Abstract
Purpose
Endometriosis is a gynecological disease influenced by multiple genetic and environmental factors. The aim of the current study was to use SNP-array technology to identify genomic aberrations that may possibly contribute to the development of endometriosis.
Methods
We performed an SNP-array genotyping of pooled DNA samples from both patients (n = 100) and controls (n = 50). Copy number variation (CNV) calling and association analyses were performed using PennCNV software. MLPA and TaqMan Copy-Number assays were used for validation of CNVs discovered.
Results
We detected 49 CNV loci that were present in patients with endometriosis and absent in the control group. After validation procedures, we confirmed six CNV loci in the subtelomeric regions, including 1p36.33, 16p13.3, 19p13.3, and 20p13, representing gains, while 17q25.3 and 20q13.33 showed losses. Among the intrachromosomal regions, our results revealed duplication at 19q13.1 within the FCGBP gene (p = 0.007).
Conclusions
We identified CNVs previously associated with endometriosis, together with six suggestive novel loci possibly involved in this disease. The intergenic locus on chromosome 19q13.1 shows strong association with endometriosis and is under further functional investigation.
Keywords: Endometriosis, Infertility, Copy number variation, DNA pooling, SNP array
Introduction
Endometriosis is a common non-malignant gynecologic disease that affects 5–10 % of women of reproductive age [1]. Pelvic pain, metastatic pattern, and infertility are the most common features of the disease. It is characterized by the presence of functioning endometrial-like tissue (epithelium and stroma) outside the uterus [1, 2]. Several models have been proposed to explain the pathogenesis of endometriosis. The most accepted model is the retrograde menstruation; however, other factors may play a role in the disease occurrence including abnormal immune response, hormonal factors, genetic predisposition, and exposure to environmental factors [1, 3, 4].
Although the increasing evidence supports a genetic component to this disease, the etiology and pathophysiology remains unclear. It is likely that endometriosis is a complex polygenic and multifactorial disease, caused by multiple genetic and environmental factors [5, 6]. Considering that endometriosis has failed to show classic Mendelian inheritance, different types of genetic approaches have been undertaken to study its molecular basis [7, 8].
Candidate genetic association studies comparing the frequency of single nucleotide polymorphisms (SNPs) in a patient vs. a control group have been widely conducted [9]. The choices of the candidate genes are based on biological mechanisms thought to contribute to the susceptibility, development, and progression of diseases, such as endometriosis [10]. Studies have demonstrated that structural variations on DNA, mainly copy number variations (CNVs), have been considered as important keys in the complex traits. The identification of disease-associated rare CNVs may help to explain some missing heritability that could not be explained by common SNPs [11].
Classical cytogenetic, molecular genetics, and molecular cytogenetic techniques applied to endometriosis have led to the identification of consistent somatic genetic alterations [7]. A linkage study that adopted a positional-cloning approach identified a significant susceptibility locus for endometriosis on chromosome 10q26 involving the genes EMX2 and PTEN [12]. Array-comparative genomic hybridization uncovered alterations at 20q13.33 that were associated with ovarian endometriosis [13]. Additionally, studies using high-density genotyping arrays have reported copy number variation loci in 6.9 % of affected women compared to 2.1 % in the general population [14]. Taken together, these findings strongly suggest the involvement of genomic aberrations in the development of the disease.
High-resolution genomic approaches, such as microarray-based genotyping, have been performed successfully in the mapping of hundreds of thousands of SNPs, and consequently, in the discovery of new genomic regions associated with multifactorial diseases [15]. In the present study, we used DNA pooling methodology and high-density SNP array as a cost-effective alternative to evaluate CNVs in endometriosis patients.
The aim of the current study was to search for genomic regions that might contribute to the development of endometriosis by identifying CNVs significantly associated to this disease compared to healthy controls.
Materials and methods
Subjects
Five hundred sixty-four infertile women with endometriosis (mean age 35.1 ± 3.9 years) from the Human Reproduction and Genetics Center of the Faculdade de Medicina do ABC (SP; in the southeast of Brazil), participated in this case-control study. The patients were diagnosed with endometriosis by laparoscopy/laparotomy and classified according to the American Society for Reproductive Medicine [16] with obligatory histological confirmation of the disease. In this group, minimal/mild (stage I and II) endometriosis was found in 229 patients (40.6 %) and moderate/severe (stage III and IV) endometriosis in 335 patients (59.4 %).
The investigation into the cause of infertility included a hormonal and biochemical profile, testing for sexually transmitted diseases, imaging examinations, investigation of genetic and/or immunological abnormalities, hysterosalpingography, hysteroscopy, laparoscopy, and semen analysis of the partner.
