Abstract
There has been growing interest in utilizing small interfering RNA (siRNA) specific to pro-inflammatory cytokines, such as tumor necrosis factor-α ( TNF-α), in chronic inflammation therapy. However, delivery systems that can increase the distribution of the siRNA in chronic inflammation sites after intravenous administration are needed. Herein we report that innovative functionalization of the surface of siRNA-incorporated poly (lactic-co-glycolic) acid (PLGA) nanoparticles significantly increases the delivery of the siRNA in the chronic inflammation sites in a mouse model. The TNF-α siRNA incorporated PLGA nanoparticles were prepared by the standard double emulsion method, but using stearoyl-hydrazone-polyethylene glycol 2000, a unique acid-sensitive surface active agent, as the emulsifying agent, which renders (i) the nanoparticles PEGylated and (ii) the PEGylation sheddable in low pH environment such as that in chronic inflammation sites. In a mouse model of lipopolysaccharide-induced chronic inflammation, the acid-sensitive sheddable PEGylated PLGA nanoparticles showed significantly higher accumulation or distribution in chronic inflammation sites than PLGA nanoparticles prepared with an acid-insensitive emulsifying agent (i.e., stearoyl-amide-polyethylene glycol 2000) and significantly increased the distribution of the TNF-α siRNA incorporated into the nanoparticles in inflamed mouse foot.
Keywords: Endo-lysosomal escape, inhibition of TNF-α release, LPS-induced chronic inflammation, macrophage uptake
Introduction
Inflammation is a part of the complex biological response of the body in reaction to tissue injury, microbial infection, or autoimmune response.1 It is a defensive attempt by the body to remove harmful stimuli and to start the healing process. If this acute inflammation response fails to resolve and becomes uncontrolled, it may develop to chronic inflammation and/or autoimmune disorders such as rheumatoid arthritis (RA).2 Many of the features of acute inflammation remain in the chronic phase, including increased capillary permeability and the accumulation of white blood cells, although the cells are predominately monocytes/macrophages and lymphocytes, instead of neutrophils as in the acute phase.3
Chronic inflammation involves the continuing induction of inflammatory mediators such as TNF-α, IL-1, and IL-12. In fact, TNF-α plays a key role in chronic inflammation, and anti-TNF-α antibodies have proven effective in treating chronic inflammation.4,5,6,7,8 Because of siRNA's advantageous properties, including high selectivity and potency, decreased immunogenicity, and the ability to be readily chemically synthesized, there is a growing interest in using small interfering RNA (siRNA), such as TNF-α siRNA, to selectively reduce the production of proinflammatory mediators to treat chronic inflammatory diseases.9,10,11 However, due to the short-half life, poor extravasation from blood vessels to target tissues, and poor cellular uptake of siRNA,12,13 TNF-α siRNA had been formulated into nanoparticles or nanocomplexes prepared with polymers (e.g., chitosan), cationic liposomes (e.g., wrapsomes), cationic poly (lactic-co-glycolic) (PLGA) nanoparticles, and PLGA microspheres, and given via various routes (e.g., intravenous, intra-articular, or intraperitoneal) in mouse models to evaluate its activity in treating collagen-induced arthritis.14,15,16,17,18,19
In the present paper, we report the preparation of novel TNF-α-siRNA-encapsulated PLGA nanoparticles by the standard double emulsion method, but using a unique acid-sensitive surface active agent, i.e., the stearoyl-hydrazone-polyethylene glycol 2000 (PHC) previously synthesized in our laboratory by conjugating polyethylene glycol 2000 (PEG2000)-aldehyde with stearic hydrazine,20 as the emulsifying agent, which renders the nanoparticles PEGylated and the PEGylation readily sheddable in low pH environment such as that in chronic inflammation sites.21 Due to the pathological features of chronic inflammation such as increased capillary permeability, angiogenesis, and inflammatory cell infiltration,22 it is expected that TNF-α siRNA carried by nanoparticles will be passively targeted to, and retained at, chronic inflammation sites via the extravasation through leaky vasculature and inflammatory cell-mediated sequestration (ELVIS) mechanism.23,24 The PEGylation of the siRNA-encapsulated PLGA nanoparticles is expected to increase the extravasation of the nanoparticles to chronic inflammation sites, by minimizing the opsonization of the nanoparticles and their subsequent distribution in the mononuclear phagocyte system such as the spleen and liver.25 Importantly, the acid-sensitive sheddable nature of the PEGylation is expected to facilitate the shedding of the PEG chains from the nanoparticles in the relatively lower pH environment in chronic inflammation sites after extravasation, and the PEG-shed nanoparticles will then be readily taken up by inflammatory cells such as macrophages and retained in the chronic inflammation sites. In a mouse model of lipopolysaccharide (LPS)-induced chronic inflammation,26 we showed that the acid-sensitive sheddable PEGylated PLGA nanoparticles significantly increase the accumulation/distribution of the TNF-α siRNA encapsulated in them in the inflamed mouse foot.
Results and discussion
Preparation and characterization of siRNA-incorporated PLGA nanoparticles
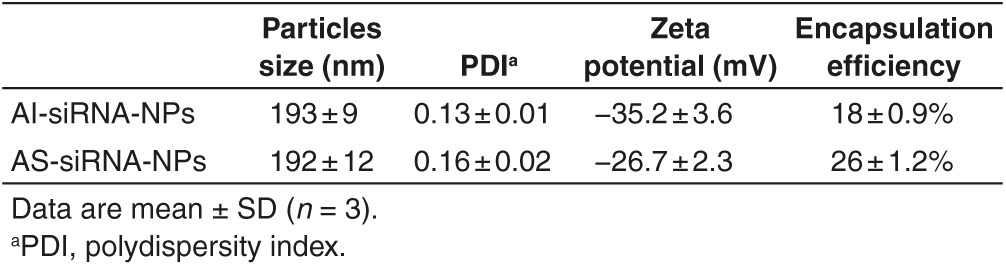
Nanoparticles prepared with PLGA are often used as an siRNA delivery system.18,27,28,29,30,31,32,33 When preparing siRNA-incorporated PLGA nanoparticles using the double emulsion method, poly (vinyl alcohol) (PVA) is usually used as the emulsifying agent to create stable and homogenous emulsions.18,27,28,29,30,31,34 In order to PEGylate the PLGA nanoparticles, copolymers synthesized by conjugating PEG molecules with PLGA or PLA are regularly used.31,32,33 In the present study, we used the acid-sensitive PEG-hydrazone-C18 (PHC) previously synthesized in our laboratory as an emulsifying agent in the second water phase to prepare siRNA-incorporated PLGA nanoparticles using the double emulsion method (i.e., AS-siRNA-NPs, where AS indicates that the nanoparticles are PEGylated with the acid-sensitive sheddable PEG). Using the acid-sensitive PHC as an emulsifying agent is expected to render the resultant nanoparticles PEGylated and the PEGylation readily sheddable in low pH environment. As a control, siRNA-incorporated PLGA nanoparticles were also prepared using the acid-insensitive PEG-amide-C18 (PAC) as an emulsifying agent (i.e., AI-siRNA-NPs, where AI indicates that the nanoparticles are PEGylated with the acid-insensitive PEG). The particle size, zeta potential, and encapsulation efficiency values of the final nanoparticles used in the present study are shown in Table 1. The particle sizes and zeta potentials of the AI-siRNA-NPs and AS-siRNA-NPs are not different from each other. However, the encapsulation efficiency of the siRNA in the AI-siRNA-NPs was lower than in the AS-siRNA-NPs, which is likely due to the differences between PAC and PHC as emulsifying agents.35
Table 1. Characterization of AS-siRNA-NPs and AI-siRNA-NPs.
