Abstract
We have evaluated the effect of heart extracellular matrix (ECM) on the cardiomyocyte differentiation of mouse embryonic stem cells (ES cells) using de-cellularized heart tissue. Several lines of evidence indicate that ECM plays significant roles in cell proliferation, cell death and differentiation, but role of ECM possessing a 3D structure in differentiation has not been studied in detail. We found that there are substantial differences in the quantitative protein profiles of ECM in SDS-treated heart tissue compared to that of liver tissue, as assessed by iTRAQ™ quantitative proteomics analysis. When mouse ES cells were cultured on thin (60 μm) sections of de-cellularized tissue, the expression of cardiac myosin heavy chain (cMHC) and cardiac troponin I (cTnI) was high in ES cells cultured on heart ECM compared with those cultured on liver ECM. In addition, the protein expression of cMHC and cTnI was detected in cells on heart ECM after 2 weeks, which was not detectable in cells on liver ECM. These results indicate that heart ECM plays a critical role in the cardiomyocyte differentiation of ES cells. We propose that tissue-specific ECM induced cell lineage specification through mechano-transduction mediated by the structure, elasticity and components of ECM.
Keywords: Embryonic stem cells, Cardiomyocyte, Differentiation, Extracellular matrix, Mechanobiology
Ischemic heart disease is a major cause of mortality in many countries. For decades, attempts have been made to show the safety, practicability and effectiveness of stem cell-based therapies for tissue regeneration in the ischemic myocardium. Regarding recent stem cell studies, an important issue is how to induce stem cell differentiation into cardiomyocytes. Various chemical factors and approaches have been tried for the differentiation of several stem cell types, such as embryonic stem (ES) cells, bone marrow mesenchymal stem cells (MSCs), and induced pluripotent stem (iPS) cells, into cardiomyocytes (1–3). Though it has been shown that these cells have the capacity to differentiate into cardiomyocytes, a complete method exhibiting a high efficiency of cardiomyocyte differentiation has not been established yet.
Recently, it has become gradually recognized that environmental factors other than chemical factors contribute to stem cell activity. The extracellular space size and shape have been shown to regulate the differentiation of human MSCs into adipocytes or osteoblasts (4). A study involving a pore-size gradient material showed that there is an optimal pore-size for chondrogenesis by adipose stem cells (5), and hematopoietic differentiation ofmouse ES cells (6). On the otherhand, the importance of matrixelasticity has been demonstrated by another group (7), who showed that MSCs specify lineage with extreme sensitivity to tissue-level elasticity. They showed that, (i) soft matrices that mimic the brain are neurogenic, (ii) stiffer matrices that mimic muscle are myogenic, and (iii) comparatively rigid matrices resemble collagenoustissues. Additionally, ithas been shownthatdifferences in ECM components have different effects through activation of individual integrin family receptors as mechanosensors (8). In fact, in cardiomyocyte differentiation, cardiomyocyte-like cells (CLCs) show enhanced GATA4 and Nkx2.5 expression when cultured on a collagen V as compared to a collagen I matrix (9).
Although a role of ECM in differentiation has been indicated, to the best of our knowledge, the effect of ECM in retaining its components and 3D structure on ES cell differentiation has not been studied in vitro. Thus, we hypothesized that the commitment of stem cells to become cardiomyocytes is mediated by heart-specific ECM due to its structure, elasticity and components. In this study, we first compared protein profiles of ECM proteins between heart and liver ECM using iTRAQ™ quantitative proteomics approach, and found that the ECM composition largely differs between these two tissues. Next, we cultured ES cells on de-cellularized heart and liver ECM, and examined whether cells are directed to heart or liver differentiation. We found that ES cells on heart ECM differentiate into cardiomyocytes, showing that tissue-specific ECM directs ES cell differentiation.