Six hundred fifty-two fertile women (mean age 39.2 ± 5.8 years), from the Family Planning Outpatient Clinic of the Faculdade de Medicina do ABC, who were among subjects evaluated for tubal ligation participated as control group. In all of them, absence of endometriosis was confirmed by inspection of the pelvic cavity during the laparoscopy.
Genomic DNA for each individual was extracted from peripheral blood according to as previously described [17].
DNA pool construction
Pooling consists of combining multiple DNA samples for the purpose of screening many subjects simultaneously [18]. The construction is based on the addition of equal amounts of DNA from each individual to either patient or control pools [19].
A total of 150 samples were analyzed according to DNA pooling methodology. To establish homogeneous pools, the samples selected from the endometriosis group included 100 patients who did not have ovulatory and endocrine disorders, Mullerian defects, or autoimmune diseases and whose partner presented any male factor associated with infertility. In relation to the control group, we selected 50 samples per pool without medical history of autoimmune diseases, pelvic pain complaints, and spontaneous miscarriage.
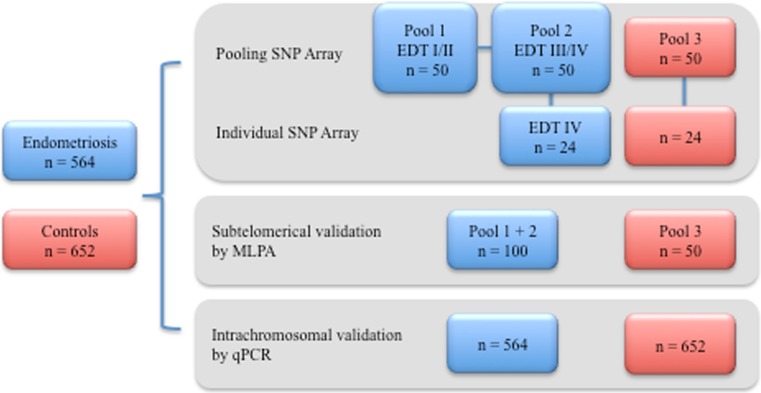
DNA concentrations in each sample were measured using the spectrophotometer NanoDrop 2000 (Thermo Scientific, CA, USA) for the quantification of double-stranded DNA. DNA samples were diluted to a final concentration of 50 ng/μL using 1x Tris–EDTA (TE). Pools were constructed by combining equal volumes (10 μL) of each DNA sample. Each pool was composed by 50 samples. A total of three pools were constructed for the group of endometriosis and subdivided according to the stages of the disease (pool 1: minimal/mild endometriosis, pool 2: moderate/severe endometriosis, and pool 3: control group). The scheme in Fig. 1 represents the distribution of individuals subjected to the SNP array experiments and the following validations.
Fig. 1.
Distribution of subjects analyzed in each step of the study. To establish homogeneous pools 50/229 patients with EDT I/II, 50/335 EDT III/IV and 50/652 controls were selected (EDT endometriosis, SNP single nucleotide polymorphism, MLPA multiplex ligation-dependent probe amplification, qPCR quantitative polymerase chain reaction)
High-density SNP genotyping
Once equimolar amounts of each sample were combined, the genomic DNA pools were assayed using the Illumina protocol for individual genotyping. Briefly, 750 ng of pooled genomic DNA was labeled and hybridized to the Illumina HumanOmni 2.5 BeadChip (Illumina, San Diego, CA, USA), which interrogated ∼2.5 million SNPs. The pools were genotyped in duplicate, serving as technical replicates.
In addition, 48 samples that composed the pools were selected to be genotyped individually and used as experiment controls. Genotyping using the Illumina Human OmniExpress BeadChip, which interrogated >715,000 SNPs was performed in 24 samples of the control group and 24 samples of the patient group (only endometriosis stage IV).
Data analysis
BeadChip data was processed using GenomeStudio Software v.3.1 (Illumina Inc.) according to the manufacturer’s recommendations. Primary data analyses, including raw data normalization, clustering, and genotype calling were performed using algorithms in the genotyping module (GT). The software provided log R ratios (LRRs) and B allele frequencies (BAFs) for each probe on the SNP array.
The Software UPDG [20] was used to perform the data analysis of the pooled DNA. Raw data were manipulated for correcting the preferential allelic amplification, normalization, and analysis of variance for genetic association test.
