Abstract
FLI1 (Friend leukemia virus integration 1) and IL6 (interleukin 6; IL-6) are associated with Leishmania braziliensis susceptibility. Cutaneous lesions show exaggerated matrix metalloproteinase 1 (MMP1). In other skin diseases, FLI1 promoter methylation reduces FLI1 expression, and low FLI1 down-regulates MMP1. IL-6 increases FLI1 expression. We hypothesized that epigenetic regulation of FLI1 in cutaneous leishmaniasis, together with IL-6, might determine MMP1 expression. While generally low (<10%), percent FLI1 promoter methylation was lower (P=0.001) in lesion biopsies than normal skin. Contrary to expectation, a strong positive correlation occurred between FLI1 methylation and gene expression in lesions (r=0.98, P=0.0005) and in IL-6-treated L. braziliensis-infected macrophages (r=0.99, P=0.0004). In silico analysis of the FLI1 promoter revealed co-occurring active H3K27ac and repressive DNA methylation marks to enhance gene expression. FLI1 expression was enhanced between 3 and 24 hours post infection in untreated (P=0.0002) and IL-6-treated (P=0.028) macrophages. MMP1 was enhanced in lesion biopsies (P=0.0002), induced (P=0.007) in infected macrophages, but strongly inhibited by IL-6. No correlations occurred between FLI1 and MMP1 expression in lesions or infected macrophages (with/without IL-6). We conclude that MMP1 is regulated by factors other than FLI1, and that the influence of IL-6 on MMP1 was independent of its effect on FLI1.
Keywords: leishmaniasis, FLI1, MMP1, IL-6, cutaneous lesions, macrophages
1. Introduction
Leishmaniasis is a disease with a wide spectrum of clinical presentations, ranging from self-limiting lesions to severe mucosal disease. The magnitude of disease severity results from environmental factors such as parasite polymorphism, phlebotomine sandfly components, as well as the host’s immune and genetic background. In particular, a number of studies (reviewed (Blackwell, 2010; Burgner et al., 2006; Castellucci et al., 2014)) have reported the role of host genetic factors in regulating the clinical disease outcome of Leishmania braziliensis infection in humans. The importance of the wound healing processes in cutaneous forms of leishmaniasis has also been demonstrated from studies mapping murine susceptibility genes (Sakthianandeswaren et al., 2010; Sakthianandeswaren et al., 2005; Sakthianandeswaren et al., 2009). In particular, fine mapping in the region of Chromosome 9 in mice (Chromosome 11q24 in humans) identified Fli1 (Friend leukemia virus integration 1; FLI1 in humans) as a novel candidate influencing both resistance to L. major and an enhanced wound healing response (Sakthianandeswaren et al., 2010). Recently we demonstrated (Castellucci et al., 2011) that polymorphism at FLI1 is associated with cutaneous leishmaniasis (CL) caused by L. braziliensis in humans. We have also shown that the C allele at the IL6-174 G/C promoter polymorphism, which determines low levels of IL-6 release from macrophages, is a risk factor for mucosal leishmaniasis that occurs in some individuals following CL disease (Castellucci et al., 2006). IL-6 has been shown by RT-PCR to be expressed in lesions from both cutaneous and mucosal forms of American cutaneous leishmaniasis caused by L. braziliensis (Caceres-Dittmar et al., 1993), as well as in experimental cutaneous lesions caused by L. major infection (Karam et al., 2011). IL-6 is known to increase expression of FLI1 (Thaler et al., 2011).
Recent RNA expression profiling (Novais et al., 2015) has demonstrated that the gene encoding matrix metalloproteinase 1 (MMP1) was amongst the most highly up-regulated genes in CL lesions caused by L. braziliensis infection in our study area. This was of interest since reduction of FLI1 expression in human fibroblasts has been shown to result in down-regulation of MMP1, the authors of this work concluding that FLI1 suppression is involved in activation of the profibrotic gene program in fibroblasts (Nakerakanti et al., 2006). Heavy methylation of the CpG islands in the FLI1 promoter region occurs in fibroblasts and skin biopsies, but not normal fibroblasts or skin, from systemic sclerosis patients (Wang et al., 2006), rendering FLI1 transcriptionally inactive. Of interest too are recent observations that infection of macrophages with live Leishmania parasites is associated with changes in DNA methylation at hundreds of CpG islands (Marr et al., 2014), suggesting that the parasite might influence such epigenetic regulation of gene expression. The aims of the present study were: (i) to examine methylation and expression of FLI1 in normal and lesion skin biopsies from patients with CL; (ii) to determine whether L. braziliensis parasites influence methylation and expression of FLI1 in macrophages infected ex vivo; and (iii) to determine whether or not changes in expression of FLI1 correlate with expression of MMP1 in lesion biopsies and/or infected macrophages.
2. Subjects and methods
2.1. Ethical statement
The study was conducted according to the principles specified in the Declaration of Helsinki and under local ethical guidelines. The study was approved by the institutional review board of the Federal University of Bahia (CEP-UFBA 22/2012) and the Brazilian National Ethical Committee (CONEP: 1258513.1.000.5537). All patients provided written informed consent for the collection of samples and subsequent analysis. Parents or guardians provided informed consent for patients less than 18 years of age. Patients between 12 and 17 years of age provided written assent.
2.2. Study site, diagnosis and sample collection
The study was conducted in the rural area of Corte de Pedra, Bahia, Brazil, where L. braziliensis is endemic. The endemic area of Corte de Pedra is characterized by isolated sites of secondary forest with agricultural activities providing the main source of income for the majority of its inhabitants. The work on farms and homes built in clearings in the woods has increased the population’s exposure to L. braziliensis infection over the last decades (Queiroz et al., 2012). Participants of the present study include CL patients recruited at the health post in Corte de Pedra, the reference center for disease treatment of twenty municipalities. As in our previous studies (Castellucci et al., 2012; Castellucci et al., 2011; Castellucci et al., 2010; Castellucci et al., 2006) CL is defined as the presence of a single chronic ulcerative lesion at a skin site without evidence of mucosal involvement, without evidence of dissemination to 10 or more sites, and confirmed by detection of parasites or two of the three following criteria: positive delayed-type hypersensitivity test, PCR positive to L. braziliensis, and a histopathology consistent with leishmaniasis. Of the 31 patients used in this study, 65% (N=20) presented with a single lesion, 16% (N=5) with 2 lesions, 16% (N=5) with 3 lesions, and 3% (N=1) with 4 lesions. Full clinical details of these 31 patients are included under Supplementary Information, which also provides details of the numbers of patients used in each type of study presented here, including the patient ID (far left column) which allows the reader to determine which pairs of samples were used in each type of study. All cases in the current study also responded to the standard anti-leishmanial therapy subsequent to samples being taken.