MATERIALS AND METHODS
De-cellularization of mouse tissues
Animal tissues were obtained via the tissue-sharing system of the National University of Singapore. De-cellularization of whole heart and liver was performed by a chemical method according to a reported protocol (10,11), and the whole de-cellularized samples were subjected to quantitative proteomics analysis. To obtain ECM sheets from heart and liver tissues, tissues were embedded in O.C.T. compound (Sakura Finetek, Japan) on dry ice, and 60 μm thick sections were made using a cryostat (CM3050-S, Leica, Wetzlar, Germany). To avoid the tissue breakage from procedure of de-cellularization, we chose the maximum thickness that can be cut by our microtome. Tissue sections were attached to deckgläser cover glasses (ϕ 25 mm) (Marienfeld, Lauda-Konigshofen, Germany), and de-cellularized by procedures as follows. First, thin tissue sections were treated with 1% SDS for 1 h, and then washed three times with PBS. Then, thin tissue sections were washed with 1% Triton X-100 in PBS for 10 min, and rinsed four times for 5 min in PBS. To remove DNA, thin tissue sections were treated with DNase (Qiagen, Hilden, Germany) for 1 h at room temperature, and then washed three times in PBS. For sterilization, de-cellularized thin tissue sections were UV-irradiated for 15 min in a clean work station.
Quantitative proteomics analysis
iTRAQ quantitative proteomics analysis was performed by the method described previously, with modifications (12). SDS-treated samples were first digested with 0.5 mg/ml Collagenase NB 8 Broad Range (SERVA Electrophoresis, Heidelberg, Germany) in Krebs Henseleit buffer (118 mM NaCl, 4.7 mM KCl, 1.64 mM MgCl2, 24.88 mM NaHCO3, 1.18 mM KH2PO4, 5.55 mM Glucose, and 2.0 mM Na-Pyruvat) at 37°C for 17 h. The samples were then centrifuged at 12,000 × g for 30 min. The supernatants containing 100 μg each of the proteins were reduced with 5 mM tris-(2-carboxyethyl) phosphine (TCEP), alkylated with 10 mM methyl methane-thiosulfonate (MMTS) and digested with trypsin (Promega, Madison, WI, USA), with a trypsin to protein ratio of 1:10 (w/w) at 37°C for 16 h. Digested peptides were then dried and reconstituted with 0.5 M triethylammonium bicarbonate (TEAB). Samples were labeled using iTRAQ Reagent 8-Plex kit (AB SCIEX, Foster City, CA, USA) based on the manufacturer’s protocol. Duplicate liver samples were labeled with 113 and 116 iTRAQ reagents, while duplicate heart samples were labeled with 114 and 118 reagents. To remove all interfering substances, strong cation exchange chromatography was carried out using the cation exchange system provided in the iTRAQ Method Development Kit (AB SCIEX). Peptide separation was performed on an Ultimate 3000 liquid chromatography (LC) system (Thermo Fisher Scientific, Waltham, MA, USA) equipped with a Probot MALDI spotting device with a 300 μm × 150 mm Symmetry C18 NanoEase™ reversed phase (RP) column (C18 SYMMETRY 300™) (Waters, Milford, MA, USA). Mobile phases A and B used for second – the separation were 100% water with 0.05% TFA and 100% ACN with 0.04% TFA respectively. The sequential gradients were 2–5% B in 10 min, 5–14% in 120 min, 14–25% in 60 min and 25–40% in 10 min, at a flow rate of 2.7 μl/min. LC fractions eluted from the RP column were then mixed directly with MALDI matrix solution (7 mg/ml α-cyano-4-hydroxycinnamic acid (CHCA) and 130 μg/ml ammonium citrate in 75% ACN) at a flow rate of 5.4 μl/min via a 25 nl mixing tee before they were spotted in 28 × 44 spot arrays on 123 × 81 mm Opti-TOF™ LC/MALDI Inserts (AB SCIEX) using a Probot Micro Precision Fraction collector (Thermo Fisher Scientific), at a frequency of one spot per 5 s.
MS/MS analyses were performed on a 4800 MALDI-TOF/TOF Analyzer (AB SCIEX) operating in MS-positive reflector mode. Protein identification and relative iTRAQ quantification were performed with ProteinPilot Software 2.0.1 (AB SCIEX) using the Paragon algorithm for the peptide identification which was further processed by Pro-Group algorithm where isoform-specific quantification was adopted to trace the differences between expressions of various isoforms.