Standard quality control
To allow cross-platform evaluation, we kept only common SNPs between Omni 2.5 and OmniExpress beadchips. Standard quality control steps were as follows: SNPs with minor allele frequency (MAFs) >0.05, missing rates <0.05, or P value >0.0001 for the Hardy–Weinberg equilibrium test were included. Principal component analysis (PCA) was applied considering all valid SNPs to calculate genetic distances and account for population stratification in subsequent association analyses. Data processing was performed using PLINK (version 1.07) [21].
CNV analysis and post-CNV calling
CNVs were called using PennCNV algorithm, which considers the total signal intensity and allelic intensity ratio at each SNP, the distances between SNPs, and the frequency of each SNP via a hidden Markov model (HMM) [22]. The software is capable of processing integer copy numbers, according to a six-state definition: state 1 = deletion of two copies (copy number 0), state 2 = deletion of one copy (copy number 1), state 3 = two-copy state (copy number 2), state 4 = two-copy state with LOH (copy number 2), state 5 = single-copy duplication (copy number 3), and state 6 = double-copy duplication (copy number 4). A copy number state of two per individual was considered normal (one copy per chromosome); CNVs with copy number >2 were defined as duplications, while those with copy number <2 were considered deletions.
The Software BEDTools [23] of genomic arithmetic integrated the CNV data from the individual and pooled sample analysis, merging the CNV regions that were common among them. The most relevant regions were saved for subsequent validation.
Defining the CNV Loci
CNV calls were grouped into loci having at least 1 kb in length. A confidence score of 10 probes minimum was used as threshold to classify reliable CNV calls [24]. To identify if the CNV loci were common or novel, we compared our results with those published in the Database of Genomic Variants (DGV) [25]. We used the gene annotation of the University of California Santa Cruz (UCSC) Genome Browser [26] to identify genes that were located within or partially overlapped with CNV loci. All annotated CNV genomic regions were based on the Human genome build 19 (hg19).
Validation of CNV calls
For validation of the selected CNVs, the loci were subdivided into subtelomeric and intrachromosomal, considering that the validation strategy for each of these findings was performed differently. The same 150 subjects (100 patients and 50 controls) used in the SNP array were analyzed in the MLPA experiments. For qPCR, all 564 endometriosis samples and 652 controls included in the study were analyzed.
Multiplex ligation-dependent probe amplification
Deletions and duplications in subtelomeric regions were analyzed using the multiplex ligation-dependent probe amplification (MLPA) technique with the kit SALSA P070 Human Telomere probe mix, according to the manufacturer’s protocol (MRC Holland, Amsterdam). The probe amplification was performed in the thermocycler MasterCycler Gradient (Eppendorf) and the fragment analysis through capillary electrophoresis using the ABI 3500 Sequencer (Applied, Life Technologies). MLPA results were analyzed through GeneMarker Software (Softgenetics®, LLC, State College, PA, USA).
Real time qPCR
Four intrachromosomal loci were selected and subjected to TaqMan CNV detection analysis. Real-time qPCR was performed to validate the candidate CNVs in the FCGBP, NR5A1, PTGES2, SLC25A25, and CXXC5 gene regions, and Taqman assays Hs02788922_cn, Hs00492415_cn, Hs01128655_cn, Hs02406760_cn and Hs06063192_cn were used for genotyping each locus, respectively. TaqMan Copy Number Reference Assays (RNaseP and TERT) were used as references (internal control). Reaction plates (384-well) with a mixture of TaqMan Fast Advanced Master Mix (Applied Biosystems, USA), target and reference TaqMan Assays (Applied Biosystems, USA) and 10 ng DNA/well were prepared using a Biomek FX (Beckman Coulter, Fullerton, CA, USA). Experiments were performed according to the manufacturer’s instructions and ViiA 7 Real-Time PCR System (Life Technologies, CA, USA) was used for the quantitative PCR reactions (qPCR). The reactions were done in triplicate and the results analyzed based on the delta-delta Ct method were imported to Copy Caller Software v.2.0 (Applied Biosystems Inc., USA) to determine copy numbers. A confidence level of 95 % and |z-score| value of <1.75 was applied to call the CNVs.
Statistical analysis
Fisher’s exact test was used to carry out the case-control association analysis of CNVs identified. The significance threshold was chosen as 0.05. For comparison of CNV sizes and mean numbers between patients and controls, two-tailed Mann–Whitney U test was used.
Results
A basic association check was performed between the SNPs and phenotype to investigate possible systematic biases in the association results, revealing a genomic inflation factor (GIF) of 1.41082. Outliers were subsequently dismissed, and the data was corrected by three components of ancestry that reduced the GIF value to 1.11251, which we considered acceptable.