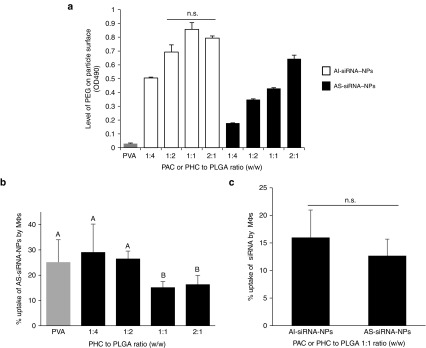
The final nanoparticles were prepared with a PHC to PLGA, or PAC to PLGA, ratio of 1:1 (w/w) based on data from experiments designed to identify the optimal ratio of PHC or PAC to PLGA. The presence of PEG on the surface of the nanoparticles was confirmed using the Lugol's iodine solution assay and a cell uptake assay.36 As shown in Figure 1a, increasing the amount of PAC or PHC, relative to PLGA, in the nanoparticle preparations in general led to the detection of more PEG on the surface of the nanoparticles. For the AI-siRNA-NPs, when the PAC to PLGA ratio reached 1:2 (w/w), the amount of PEG on the surface of the resultant nanoparticles appeared to reach the maximum, because increasing the PAC to PLGA ratio from 1:2 to 1:1 and 2:1 did not further increase the amount of the PEG on the nanoparticles (Figure 1a). However, for the AS-siRNA-NPs, increasing the ratio of PHC to PLGA from 1:1 to 2:1 continued to increase the amount of PEG on the nanoparticles as measured using the Lugol's iodine solution (Figure 1a), likely because differences in the physical and chemical properties of the PAC and PHC.20,37 The differences in the physical and chemical properties of the PAC and PHC are likely also responsible for the overall lower PEG contents in the nanoparticles prepared with the PHC, as compared to those prepared with the same amount of PAC (Figure 1a). To further identify the optimal ratio of the PHC to PLGA, the effect of PEGylation on the cellular uptake of the AS-siRNA-NPs was studied using the murine J774A.1 macrophage cells. As shown in Figure 1b, overall, increasing the ratio of the PHC to PLGA in the nanoparticle preparation decreased the cellular uptake of AS-siRNA-NPs. The PHC to PLGA ratio of 1:1 (w/w) was chosen for further studies, because increasing their ratio from 1:1 to 2:1 (w/w) did not further decrease the uptake of the resultant nanoparticles, although the PEG content in the nanoparticles prepared with the PHC to PLGA ratio of 2:1 was significantly higher than that in the nanoparticles prepared with the PHC to PLGA ratio of 1:1 (Figure 1a), likely because when the level of PEG on the surface of nanoparticles reaches a certain level, further increasing it will not necessarily leads to more inhibition of cell uptake.38
Figure 1.
Confirmation of the presence of PEG on the surface of AS-siRNA-NPs and AI-siRNA-NPs. (a) levels of PEG on the surface of siRNA-incorporated PLGA nanoparticles prepared with various amounts of PHC or PAC. Shown are OD490 values after samples were reacted with Lugol's solution. The numbers in X-axis indicate the ratios of PAC to PLGA, or PHC to PLGA, in the nanoparticle preparations (n.s., no significant difference). (b) The effect of the amount of PHC used in preparing the AS-siRNA-NPs on the cellular uptake of the AS-siRNA-NPs by J774A.1 macrophages. As a control, nanoparticles were also prepared using PVA as an emulsifying agent (A–B, P < 0.05). (c) Cellular uptake of AI-siRNA-NPs and AS-siRNA-NPs prepared with a PAC or PHC to PLGA ratio of 1:1 (w/w) by J774A.1 macrophages. Data shown are mean ± SE (n ≥ 3).
The AI-siRNA-NPs prepared with a PAC to PLGA ratio of 1:1 (w/w) were also chosen for further studies, which allowed the PAC to PLGA and PHC to PLGA ratios identical in the AI-siRNA-NPs and the AS-siRNA-NPs, respectively. Moreover, data from a cell uptake study showed that the extent to which the resultant AI-siRNA-NPs were taken up by J774A.1 cells in culture was not different from that of the AS-siRNA-NPs (Figure 1c).
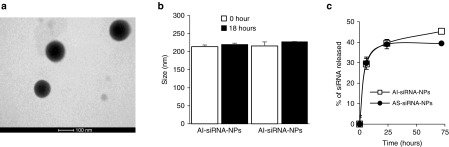
The AS-siRNA-NPs and AI-siRNA-NPs are spherical (Figure 2a and data not shown). Both AS-siRNA-NPs and AI-siRNA-NPs are stable in a simulated biological medium, as their sizes did not significantly change after 18 hours incubation in phosphate-buffered saline (PBS, pH 7.4, 10 mM) supplemented with 10% fetal bovine serum (Figure 2b). The release of siRNA from the nanoparticles is characterized by a typical initial burst release phase, followed by a relatively slower release phase (Figure 2c).
Figure 2.
Physical characterization of the AS-siRNA-NPs. (a) A representative transmission electron microscopy (TEM) image of AS-siRNA-NPs. (b) The stability of AS-siRNA-NPs and AI-siRNA-NPs in a simulated biological medium (i.e., 10% fetal bovine serum (FBS) in PBS, v/v). (c) The in vitro release profiles of siRNA from AI-siRNA-NPs and AS-siRNA-NPs. Data are mean ± SE (n = 3).