Long time culture of ES cell on de-cellularized tissue ECM
ES cells were maintained in high glucose DMEM (Gibco, Gaithersburg, MD, USA) containing 10% FBS (Sigma–Aldrich, St. Louis, MO), 1% penicillin (Sigma–Aldrich), 1% streptomycin (Sigma–Aldrich), 1% GlutaMAX-1 (Gibco), 1% non-essential amino acid (Gibco), 1% nucleosides (Millipore), 1% sodium pyruvate (Sigma–Aldrich), 0.1% 2-mercaptoethanol (Sigma–Aldrich), and 0.1% leukemia inhibitory factor (LIF) (Sigma–Aldrich). For prior differentiation into cardiomyocyte, ES cells were treated with 5-azacytidine (10 μM) (Sigma–Aldrich) in ES culture medium without LIF. After 24 h, the medium was changed to ES culture medium without LIF, and the cells were cultured for another 24 h. Then, de-cellularized ECM placed on ϕ 25 mm cover glass was placed in a 6-well culture plate, and ES cells were seeded at 2 × 104 cells/well, and cultured in ES cell medium without LIF at 37°C under 5% CO2. The medium was changed every day.
Immunohistochemistry
Cultured cells were washed once with PBS, and then fixed with 4% paraformaldehyde (PFA) for 20 min at 4°C. Samples were washed three times, each for 5 min, in PBS and then treated with CAS-BLOCK (Invitrogen, Carlsbad, CA, USA) solution for 30 min at room temperature. Primary antibodies were diluted with CAS-BLOCK, and then applied to samples at 4°C overnight. We used following antibodies at 1:100 dilutions: collagen I antibody (abcam, Cambridge, UK), collagen III antibody (abcam), collagen V antibody (abcam), cardiac myosin heavy chain (cMHC) antibody, and myosin light chain 3 (MLC3) antibody (abcam). After rinsing four times, each for 5 min in PBS, samples were incubated at the room temperature for 1.5 h with fluorescent secondary antibodies (Invitrogen). Samples were washed two times, each for 5 min in PBS, and then stained with 4′,6-diamidino-2-phenylindole (DAPI) (0.1 μg/ml). Samples were observed under a fluorescence microscope after final rinsing three times, each for 5 min, in PBS.
Western blotting
Samples were prepared using lysis buffer as previously described (13). Samples containing 25 μg of total protein were subjected to electrophoresis in 12.5% SDS-containing polyacrylamide gels. After blotting onto polyvinylidenedifluoride (PVDF)-membranes, they were incubated for 1 h with the blocking buffer containing 6% milk powder in Tris-buffered saline with 0.1% Tween-20 (TBST), and then incubated overnight at 4°C with primary antibodies in 1% milk powder in TBST. We used following antibodies: cardiac troponin I (cTnI) antibody (abcam, 1:2000) and α-tubulin antibody (Sigma, 1:1000). After rinsing five times, each for 10 min, in TBST, samples were incubated with secondary antibodies for 1.5 h at room temperature. Samples were rinsed five times, each for 10 min, in TBST, and proteins were detected by using Western blotting detection reagents (Thermo Fisher Scientific).
Quantitative real-time PCR
RNA was isolated using an RNeasy mini kit (Qiagen), and cDNA was synthesized using an Omniscript RT Kit (Qiagen). cDNA was used for quantitative real-time polymerase chain reaction (PCR) analysis. Analyses were performed using following primers corresponding to mus musculus sequences: ubiquitin forward, 5′-CGAGCCCAGTGACACCATT-3′; reverse, 5′-TGCCTTGACAT TCTCTATGGT-3′. cTnI forward, 5′-CTGCCAACTACCGAGCCTAT-3′; reverse, 5′-CTC GTTCCATCTCCTGCTTC-3′. cMHC forward, 5′-GAAGGGCATGAGGAAGAGCG-3′; reverse, 5′-CAGCTTGTTGACCTGGGACT-3′. Eight microliters of a real-time PCR mixture including 1× Quantities Eva Green Super Mix (Bio-Rad), gene-specific primers (0.4 μM), and cDNA template (300 ng) was reacted in a CFX96 real-time PCR analysis system (Bio-Rad, Hercules, CA, USA). PCR conditions were as follows: 98°C for 30 s, 40 cycles at 98°C for 5 s, 59.7°C (ubiquitin), 50.0°C (cTnI) or 60.3°C (cMHC) for 5 s, 50°C for 5 s, and 95°C for 5 s.