The combined analysis of individual and pooled samples using BedTools Software was performed to integrate the CNV regions that were common among them, revealing 49 CNV loci present in patients with endometriosis and absent in the control group. Of these, 33 were kept and 16 were excluded from the study—6 due to size smaller than 1 kb and 10 for absence of the minimum number of probes required to be considered a reliable CNV.
The CNV sizes ranged from 4 to 61 kb with a median size of 19 kb. These regions were studied and screened according to length, presence of overlapping regions in DGV, chromosomal location, and overlapping of genes and its functions. The results were divided into subtelomeric and intrachromosomal CNVs.
MLPA was used to confirm the CNVs located at subtelomeric regions that represented 48 % (16/33) of the CNVs of interest (Table 1). Of these, six showed agreement between the MLPA and SNP Array analysis. The regions 1p36.33, 16p13.3, 19p13.3, and 20p13 represented gains while 17q25.3 and 20q13.33 showed losses. The non-validated regions were considered to be false positives due to the limited specificity of the DNA-pooling methodology that tends to overestimate the effect size and were not pursued any further [27].
Table 1.
Characteristics of the loci located in the subtelomeric regions
| CN size (bp) | Chromosomal region | CN region | CN type | Gene |
|---|---|---|---|---|
| 9444 | 1543311-1552755 | 1p36.33 | Gain | |
| 12899 | 194875096-194887995 | 3q29 | Gain | |
| 36637 | 3806638-3843275 | 4q35.2 | Loss | |
| 14796 | 1765278-1780074 | 5q35.3 | Loss | |
| 35622 | 143813581-143849203 | 8q24.3 | Gain | LYNX1 |
| 1046 | 143614478-143615524 | 8q24.3 | Gain | BAI1 |
| 18167 | 138357705-138375872 | 9q34.3 | Gain | PPP1R26 |
| 6230 | 138149166-138155396 | 9q34.3 | Gain | |
| 19047 | 131553189-131572236 | 12q24.33 | Loss | |
| 10696 | 2803994-2814690 | 16p13.3 | Gain | SRRM2 |
| 27545 | 76948501-76976046 | 17q25.3 | Loss | LGALS3BP |
| 12337 | 1401668-1414005 | 18p11.32 | Gain | |
| 30028 | 3308908-3338936 | 19p13.3 | Gain | |
| 11593 | 4279824-4291417 | 19p13.3 | Gain | SHD |
| 9406 | 2524509-2533915 | 20p13 | Gain | |
| 3030 | 60961271-60964301 | 20q13.33 | Loss | CABLES2 |
In bold are represented the loci that were validated by MLPA
Among the 33 CNV loci, 17 (52 %) were intrachromosomal. Two of them were not involved with any gene, while 15 were located within intergenic and gene coding regions. All investigated regions were previously reported on DGV (Table 2).
Table 2.
Detailed CNV findings and characteristics of intrachromosomal regions
| Genes | CN size | Chromosomal region | CN region | N probes | CN type | Validation | No. of individuals confirmed |
|---|---|---|---|---|---|---|---|
| NUP93 and SLC12A3 | 61 kb | 16q13 | 56851548–56912903 | 46 | Gain | Not tested | |
| (None) | 58 kb | 11p11.2 | 45413385–45471582 | 59 | Loss | Not tested | |
| SLC25A25 and PTGES2 | 38 kb | 9q34.11 | 130850879–130889600 | 30 | Loss | Tested—not confirmed | 4 cases/6 controls |
| PARVG | 34 kb | 22q13.31 | 44582820–44617272 | 69 | Gain | Not tested | |
| GPR144 and NR5A1 | 26 kb | 9q33.3 | 127229652–127256223 | 30 | Loss | Tested—not confirmed | 0 cases/0 controls |
| CCDC64 and RAB35 | 23 kb | 12q24.23 | 120525746–120548898 | 25 | Loss | Not tested | |
| PCBD1 | 22 kb | 10q22.1 | 72642702–72665347 | 32 | Loss | Not tested | |
| CXXC5 | 17 kb | 5q31.2 | 139020091–139037511 | 12 | Loss | Tested—not confirmed | 0 cases/0 controls |
| TMEM119 | 14 kb | 12q23.3 | 108981642–108996162 | 22 | Loss | Not tested | |
| VAV2 | 14 kb | 9q34.2 | 136664428–136678685 | 20 | Loss | Not tested | |
| SARDH | 14 kb | 9q34.2 | 136583939–136598128 | 22 | Loss | Not tested | |
| (None) | 12 kb | 1p35.2 | 30816458–30828841 | 10 | Loss | Not tested | |
| HERPUD1 | 9 kb | 16q13 | 56965707–56975208 | 14 | Loss | Not tested | |
| JPH3 | 8 kb | 16q24.2 | 87702670–87711308 | 12 | Loss | Not tested | |
| GLP1R | 6 kb | 6p21.2 | 39047753–39054138 | 21 | Loss | Not tested | |
| PAX5 | 6 kb | 9p13.2 | 36834277–36840446 | 10 | Loss | Not tested | |
| FCGBP | 4 kb | 19q13.2 | 40360845 –40365523 | 10 | Gain | Tested—confirmed | 8 cases/0 controls |
In bold are represented the loci that were selected for validation by qPCR
Of note, a 4678-bp duplication (chr19 40,360,845–40,365,523) involving the FCGBP gene (Fig. 2) was observed in 8 patients and was absent in controls, thus conferring significantly increased endometriosis risk (p = 0.007).
