans. Thirdly, mutation at these BNAb epitope sites may be associated with a fitness cost that hinders viral replication. For example, it has been demonstrated that W420A mutation confers resistance to the murine BNAb AP33. Yet, no variations of W420 were seen in human sequences. Tarr et al. reported that only about 2.5% of chronic HCV carriers produce antibodies that recognise the linear epitope targeted by AP33 (Tarr et al., 2007). Nevertheless, epitopes targeted by such murine antibodies remain an area of interest for vaccine development. If the residues are highly conserved in the host population, this implies a selection pressure to constrain mutation. Indeed, such heterologous approaches are commonly used in passive immunisation against rabies (Both et al., 2012), and are being utilised for products in development against HIV (Heydarchi et al., 2016). Finally, the resistant mutations may occur as the dominant variants when put under appropriate immune pressure. Chung et al. described a randomized controlled trial where monoclonal BNAb HCV-1 (binding epitope: 412–423) was administered to 6 patients with hepatitis C requiring liver transplantation (the placebo arm included 5 patients) (Chung et al., 2013). In all patients who received the antibody, the resistant variants emerged at positions 415 and 417 (N415D/S/K, N417S) and dominated over the wild type starting from day 7 post-transplantation. It was also demonstrated that either the 415 or 417 mutation was sufficient to confer resistance. In the placebo group, the sensitive variant remained except in one patient who had the resistant N417S as the dominant type prior to transplantation (Babcock et al., 2014; Chung et al., 2013). It is evident that the data in table 2 should be interpreted in this context, where the low frequency of occurrence may be due to the lack of a selection pressure by naturally occurring antibodies. Should such antibodies be used therapeutically, or an Envelope-containing vaccine be deployed the prevalence of resistant variants may increase.
A recent study has shown that two mutations (A475T, T563V) which occur at a frequency of greater than 10% in the host population, but sit outside the epitope regions defined here can also confer resistance to several BNAb (Bailey et al., 2015). In the previously cited sequence analysis following HCV-1 administration to patients undergoing liver transplantation, in addition to the key mutations at positions 415 and 417, several other sites also mutated between pre and post transplantation between the treatment and placebo groups (Babcock et al., 2014). Not all these sites were within the defined epitope regions. This suggests that critical binding residues can either occur outside the recognised epitope regions, or that co-occurring mutations can balance the survival disadvantage conferred by a non-fit resistance mutation. The interactions between binding residues (or their variants) with other residues may not be simplified to a pairwise interaction. In fact, epistatic interactions between residues may well happen in a more complex network, in which multiple residues influence each other across the genome, potentially including residues outside the envelope proteins. This has been shown to be true of other rapidly mutating viruses such as influenza (Wu et al., 2016).
Assessment of the consensus level differences between the acute and chronic infections, revealed two significant differences at positions 610 and 655. Experimental evidence shows these positions to be important for neutralization by antibodies, and in the case of 610, for viral replication (Kawaguchi et al., 2011). At position 655, E and D are chemically equivalent amino acids and a switch may not affect the protein folding or the chemical environment of the surrounding amino acids. However, other mutations at position 655 have been shown to confer resistance to the BNAbs AP33 and 3/11, in conjunction with a mutation at position 415 (Gal-Tanamy et al., 2008). In particular, N415Y together with E655G resulted in a more potent viral escape than with either N415Y or E655G by itself, whereas the E655G mutation by itself conferred partial resistance against AP33 (Gal-Tanamy et al., 2008). Position 610 is located at the centre of the third epitope region, which sits to the side of the broadly neutralising face, which encompasses the first and second epitope regions, and is often referred to as the non- or weak-neutralising face. This region includes residues (613, 617 and 618) that have also been reported to influence the interaction with CD81 (Wang et al., 2011). Despite lying outside of the broadly neutralising face of E2, position 610 has been suggested to confer resistance via the K610R mutation to the human BNAb, HC33.1, that targets residues within the first epitope region (Keck et al., 2014). However, neither the sensitive, nor the resistant variant, was observed in the sequences derived from natural infection described here. In addition, a D610N mutation has been shown to emerge, along with co-occurring mutations at positions E431D and N434T, after passaging patient-derived E1 and E2 JFH-1 chimeras in vitro (Kawaguchi et al., 2011). These three mutations synergistically reduced the neutralization capacity of LMF87, an anti-HVR1 antibody, despite lying outside the binding domain of the antibody. Interestingly, the mutations also enhanced the replicative capacity of the chimeric virus in cell culture. More recently, the naturally occurring residues at position 610 have been shown to be directly involved in resistance to a key BNAb; HC84.26 with variants carrying H610 (instead of D) being more sensitive to neutralization (Prentoe et al., 2016). It should be noted that this was demonstrated with a genotype 5a, HVR-deleted, chimeric virus, while our analysis between acute and chronic samples was restricted to genotype 1a.