2.3. Biopsy specimens and DNA/RNA extraction
Biopsies of CL lesions (taken from the active leading edge at the lesion border) and normal skin (taken from a site remote to the lesion, usually another limb) from the same patient were taken using a 4-mm punch and tissues preserved in RNAlater Solution (Ambion) until the extraction of RNA and DNA. After intensive mechanical maceration using a syringe plunger, RNA and DNA were extracted for the gene expression and epigenetic regulation studies, respectively. RNA was extracted by using the PureLink RNA Mini Kit (Ambion) and DNA extraction was performed using the QIAamp DNA Mini Kit (Qiagen). The protocols recommended by the kit manufacturers were followed precisely. Both, RNA and DNA concentration and integrity were determined by spectrophotometric optical density measurement (260 and 280 nm) and samples stored at −70°C.
2.4. Macrophages cultures and DNA/RNA extraction
Monocyte-derived macrophages (hereinafter referred to as macrophages) were prepared following a method previously shown (Giudice et al., 2012) by our laboratory to yield >99% macrophages characterized by flow cytometry as CD14-positive, CD3-negative, CD19-negative. Briefly, peripheral blood mononuclear cells were separated from blood over Ficoll hypaque (GE Healthcare Bio Sciences AB), and monocytes separated by adherence to plastic. Cells were maintained in Teflon vials in RPMI-1640 (Gibco) supplemented with 10% heat-inactivated fetal calf serum (Sigma), 100 IU/mL penicillin and 100 μg/mL streptomycin (RP-10). After six days culture at 37°C and 5% CO2, differentiated macrophages were harvested by centrifugation, resuspended and allowed to adhere to glass coverslips for 24 hours at 37°C and 5% CO2, in the presence or absence of recombinant human IL-6 (20 ng/mL) (Gibco). Following this incubation period, non-adherent cells were removed by rinsing, and adherent macrophages were cultivated in RP-10, and incubated at 37°C, 5% CO2.
Macrophage cultures were infected with L. braziliensis stationary phase promastigotes at a 5:1 ratio. Uninfected macrophages were used as controls. Infected macrophages were incubated for 2 hours, after which time remaining extracellular parasites were removed by washing in HBSS. At each time point (3 and 24 hours after infection), cells were harvested for DNA and RNA extraction. Cells used for DNA extraction were harvested in 50% fetal bovine serum, 40% RPMI-1640 (Gibco) and 10% DMSO and maintained at −70°C. The remaining cells to be used for matched RNA extraction were harvested in TRIzol Reagent (Invitrogen) and also stored at −70°C until use. DNA and RNA were extracted using the same kits and protocols described above.
2.5. Gene expression of FLI1 and MMP1
Reverse transcription reactions were performed using the commercially available High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), starting from 10μl of total RNA and using the MultiScribe Reverse Transcriptase enzyme, according to the manufacturer’s instructions. cDNA synthesis was carried out using the GeneAmp PCR System 9700 as follows: 25°C for 10 minutes, 37°C for 120 minutes, 85°C for 5 minutes, finally cooling at 4°C; and adjusted to 5ng/μl. Assays containing specific primers and probes for FLI1 (Hs00956709_m1) and MMP1 (Hs00899658_m1) genes were pre-designed by Applied Biosystems®. The mRNA levels within each sample were normalized to the level of the house-keeping gene Beta-actin (Hs01060665_g1). The quantitative RT-PCR (qRT-PCR) individual reactions were prepared from 2 μl of cDNA, 0.5 μl of each assay, 5 μl of TaqMan Universal Mastermix® and 2.5 μl of RNase-DNase free water in a final volume of 10 μl. All samples were run in duplicate and the output level reported as the average of the two samples. Amplification conditions included: 50°C for 2 minutes, 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds and 60°C for 1 minute on the 7500 Real Time PCR System (Applied Biosystems). The data were analyzed by comparing the threshold cycle (Ct), according to the equation 2−ΔΔCT where ΔCT is the Ct value of the target gene subtracted from the Ct of the endogenous house-keeping gene, and ΔΔCT is the ΔCT value of each individual less the median ΔCT of the control group.
2.6. Methylation of FLI1 gene
Analysis of DNA methylation was performed using an EpiTect Methyl II PCR assay specific for the FLI1 promoter CpG island (Reference EPHS102855-1A; Qiagen). This assay is based on the differential cleavage of methylated versus unmethylated genomic DNA when using methylation sensitive or dependent restriction enzymes. Briefly, prior to performing the PCR reactions the DNAs were digested using the EpiTect Methyl II DNA Restriction Kit (Qiagen) according to the manufacturer’s instructions. The percentage of DNA that is fully methylated or fully unmethylated is determined by quantitative PCR (q-PCR) of digestion products. The q-PCR was set up by preparing individual reactions for each of the digestions following the protocol provided with the assay. Cycling was performed by the 7500 Real Time PCR System programmed according to the manufacturer’s instructions. The data were analyzed using the EpiTect Methyl II PCR Array Microsoft Excel template, available at www.sabiosciences.com/dna_methylation_data_analysis.php. This template automatically performs all ΔCt based calculations from the raw threshold cycle (Ct) values to determine gene specific DNA methylation status. The Excel template normalizes the Ct values of both digestions against mock digestion values to calculate and report the percentage of the DNA that is methylated and unmethylated.