Confirmation of the acid-sensitive sheddable PEGylation of the AS-siRNA-NPs by determining the uptake of the siRNA-incorporated PLGA nanoparticles by macrophages in culture
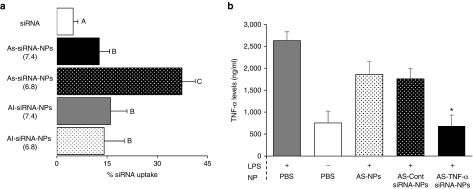
The siRNA was incorporated into PLGA nanoparticles by the traditional double emulsion method, but the acid-sensitive PHC was used as an emulsifying agent. To confirm the acid-sensitive sheddable PEGylation of the nanoparticles, the uptake of the nanoparticles, prepared with fluorescein-labeled siRNA, by mouse J774A.1 macrophages was evaluated after the nanoparticles were pre-incubated in pH 6.8 PBS (10 mM, and pH 7.4 as a control) for 6 hours. It is known that PEGylation inhibits the cellular uptake of nanoparticles.36,39 Because the PHC is acid-sensitive, pre-incubation of the AS-siRNA-NPs in pH 6.8 is expected to facilitate the hydrolysis of the PEG chains from the AS-siRNA-NPs and thus, as shown in Figure 3a, increase their uptake by J774A.1 cells. In contrast, PAC is not more sensitive in lower pH, and the uptake of AI-siRNA-NPs was not affected by pre-incubation of the nanoparticles at pH 6.8 or pH 7.4 (Figure 3a). The increased cellular uptake of the AS-siRNA-NPs, but not the AI-siRNA-NPs, by J774A.1 cells after 6 hours of pre-incubation of the nanoparticles at pH 6.8 indicated the proper PEGylation of the nanoparticles.
Figure 3.
The uptake of the TNF-α siRNA by J774A.1 cells and the down-regulation of TNF-α release by AS-siRNA-NPs. (a) J774A.1 cells (2.5 × 105) were seeded in 24-well plates. After 20 hours, the medium was replaced with serum-free DMEM containing fluorescein-labeled siRNA-AS-NPs or siRNA-AI-NPs that were pre-incubated at pH 6.8 or pH 7.4 for 6 hours. The cells were washed after 45 minutes of incubation and lysed, and the fluorescence intensity was measured (A–C, P < 0.05). (b) J774A.1 cells (5 × 105) were seeded in 12-well plates. After 20 hours, the medium was replaced with serum-free DMEM containing AS-siRNA-NPs prepared with TNF-α siRNA (siRNA = 47.25 ng/ml). After 4 hours of incubation, the medium was replaced with fresh medium containing 10% fetal bovine serum (FBS). After 19 hours, LPS (100 ng/ml) was added, and the cells were incubated for 5 additional hours. The TNF-α levels in the medium were then measured. * The value of the AS-TNF-α-siRNA-NPs is different from that of the siRNA-free AS-NPs and the AS-Cont siRNA-NPs (P < 0.05), but is not different from that of the cells that were not stimulated with LPS.
Inhibition of TNF-α release by TNF-α siRNA-incorporated PLGA nanoparticles
To validate the function of the TNF-α siRNA after it is incorporated into the nanoparticles, J774A.1 cells were treated with AS-TNF-α-siRNA-NPs and stimulated with LPS to evaluate the nanoparticles' ability to downregulate TNF-α expression. As controls, J774A.1 cells were treated with sterile PBS, siRNA-free AS-NPs, or AS-NPs incorporated with a negative control siRNA (i.e., Medium GC Duplex) before they were stimulated with LPS. Cells that were not stimulated with LPS were used as a control as well. As shown in Figure 3b, the AS-TNF-α-siRNA-NPs were able to significantly decrease TNF-α release by J774A.1 cells, to a level that was not significantly different from that of cells not stimulated with LPS, demonstrating that the siRNA in the AS-siRNA-NPs was functional.
Escaping of the siRNA-incorporated PLGA nanoparticles from endo-lysosomes
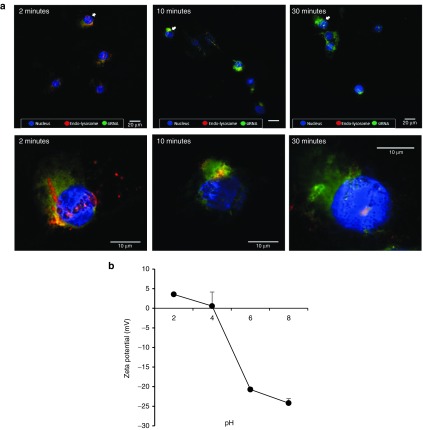
The TNF-α inhibition data shown in Figure 3b indicate that the TNF-α siRNA was successfully released into the cytoplasm of the J774A.1 cells after the uptake of the AS-TNF-α-siRNA-NPs. To confirm that the AS-TNF-α-siRNA-NPs escape the endo-lysosomes after being internalized, J774A.1 cells were stained with the LysoTracker Red DND-99 to identify secondary endosomes and lysosomes, Hoechst 33342 to identify cell nucleus, and then incubated with AS-siRNA-NPs prepared with fluorescein-labeled siRNA for an additional 2, 10 or 30 minutes before they were examined under a confocal microscope. As shown in Figure 4a, the appearance of orange to yellow fluorescence signals in cytoplasm within 2 minutes of incubation indicates the presence of AS-siRNA-NPs in the endo-lysosomal compartment. As the incubation time was increased (to 10 minutes), more green fluorescence and less yellow fluorescence were observed around the nucleus, indicating the escaping of the AS-siRNA-NPs out of the endo-lysosomes. After 30 minutes of incubation, the AS-siRNA-NPs appeared to have completely escaped from the endo-lysosomes (Figure 4a).
Figure 4.
The escape of AS-siRNA-NPs from endo-lysosomes in J774A.1 cells. (a) J774A.1 cells (1 × 105) were seeded on sterile coverslides placed in 6-well plates. Cells were stained with Hoechst 33342 (blue) and a LysoTracker (red) for 30 minutes and then incubated with AS-siRNA-NPs (green) for 2, 10, and 30 minutes before examining under a confocal microscope. White arrows in the upper row indicate cells that are enlarged in the lower row. (b) Zeta potential of AS-siRNA-NPs at a nanoparticle concentration of 100 μg/ml in HEPES (10 mM) buffer of different pH values. Data are mean ± SE (n = 3).