Fig. 2.
Graphical representation of the duplication located within the FCGBP gene visualized at the UCSC genome browser view (version hg19)
We detected a 38,721-bp deletion, involving the SLC25A25 and PTGES2 genes (chr9 130,850,879–130,889,600), impacting 4 patients and in 6 controls. Thus, we could not validate this locus (p = 0.92). The results of NR5A1 and CXXC5 were also not validated by qPCR.
Discussion
Large-scale genotyping using SNP microarrays has been performed successfully in the discovery of new genomic regions that are associated with multifactorial diseases [15, 28]. The methodology assesses variation across the genome using computational models to compare the genotypes of people with and without disease to identify variants that are associated with disease.
Even though the cost of SNP arrays was significantly reduced in the last years, it remains prohibitive for numerous research centers to genotype a large number of samples [18, 19]. One way to reduce the cost is to use the DNA pooling strategy, an alternative and attractive method that can unveil loci that are highly informative. However, DNA pooling has limitations and the most common is the difficulty to access genotyping frequencies. Instead, this methodology is used to estimate allele frequencies, a process referred as “allelotyping” [29].
DNA pooling has been originally used as an economic and alternative tool for genotyping and GWAS; therefore, it can be applied to discover common CNVs in certain studies [30, 31]. However, the copy number estimation represents a challenge in terms of false-positive rates, so it has been suggested that CNVs identified from SNP genotyping data must always be validated with an alternative method to avoid erroneous calls [31, 32]. A past study with schizophrenia in Brazilian samples also used pooling strategy to evaluate the CNVs and the possible role in the disease. As in our study, they were able to identify few CNVs with relevant association with the phenotype, but they also revealed the limitations of pooling in admixture population and the risk of false-positive calls [33].
In the present study, we have carried out a SNP array analysis through a pooled-sample strategy in a Brazilian cohort with endometriosis. The high-resolution SNP array used was able to produce enough data to support CNV detection, thus enabling integrated analysis of SNPs and CNVs in the same patients.
Our findings revealed 33 CNV loci in common between-individual analysis (1 array per individual) and pooled samples, which were present in patients with endometriosis and absent in the control group. Of these, 16 were located in the subtelomeric regions with the 1p36.33, 16p13.3, 17q25.3, 19p13.3, 20p13, and 20q13.33 loci confirmed through MLPA validation.
Our results are in agreement with previous studies that demonstrated an association between 1p36.33 and endometriosis [34–36]. This locus contains markers located in or near the WNT4 gene, which plays an important role on the development of the female reproductive tract and steroidogenesis [37, 38]. The literature emphasizes the role of the WNT4 as a regulator of cell proliferation and differentiation, in which the signaling pathway involves proteins that directly participate in both gene transcription and cell adhesion [39]. Based on these biological functions, the duplication involving the 1p36.33 locus is likely to be involved on endometriosis development.
Genomic imbalance at the 17q25 and 19p13 loci was also found to be associated with endometriosis by Veiga-Castelli et al. [40] in a comparative genomic hybridization (CGH) study conducted in Brazilian women. In another study, using the same samples, the authors observed a differential expression of the genes MXRA7 and UBA52, located respectively in 17q25 and 19p13, suggesting that these alterations could lead to the development, establishment, and maintenance of the ovarian endometriomas [41].