While there were no differences between acute and chronic infection datasets, resistance mutations at positions 424, 434, 438, 538 and 546 did occur at a frequency greater than 10% in the total sequence dataset. Of these mutations, N434D is of particular interest. In cell culture, virions carrying this mutation became dominant through cell passage (Keck et al., 2014). N434D may be a fitness mutation that can undermine the efficacy of a BNAb that is rendered ineffective by this mutation.
Calculation of significant entropy differences between acute and chronic infection samples allows identification of sites that are changing, but may not be evident at a consensus level analysis. Using a moderately conservative significance level, only one such site was observed at position 422 with higher variability (i.e higher entropy) found in the acute infection dataset. This is likely to reflect a population of viruses undergoing fixation at this position towards a more stable phenotype observed among chronic infections (from valine to isoleucine). However, confirmation of this hypothesis would require sequence analysis from samples collected during longitudinal follow up of individual subjects from acute to chronic infection. The I422 residue is highly conserved in the population and has not been shown to bind any BNAbs reported to date, but does sit adjacent to a glycosylation site at N423 which is 100% conserved. Glycan shielding is a well-recognized method of conferring resistance to BNAbs (Pantua et al., 2013) and in combination both these residues may form an important structural motif. Isoleucine is larger than valine which may make the glycosylation site protrude further from the tertiary structure.
This analysis sought to link the experimental observations on BNAb epitopes with sequence data from the host population. It should be acknowledged that the dataset referred to here was restricted to currently available sequencing data, which is biased towards the more common subtypes (1a, 1b, 2a, 2b, 3a). This dataset also does not capture the complexity of the viral quasi-species within each infected person. Next generation sequencing will help to overcome this limitation. Similarly, comparison of acute and chronic infections should ideally be conducted within individual subjects followed from acute to chronic infection. The InC3 collaborative is facilitating further analysis in this regard. Another limitation is that available data on BNAb binding sites identified by alanine scanning may be restricted to conserved areas. This will bias the currently available evidence to such sites. This limitation cannot be overcome until more comprehensive characterization of binding sites is available.
4. Conclusions
The recognised HCV BNAbs have overlapping epitopes in three regions when mapped onto the linear E2 protein. A population level analysis of these regions from publicly available E2 sequences showed that many residues with structurally important links to BNAbs (glycosylation sites, CD81 binding sites) are highly conserved across genotypes. In addition, many experimentally-proven escape mutations with resistance to BNAbs are absent or rare in the host population, suggesting a possible fitness cost of these mutations. A comparison of epitope regions between acute and chronic infection has identified a limited number of differences in subtype 1a envelope sequences. These findings will guide further BNAb studies, and HCV vaccine design, by linking experimental observations with sequencing data.
Supplementary Material
Highlights.