It was suggested by Panyam et al.40 that PLGA nanoparticles exhibit a surface cationization phenomenon at low pH due to hydrogen bonding between the carboxyl groups on PLGA molecules and hydronium molecules. The pH in the late endo-lysosomes is known to be low (i.e., around 4).40,41 As shown in Figure 4b, the AS-siRNA-NPs exhibits neutral to slightly positive zeta potential values when measured at pH 4, which may have caused the swelling and rupture of the lysosomes due to proton-sponge effect and osmotic imbalance.40
Accumulation of siRNA-incorporated PLGA nanoparticles in inflamed mouse foot
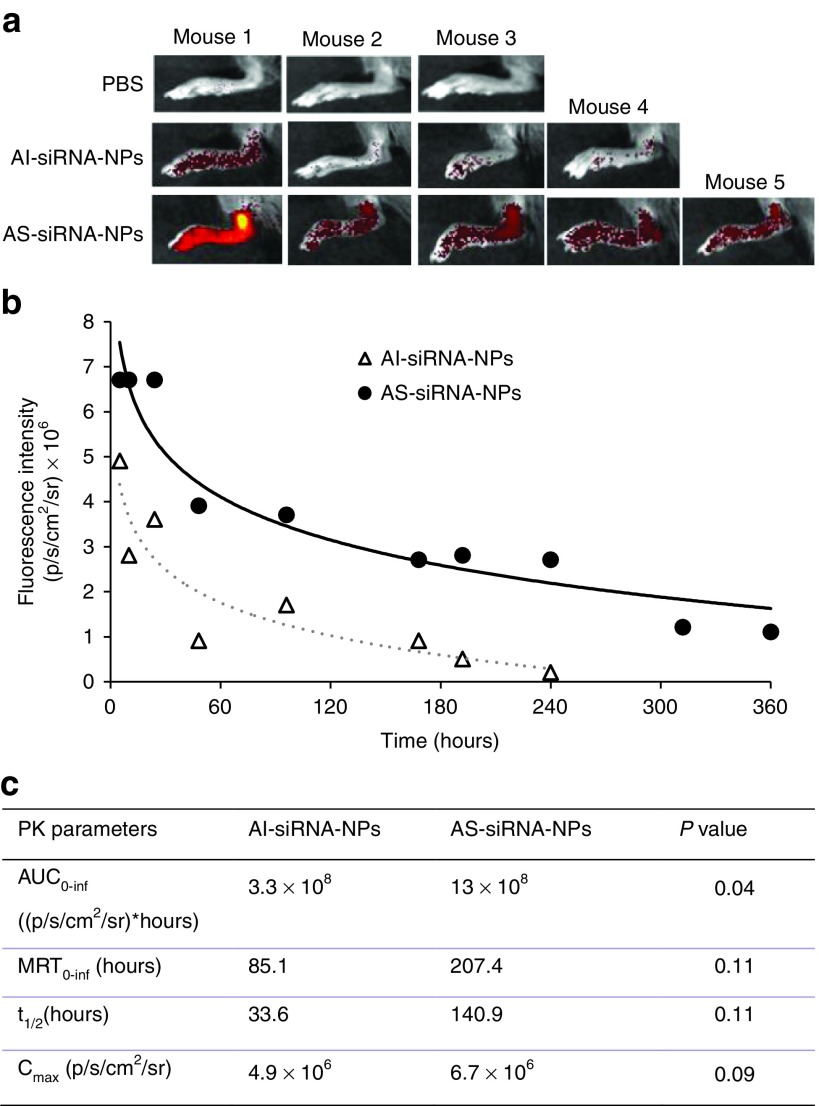
To test the AS-siRNA-NPs' ability to target chronic inflammation sites, the accumulation and elimination of the AI-siRNA-NPs and AS-siRNA-NPs in a mouse foot with LPS-induced chronic inflammation were evaluated. Following Tseng and Kung's report,26 we confirmed that after the injection of LPS in the right hind foot of mice, chronic inflammation, as visualized after intraperitoneal (i.p.) injection of lucigenin, became apparent around 8–9 days later and remained strong when monitored 25 days later (data not shown).
AI-siRNA-NPs or AS-siRNA-NPs, labeled with Cy7.5, were intravenously (i.v.) injected in mice with LPS-induced chronic inflammation in the right hind foot, and the fluorescence intensity of the inflamed foot was measured using an IVIS Spectrum in vivo imaging system at various time points. As shown in Figure 5a, 10 hours after i.v. injection, the fluorescence intensity was significantly higher in the inflamed foot in mice i.v. injected with AS-siRNA-NPs than that in mice i.v. injected with AI-siRNA-NPs; and overall the same trend holds for up to 360 hours (Figure 4b). For example, on day 13 after i.v. injection of the nanoparticles, fluorescence signal cannot be detected in the inflamed foot of mice injected with the C7.5-AI-siRNA-NPs, but was still detectable in the inflamed foot of mice injected with the C7.5-AS-siRNA-NPs. Shown in Figure 5c are selected tissue pharmacokinetic parameters of the AI-siRNA-NPs and AS-siRNA-NPs in inflamed mouse foot. The area under the curve (AUC0-inf) value of the Cy7.5-loaded-AS-siRNA-NPs is approximately fourfold higher than that of the Cy7.5-loaded-AI-siRNA-NPs (AUC values were normalized to that of the PBS group) (P = 0.04).
Figure 5.
The distribution of AI-siRNA-NPs and AS-siRNA-NPs in inflamed mouse foot. (a) In vivo fluorescence images of mouse feet at 10 hours after i.v. injection of PBS, AI-siRNA-NPs or AS-siRNA-NPs. The nanoparticles were labeled with Cy7.5. (b) Fluorescence intensity-time profiles of AI-siRNA-NPs or AS-siRNA-NPs in inflamed mouse feet. (c) A comparison of selected tissue pharmacokinetic parameters of AI-siRNA-NPs and AS-siRNA-NPs in inflamed mouse foot after i.v. injection. Data are mean ± SE (n ≥ 3).
The extravasation through leaky vasculature and subsequent inflammatory cell-mediated sequestration (i.e., ELVIS) phenomenon in inflamed tissues is likely related to the enhanced accumulation and retention of the AS-siRNA-NPs, as compared to the AI-siRNA-NPs.24,42,43 The AS-siRNA-NPs and AI-siRNA-NPs are similar in their particle sizes and zeta potentials (Table 1), and thus they may not be significantly different in their ability to extravasate through the leaky vasculature in the inflamed mouse foot. However, once the nanoparticles extravasate into inflamed tissues, the low pH environment is expected to facilitate the shedding of the PEG chains on the surface of the AS-siRNA-NPs, but not that on the AI-siRNA-NPs, and enable inflammatory cells, such as macrophages, in the inflamed tissues to readily take up the PEG-shed AS-siRNA-NPs, which may explain the enhanced accumulation or “sequestration” of the AS-siRNA-NPs in the inflamed mouse foot, as compared to the AI-siRNA-NPs.