A Chinese study by Yang et al. [13] revealed an association between 20q13.33 duplication and ovarian endometriosis. Their array-CGH results were validated through qPCR, confirming association of the genes GATA5 and SLCO4A1 in the gain region. In the present study, we also reported an evident contribution of this locus, but instead of duplication, our subjects presented a 3-kb deletion (chr20:60,961,271–60,964,301) involving both the exonic and intronic parts of RPS21, a gene responsible for encoding ribosomal proteins. In Drosophila, the complete absence of the RPS21 gene leads to excessive cell proliferation in specific tissues [42].
The 16p13.3 and 20p13 loci have never been associated with endometriosis prior to our study. A report that evaluated a 16p13.3 duplication in 12 patients reported normal to moderate mental retardation, mild arthrogryposis-like anomalies of the musculoskeletal system, mild facial dysmorphism, and occasional anomalies of the heart. In 2 patients, the duplication was inherited from an apparently normal parent, with de novo occurrence found in 10 of 12 patients, indicating that this duplication is associated in most patients with a reduced reproductive fitness [43]. In relation to 20p13, a significant linkage was previously reported at this locus and associated with intellectual and developmental disabilities [44].
Between the 17 CNV loci located within intergenic and gene coding regions, 3 represented gains, whereas 14 were losses. From these 17 loci, 4 were selected and individually checked by qPCR. The selection parameters were based on the respective gene functions of the gene candidates present in each locus and their possible correlation with the development and progression of endometriosis.
The CN gain on chromosome 19q13.2 spans 4 kb and contained ten SNP probes. It was observed in eight of the endometriosis patients, while none of the controls displayed a gain in this region (p = 0.007), suggesting that this locus is a promising risk-conferring candidate. According to the UCSC Genome Browser and Database of Genomic Variants [25, 26], this duplicated region is located within the FCGBP gene. Characterized by the production of a large (encoded by a 17-kb mRNA) mucin-like protein that binds the Fc portion of IgG molecules, this gene has been reported as differentially expressed in gallbladder [45], prostate [46], thyroid [47], lung [48], colon [49], and ovarian cancer [50]. Present in serum, the protein can be found in higher levels in patients with autoimmune diseases [51]. It has been suggested based on its IgG Fc binding property and tissue distribution that FCGBP might play a role in cell protection and anti-inflammation in tissues. Our data suggest for the first time that the duplication on the FCGBP gene could predispose to the progression of endometriosis. Thus, functional studies will be required to elucidate the exact contribution of this variant to the disease risk.
Population stratification was a concern, particularly in the current study, because the Brazilian population is one of the most heterogeneous and admixed populations in the world, formed mainly by the admixture between European, African, and Native American populations [52, 53]. After removal of SNPs through the sequential QC processes and correction by three components of ancestry, we observed genomic inflation factor of 1.11251. Thus, the inflation of false-positive rates on genetic association is within an acceptable level (genomic inflation factor <1.1) [54] for a case–control association study, indicating that the current QC processes are successful.
It has been suggested that CNVs identified from SNP genotyping data must be validated to avoid false-positive results [32]. Therefore, the identified CNVs were further validated by MLPA and qPCR in an extended sample set. The discovery of how these CNVs act together can be an important step in discovering the susceptibility, establishment, and progression of endometriosis.
Despite extensive research, the varied clinical presentations among patients, such as staging, pain, and infertility remain unclear. Genetic modifying factors are thought to underlie this variability. This study represents an effort to enlarge our knowledge about endometriosis risk genes through a genome-wide copy number variation analysis in the Brazilian population. While requiring independent validation, our novel findings contribute to the understanding of the complex pathways leading to endometriosis.
Conclusion
In summary, we identified, CNVs previously associated with endometriosis and we have uncovered novel chromosome regions that may contribute to the pathogenesis of this disease. Further large-scale discovery and replication studies will help determine the biological impact of these CNVs and eventually provide clues for the underlying mechanisms of endometriosis.
Compliance with ethical standards
Clinical data and peripheral blood samples were collected following signed informed consent, as approved by the local Research Ethics Committee (CEP FMABC n. 310.094).
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
The work was supported by a grant from Fundação de Amparo a Pesquisa do Estado de Sao Paulo (FAPESP) no. 2011/01363-7. F.M. was supported by FAPESP with two different scholarships, a PhD scholarship no. 2012/22394-8 and a scholarship for internship abroad No. 2014/07136-8, which were realized in collaboration with the Center for Applied Genomics at The Children’s Hospital of Philadelphia.
Footnotes
Capsule SNP array genotyping of pooled DNA samples revealed CNVs previously associated with endometriosis and also novel chromosome regions that may contribute to the pathogenesis of this disease.