RESULTS
Comparison of ECM molecules in heart and liver ECM
To examine protein compositions of heart and liver ECM, quantitative proteomics was performed on de-cellularized ECM. Components and the quantities were compared between the heart ECM and the liver ECM using the iTRAQ method. Results showed that ECM components were essentially same with both tissues, and no tissue-specific component in these tissue ECMs was detected. On the other hand, relative quantities of the components largely differed among tissues. In heart ECM, fibrillin-1, microfibrillar-associated protein 2 (Mfap2), microfibrillar-associated protein 5 (Mfap5), and lysyl oxidase homolog 1 were more abundant whereas collagen III and arginase-1 were less, as compared to the liver ECM. Components in which the difference was more than double/half are summarized in Table 1. The result showed that, although major components existed in both ECMs, differences in the quantities of each ECM molecules could be important for tissue-specific differentiation of stem cells.
TABLE 1.
Comparison of ECM molecules in SDS-treated heart and liver.
| Name of molecules | Ratio (heart/liver) | p-value | Function |
|---|---|---|---|
| Collagen III | 0.276 | 0.011 | ECM component |
| Fibrillin-1 | 3.037 | <0.001 | ECM component |
| Mfap2 | 2.309 | 0.007 | ECM component |
| Mfap5 | 3.632 | <0.001 | ECM component |
| Arginase-1 | 0.425 | 0.118 | ECM organization |
| Lysyl oxidase homolog 1 | 2.324 | 0.033 | ECM organization |
Mfap2: microfibrillar-associated protein 2; Mfap5: microfibrillar-associated protein 5.
Confirmation of de-cellularization and ES cell culture on different matrices
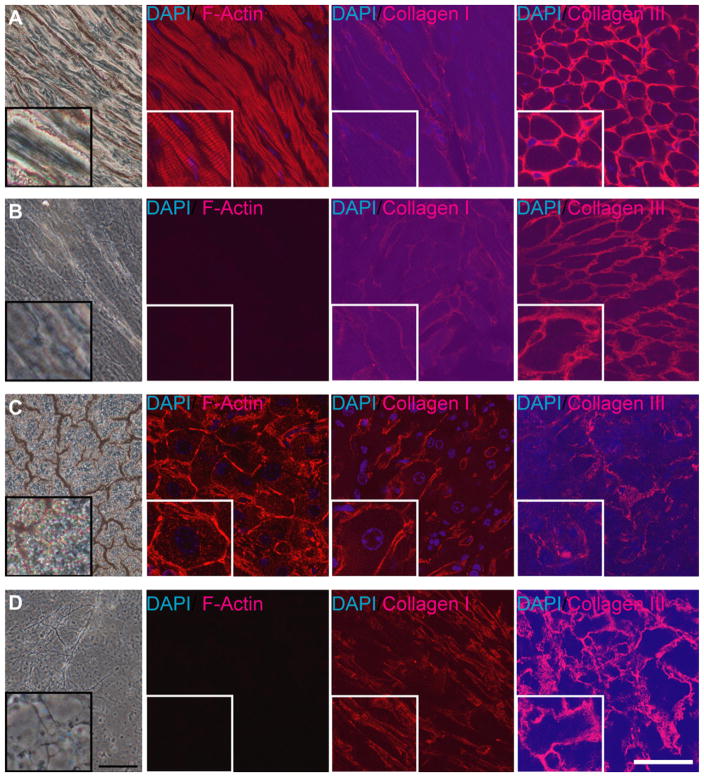
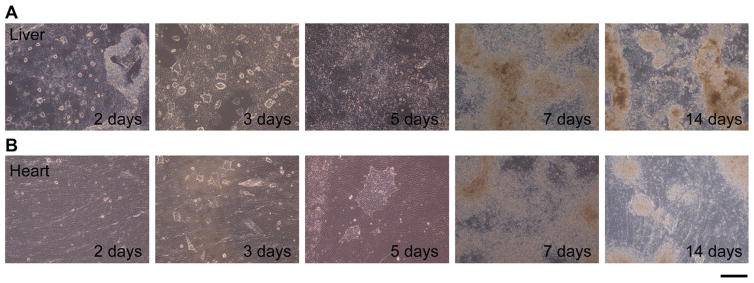
To examine the contribution of heart ECM maintaining a 3D structure to the cell fate decision to differentiate into cardiomyocytes, we cultured ES cells on de-cellularized heart and liver ECM. As described under Materials and methods, we obtained ECM slices of 60 μm thickness by de-cellularization using SDS. First, to assess the de-cellularization of cells in tissue sections, de-cellularized tissue slices were stained with Alexa-546-phalloidin (Invitrogen) and DAPI to visualize actin filaments and nuclei, and the complete removal of cells was clearly shown by the absence of both signals. ECM in de-cellularized slices remained even after the de-cellularization process, as was shown by immunostaining of collagen I and collagen III (Fig. 1A and B). We also prepared liver ECM of the same thickness via the same de-cellularization procedure (Fig. 1C and D). The liver ECM was used as a negative control so that