Broadly neutralizing antibody binding residues are clustered on three areas of HCV E2
The majority of experimentally-proven escape variants are rare in nature
Positions 610 and 655 commonly differ at the consensus level in acute and chronic infections
Acknowledgments
The infrastructure for sharing of data and specimens in InC3 was funded by the National Institute on Drug Abuse (NIDA) R01DA031056. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of NIDA or the National Institutes of Health (NIH). Research support for the InC3 cohorts includes: the Netherlands National Institute for Public Health and the Environment to the Amsterdam Cohort Study (ACS); Baltimore Before and After Study (BBA) - National Institutes of Health (NIH U19 AI088791); Boston Acute HCV Study: transmission, immunity, outcomes network (BAHSTION) is funded under NIH NIAID U19 U19 AI066345; ATAHC–NIDA RO1 DA 15999-01; HITS-p - National Health and Medical Research Council of Australia (NHMRC) - Project No. 222887, Partnership No. 1016351, Program Nos. 510488 and 1053206; HITS-c – UNSW Hepatitis C Vaccine Initiative and NHMRC Project Grant No. 630483; Networks/MIX – NHMRC Project Grants Nos. 331312 and 545891 and the Victorian Operational Infrastructure Support Programme (Department of Health, Victoria, Australia); HepCo - the Canadian Institutes of Health Research (MOP-103138 and MOP- MOP-133680); UFO – NIH R01 DA016017. The following InC3 investigators are funded by research fellowships: NHMRC, Australia – JG, MH, LM, GD, ARL; Fonds de la Recherche du Québec-Santé, Canada– JB and NHS.
Abbreviations
- T/F
Transmitted founder
- HCV
Hepatitis C virus
- NS5B
Non-structural protein 5B
- BNAbs
Broadly neutralizing antibodies
- AR
Antigentic region
- HVR
Hypervariable region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alhammad Y, Gu J, Boo I, Harrison D, McCaffrey K, Vietheer PT, Edwards S, Quinn C, Coulibaly F, Poumbourios P, Drummer HE. Monoclonal Antibodies Directed toward the Hepatitis C Virus Glycoprotein E2 Detect Antigenic Differences Modulated by the N-Terminal Hypervariable Region 1 (HVR1), HVR2, and Intergenotypic Variable Region. Journal of virology. 2015;89:12245–12261. doi: 10.1128/JVI.02070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfaq UA, Khan SN, Nawaz Z, Riazuddin S. In-vitro model systems to study Hepatitis C Virus. Genetic vaccines and therapy. 2011;9:7. doi: 10.1186/1479-0556-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock GJ, Iyer S, Smith HL, Wang Y, Rowley K, Ambrosino DM, Zamore PD, Pierce BG, Molrine DC, Weng Z. High-throughput sequencing analysis of post-liver transplantation HCV E2 glycoprotein evolution in the presence and absence of neutralizing monoclonal antibody. PloS one. 2014;9:e100325. doi: 10.1371/journal.pone.0100325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JR, Wasilewski LN, Snider AE, El-Diwany R, Osburn WO, Keck Z, Foung SK, Ray SC. Naturally selected hepatitis C virus polymorphisms confer broad neutralizing antibody resistance. The Journal of clinical investigation. 2015;125:437–447. doi: 10.1172/JCI78794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeras DI, Hraber PT, Hurlston M, Evans-Strickfaden T, Bhattacharya T, Giorgi EE, Mulenga J, Karita E, Korber BT, Allen S, Hart CE, Derdeyn CA, Hunter E. Role of donor genital tract HIV-1 diversity in the transmission bottleneck. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E1156–1163. doi: 10.1073/pnas.1103764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both L, Banyard AC, van Dolleweerd C, Horton DL, Ma JK, Fooks AR. Passive immunity in the prevention of rabies. The Lancet Infectious diseases. 2012;12:397–407. doi: 10.1016/S1473-3099(11)70340-1. [DOI] [PubMed] [Google Scholar]