References
- 1.Bulun SE. Endometriosis. N Engl J Med. 2009;360(3):268–79. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- 2.Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364(9447):1789–99. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- 3.Sasson IE, Taylor HS. Stem cells and the pathogenesis of endometriosis. Ann N Y Acad Sci. 2008;1127:106–15. doi: 10.1196/annals.1434.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Oliveira R et al. Causes of endometriosis and prevalent infertility in patients undergoing laparoscopy without achieving pregnancy. 2015. Minerva Ginecol. [PubMed]
- 5.Bischoff F, Simpson JL. Genetic basis of endometriosis. Ann N Y Acad Sci. 2004;1034:284–99. doi: 10.1196/annals.1335.030. [DOI] [PubMed] [Google Scholar]
- 6.Stefansson H, et al. Genetic factors contribute to the risk of developing endometriosis. Hum Reprod. 2002;17(3):555–9. doi: 10.1093/humrep/17.3.555. [DOI] [PubMed] [Google Scholar]
- 7.Bouquet De Joliniere J, et al. Endometriosis: a new cellular and molecular genetic approach for understanding the pathogenesis and evolutivity. Front Surg. 2014;1:16. doi: 10.3389/fsurg.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi H, et al. Understanding the role of epigenomic, genomic and genetic alterations in the development of endometriosis (review) Mol Med Rep. 2014;9(5):1483–505. doi: 10.3892/mmr.2014.2057. [DOI] [PubMed] [Google Scholar]
- 9.Spielman RS, Ewens WJ. The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet. 1996;59(5):983–9. [PMC free article] [PubMed] [Google Scholar]
- 10.Montgomery GW, et al. The search for genes contributing to endometriosis risk. Hum Reprod Update. 2008;14(5):447–57. doi: 10.1093/humupd/dmn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, et al. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–81. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Treloar SA, et al. Genomewide linkage study in 1,176 affected sister pair families identifies a significant susceptibility locus for endometriosis on chromosome 10q26. Am J Hum Genet. 2005;77(3):365–76. doi: 10.1086/432960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, et al. High-resolution array-comparative genomic hybridization profiling reveals 20q13.33 alterations associated with ovarian endometriosis. Gynecol Endocrinol. 2013;29(6):603–7. doi: 10.3109/09513590.2013.788632. [DOI] [PubMed] [Google Scholar]
- 14.Chettier R, Ward K, Albertsen HM. Endometriosis is associated with rare copy number variants. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0103968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welter D, et al. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ASRM Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril. 1997;67(5):817–21. doi: 10.1016/S0015-0282(97)81391-X. [DOI] [PubMed] [Google Scholar]
- 17.Lahiri DK, Nurnberger JI., Jr A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991;19(19):5444. doi: 10.1093/nar/19.19.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Docherty SJ, et al. Applicability of DNA pools on 500 K SNP microarrays for cost-effective initial screens in genomewide association studies. BMC Genomics. 2007;8:214. doi: 10.1186/1471-2164-8-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macgregor S, et al. Highly cost-efficient genome-wide association studies using DNA pools and dense SNP arrays. Nucleic Acids Res. 2008;36(6) doi: 10.1093/nar/gkm1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho DW, Yap MK, Yip SP. UPDG: utilities package for data analysis of pooled DNA GWAS. BMC Genet. 2012;13:1. doi: 10.1186/1471-2156-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17(11):1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glessner JT, Li J, Hakonarson H. ParseCNV integrative copy number variation association software with quality tracking. Nucleic Acids Res. 2013;41(5) doi: 10.1093/nar/gks1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacDonald JR, et al. The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42(Database issue):D986–92. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abraham R, et al. A genome-wide association study for late-onset Alzheimer’s disease using DNA pooling. BMC Med Genomics. 2008;1:44. doi: 10.1186/1755-8794-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118(5):1590–605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meaburn E, et al. Genotyping DNA pools on microarrays: tackling the QTL problem of large samples and large numbers of SNPs. BMC Genomics. 2005;6:52. doi: 10.1186/1471-2164-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin CH, et al. Genome-wide copy number analysis using copy number inferring tool (CNIT) and DNA pooling. Hum Mutat. 2008;29(8):1055–62. doi: 10.1002/humu.20760. [DOI] [PubMed] [Google Scholar]
- 31.Bosse Y, et al. Identification of susceptibility genes for complex diseases using pooling-based genome-wide association scans. Hum Genet. 2009;125(3):305–18. doi: 10.1007/s00439-009-0626-9. [DOI] [PubMed] [Google Scholar]