Biodistribution of TNF-α siRNA in AS-siRNA-NPs in mice with LPS-induced chronic inflammation
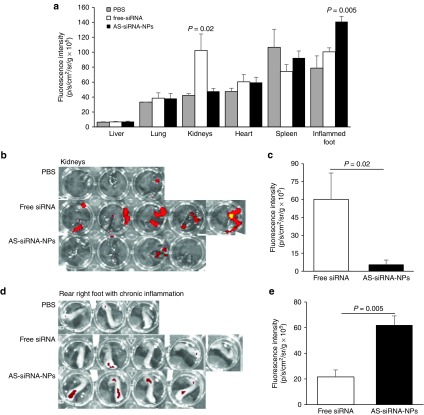
To directly evaluate the extent to which the AS-siRNA-NPs can deliver TNF-α siRNA into chronic inflammation site, while minimizing its distribution in other major organs, the distribution of AS-siRNA-NPs incorporated with fluorescently-labeled TNF-α siRNA in the inflamed foot and major organs in mice with LPS-induced chronic inflammation was analyzed 24 hours after mice were i.v. injected with AS-TNF-α-siRNA-NPs or free TNF-α siRNA. As shown in Figure 6a, significant fluorescent signals were detected only in the kidneys and the inflamed foot of the mice. The fluorescence intensity values in the kidneys were higher in mice injected with free siRNA than in mice injected with the AS-siRNA-NPs (Figure 6a–c), which is expected and has been reported by others,14,15,16,17 because of the high clearance of free siRNA through the kidneys, in addition to its degradation by endogenous enzymes. However, in the inflamed foot, the fluorescence intensity values were higher in mice i.v. injected with the AS-siRNA-NPs than in mice i.v. injected with free siRNA (Figure 6a,d,e), demonstrating that the AS-siRNA-NPs reduced the clearance of the siRNA through the kidneys, but increased the accumulation of siRNA into the inflamed foot. The proper PEGylation is critical for the accumulation of the AS-siRNA-NPs in the inflamed foot, because when AS-siRNA-NPs prepared with an insufficient amount of PHC (i.e., PHC to PLGA ratio of 1:4, instead of 1:1, w/w) were i.v. injected into mice with LPS-induced chronically inflamed foot, the siRNA was mainly accumulated in the liver, and fluorescence signal was not detected in the inflamed foot (Supplementary Figure S1).
Figure 6.
Biodistribution of fluorescently-labeled siRNA, free or in AS-siRNA-NPs, in a mouse model of LPS-induced chronic inflammation. (a) Normalized fluorescence intensities in major organs and inflamed foot in mice with LPS-induced chronic inflammation 24 hours after they were i.v. injected with free siRNA or AS-siRNA-NPs. Data are mean ± SE (n = 3–5). Data are mean ± S.E. (n = 3–5). P values shown are between free siRNA and AS-siRNA-NPs. (b–c) Ex vivo images of kidneys (b) and mean fluorescence intensity in kidneys (c) 24 hours after mice were i.v. injected with free siRNA or AS-siRNA-NPs. (d–e) Ex vivo images of inflamed foot (d) and mean fluorescence intensity in kidneys (e) 24 hours after mice were i.v. injected with free siRNA and AS-siRNA-NPs. Shown in c and e are after the mean fluorescence intensity in the kidneys or the inflamed foot of mice injected with sterile PBS was subtracted.
In conclusion, we prepared PLGA nanoparticles incorporated with siRNA using the standard double emulsion method with a new acid-sensitive stearoyl-PEG molecule (i.e., PHC) as an emulsifying agent. Due to the acid-sensitive nature of the PHC, the AS-siRNA-NPs prepared using the PHC target the siRNA into chronic inflammation sites by increasing the accumulation and retention of the nanoparticles in the inflammation sites. Delivery of TNF-α siRNA using the AS-siRNA-NPs is expected to increase its accumulation and retention in chronic inflammation sites, enhance its uptake by TNF-α producing macrophages in the inflammation sites, and thus help more effectively resolve chronic inflammation, which is currently being tested in a mouse model of collagen-induced arthritis.
Materials and methods
Materials. PHC and PAC were synthesized following our previously published methods.20 PLGA (752H), dichloromethane (DCM), tetrahydrofuran (THF), Lugol's solution, polyvinyl alcohol (PVA) 99+% hydrolyzed, Tris-EDTA (TE), sodium dodecyl sulfate, Triton X-100, N,N-dimethyl-9,9-biacridinium dinitrate (Lucigenin), LPS from Salmonella enterica serotype enteritidis were from Sigma-Aldrich (St. Louis, MO). BLOCK-iT Fluorescent Oligo siRNA was from Life Technologies (Grand Island, NY). Negative control siRNA (Medium GC Duplex), LysoTracker Red DND-99, Hoechst 33342, Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum, and streptomycin/penicillin were from Invitrogen (Carlsbad, CA). TNF-α siRNA (5′-GUCUCAGCCUCUUCUCAUUCCUGCT-3′) was synthesized by Integrated DNA Technologies (Coralville, IA).
Preparation of siRNA-incorporated nanoparticles. The siRNA-incorporated PLGA nanoparticles were prepared using the standard double emulsion method with modifications.44,45 Briefly, a 200 µl solution of 20 µM of siRNA in TE buffer (10 mM Tris–HCl and 1 mM EDTA in water, pH 7.5) was added drop-wise to 800 µl of DCM containing 25 mg of PLGA, and the mixture was emulsified by sonication using an ultrasonic processor (QSONICA Ultrasonicator, Newtown, CT) into a primary W1/O emulsion. PHC or PAC (25 mg) as an emulsifying agent was dissolved in 2.5 ml of water to form the second water phase (W2). The primary emulsion was added drop-wise into the second water phase and further emulsified by sonication for 2 minutes to form a W1/O/W2 double emulsion. The resultant emulsion was diluted with 6 ml of water and stirred for 3 hours at room temperature to evaporate the DCM. The nanoparticles were collected by centrifugation at 17,500× g for 30 minutes at 4 °C, washed, and re-suspended in diethylpyrocarbonate (DEPC)-treated water (Invitrogen, Carlsbad, CA). Nanoparticles prepared with PHC are named AS-siRNA-NPs, where AS indicates that the nanoparticles are PEGylated with the acid-sensitive sheddable PEG (i.e., PHC). Nanoparticles prepared with PAC are named AI-siRNA-NPs, where AI indicates that the nanoparticles were PEGylated with PAC, which is not acid-sensitive. Fluorescently labeled nanoparticles were prepared by using fluorescein-labeled siRNA or by including 200 µg of Cy7.5 in the TE solution that contained siRNA (i.e., W1).
Characterization of siRNA-incorporated nanoparticles. The particle size, polydispersity index (PDI), and zeta potential of the siRNA-incorporated nanoparticles were determined using a Malvern Zeta Sizer Nano ZS (Westborough, MA). Zeta potential was also measured at an AS-siRNA-NPs concentration of 100 μg/ml in 0.001 M HEPES buffers adjusted to pH 2, 4, 6 and 8 with either 0.1 N sodium hydroxide or 0.1 N hydrochloric acid. To determine the encapsulation efficiency of siRNA in the nanoparticles, nanoparticles prepared with fluorescein-labeled siRNA were centrifuged at 15,000 rpm for 30 minutes, and the amounts of fluorescein-labeled siRNA in the supernatant and extracted from the nanoparticle pellet were measured. To extract siRNA, the pellet from the centrifugation was dissolved in 200 μl of THF and then mixed with 1 ml of TE buffer. THF (200 μl) was also added to 1 ml of the supernatant. The fluorescence intensity was measured using a BioTek Synergy HT Multi-Mode Microplate Reader (Winooski, VT, Ex = 485 nm, Em = 528 nm).