- Broering TJ, Garrity KA, Boatright NK, Sloan SE, Sandor F, Thomas WD, Jr, Szabo G, Finberg RW, Ambrosino DM, Babcock GJ. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. Journal of virology. 2009;83:12473–12482. doi: 10.1128/JVI.01138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RA, Luciani F, McElroy K, Gaudieri S, Pham ST, Chopra A, Cameron B, Maher L, Dore GJ, White PA, Lloyd AR. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS pathogens. 2011;7:e1002243. doi: 10.1371/journal.ppat.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RT, Gordon FD, Curry MP, Schiano TD, Emre S, Corey K, Markmann JF, Hertl M, Pomposelli JJ, Pomfret EA, Florman S, Schilsky M, Broering TJ, Finberg RW, Szabo G, Zamore PD, Khettry U, Babcock GJ, Ambrosino DM, Leav B, Leney M, Smith HL, Molrine DC. Human monoclonal antibody MBL-HCV1 delays HCV viral rebound following liver transplantation: a randomized controlled study. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2013;13:1047–1054. doi: 10.1111/ajt.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Lanzavecchia A. Broadly Neutralizing Antiviral Antibodies. Annu Rev Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- Dhillon S, Witteveldt J, Gatherer D, Owsianka AM, Zeisel MB, Zahid MN, Rychlowska M, Foung SK, Baumert TF, Angus AG, Patel AH. Mutations within a conserved region of the hepatitis C virus E2 glycoprotein that influence virus-receptor interactions and sensitivity to neutralizing antibodies. Journal of virology. 2010;84:5494–5507. doi: 10.1128/JVI.02153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fofana I, Fafi-Kremer S, Carolla P, Fauvelle C, Zahid MN, Turek M, Heydmann L, Cury K, Hayer J, Combet C, Cosset FL, Pietschmann T, Hiet MS, Bartenschlager R, Habersetzer F, Doffoel M, Keck ZY, Foung SK, Zeisel MB, Stoll-Keller F, Baumert TF. Mutations that alter use of hepatitis C virus cell entry factors mediate escape from neutralizing antibodies. Gastroenterology. 2012;143:223–233. e229. doi: 10.1053/j.gastro.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Tanamy M, Keck ZY, Yi M, McKeating JA, Patel AH, Foung SK, Lemon SM. In vitro selection of a neutralization-resistant hepatitis C virus escape mutant. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19450–19455. doi: 10.1073/pnas.0809879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, Rice CM, Ploss A, Burton DR, Law M. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. Journal of hepatology. 2014;61:S45–57. doi: 10.1016/j.jhep.2014.07.027. [DOI] [PubMed] [Google Scholar]
- Grebely J, Morris MD, Rice TM, Bruneau J, Cox AL, Kim AY, McGovern BH, Shoukry NH, Lauer G, Maher L, Lloyd AR, Hellard M, Prins M, Dore GJ, Page K. Cohort profile: the international collaboration of incident HIV and hepatitis C in injecting cohorts (InC3) study. International journal of epidemiology. 2013;42:1649–1659. doi: 10.1093/ije/dys167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebely J, Page K, Sacks-Davis R, van der Loeff MS, Rice TM, Bruneau J, Morris MD, Hajarizadeh B, Amin J, Cox AL, Kim AY, McGovern BH, Schinkel J, George J, Shoukry NH, Lauer GM, Maher L, Lloyd AR, Hellard M, Dore GJ, Prins M. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology. 2014;59:109–120. doi: 10.1002/hep.26639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nature reviews Gastroenterology & hepatology. 2013;10:553–562. doi: 10.1038/nrgastro.2013.107. [DOI] [PubMed] [Google Scholar]