results could be compared with those for ECMs from tissues other than the heart tissue. We chose liver ECM because it is prominently different from heart tissue, where liver arose from endoderm whereas the heart is from mesoderm. At earlier points in culture, within the first week, slow cell growth was observed for cells cultured on the heart ECM, as compared to those cultured on the liver ECM (Fig. 2). Five days after seeding, the cells on liver ECM were shown to spread across the ECM. On the other hand, some of the cells on the heart ECM still had a colony-like morphology. These results show that different matrices had different effects on the cell proliferation of ES cells.
FIG. 1.
Intact and de-cellularized sections of mouse heart and liver. Tissue sections were stained with phalloidin, DAPI, collagen I and collagen III antibodies. From left to right, bright field, phalloidin staining, collagen I staining, and collagen III staining. The inserted square area in the left bottom of each photographs are 140% magnification views of the center part of each photograph. (A) Intact heart sections. (B) De-cellularized heart sections. (C) Intact liver sections. (D) De-cellularized liver sections. Scale bars: 80 μm for bright field and 50 μm for fluorescence, respectively.
FIG. 2.
ES cells cultured on different matrices for 2 weeks. (A) ES cells on liver ECM. (B) ES cells on heart ECM. Days denote period of cultured days after seeding. Scale bar: 250 μm.
Heart ECM supports cardiomyocyte differentiation
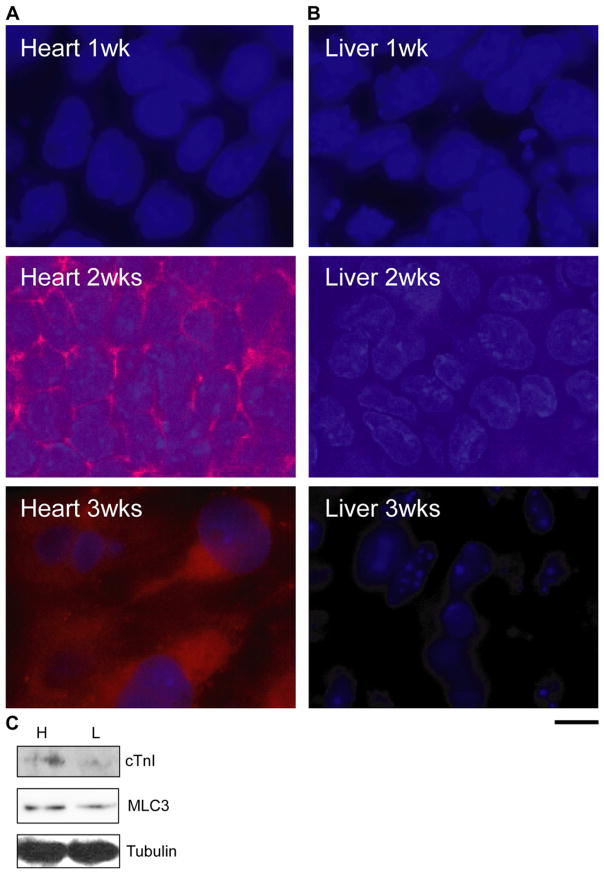
Because we observed a difference in the behavior of ES cells between heart and liver ECMs, we evaluated effects of heart and liver ECMs on the cell fate decision by quantitative real-time PCR (RT-PCR) and Western blot analyses. At 1, 2 and 3 weeks after seeding of ES cells, expression levels of cardiac genes were examined by quantitative RT-PCR analysis. Amazingly, cMHC (p < 0.1) and cTnI (p < 0.05) were highly expressed in ES cells cultured on heart ECM for 2 weeks, as compared with cells on liver ECM (Fig. 3A and B). There were no significant differences at other time points. Additionally, increases in the protein expression of cMHC and cTnI were detected in cells cultured on the heart ECM after 2 weeks from seeding (Fig. 4A and C). On the other hand, ES cells cultured on the liver ECM did not express cMHC, and the expression of cTnI was very low, as compared to cells on the heart ECM (Fig. 4B and C). Also, we detected a high expression of MLC3 in cells seeded on the heart ECM (Fig. 4C). These results show that the heart ECM induced cardiomyocyte differentiation of ES cells.