2.7. Statistical analysis
Statistical significance was assessed by Mann-Whitney for comparison between two groups, using paired test where appropriate. P<0.05 were considered statistically significant results. Pearson’s correlation coefficients (r) were used to evaluate correlations between gene-gene expression and/or gene-methylation and/or gene-lesion sizes, and lines fitted using linear regression. All statistical analyses were carried out within GraphPad Prism5.
2.8. In silico analysis
We examined regulatory regions in the 5’ and upstream regions of the FLI1 gene using standard and custom tracks in the UCSC Genome Browser (http://genome.ucsc.edu), which brings together public domain data including data from the ENCODE project (2004) to throw light on the regulatory elements that control gene expression.
3. Results
3.1. FLI1-specific methylation and FLI1 expression in normal skin and lesion biopsies from CL patients
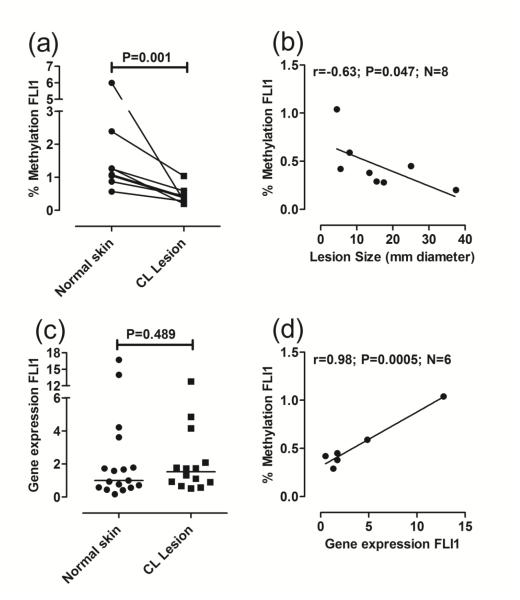
Previous studies observed heavy methylation of upstream CpGs associated with low FLI1 expression in skin biopsies from systemic sclerosis patients compared to normal skin (Wang et al., 2006). Here we find the converse. While the percentage of FLI1 promoter-specific methylated DNA in biopsies was generally low (<10%), we observed a significant reduction in FLI1 methylation in DNA from biopsies of CL lesions caused by L. braziliensis infection compared to paired normal skin from the same donors (Fig. 1a). This suggests that pathology associated with CL results in de-methylation of CpGs in the FLI1 promoter. This is supported too by the observation that the larger the lesion (mm diameter of the primary lesion biopsied), which is positively correlated (r=0.71, P=0.025, N=8 paired observations; data not shown) with lesion duration in days, the lower the percent of FLI1-specific methylation (Fig. 1b; r=−0.63, P=0.047, N=8 paired observations). However, the reduced methylation was not associated with a concomitant increase in FLI1 gene expression in lesion biopsies compared to normal skin (Fig. 1c). Indeed, where paired observations were available, there was a strong positive correlation in lesions between percent methylation of FLI1 and expression of FLI1 at the mRNA level (Fig. 1d; r=0.98, P=0.0005, N=6). Thus our results differ from the conventional interpretation of methylation data, where increased CpG methylation is generally associated with decreased gene expression.
Fig 1. FLI1 promoter-specific DNA methylation and gene expression in lesion compared to normal skin biopsies.
(a) Compares percent methylation in DNA from paired biopsies of normal skin and cutaneous lesions for each individual (N=8 paired observations). DNA methylation was measured using an EpiTect Methyl II PCR assay specific for the FLI1 promoter CpG island (Reference EPHS102855-1A; Qiagen). Following real-time PCR, the data are expressed as percentage of methylated DNA. (b) Plots percent methylation against lesion size expressed as the mean diameter (mm) between two readings taken at right angles to each other. (c) Compares gene expression, represented as 2−ΔΔCT as described in the methods, following qRT/PCR of RNA from biopsies of normal skin (N=17) and cutaneous lesions (N=14). (d) Plots the correlation between percent FLI1 methylation and FLI1 gene expression for paired observations in lesion biopsies. In (a) and (c) P-values are for the non-parametric Mann-Whitney tests comparing data for normal skin biopsies with lesion biopsies. In (b) and (d) results are shown for Pearson’s correlation coefficients (r) and associated P-values are indicated for the number (N) of paired observations available. Lines were fitted using linear regression.
3.2. Influence of L. braziliensis infection on FLI1 methylation and gene expression in macrophages
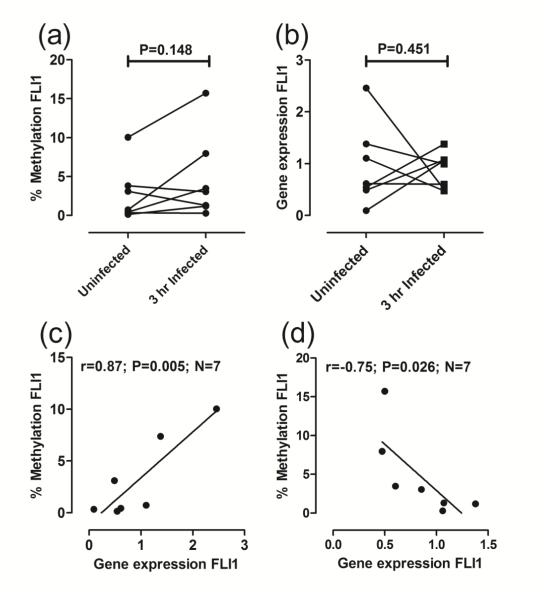
To determine to what extent the reduction in methylation of the FLI1 promoter observed ex vivo in CL lesion samples was due to direct effects of the parasite on its preferred host cell, we also evaluated the percentage of FLI1 methylated DNA in uninfected macrophages compared to macrophages 3 hours following infection with L. braziliensis. Although there was a trend for increased methylation in the FLI-specific CpG with infection (Fig. 2a), this did not achieve statistical significance. Likewise, no significant difference in FLI1 expression was observed between uninfected macrophages and macrophages harvested 3 hours following infection (Fig. 2b). The results for both FLI1 methylation and FLI1 gene expression again showed high individual variability across donors. Where paired observations were available, we again saw a positive correlation between FLI1 methylation and FLI1 gene expression in uninfected macrophages (Fig. 2c; r=0.87, P=0.005, N=7 paired observations). However, following infection (3 hours), a negative correlation between FLI1 methylation and FLI1 gene expression (Fig. 2d; r=−0.75, P=0.043, N=6 paired observations), consistent with the more usual observation that increased methylation of CpGs in a gene promoter results in decreased gene expression.