- Heydarchi B, Center RJ, Gonelli C, Muller B, Mackenzie C, Khoury G, Lichtfuss M, Rawlin G, Purcell DF. Repeated Vaccination of Cows with HIV Env gp140 during Subsequent Pregnancies Elicits and Sustains an Enduring Strong Env-Binding and Neutralising Antibody Response. PloS one. 2016;11:e0157353. doi: 10.1371/journal.pone.0157353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUPAC; IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN) Nomenclature and symbolism for amino acids and peptides. Recommendations 1983. The Biochemical journal. 1984;219:345–373. doi: 10.1042/bj2190345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson DX, Voisset C, Tarr AW, Aung M, Ball JK, Dubuisson J, Persson MA. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16269–16274. doi: 10.1073/pnas.0705522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi K, Faulk K, Purcell RH, Emerson SU. Reproduction in vitro of a quasispecies from a hepatitis C virus-infected patient and determination of factors that influence selection of a dominant species. Journal of virology. 2011;85:3408–3414. doi: 10.1128/JVI.02554-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck Z, Wang W, Wang Y, Lau P, Carlsen TH, Prentoe J, Xia J, Patel AH, Bukh J, Foung SK. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. Journal of virology. 2013;87:37–51. doi: 10.1128/JVI.01941-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck ZY, Angus AG, Wang W, Lau P, Wang Y, Gatherer D, Patel AH, Foung SK. Non-random escape pathways from a broadly neutralizing human monoclonal antibody map to a highly conserved region on the hepatitis C virus E2 glycoprotein encompassing amino acids 412–423. PLoS pathogens. 2014;10:e1004297. doi: 10.1371/journal.ppat.1004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck ZY, Olson O, Gal-Tanamy M, Xia J, Patel AH, Dreux M, Cosset FL, Lemon SM, Foung SK. A point mutation leading to hepatitis C virus escape from neutralization by a monoclonal antibody to a conserved conformational epitope. Journal of virology. 2008;82:6067–6072. doi: 10.1128/JVI.00252-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck ZY, Saha A, Xia J, Wang Y, Lau P, Krey T, Rey FA, Foung SK. Mapping a region of hepatitis C virus E2 that is responsible for escape from neutralizing antibodies and a core CD81-binding region that does not tolerate neutralization escape mutations. Journal of virology. 2011;85:10451–10463. doi: 10.1128/JVI.05259-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck ZY, Xia J, Cai Z, Li TK, Owsianka AM, Patel AH, Luo G, Foung SK. Immunogenic and functional organization of hepatitis C virus (HCV) glycoprotein E2 on infectious HCV virions. Journal of virology. 2007;81:1043–1047. doi: 10.1128/JVI.01710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, Grakoui A, Marcotrigiano J. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, Wilson IA, Law M. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken C, Hraber P, Thurmond J, Yusim K. The hepatitis C sequence database in Los Alamos. Nucleic acids research. 2008;36:D512–516. doi: 10.1093/nar/gkm962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntzen T, Timm J, Berical A, Lennon N, Berlin AM, Young SK, Lee B, Heckerman D, Carlson J, Reyor LL, Kleyman M, McMahon CM, Birch C, Schulze Zur Wiesch J, Ledlie T, Koehrsen M, Kodira C, Roberts AD, Lauer GM, Rosen HR, Bihl F, Cerny A, Spengler U, Liu Z, Kim AY, Xing Y, Schneidewind A, Madey MA, Fleckenstein JF, Park VM, Galagan JE, Nusbaum C, Walker BD, Lake-Bakaar GV, Daar ES, Jacobson IM, Gomperts ED, Edlin BR, Donfield SM, Chung RT, Talal AH, Marion T, Birren BW, Henn MR, Allen TM. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology. 2008;48:1769–1778. doi: 10.1002/hep.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, Ball JK, McKeating JA, Kneteman NM, Burton DR. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nature medicine. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- Liang TJ. Current progress in development of hepatitis C virus vaccines. Nat Med. 2013;19:869–878. doi: 10.1038/nm.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani F, Bretana NA, Teutsch S, Amin J, Topp L, Dore GJ, Maher L, Dolan K, Lloyd AR. A prospective study of hepatitis C incidence in Australian prisoners. Addiction (Abingdon, England) 2014;109:1695–1706. doi: 10.1111/add.12643. [DOI] [PubMed] [Google Scholar]