- 32.Kim SY, Kim JH, Chung YJ. Effect of combining multiple CNV defining algorithms on the reliability of CNV calls from SNP genotyping data. Genomics Inform. 2012;10(3):194–9. doi: 10.5808/GI.2012.10.3.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ota VK, et al. Candidate genes for schizophrenia in a mixed Brazilian population using pooled DNA. Psychiatry Res. 2013;208(2):201–2. doi: 10.1016/j.psychres.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Albertsen HM, et al. Genome-wide association study link novel loci to endometriosis. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0058257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyholt DR, et al. Genome-wide association meta-analysis identifies new endometriosis risk loci. Nat Genet. 2012;44(12):1355–9. doi: 10.1038/ng.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uno S, et al. A genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat Genet. 2010;42(8):707–10. doi: 10.1038/ng.612. [DOI] [PubMed] [Google Scholar]
- 37.Boyer A, et al. WNT4 is required for normal ovarian follicle development and female fertility. FASEB J. 2010;24(8):3010–25. doi: 10.1096/fj.09-145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vainio S, et al. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397(6718):405–9. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- 39.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303(5663):1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veiga-Castelli LC, et al. Genomic alterations detected by comparative genomic hybridization in ovarian endometriomas. Braz J Med Biol Res. 2010;43(8):799–805. doi: 10.1590/S0100-879X2010007500072. [DOI] [PubMed] [Google Scholar]
- 41.Meola J, et al. Differentially expressed genes in eutopic and ectopic endometrium of women with endometriosis. Fertil Steril. 2010;93(6):1750–73. doi: 10.1016/j.fertnstert.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 42.Torok I, et al. Down-regulation of RpS21, a putative translation initiation factor interacting with P40, produces viable minute imagos and larval lethality with overgrown hematopoietic organs and imaginal discs. Mol Cell Biol. 1999;19(3):2308–21. doi: 10.1128/MCB.19.3.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thienpont B, et al. Duplications of the critical Rubinstein-Taybi deletion region on chromosome 16p13.3 cause a novel recognisable syndrome. J Med Genet. 2010;47(3):155–61. doi: 10.1136/jmg.2009.070573. [DOI] [PubMed] [Google Scholar]
- 44.Weiss LA, et al. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461(7265):802–8. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiong L, et al. NT5E and FcGBP as key regulators of TGF-1-induced epithelial-mesenchymal transition (EMT) are associated with tumor progression and survival of patients with gallbladder cancer. Cell Tissue Res. 2014;355(2):365–74. doi: 10.1007/s00441-013-1752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gazi MH, et al. Downregulation of IgG Fc binding protein (Fc gammaBP) in prostate cancer. Cancer Biol Ther. 2008;7(1):70–5. doi: 10.4161/cbt.7.1.5131. [DOI] [PubMed] [Google Scholar]
- 47.O’Donovan N, et al. Differential expression of IgG Fc binding protein (FcgammaBP) in human normal thyroid tissue, thyroid adenomas and thyroid carcinomas. J Endocrinol. 2002;174(3):517–24. doi: 10.1677/joe.0.1740517. [DOI] [PubMed] [Google Scholar]
- 48.Zhou C, et al. Screening of genes related to lung cancer caused by smoking with RNA-Seq. Eur Rev Med Pharmacol Sci. 2014;18(1):117–25. [PubMed] [Google Scholar]
- 49.Zhu H, et al. Screening for differentially expressed genes between left- and right-sided colon carcinoma by microarray analysis. Oncol Lett. 2013;6(2):353–8. doi: 10.3892/ol.2013.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi CH, et al. Identification of differentially expressed genes according to chemosensitivity in advanced ovarian serous adenocarcinomas: expression of GRIA2 predicts better survival. Br J Cancer. 2012;107(1):91–9. doi: 10.1038/bjc.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi K, et al. Detection of Fcgamma binding protein antigen in human sera and its relation with autoimmune diseases. Immunol Lett. 2001;79(3):229–35. doi: 10.1016/S0165-2478(01)00288-7. [DOI] [PubMed] [Google Scholar]
- 52.Cordeiro Q, et al. A review of psychiatric genetics research in the Brazilian population. Rev Bras Psiquiatr. 2009;31(2):154–62. doi: 10.1590/S1516-44462009000200013. [DOI] [PubMed] [Google Scholar]
- 53.Giolo SR, et al. Brazilian urban population genetic structure reveals a high degree of admixture. Eur J Hum Genet. 2012;20(1):111–6. doi: 10.1038/ejhg.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaguchi-Kabata Y, et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet. 2008;83(4):445–56. doi: 10.1016/j.ajhg.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]