Fig 2. FLI1 promoter-specific DNA methylation and gene expression in monocyte-derived macrophages.
(a) Compares percent FLI1 methylation and (b) FLI1 gene expression, in DNA from paired samples prepared from uninfected macrophages and at 3 hours following infection with L. braziliensis in vitro. P-values are for the non-parametric Mann-Whitney tests comparing uninfected macrophages with 3 hour infected macrophages. (c) and (d) plot percent methylation against FLI1 gene expression for uninfected and 3 hour infected macrophages, respectively. Results are shown for Pearson’s correlation coefficients (r) and associated P-values are indicated for the number (N) of paired observations available. Lines were fitted using linear regression.
3.3. Influence of IL-6 on FLI1 methylation and gene expression in L. braziliensis infected macrophages
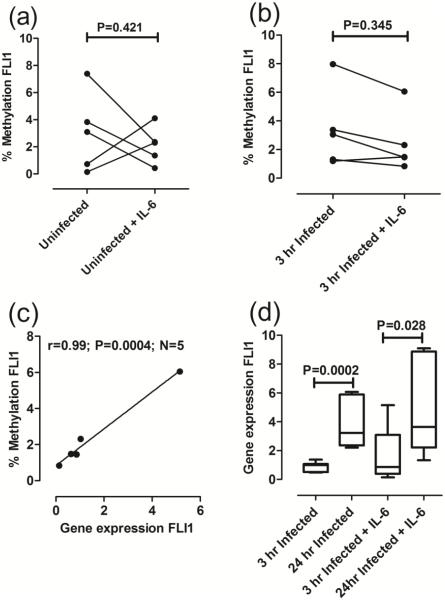
Results from lesion biopsies examined ex vivo (Fig. 1d), and macrophages infected in vitro (Fig. 2d), provide apparently conflicting results, i.e. a positive correlation between FLI1 methylation and FLI1 gene expression in lesions versus a negative correlation observed at 3 hours post infection in macrophages. This likely reflects the more complex set of cellular and cytokine interactions occurring in lesions compared to the isolated interaction between parasites and macrophages in vitro. In particular, the cytokine IL-6 is known to be present in CL lesions (Caceres-Dittmar et al., 1993), and to increase FLI1 gene expression (Thaler et al., 2011), and is likely to impact on interactions between parasites and macrophages. We therefore looked to see whether the presence of IL-6 in infected macrophage cultures would influence percent methylation of CpGs in the FLI1 promoter and FLI1 gene expression. The effect of IL-6 on FLI1 methylation in uninfected cells was variable (Fig. 3a). Of interest, while infection had caused an upward trend in FLI1 methylation (Fig. 2a), IL-6 was associated with a downward trend in FLI1 methylation in 3 hour infected macrophages compared to infection alone (Fig. 3b). While there was no significant correlation between percent FLI1 methylation and FLI1 gene expression in uninfected macrophages (r=−0.67, P=0.109, N=5; data not shown), the combination of infection plus IL-6 resulted in a strong positive correlation (Fig. 3c: r=0.99, P=0.0004, N=5 paired observations) where paired observations were available. Hence, the combination of parasites plus IL-6 provided strong support for the highly significant correlation (Fig. 1d) we had observed between FLI1 methylation and FLI1 gene expression in lesions ex vivo. The downstream effect of infection with/without IL-6 (Fig. 3d) was to enhance FLI1 gene expression at 24 hours compared to 3 hours post infection (P=0.0002 infection only; P=028 infection plus IL-6).
Fig 3. Influence of IL-6 on FLI1 methylation and gene expression in macrophages.
(a) and (b) compare percent FLI1 methylation in DNA from paired samples prepared from uninfected and 3 hour infected macrophages, respectively, with/without IL-6 as indicated. P-values are for the non-parametric Mann-Whitney tests comparing untreated and IL-6 treated macrophages. (c) Plots percent methylation against FLI1 gene expression for 3 hour infected macrophages treated with IL-6. Results are shown for Pearson’s correlation coefficients (r) and associated P-values are indicated for the number (N) of paired observations available. Lines were fitted using linear regression. (d) Compares FLI1 gene expression at 3 and 24 hours after infection, with/without IL-6 treatment as indicated. Results are plotted using Tukey Box and Whiskers plots. The band within the box shows the second quartile or median value. P-values are for the non-parametric Mann-Whitney tests.
3.4. No evidence for an influence of FLI1 on MMP1 expression
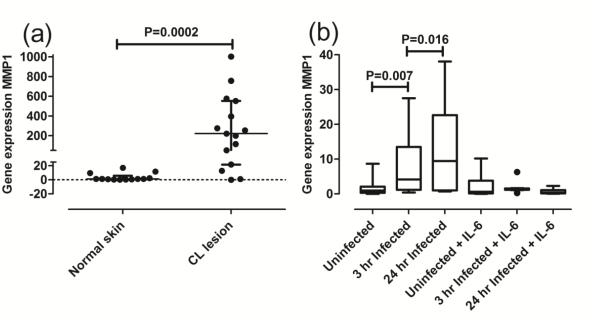
Previous studies suggest that a reduction in FLI1 expression results in a concomitant reduction in MMP1 expression (Nakerakanti et al., 2006). As in the earlier expression profiling studies (Novais et al., 2015), we here find a highly significant increase in MMP1 expression at the mRNA level in lesions of CL patients compared to paired normal skin from the same donors (Fig. 4a; P=0.0002) and in macrophages infected for 3 hours (P=0.007) and 24 hours (P=0.016) in vitro (Fig. 4b). The effect of IL-6 was to inhibit MMP1 expression in macrophages at 3 hours and 24 hours post infection in vitro (Fig. 4b), despite the increase in FLI1 expression (Fig. 3d). Where paired data were available, there were no significant correlations in lesion biopsies between percent methylation of FLI1 and MMP1 gene expression (r=0.33, P=0.237, N=6 paired observations; data not shown), or between FLI gene expression and MMP1 gene expression (r=0.37, P=0.233, N=6 paired observations; data not shown). There was also no correlation between MMP1 gene expression and lesion size (r=0.52, P=293, N=6 paired observations; data not shown). There were no correlations between FLI1 gene expression and MMP1 gene expression at either 3 hours (r=−0.21, P=0.196, N=19 paired observations; data not shown) or 24 hours (r=−0.16, P=0.341, N=9 paired observations; data not shown) post infection. Hence there was no suggestion of any relationship between FLI1 and regulation of MMP1 expression, at least as could be detected at the level of mRNA and DNA isolated from complex CL lesions and studied ex vivo or from macrophages infected with L. braziliensis in vitro.