The presence of PEG on the surface of the nanoparticles was confirmed using an iodide staining method.4 Briefly, about 5 mg of nanoparticles were re-suspended in 200 µl of water to make a concentrated solution of nanoparticles, and 50 µl of the concentrated nanoparticles were added to a solution that contained 950 µl of PBS (pH 7.4, 10 mM) and 68 µl of Lugol's solution. After 5 minutes of incubation at room temperature, the absorbance (OD490 nm) was measured using a BioTek Synergy HT Multi-Mode Microplate Reader.
Transmission electron microscopy. The morphology of the AS-siRNA-NPs and AI-siRNA-NPs was examined using an FEI Tecnai Transmission Electron Microscope in the Institute for Cellular and Molecular Biology (ICMB) Microscopy and Imaging Facility at The University of Texas at Austin. Carbon-coated 400-mesh grids were activated for 1–2 minutes. One drop of the nanoparticle suspension was deposited on the grids and incubated overnight at room temperature before examination.
In vitro release of siRNA from the nanoparticles. The release of siRNA from the nanoparticles was measured using nanoparticles prepared with fluorescein-labeled siRNA. Briefly, 5 mg of AS-siRNA-NPs or AI-siRNA-NPs were suspended in 1 ml TE buffer in RNase-free Eppendorf tubes, which were then placed in shaker incubator (MAQ 5000, MODEL 4350, Thermo Fisher Scientific, Waltham, MA) (100 rpm, 37 °C). At given time points (6, 24, 48, and 72 hours), the tubes were centrifuged (17,500× g, 30 minutes), and the amount of labeled siRNAs was measured in both the supernatant and the precipitate using a BioTek Synergy HT Multi-Mode Microplate Reader as mentioned above. The percent of siRNA released was calculated using the following equation: % released = 100 X fluorescence intensity in supernatant / (fluorescence intensity in precipitate + fluorescence intensity in supernatant).
In vitro uptake of siRNA-incorporated nanoparticles by macrophages. Murine macrophage J774A.1 cells (American Type Culture Collection, Manassas, VA) were seeded in a 24-well plate (2.5 × 105 cells/well). To study the effect of the acid-sensitive sheddable PEGylation of the nanoparticles on the uptake of the nanoparticles, the AS-siRNA-NPs or AI-siRNA-NPs were pre-incubated in pH 6.8 PBS, or 7.4 as a control, for 6 hours to facilitate the shedding of the PEG before the nanoparticles were added into the cell culture medium. After 45 minutes of co-incubation, the cells were washed with PBS (10 mM, pH 7.4) and lysed with 2% (v/v) sodium dodecyl sulfate and 1% Triton X-100. The fluorescence intensity in the cell lysates was measured (Ex = 485 nm, Em = 528 nm).
Confocal microscopy. Murine macrophage J774A.1 cells were plated 18 hours prior to the experiment on sterile coverslips placed in 6-well plates (1 × 105 cells/well) in 1 ml of growth medium. To study the intracellular distribution of AS-siRNA-NPs, the cells were first pre-incubated for 30 minutes with serum-free growth medium containing of LysoTracker Red DND-99 (10 μg/ml) and Hoechst 33342 (100 μg/ml), and then incubated for 2, 10 or 30 additional minutes with a suspension of the AS-siRNA-NPs (100 μg/ml) containing 224 ng of fluorescein-labeled siRNA. Cells were washed three times with PBS and incubated in HEPES buffer (pH 8.0) and then visualized under a Leica SP5 AOBS WLL confocal microscope (Lecia, Buffalo Grove). Images were captured using Ex = 460 nm, Em = 490 nm for nucleus (Hoechst 33342), Ex = 485 nm, Em = 528 nm for siRNA (fluorescein), and Ex = 577 nm, Em = 590 nm for Lysosomes (LysoTracker Red DND-99). All images were obtained with the same gain and offset. Cells were randomly selected and analyzed using the ImageJ software (NIH, Bethesda, MD).
TNF-α release from J774A.1 macrophages in culture. J774A.1 cells were seeded in 12-well plates (500,000 cells per well). After 20 hours incubation at 37 °C, 5% CO2, the culture medium was replaced with serum-free DMEM and TNF-α–siRNA incorporated AS-NPs to a final siRNA concentration of 50 ng/ml. The culture medium was replaced 4 hours later with fresh DMEM containing 10% fetal bovine serum. Nineteen hours later, LPS was added into the cell culture medium to a final concentration of 100 ng/ml. The cell culture medium was harvested after five additional hours of incubation to measure TNF-α concentration using a mouse TNF-α ELISA Kit (Thermo Fisher Scientific).
LPS-induced mouse model of chronic inflammation. All animal studies were conducted in accordance with the US National Research Council guidelines for the care and use of laboratory animals. The animal protocol was approved by the Institutional Animal Care and Use Committee at The University of Texas at Austin. Female C57BL/6 mice (6–8 weeks) were from Charles River Laboratories (Wilmington, MA). For imaging, mice were fed with alfalfa-free diet (Harlan, Indianapolis, IN) to minimize unwanted background signals. An LPS-induced mouse model of chronic inflammation was established as previously described.26 Briefly, LPS was dissolved in sterile PBS (pH 7.4, 10 mM) at a concentration of 1 mg/ml. A 50 μl of the solution was injected into the right hind footpad of the mice on day 0. On day 8, chronic inflammation was confirmed using an IVIS Spectrum (Caliper, Hopkinton, MA) with a bioluminescence imaging system 20 minutes following i.p. injection of 15 mg/kg of lucigenin (exposure time 60 seconds, large binning, field B). Lucigenin is known to react with the superoxide produced by macrophages during chronic inflammation.26 Mice that did not show significant chronic inflammation were excluded.
Biodistribution studies. To evaluate the kinetics of the accumulation and elimination of the AI-siRNA-NPs and AS-siRNA-NPs in LPS-induced inflamed mouse feet, mice were i.v. injected with AI-siRNA-NPs or AS-siRNA-NPs (NPs were labeled by Cy7.5, 0.2 mg/kg),28 and the inflamed foot (i.e., right hind) was imaged using IVIS Spectrum 5 hours, 10 hours, 24 hours, and 2, 4, 7, 8, 10, 13 and 15 days after the injection. As controls, mice were i.v. injected with sterile PBS. The kinetics data were analyzed using PK Solver.46
In another study in mice with LPS-induced inflamed foot, mice were i.v. injected with PBS, free siRNA, or AS-siRNA-NPs (siRNA was fluorescently-labeled, 0.5 mg/kg). Mice were euthanized 24 hours later to collect inflamed foot and major organs (i.e., heart, kidneys, liver, spleen, and lung). All samples were then imaged using an IVIS Spectrum (Ex = 485 nm, Em = 528 nm). All the fluorescent units are in photons per second per centimeter square per steradian (p/s/cm2/sr).