FIG. 3.
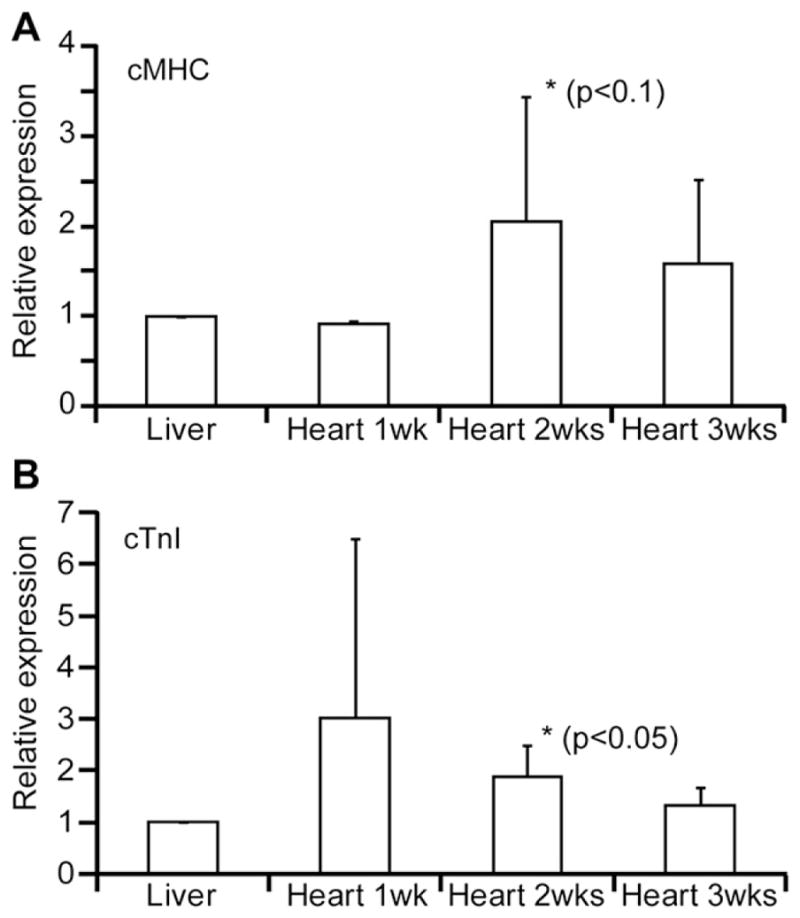
Expression levels of cardiac genes determined by quantitative real-time PCR analysis after 1, 2, and 3 weeks of cultures, relative to levels in cells cultured on the liver ECM. (A) The expression level of cMHC. (B) The expression level of cTnI. Each value represents the average of four experiments. Error bars: SD.
FIG. 4.
Protein expression of cardiac genes examined in cells cultured on different matrices. (A) ES cells cultured on the heart ECM for 1, 2, and 3 weeks. (B) ES cells cultured on the liver ECM for 1, 2, and 3 weeks. Blue, DAPI; red, cMHC. Scale bar: 10 μm. (C) Detection of cTnI and MLC3 expression by Western blotting at 2 weeks. Abbreviations: “H”, cells on heart ECM; “L”, cells on liver ECM. Representative immunoblots from three independent experiments are shown.