Fig 4. MMP1 gene expression in biopsies and macrophages.
(a) MMP1 gene expression, represented as 2−ΔΔCT as described in the methods, following qRT/PCR of RNA from biopsies of normal skin (N=13) and cutaneous lesions (N=15). (b) MMP1 gene expression in uninfected macrophages and at 3 and 24 hours after infection, with/without IL-6 treatment as indicated. Results are plotted using Tukey Box and Whiskers plots. The band within the box shows the second quartile or median value. P-values in (a) and (b) are for the non-parametric Mann-Whitney tests.
3.5. In silico analysis of the FLI1 promoter
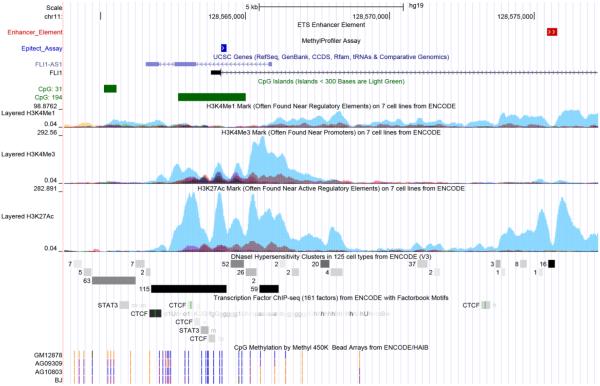
Results presented here showed an apparent contradiction with current convention in showing a positive correlation between FLI1 promoter methylation and FLI1 gene expression, both ex vivo in lesion biopsies (Fig. 1d), and in L. braziliensis infected macrophages treated with IL-6 (Fig. 3b). To try to understand this further, we carried out an in silico analysis of transcription factor binding sites in and around the region of the CpG island in the FLI1 gene promoter that contains the probe used to amplify methylated DNA (Fig. 5). In figure 5, the region of the larger CpG island (CpG:194; annotated in green) covered by the methylation assay used in this study is annotated as “Epitect Assay” in blue. The H3K4Me1 and H3K4Me3 tracks for 7 cell lines from the ENCODE (2004) project provide markers for regulatory elements found near promoter regions, with the strongest signal (turquoise) found in the HUVEC cell line which has been shown to express both FLI1 and the FLI1-AS1 (SENCR) gene encoded as anti-sense on the reverse strand to FLI1 (Bell et al., 2014). There are numerous transcription factor binding sites aligning directly beneath and within the region of CpG:194, including the region covered by our methylation assay. Notably there are relevant transcription factor binding sites that lie outside the region of CpG:194 that may be used when this CpG is methylated, including an alternative STAT3 site that would be influenced by IL-6 signalling (Heinrich et al., 1998). Note the presence too of DNase hypersensitivity clusters that provide further markers of DNA regulatory regions, and the presence of a number of binding sites for the insulator protein CTCF that lie within and outside the region of CpG:194, including one that lies near an ETS enhancer (Donaldson et al., 2005) element (white arrows on red) within a tract of active H3K27ac marks. Recent work has highlighted enhancers that show active H3K27ac marks coexisting with DNA methylation to provide a positive signal for enhanced gene expression (Charlet et al., 2016). Of interest too are tracts showing results of CpG methylation based on ENCODE Illumina Infinium HumanMethylation450 Bead Chip data for 3 fibroblast lines (AG09309, AG10803, BJ) and one B lymphoblastoid cell line (GM12878) that show low methylation (blue vertical lines) across the region of CpG:194 within which both our assay, and that of Wang et al. (Wang et al., 2006), were targeted. Overall, the results of our study suggest that methylation in the FLI1 promoter in CL lesions is generally low, and when present may alter the balance and interaction between potential regulatory regions that lie within or outside the CpG Island in the FLI1 promoter, leading to the novel observation that increased methylation of the CpG is associated with increased rather than decreased FLI1 gene expression.
Fig 5. In silico analysis of the 5’ promoter and intron 1 enhancer regions of the FLI1 gene.
Putative regulatory regions in the 5’ and upstream regions of the FLI1 gene were annotated using standard and custom tracks in the UCSC Genome Browser (http://genome.ucsc.edu). The image is based on GRCh37/hg19 assembly. FLI1 is encoded on the forward strand (only exon 1 is shown), and FLI1-AS1 (SENCR) as anti-sense on the reverse strand. The region of the larger CpG island (CpG:194; annotated in green) covered by the methylation assay used in this study is annotated as “Epitect Assay” in blue. An ETS enhancer element in intron 1 of the FLI1 gene is annotated as white arrows on red. The H3K4Me1, H3K4Me3 and H3K27ac tracks are shown for 7 cell lines from the ENCODE (2004) project, the strongest signal (turquoise) is for the HUVEC cell line. DNase hypersensitivity clusters and individual (STAT3; CTCF) or condensed tracts for transcription factor binding sites are annotated in grey scale (light grey to black). ENCODE results for CpG methylation determined using the Illumina Infinium HumanMethylation450 Bead Chip are shown for one B lymphoblastoid cell line (GM12878) and 3 fibroblast cell lines (AG09309, AG10803, BJ). In these tracts blue vertical lines indicate that sites are unmethylated, purple vertical lines that sites are partially methyled, and orange vertical lines that sites are fully methylated.