Statistical analysis. Statistical analyses were completed by performing analysis of variance followed by Fisher's protected least significant difference procedure. A P value of ≤0.05 (two-tail) was considered significant.
SUPPLEMENTARY MATERIAL Figure S1. Representative ex vivo images of major organs 24 h after C57BL/6 mice with LPS-induced chronic inflammation in the right hind foot were i.v. injected with free siRNA or AS-siRNA-NPs that were prepared with a PHC to PLGA ratio of 1:4 (w/w). The siRNA was fluorescently labeled.
Acknowledgments
This work was supported in part by a grant from the US National Institutes of Health (CA135274 to Z.C.) and the Alfred and Dorothy Mannino Fellowship in Pharmacy at UT-Austin (to Z.C). A.M.A. is a King Abdullah International Medical Research Center (KAIMRC) scholar and is supported by the KAIMRC scholarship program. The authors would like to thank Yue Li for her help in acquiring confocal microscopic images.
Supplementary Material
References
- Jahoor, A, Patel, R, Bryan, A, Do, C, Krier, J, Watters, C et al. (2008). Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J Bacteriol 190: 4408–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy, EH and Panayi, GS (2001). Cytokine pathways and joint inflammation in rheumatoid arthritis. N Engl J Med 344: 907–916. [DOI] [PubMed] [Google Scholar]
- Fujiwara, N and Kobayashi, K (2005). Macrophages in inflammation. Curr Drug Targets Inflamm Allergy 4: 281–286. [DOI] [PubMed] [Google Scholar]
- van de Putte, LB, Rau, R, Breedveld, FC, Kalden, JR, Malaise, MG, van Riel, PL et al. (2003) Efficacy and safety of the fully human anti-tumour necrosis factor alpha monoclonal antibody adalimumab (D2E7) in DMARD refractory patients with rheumatoid arthritis: a 12 week, phase II study. Ann Rheum Dis, 62, 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff, MH, Burmester, GR, Kent, JD, Pangan, AL, Kupper, H, Fitzpatrick, SB and Donovan, C(2006)Safety analyses of adalimumab (HUMIRA) in global clinical trials and US postmarketing surveillance of patients with rheumatoid arthritis. Ann Rheum Dis 65: 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber, S. (2011). Certolizumab pegol for the treatment of Crohn's disease.Therap Adv Gastroenterol 4: 375–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Haens, GR.(1999). Infliximab (Remicade), a new biological treatment for Crohn's disease. Ital J Gastroenterol Hepatol 31: 519–520. [PubMed] [Google Scholar]
- Weinblatt, ME, Bingham, CO, 3rd, Mendelsohn, AM, Kim, L, Mack, M, Lu, J, Baker, D and Westhovens, R.(2013). Intravenous golimumab is effective in patients with active rheumatoid arthritis despite methotrexate therapy with responses as early as week 2: results of the phase 3, randomised, multicentre, double-blind, placebo-controlled GO-FURTHER trial. Ann Rheum Dis 72: 381–389. [DOI] [PubMed] [Google Scholar]
- Fujita, Y, Takeshita, F, Kuwano, K and Ochiya, T (2013). RNAi therapeutic platforms for lung diseases. Pharmaceuticals (Basel) 6: 223–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet, DW, Nelson, J, Tucker, CE, Zack, PM, Nixon, K, Bolin, R et al. (2000). Pharmacokinetics and safety of an anti-vascular endothelial growth factor aptamer (NX1838) following injection into the vitreous humor of rhesus monkeys. Pharm Res 17: 1503–1510. [DOI] [PubMed] [Google Scholar]
- Wang, J, Lu, Z, Wientjes, MG and Au, JL (2010). Delivery of siRNA therapeutics: barriers and carriers. AAPS J 12: 492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, S, Dagnaes-Hansen, F, Nielsen, EJ, Wengel, J, Besenbacher, F, Howard, KA et al. (2009). The effect of chemical modification and nanoparticle formulation on stability and biodistribution of siRNA in mice. Mol Ther 17: 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng, Q, Woodle, MC, Lu, PY and Mixson, AJ (2009). Advances in Systemic siRNA Delivery. Drugs Future 34: 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, KA, Paludan, SR, Behlke, MA, Besenbacher, F, Deleuran, B and Kjems, J (2009). Chitosan/siRNA nanoparticle-mediated TNF-alpha knockdown in peritoneal macrophages for anti-inflammatory treatment in a murine arthritis model. Mol Ther 17: 162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, SS, Ye, C, Kumar, P, Chiu, I, Subramanya, S, Wu, H et al. (2010). Targeted delivery of siRNA to macrophages for anti-inflammatory treatment. Mol Ther 18: 993–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano, Y, Yagi, N, Onoue, I, Kaneko, K, Miyasaka, N and Nanki, T (2012). Arthritic joint-targeting small interfering RNA-encapsulated liposome: implication for treatment strategy for rheumatoid arthritis. J Pharmacol Exp Ther 340: 109–113. [DOI] [PubMed] [Google Scholar]
- Présumey, J, Salzano, G, Courties, G, Shires, M, Ponchel, F, Jorgensen, C et al. (2012). PLGA microspheres encapsulating siRNA anti-TNFalpha: efficient RNAi-mediated treatment of arthritic joints. Eur J Pharm Biopharm 82: 457–464. [DOI] [PubMed] [Google Scholar]
- te Boekhorst, BC, Jensen, LB, Colombo, S, Varkouhi, AK, Schiffelers, RM, Lammers, T et al. (2012). MRI-assessed therapeutic effects of locally administered PLGA nanoparticles loaded with anti-inflammatory siRNA in a murine arthritis model. J Control Release 161: 772–780. [DOI] [PubMed] [Google Scholar]
- Lee, SJ, Lee, A, Hwang, SR, Park, JS, Jang, J, Huh, MS et al. (2014). TNF-α gene silencing using polymerized siRNA/thiolated glycol chitosan nanoparticles for rheumatoid arthritis. Mol Ther 22: 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S, Wonganan, P, Lansakara-P, DS, O'Mary, HL, Li, Y and Cui, Z (2013). The effect of the acid-sensitivity of 4-(N)-stearoyl gemcitabine-loaded micelles on drug resistance caused by RRM1 overexpression. Biomaterials 34: 2327–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr, M, Garvey, K, Bold, AM, Kendall, MJ and Bacon, PA (1985). Significance of the hydrogen ion concentration in synovial fluid in rheumatoid arthritis. Clin Exp Rheumatol 3: 99–104. [PubMed] [Google Scholar]