DISCUSSION
Recently, it was reported that environmental factors contribute to stem cell activity (14). In this study, we clearly demonstrated that the heart ECM contributes to the cell fate decision of stem cells to differentiate into cardiomyocytes. Previously, it was reported that cardiomyocyte-like cells (CLCs) showed enhanced GATA4 and Nkx2.5 expression when cultured on a collagen V, as compared to on a collagen I matrix (9). Our study differs from this previous study in that we demonstrated cardiomyocyte differentiation from undifferentiated stem cells, which were not committed to differentiate into cells such as CLC. In this study, we used heart ECM derived from tissue as a substrate for stem cell culture. There could be two possible reasons why heart ECM was effective as to cardiomyocyte differentiation.
First, heart ECM contains several components other than collagen V that may assist cardiomyocyte differentiation. According to the results of the mass-spectrometric study, heart ECM contained many protein components other than collagen V. Recently, it was shown that differences in ECM components have different effects on intercellular dynamics through activation of each integrin family receptor. So far, 24 kinds of integrins have been reported, and selective binding between these integrin families and ECM molecules has been revealed (8). For example, it is known that four forms of integrin, α1β1, α2β1, α10β1 and α11β1, bind to collagen, whereas α1β1, α2β1 and α10β1 integrin receptors bind to laminin. In addition to the difference in the ECM components, the differences in the relative quantities of ECM components could also be important. In our study, major components that exist in heart ECM were found to also be present in liver ECM. Thus, although the components may not differ significantly, there could be a specific ratio of ECM proteins that promotes cardiomyocyte differentiation.
Secondly, the contribution of the 3D structure of tissue-specific ECM of suitable size and shape may occur in de-cellularized tissue. It is well known that pore-size is one of the factors that are important for the cell fate decision (4). The nanotube dimension was shown to control the differentiation of adipose stem cells (15). Thus, our results show that heart ECM had a suitable pore-size for cardiomyocyte differentiation. Also, it seems that the 3D structure of the heart ECM, which is like a sponge, has a suitable elasticity for cardiomyocyte differentiation. According to our preliminary data, heart and liver ECM exhibit different elasticities (data not shown). Overall, we hypothesize that ES cells could become committed to differentiate into cardiomyocytes due to environmental cues from heart ECM with suitable ECM molecules, pore-size and elasticity. Previous study reported the cell culture protocol on tissue/organ sections for histopathology (TOSHI) substratum (16). Their culture system and concept, especially using of acellular type of TOSHI, are very similar to our culturing system. By using the general type of TOSHI substratum prepared from livers, they showed that the substratum had a high potential to efficiently differentiate ES cells into hepatocyte-like colonies with functional and morphological characteristics of mature parenchyma hepatocytes (17). Our finding, together with study using TOSHI substrate, shows that thin tissue section can be applicable for directing stem cells to specific linage.
In this study, though we showed the contribution of the extracellular environment for cardiomyocyte differentiation, we should point out that the commitment due to chemical factors is also essential. In our experiment, cells seeded on a de-cellularized ECM were cultured in the absence of leukemia inhibitory factor (LIF) for induction into mesodermal cells. In 2000, Yamashita and colleagues discovered that Flk1+ mesoderm cells were obtained from undifferentiated ES cells in the absence of LIF (18,19). In addition, ES cells were pretreated with 5-azacytidine, which had been reported to be a drug inducing cardiomyocyte differentiation in mesenchymal stem cells and ES cells (20,21). In our experiments, it was difficult to induce cardiomyocyte differentiation by only stimulation with heart ECM without such chemical treatment (data not shown). However, chemical factors alone were also not sufficient for the differentiation of ES cells into cardiomyocytes, because eventually the expression of cardiac genes was induced by additional stimulation by the heart ECM. Thus, this study showed that both mechanical factors in ECM as well as chemical factors are necessary for cardiomyocyte differentiation.
During ontogenesis, the mammalian heart is derived from the anterior portion of the lateral plate mesoderm. It has been suggested that both ECM and growth factors are necessary for cardiovascular development (22). To obtain a complete protocol of cardiomyocyte differentiation for safe stem cell therapy, a detailed understanding of the mechanism of mechano-transduction through stimulation by ECM in addition to chemical transduction is needed.