4. Discussion
Previous studies have demonstrated a role for wound healing genes in resolution of cutaneous lesions caused by Leishmania spp. (Castellucci et al., 2014), including the Fli1/FLI1 gene in both mice (Sakthianandeswaren et al., 2010) and humans (Castellucci et al., 2011). Skin diseases can share some characteristics (Cho and Blaser, 2012; Naik et al., 2012; Novais et al., 2015), and we were interested in the observation that methylation of CpG islands in the FLI1 promoter reduced FLI1 gene expression in fibroblasts and skin biopsies from systemic sclerosis patients, but not normal fibroblasts or skin (Wang et al., 2006). Reduction of FLI1 expression has been shown to down-regulate MMP1 in human fibroblasts (Nakerakanti et al., 2006) and in LPS treated macrophages (Ho and Ivashkiv, 2010), prompting speculation that FLI1 might be regulating the exaggerated expression of MMP1 that we and others (Novais et al., 2015) have observed in cutaneous lesions caused by L. braziliensis. Our expectation was that the reduced FLI1 methylation we observed in lesions would translate into high FLI1 and high MMP1 gene expression. In the event, the reduction in FLI1 methylation we observed in lesion biopsies ex vivo compared to biopsies from normal skin did not translate into enhanced FLI1 gene expression, and there were no correlations between FLI1 gene expression and MMP1 gene expression ex vivo or in vitro. Hence, we conclude that FLI1 is unlikely to be a major regulator of MMP1 in lesions caused by L. braziliensis infection.
A second factor that we hypothesized might contribute to regulation of MMP1 in lesions was the cytokine IL-6, which has been shown to be present in both cutaneous and mucosal lesions associated with L. braziliensis infection (Caceres-Dittmar et al., 1993) and is known to upregulate FLI1 (Thaler et al., 2011). Initially we proposed that the influence of IL-6 on MMP1 might be secondary to its influence on FLI1 expression (Thaler et al., 2011). However, while FLI1 expression was enhanced in the presence of IL-6 in macrophages infected in vitro, MMP1 was strongly reduced. Hence we conclude that the influence of IL-6 on MMP1 expression in macrophages in vitro was independent of its effect on FLI1. Understanding its role in modulating MMP1 expression in lesions in vivo will require further investigation, but our observations in macrophages infected in vitro suggest that it would be involved in down-regulating, rather than exaggerating, MMP1 expression.
The situation in the CL lesion is complex, with many cell types contributing to tissue injury and ultimately tissue repair and wound healing. Our biopsy samples were taken when patients presented with active lesions, before treatment had commenced. At this stage lesions are characterized by tissue damage and ulceration. MMP1 is a matrix metalloproteinase that breaks down interstitial type I collagens (Brinckerhoff and Matrisian, 2002). Normally in injured human skin, re-epithelialization occurs when keratinocytes migrate along type I collagen, but this can only occur when MMP1 is present to cleave type I collagens and loosen the tight contacts keratinocytes establish with the dermal matrix (Rohani and Parks, 2015). However, the very high levels of MMP1 observed in cutaneous lesions caused by L. braziliensis suggest that the enzyme may be mediating tissue damage rather than repair. Such exaggerated expression of MMP1 has been observed to contribute to tissue destruction and disease progression in a number of other clinical contexts, including progression of rheumatoid arthritis (Xu et al., 2015), chronic obstructive pulmonary disease, acute respiratory distress syndrome, sarcoidosis, and tuberculosis (Elkington and Friedland, 2006). The pro-inflammatory cytokine tumor necrosis factor (TNF), which characterizes human CL caused by L. braziliensis (Castes et al., 1993; Ribeiro-de-Jesus et al., 1998), could be contributing to exaggerated expression of matrix metalloproteinases (Campos et al., 2014; Lehmann et al., 2005). TNF is known to accelerate degradation of type I collagens by MMP-1 leading to their gradual loss in human skin (Agren et al., 2015). As we observed in CL lesions, there was an exaggerated expression of MMP1 following infection of macrophages in vitro. High levels of TNF are produced following infection of monocytes with L. braziliensis (Campos et al., 2014). Thus it seems likely that high levels of MMP1 associated with CL pathogenesis may be contributed to by a direct effect of the parasite on macrophage function. This is also similar to the situation in tuberculosis infection, where exaggerated MMP1 responses are observed in infected macrophages (Elkington et al., 2005), leading to tissue destruction and highlighting MMP1 inhibition as a potential therapeutic target (Elkington and Friedland, 2006).
An unusual finding of our study was that, at the level of individual responses, we observed a strong positive correlation between the degree of FLI1 methylation and FLI1 gene expression, both in lesions ex vivo and in IL-6 treated macrophages infected with L. braziliensis in vitro. This was at variance with the more conventional observation that higher methylation of CpGs in gene promoters is usually associated with lower gene expression, and vice versa. That is, we expect a negative correlation between FLI1 methylation and FLI1 gene expression. One caveat is that the overall level of methylation in our studies of lesion biopsies and macrophages was low (generally <10% methylated). A number of studies now recognize the complexity of interactions between enhancer and promoter elements, and the role of epigenetic modifications in determining gene expression. In particular, that enhancers can show active H3K27ac and repressive DNA methylation marks on the same nucleosomes to provide a positive signal for enhanced gene expression (Charlet et al., 2016), and that the insulator protein CTCF can bind to these enhancers characterized by active histone marks, DNase hypersensitivity, and low methylation (Stadler et al., 2011). Our in silico analysis demonstrated the presence of these enhancer elements in the regulatory regions upstream and in intron 1 of the FLI1 gene. Further research is required to determine how these elements regulate transcription of FLI1 and the overlapping FLI1-AS1 (SENCR) gene encoded on the reverse strand. FLI1-AS1 is known to be co-expressed with FLI1 but does not itself have any cis-acting effect on FLI1 gene expression (Bell et al., 2014). Of interest too are recent observations that infection of macrophages with live Leishmania parasites is associated with changes in DNA methylation at hundreds of CpG islands (Marr et al., 2014), suggesting that the parasite might influence such epigenetic regulation of gene expression. The only conditions under which we observed the expected negative correlation between percent FLI1 promoter methylation and FLI1 gene expression was in macrophages 3 hours after infection in vitro. It will be of interest to determine whether this is a direct or indirect effect of the parasite on macrophage function.