- Mancini N, Diotti RA, Perotti M, Sautto G, Clementi N, Nitti G, Patel AH, Ball JK, Clementi M, Burioni R. Hepatitis C virus (HCV) infection may elicit neutralizing antibodies targeting epitopes conserved in all viral genotypes. PloS one. 2009;4:e8254. doi: 10.1371/journal.pone.0008254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May S, Ngui SL, Collins S, Lattimore S, Ramsay M, Tedder RS, Ijaz S. Molecular epidemiology of newly acquired hepatitis C infections in England 2008–2011: genotype, phylogeny and mutation analysis. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2015;64:6–11. doi: 10.1016/j.jcv.2014.12.014. [DOI] [PubMed] [Google Scholar]
- Meunier JC, Russell RS, Goossens V, Priem S, Walter H, Depla E, Union A, Faulk KN, Bukh J, Emerson SU, Purcell RH. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the e1 glycoprotein of hepatitis C virus. Journal of virology. 2008;82:966–973. doi: 10.1128/JVI.01872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osburn WO, Fisher BE, Dowd KA, Urban G, Liu L, Ray SC, Thomas DL, Cox AL. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology. 2010;138:315–324. doi: 10.1053/j.gastro.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osburn WO, Snider AE, Wells BL, Latanich R, Bailey JR, Thomas DL, Cox AL, Ray SC. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology. 2014;59:2140–2151. doi: 10.1002/hep.27013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owsianka A, Tarr AW, Juttla VS, Lavillette D, Bartosch B, Cosset FL, Ball JK, Patel AH. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. Journal of virology. 2005;79:11095–11104. doi: 10.1128/JVI.79.17.11095-11104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owsianka AM, Tarr AW, Keck ZY, Li TK, Witteveldt J, Adair R, Foung SK, Ball JK, Patel AH. Broadly neutralizing human monoclonal antibodies to the hepatitis C virus E2 glycoprotein. The Journal of general virology. 2008;89:653–659. doi: 10.1099/vir.0.83386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantua H, Diao J, Ultsch M, Hazen M, Mathieu M, McCutcheon K, Takeda K, Date S, Cheung TK, Phung Q, Hass P, Arnott D, Hongo JA, Matthews DJ, Brown A, Patel AH, Kelley RF, Eigenbrot C, Kapadia SB. Glycan shifting on hepatitis C virus (HCV) E2 glycoprotein is a mechanism for escape from broadly neutralizing antibodies. Journal of molecular biology. 2013;425:1899–1914. doi: 10.1016/j.jmb.2013.02.025. [DOI] [PubMed] [Google Scholar]
- Perotti M, Mancini N, Diotti RA, Tarr AW, Ball JK, Owsianka A, Adair R, Patel AH, Clementi M, Burioni R. Identification of a broadly cross-reacting and neutralizing human monoclonal antibody directed against the hepatitis C virus E2 protein. Journal of virology. 2008;82:1047–1052. doi: 10.1128/JVI.01986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham ST, Bull RA, Bennett JM, Rawlinson WD, Dore GJ, Lloyd AR, White PA. Frequent multiple hepatitis C virus infections among injection drug users in a prison setting. Hepatology. 2010;52:1564–1572. doi: 10.1002/hep.23885. [DOI] [PubMed] [Google Scholar]
- Prentoe J, Velazquez-Moctezuma R, Foung SK, Law M, Bukh J. Hypervariable region 1 shielding of hepatitis C virus is a main contributor to genotypic differences in neutralization sensitivity. Hepatology. 2016 doi: 10.1002/hep.28705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo CEA, Bull R, Luciani F, Page K, Kim AY, Lauer G, Cox AL, Shoukry N, Bruneau J, Morris M, Schinkel J, Applegate T, Maher L, Hellard M, Prins M, Dore GJ, Grebely J, Lloyd AR. The Australian Centre for HIV and Hepatitis Virology Research workshop. The Australian Centre for HIV and Hepatitis Virology Research; Hunter Valley, NSW, Australia: 2015. Global molecular epidemiology of acute hepatitis C infection via phylogenetic analysis of full length genomes across nine international cohorts. [Google Scholar]