- Pène, J, Chevalier, S, Preisser, L, Vénéreau, E, Guilleux, MH, Ghannam, S et al. (2008). Chronically inflamed human tissues are infiltrated by highly differentiated Th17 lymphocytes. J Immunol 180: 7423–7430. [DOI] [PubMed] [Google Scholar]
- Wang, D and Goldring, SR (2011). The bone, the joints and the Balm of Gilead. Mol Pharm 8: 991–993. [DOI] [PubMed] [Google Scholar]
- Yuan, F, Quan, LD, Cui, L, Goldring, SR and Wang, D (2012). Development of macromolecular prodrug for rheumatoid arthritis. Adv Drug Deliv Rev 64: 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens, DE 3rd and Peppas, NA (2006). Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm 307: 93–102. [DOI] [PubMed] [Google Scholar]
- Tseng, JC and Kung, AL (2013). In vivo imaging method to distinguish acute and chronic inflammation. J Vis Exp 78: e50690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cun, D, Jensen, DK, Maltesen, MJ, Bunker, M, Whiteside, P, Scurr, D et al. (2011). High loading efficiency and sustained release of siRNA encapsulated in PLGA nanoparticles: quality by design optimization and characterization. Eur J Pharm Biopharm 77: 26–35. [DOI] [PubMed] [Google Scholar]
- Patil, Y and Panyam, J (2009). Polymeric nanoparticles for siRNA delivery and gene silencing. Int J Pharm 367: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C, Mei, H, Shi, W, Deng, J, Zhang, B, Guo, T et al. (2013). EGFP-EGF1-conjugated PLGA nanoparticles for targeted delivery of siRNA into injured brain microvascular endothelial cells for efficient RNA interference. PLoS One 8: e60860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, J, Sun, Y, Shi, QS, Liu, PF, Zhu, MJ, Wang, CH et al. (2012). Biodegradable nanoparticles of mPEG-PLGA-PLL triblock copolymers as novel non-viral vectors for improving siRNA delivery and gene silencing. Int J Mol Sci 13: 516–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, XZ, Dou, S, Sun, TM, Mao, CQ, Wang, HX and Wang, J (2011). Systemic delivery of siRNA with cationic lipid assisted PEG-PLA nanoparticles for cancer therapy. J Control Release 156: 203–211. [DOI] [PubMed] [Google Scholar]
- Zhu, X, Xu, Y, Solis, LM, Tao, W, Wang, L, Behrens, C et al. (2015). Long-circulating siRNA nanoparticles for validating Prohibitin1-targeted non-small cell lung cancer treatment. Proc Natl Acad Sci U S A 112: 7779–7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson, S, Case, RI, Cole, RO, Hwang, J, Kabir, SR, Lazarus, D et al. (2016). Tumor selective silencing using an RNAi-conjugated polymeric nanopharmaceutical. Mol Pharm 13: 737–747. [DOI] [PubMed] [Google Scholar]
- Wischke, C and Schwendeman, SP (2008). Principles of encapsulating hydrophobic drugs in PLA/PLGA microparticles. Int J Pharm 364: 298–327. [DOI] [PubMed] [Google Scholar]
- Zhu, S, Niu, M, O'Mary, H and Cui, Z (2013). Targeting of tumor-associated macrophages made possible by PEG-sheddable, mannose-modified nanoparticles. Mol Pharm 10: 3525–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazile, D, Prud'homme, C, Bassoullet, MT, Marlard, M, Spenlehauer, G and Veillard, M (1995). Stealth Me.PEG-PLA nanoparticles avoid uptake by the mononuclear phagocytes system. J Pharm Sci 84: 493–498. [DOI] [PubMed] [Google Scholar]
- Zhu, S, Lansakara-P, DS, Li, X and Cui, Z (2012). Lysosomal delivery of a lipophilic gemcitabine prodrug using novel acid-sensitive micelles improved its antitumor activity. Bioconjug Chem 23: 966–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hak, S, Helgesen, E, Hektoen, HH, Huuse, EM, Jarzyna, PA, Mulder, WJ et al. (2012). The effect of nanoparticle polyethylene glycol surface density on ligand-directed tumor targeting studied in vivo by dual modality imaging. ACS Nano 6: 5648–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok, H, Park, JW and Park, TG (2008). Enhanced intracellular delivery of quantum dot and adenovirus nanoparticles triggered by acidic pH via surface charge reversal. Bioconjug Chem 19: 797–801. [DOI] [PubMed] [Google Scholar]
- Panyam, J, Zhou, WZ, Prabha, S, Sahoo, SK and Labhasetwar, V (2002). Rapid endo-lysosomal escape of poly(DL-lactide-co-glycolide) nanoparticles: implications for drug and gene delivery. FASEB J 16: 1217–1226. [DOI] [PubMed] [Google Scholar]
- Mukherjee, S, Ghosh, RN and Maxfield, FR (1997). Endocytosis. Physiol Rev 77: 759–803. [DOI] [PubMed] [Google Scholar]
- Baluk, P, Hirata, A, Thurston, G, Fujiwara, T, Neal, CR, Michel, CC et al. (1997). Endothelial gaps: time course of formation and closure in inflamed venules of rats. Am J Physiol 272(1 Pt 1): L155–L170. [DOI] [PubMed] [Google Scholar]
- van den Hoven, JM, Van Tomme, SR, Metselaar, JM, Nuijen, B, Beijnen, JH and Storm, G (2011). Liposomal drug formulations in the treatment of rheumatoid arthritis. Mol Pharm 8: 1002–1015. [DOI] [PubMed] [Google Scholar]
- Cun, D, Jensen, DK, Maltesen, MJ, Bunker, M, Whiteside, P, Scurr, D et al. (2011). High loading efficiency and sustained release of siRNA encapsulated in PLGA nanoparticles: quality by design optimization and characterization. Eur J Pharm Biopharm 77: 26–35. [DOI] [PubMed] [Google Scholar]
- Leach, JK, Patterson, E and O'Rear, EA (2004). Encapsulation of a plasminogen activator speeds reperfusion, lessens infarct and reduces blood loss in a canine model of coronary artery thrombosis. Thromb Haemost 91: 1213–1218. [DOI] [PubMed] [Google Scholar]
- Zhang, Y, Huo, M, Zhou, J and Xie, S (2010). PKSolver: an add-in program for pharmacokinetic and pharmacodynamic data analysis in Microsoft Excel. Comput Methods Programs Biomed 99: 306–314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.