In summary, our work suggests that the exaggerated MMP1 gene expression may play a key role in tissue destruction presented in CL, but that this is more likely through the independent influence of cytokines like TNF than through the action of FLI1. Our data, like others (Novais et al., 2015), points to the possibility that modulation of this pathway through therapeutic intervention might be effective in resolving lesions in CL. Novel pathways to therapy would be welcome in severe forms of disease that are less responsive to treatments currently available.
Supplementary Material
Highlights.
FLI1 and MMP1 were shown by us to important in L. braziliensis infection
Heavy FLI1 promoter methylation is associated with low FLI1 and MMP1 in sclerosis
Conversely, we find FLI1 promoter methylation is reduced in L. braziliensis lesions
Unusually, we find a positive correlation between FLI1 methylation and expression
We find no correlation between FLI1 and MMP1 in lesions or infected macrophages
Acknowledgements
This work was supported by the NIH Grant AI 30639 and CNPq/INCT. We thank the patients for participating in this study. We also acknowledge Ednaldo Lago for the support in the field work in Brazil.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ contributions
LA carried out the field collection and preparation of the samples, as well as the laboratory investigations. LC supervised the laboratory work. LA, LCC and JMB undertook the statistical analyses, interpretation of the data, and preparation of the draft manuscript. JAS, VMA, PM participated in clinical and/or data collection in the field. SEJ helped conceive the study and provided expert guidance on epigenetics methodology and data interpretation. EMC helped conceive the study, initial selection of cases from the health post, and provided the logistical support to make the study possible. JMB participated in the design of the study, made the final interpretation of the data, and finalized preparation of the manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors declare no conflicts of interests.
References
- The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Agren MS, Schnabel R, Christensen LH, Mirastschijski U. Tumor necrosis factor-alpha-accelerated degradation of type I collagen in human skin is associated with elevated matrix metalloproteinase (MMP)-1 and MMP-3 ex vivo. Eur J Cell Biol. 2015;94:12–21. doi: 10.1016/j.ejcb.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, Miano JM. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol. 2014;34:1249–1259. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell JM. Chapter 35: Immunogenetics of host response to parasites in humans. In: Kaufmann SHE, Rouse B, Sacks D, editors. Immunology of Infectious Diseases. ASM Publications; Washington: 2010. pp. 483–490. [Google Scholar]
- Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: a tail of a frog that became a prince. Nat Rev Mol Cell Biol. 2002;3:207–214. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- Burgner D, Jamieson SE, Blackwell JM. Genetic susceptibility to infectious diseases: big is beautiful, but will bigger be even better? Lancet Infect Dis. 2006;6:653–663. doi: 10.1016/S1473-3099(06)70601-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres-Dittmar G, Tapia FJ, Sanchez MA, Yamamura M, Uyemura K, Modlin RL, Bloom BR, Convit J. Determination of the cytokine profile in American cutaneous leishmaniasis using the polymerase chain reaction. 1993;91:500–505. doi: 10.1111/j.1365-2249.1993.tb05931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos TM, Passos ST, Novais FO, Beiting DP, Costa RS, Queiroz A, Mosser D, Scott P, Carvalho EM, Carvalho LP. Matrix metalloproteinase 9 production by monocytes is enhanced by TNF and participates in the pathology of human cutaneous Leishmaniasis. PLoS Negl Trop Dis. 2014;8:e3282. doi: 10.1371/journal.pntd.0003282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci L, Jamieson SE, Almeida L, Oliveira J, Guimaraes LH, Lessa M, Fakiola M, Jesus AR, Nancy Miller E, Carvalho EM, Blackwell JM. Wound healing genes and susceptibility to cutaneous leishmaniasis in Brazil. Infect Genet Evol. 2012;12:1102–1110. doi: 10.1016/j.meegid.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci L, Jamieson SE, Miller EN, de Almeida LF, Oliveira J, Magalhaes A, Guimaraes LH, Lessa M, Lago E, de Jesus AR, Carvalho EM, Blackwell JM. FLI1 polymorphism affects susceptibility to cutaneous leishmaniasis in Brazil. Genes Immun. 2011;12:589–594. doi: 10.1038/gene.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci L, Jamieson SE, Miller EN, Menezes E, Oliveira J, Magalhaes A, Guimaraes LH, Lessa M, de Jesus AR, Carvalho EM, Blackwell JM. CXCR1 and SLC11A1 polymorphisms affect susceptibility to cutaneous leishmaniasis in Brazil: a case-control and family-based study. BMC Med Genet. 2010;11:10. doi: 10.1186/1471-2350-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci L, Menezes E, Oliveira J, Magalhaes A, Guimaraes LH, Lessa M, Ribeiro S, Reale J, Noronha EF, Wilson ME, Duggal P, Beaty TH, Jeronimo S, Jamieson SE, Bales A, Blackwell JM, de Jesus AR, Carvalho EM. IL6 -174 G/C promoter polymorphism influences susceptibility to mucosal but not localized cutaneous leishmaniasis in Brazil. J Infect Dis. 2006;194:519–527. doi: 10.1086/505504. [DOI] [PubMed] [Google Scholar]
- Castellucci LC, Almeida LF, Jamieson SE, Fakiola M, Carvalho EM, Blackwell JM. Host genetic factors in American cutaneous leishmaniasis: a critical appraisal of studies conducted in an endemic area of Brazil. Mem Inst Oswaldo Cruz. 2014;109:279–288. doi: 10.1590/0074-0276140028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castes M, Trujillo D, Rojas ME, Fernandez CT, Araya L, Cabrera M, Blackwell J, Convit J. Serum levels of tumor necrosis factor in patients with American cutaneous leishmaniasis. Biol. Res. 1993;26:233–238. [PubMed] [Google Scholar]