Acknowledgments
Authors thank Yuliansa Sudibyo for technical assistance.
References
- 1.Heng BC, Haider H, Sim EK, Cao T, Ng SC. Strategies for directing the differentiation of stem cells into the cardiomyogenic lineage in vitro, Cardiovasc. Res. 2004;62:34–42. doi: 10.1016/j.cardiores.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 2.Singla DK, Long X, Glass C, Singla RD, Yan B. Induced pluripotent stem (iPS) cells repair and regenerate infarcted myocardium, Mol. Pharm. 2011;8:1573–1581. doi: 10.1021/mp2001704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells, Circ. Res. 2009;104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment, Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 5.Oh SH, Kim TH, Im GI, Lee JH. Investigation of pore size effect on chondrogenic differentiation of adipose stem cells using a pore size gradient scaffold. Biomacromolecules. 2010;11:1948–1955. doi: 10.1021/bm100199m. [DOI] [PubMed] [Google Scholar]
- 6.Taqvi S, Roy K. Influence of scaffold physical properties and stromal cell coculture on hematopoietic differentiation of mouse embryonic stem cells. Biomaterials. 2006;27:6024–6031. doi: 10.1016/j.biomaterials.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 7.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 8.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 9.Tan G, Shim W, Gu Y, Qian L, Chung YY, Lim SY, Yong P, Sim E, Wong P. Differential effect of myocardial matrix and integrins on cardiac differentiation of human mesenchymal stem cells. Differentiation. 2010;79:260–271. doi: 10.1016/j.diff.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, Taylor DA. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart, Nat. Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 11.Uygun BE, Soto-Gutierrez A, Yagi H, Izamis ML, Guzzardi MA, Shulman C, Milwid J, Kobayashi N, Tilles A, Berthiaume F, et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med. 2010;16:814–820. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh D, Yu H, Tan XF, Lim TK, Zubaidah RM, Tan HT, Chung MC, Lin Q. Identification of key players for colorectal cancer metastasis by iTRAQ quantitative proteomics profiling of isogenic SW480 and SW620 cell lines. J Proteome Res. 2011;10:4373–4387. doi: 10.1021/pr2005617. [DOI] [PubMed] [Google Scholar]
- 13.Fujita H, Shimizu K, Nagamori E. Novel method for measuring active tension generation by C2C12 myotube using UV-crosslinked collagen film, Biotechnol. Bioeng. 2010;106:482–489. doi: 10.1002/bit.22705. [DOI] [PubMed] [Google Scholar]
- 14.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh S, Brammer KS, Li YS, Teng D, Engler AJ, Chien S, Jin S. Stem cell fate dictated solely by altered nanotube dimension, Proc. Natl Acad Sci USA. 2009;106:2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takezawa T, Takenouchi T, Imai K, Takahashi T, Hashizume K. Cell culture on thin tissue sections commonly prepared for histopathology. FASEB J. 2002;16:1847–1849. doi: 10.1096/fj.02-0405fje. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi T, Ochiya T, Takezawa T. Tissue array substratum composed of histological sections: a new platform for orienting differentiation of embryonic stem cells towards hepatic lineage, Tissue Eng. Part A. 2008;14:267–274. doi: 10.1089/tea.2007.0188. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita J, Itoh H, Hirashima M, Ogawa M, Nishikawa S, Yurugi T, Naito M, Nakao K. Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature. 2000;408:92–96. doi: 10.1038/35040568. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita JK. ES and iPS cell research for cardiovascular regeneration, Exp. Cell Res. 2010;316:2555–2559. doi: 10.1016/j.yexcr.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells, Circ. Res. 2002;91:501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 21.Fukuda K. Reprogramming of bone marrow mesenchymal stem cells into cardiomyocytes, C. R Biol. 2002;325:1027–1038. doi: 10.1016/s1631-0691(02)01524-x. [DOI] [PubMed] [Google Scholar]
- 22.Corda S, Samuel JL, Rappaport L. Extracellular matrix and growth factors during heart growth, Heart Fail. Rev. 2000;5:119–130. doi: 10.1023/A:1009806403194. [DOI] [PubMed] [Google Scholar]