- Rothwangl KB, Manicassamy B, Uprichard SL, Rong L. Dissecting the role of putative CD81 binding regions of E2 in mediating HCV entry: putative CD81 binding region 1 is not involved in CD81 binding. Virology journal. 2008;5:46. doi: 10.1186/1743-422X-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks-Davis R, Grebely J, Dore GJ, Osburn W, Cox AL, Rice TM, Spelman T, Bruneau J, Prins M, Kim AY, McGovern BH, Shoukry NH, Schinkel J, Allen TM, Morris M, Hajarizadeh B, Maher L, Lloyd AR, Page K, Hellard M. Hepatitis C Virus Reinfection and Spontaneous Clearance of Reinfection-the InC3 Study. The Journal of infectious diseases. 2015;212:1407–1419. doi: 10.1093/infdis/jiv220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautto G, Tarr AW, Mancini N, Clementi M. Structural and antigenic definition of hepatitis C virus E2 glycoprotein epitopes targeted by monoclonal antibodies. Clinical & developmental immunology. 2013;2013:450963. doi: 10.1155/2013/450963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautto GA, Diotti RA, Clementi M. New therapeutic options for HCV infection in the monoclonal antibody era. The new microbiologica. 2012;35:387–397. [PubMed] [Google Scholar]
- Shimizu YK, Hijikata M, Oshima M, Shimizu K, Alter HJ, Purcell RH, Yoshikura H, Hotta H. Isolation of human monoclonal antibodies to the envelope e2 protein of hepatitis C virus and their characterization. PloS one. 2013;8:e55874. doi: 10.1371/journal.pone.0055874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr A, Lafaye P, Meredith L, Damier-Piolle L, Urbanowicz R, Meola A, Jestin JL, Brown R, McKeating J, Rey F, Ball J, Krey T. An alpaca nanobody inhibits hepatitis C virus entry and cell-to-cell transmission. Hepatology. 2013;58:932–939. doi: 10.1002/hep.26430. [DOI] [PubMed] [Google Scholar]
- Tarr AW, Owsianka AM, Jayaraj D, Brown RJ, Hickling TP, Irving WL, Patel AH, Ball JK. Determination of the human antibody response to the epitope defined by the hepatitis C virus-neutralizing monoclonal antibody AP33. The Journal of general virology. 2007;88:2991–3001. doi: 10.1099/vir.0.83065-0. [DOI] [PubMed] [Google Scholar]
- Tarr AW, Owsianka AM, Timms JM, McClure CP, Brown RJP, Hickling TP, Pietschmann T, Bartenschlager R, Patel AH, Ball JK. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology. 2006;43:592–601. doi: 10.1002/hep.21088. [DOI] [PubMed] [Google Scholar]
- Wang Y, Keck ZY, Foung SK. Neutralizing antibody response to hepatitis C virus. Viruses. 2011;3:2127–2145. doi: 10.3390/v3112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewski LN, El-Diwany R, Munshaw S, Snider AE, Brady JK, Osburn WO, Ray SC, Bailey JR. A Hepatitis C Virus Envelope Polymorphism Confers Resistance to Neutralization by Polyclonal Sera and Broadly Neutralizing Monoclonal Antibodies. Journal of virology. 2016;90:3773–3782. doi: 10.1128/JVI.02837-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Du Y, Le S, Young AP, Zhang TH, Wang Y, Zhou J, Yoshizawa JM, Dong L, Li X, Wu TT, Sun R. Coupling high-throughput genetics with phylogenetic information reveals an epistatic interaction on the influenza A virus M segment. BMC genomics. 2016;17:46. doi: 10.1186/s12864-015-2358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel MB, Fofana I, Fafi-Kremer S, Baumert TF. Hepatitis C virus entry into hepatocytes: Molecular mechanisms and targets for antiviral therapies. J Hepatol. 2011;54:566–576. doi: 10.1016/j.jhep.2010.10.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.