- Charlet J, Duymich CE, Lay FD, Mundbjerg K, Dalsgaard Sorensen K, Liang G, Jones PA. Bivalent Regions of Cytosine Methylation and H3K27 Acetylation Suggest an Active Role for DNA Methylation at Enhancers. Molecular cell. 2016;62:422–431. doi: 10.1016/j.molcel.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson IJ, Chapman M, Kinston S, Landry JR, Knezevic K, Piltz S, Buckley N, Green AR, Gottgens B. Genome-wide identification of cis-regulatory sequences controlling blood and endothelial development. Hum Mol Genet. 2005;14:595–601. doi: 10.1093/hmg/ddi056. [DOI] [PubMed] [Google Scholar]
- Elkington PT, Friedland JS. Matrix metalloproteinases in destructive pulmonary pathology. Thorax. 2006;61:259–266. doi: 10.1136/thx.2005.051979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkington PT, Nuttall RK, Boyle JJ, O'Kane CM, Horncastle DE, Edwards DR, Friedland JS. Mycobacterium tuberculosis, but not vaccine BCG, specifically upregulates matrix metalloproteinase-1. Am J Respir Crit Care Med. 2005;172:1596–1604. doi: 10.1164/rccm.200505-753OC. [DOI] [PubMed] [Google Scholar]
- Giudice A, Vendrame C, Bezerra C, Carvalho LP, Delavechia T, Carvalho EM, Bacellar O. Macrophages participate in host protection and the disease pathology associated with Leishmania braziliensis infection. BMC Infect Dis. 2012;12:75. doi: 10.1186/1471-2334-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Muller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334:297–314. doi: 10.1042/bj3340297. Pt 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HH, Ivashkiv LB. Downregulation of Friend leukemia virus integration 1 as a feedback mechanism that restrains lipopolysaccharide induction of matrix metalloproteases and interleukin-10 in human macrophages. J Interferon Cytokine Res. 2010;30:893–900. doi: 10.1089/jir.2010.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam MC, Al-Kouba JE, Bazzi SI, Smith CB, Leung L. Interleukin-13 reduces hyperalgesia and the level of interleukin-1beta in BALB/c mice infected with Leishmania major with an up-regulation of interleukin-6. J Neuroimmunol. 2011;234:49–54. doi: 10.1016/j.jneuroim.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Lehmann W, Edgar CM, Wang K, Cho TJ, Barnes GL, Kakar S, Graves DT, Rueger JM, Gerstenfeld LC, Einhorn TA. Tumor necrosis factor alpha (TNF-alpha) coordinately regulates the expression of specific matrix metalloproteinases (MMPS) and angiogenic factors during fracture healing. Bone. 2005;36:300–310. doi: 10.1016/j.bone.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Marr AK, MacIsaac JL, Jiang R, Airo AM, Kobor MS, McMaster WR. Leishmania donovani infection causes distinct epigenetic DNA methylation changes in host macrophages. PLoS Pathog. 2014;10:e1004419. doi: 10.1371/journal.ppat.1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, Spencer S, Hall JA, Dzutsev A, Kong H, Campbell DJ, Trinchieri G, Segre JA, Belkaid Y. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakerakanti SS, Kapanadze B, Yamasaki M, Markiewicz M, Trojanowska M. Fli1 and Ets1 have distinct roles in connective tissue growth factor/CCN2 gene regulation and induction of the profibrotic gene program. J Biol Chem. 2006;281:25259–25269. doi: 10.1074/jbc.M600466200. [DOI] [PubMed] [Google Scholar]
- Novais FO, Carvalho LP, Passos S, Roos DS, Carvalho EM, Scott P, Beiting DP. Genomic profiling of human Leishmania braziliensis lesions identifies transcriptional modules associated with cutaneous immunopathology. J Invest Dermatol. 2015;135:94–101. doi: 10.1038/jid.2014.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz A, Sousa R, Heine C, Cardoso M, Guimaraes LH, Machado PR, Carvalho EM, Riley LW, Wilson ME, Schriefer A. Association between an emerging disseminated form of leishmaniasis and Leishmania (Viannia) braziliensis strain polymorphisms. J Clin Microbiol. 2012;50:4028–4034. doi: 10.1128/JCM.02064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-de-Jesus A, Almeida RP, Lessa H, Bacellar O, Carvalho EM. Cytokine profile and pathology in human leishmaniasis. Braz J Med Biol Res. 1998;31:143–148. doi: 10.1590/s0100-879x1998000100020. [DOI] [PubMed] [Google Scholar]
- Rohani MG, Parks WC. Matrix remodeling by MMPs during wound repair. Matrix biology : journal of the International Society for Matrix Biology. 2015;44-46:113–121. doi: 10.1016/j.matbio.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Sakthianandeswaren A, Curtis JM, Elso C, Kumar B, Baldwin TM, Lopaticki S, Kedzierski L, Smyth GK, Foote SJ, Handman E. Fine mapping of Leishmania major susceptibility Locus lmr2 and evidence of a role for Fli1 in disease and wound healing. Infect Immun. 2010;78:2734–2744. doi: 10.1128/IAI.00126-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakthianandeswaren A, Elso CM, Simpson K, Curtis JM, Kumar B, Speed TP, Handman E, Foote SJ. The wound repair response controls outcome to cutaneous leishmaniasis. Proc Natl Acad Sci U S A. 2005;102:15551–15556. doi: 10.1073/pnas.0505630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakthianandeswaren A, Foote SJ, Handman E. The role of host genetics in leishmaniasis. Trends Parasitol. 2009;25:383–391. doi: 10.1016/j.pt.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schubeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Thaler R, Agsten M, Spitzer S, Paschalis EP, Karlic H, Klaushofer K, Varga F. Homocysteine suppresses the expression of the collagen cross-linker lysyl oxidase involving IL-6, Fli1, and epigenetic DNA methylation. J Biol Chem. 2011;286:5578–5588. doi: 10.1074/jbc.M110.166181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006;54:2271–2279. doi: 10.1002/art.21948. [DOI] [PubMed] [Google Scholar]
- Xu Y, Huang Y, Cai D, Liu J, Cao X. Analysis of differences in the molecular mechanism of rheumatoid arthritis and osteoarthritis based on integration of gene expression profiles. Immunol Lett. 2015;168:246–253. doi: 10.1016/j.imlet.2015.09.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.