Abstract
Pseudomonas aeruginosa is capable of establishing airway infections. Human airway mucus contains a large amount of lysozyme, which hydrolyzes bacterial cell walls. P. aeruginosa, however, is known to be resistant to lysozyme. Here, we performed a genetic screen using a mutant library of PAO1, a prototype P. aeruginosa strain, and identified two mutants (ΔbamB and ΔfabY) that exhibited decrease in survival after lysozyme treatment. The bamB and fabY genes encode an outer membrane assembly protein and a fatty acid synthesis enzyme, respectively. These two mutants displayed retarded growth in the airway mucus secretion (AMS). In addition, these mutants exhibited reduced virulence and compromised survival fitness in two different in vivo infection models. The mutants also showed susceptibility to several antibiotics. Especially, ΔbamB mutant was very sensitive to vancomycin, ampicillin, and ceftazidime that target cell wall synthesis. The ΔfabY displayed compromised membrane integrity. In conclusion, this study uncovered a common aspect of two different P. aeruginosa mutants with pleiotropic phenotypes, and suggests that BamB and FabY could be novel potential drug targets for the treatment of P. aeruginosa infection.
Keywords: Pseudomonas aeruginosa, lysozyme, treatment regimen, airway infection, multi-drug resistance
Introduction
Antibiotic-resistant P. aeruginosa strains are emerging at a significantly faster rate than the introduction of new anti-Pseudomonal agents (Pendleton et al., 2013). Furthermore, prolonged use of antibiotics has been proven to be a critical risk factor in the selection of multi-drug resistant variants, especially in the case of P. aeruginosa infection (Merlo et al., 2007). It is, therefore, crucial to devise a revolutionary strategy to combat P. aeruginosa infection. One such way would be to take advantage of the host innate defense molecules. Since the early discovery of the inherent lysozyme resistance of P. aeruginosa (Warren et al., 1955), no significant efforts have been made to increase the susceptibility of P. aeruginosa to lysozyme.
Lysozyme is an important antibacterial protein that is abundantly present in the human airway (Duszyk, 2001; Dubin et al., 2004; Dajani et al., 2005). Its enzymatic activity is to cleave β-1,4-glycosidic linkages on the N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) polysaccharide chains within the bacterial peptidoglycan layer (Vanderwinkel et al., 1995). Lysozyme found in the human airway is more active than egg-white lysozyme due to distinct structural differences between the two molecules (Marx et al., 1986). Although lysozyme has been proposed to play a crucial role in host defense, persistent P. aeruginosa airway infection is a major cause of morbidity and mortality in cystic fibrosis (CF) patients. Moreover, higher lysozyme activity has been detected in the serum and saliva of CF patients compared with normal individuals (Hughes et al., 1982). Likewise, lysozyme activity was also higher in bronchoalveolar lavage fluid (BALF) collected from CF patients (Sagel et al., 2009). Together, these results suggest that human lysozyme is ineffective in killing P. aeruginosa during infection. The lack of lysozyme effectiveness against P. aeruginosa has also been attributed to its active production of elastase, a major metalloprotease that can cleave human lysozyme (Jacquot et al., 1985).
Besides the infection in CF patients, P. aeruginosa is one of the major pathogens that cause ventilator-associated pneumonia (VAP). Especially, the pneumonia and sepsis by P. aeruginosa are the serious threats to the cardiac or thoracic surgical patients and trauma patients in the Intensive Care Unit (ICU; Berra et al., 2010). P. aeruginosa can also cause acute lung injury by injecting the ExoU toxin into the cytosol of eukaryotic cells using Type III Secretion System (TTSS; Pankhaniya et al., 2004). The acute P. aeruginosa infection could disrupt the alveolar epithelial barrier to promote necrosis of lung epithelial cells and sepsis, leading rapid increase in mortality (Sawa, 2014).
In this study, we aimed to identify novel interventional approaches that increase the sensitivity of P. aeruginosa to lysozyme. To this end, we isolated P. aeruginosa mutants that lost their viability when treated with lysozyme and sought to describe the genetic and phenotypic bases of their lysozyme sensitivity. Such defective mutants were found to be incapable of propagating in airway mucus secretions (AMSs) or establishing infection in the mouse airway, two locations where lysozyme is present in high concentrations. Results provided here will stimulate future works to propose novel approaches for inhibiting P. aeruginosa growth under lysozyme-rich environments.
Materials and methods
Experimental ethics
Experiments using human subjects and experimental animals were performed in strict accordance with guidelines provided by Yonsei University. Protocols were reviewed and approved by Institutional Review Board of Yonsei University College of Medicine. Permit numbers for primary culture of human tissues and mouse infection experiment were 2014-1842-001 and 2013-0369-5, respectively.
Bacterial strains and growth conditions
A prototype strain of P. aeruginosa called PAO1 was used in this study (Yoon et al., 2006). The bacterial strains of the other species, as listed in Figure 1, were obtained from our laboratory stock and were previously reported (Gi et al., 2015). Bacterial cultures were grown in Luria-Bertani medium (LB; 10 g tryptone, 5 g yeast extract, and 10 g NaCl/l), unless otherwise stated. Bacterial cells were cultured at 37°C for 16 h and shaken at 200 rpm. Bacterial growth in AMS and survival following the treatments with diverse antibiotics were assessed by quantifying colony forming units (CFUs).
Figure 1.
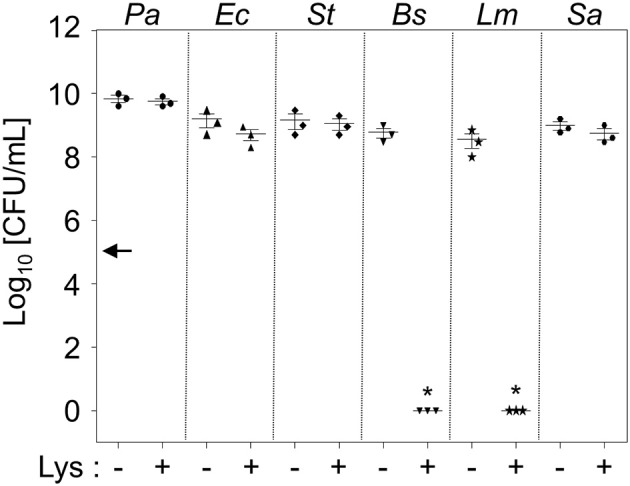
Intrinsic resistance of P. aeruginosa to lysozyme. Six different bacterial species were treated with 1 mg/mL lysozyme in 0.5 × LB for 16 h. Bacterial cells were enumerated by plating serially-diluted cells on LB agar plates. Each culture contained an initial inoculum of 1 × 105 cells. Bacterial cultures were repeated in triplicate, and the mean ± SD values are displayed. *P < 0.05 vs. CFU of untreated cultures.
Mutant library screen for lysozyme-sensitive mutants and preparation of airway mucus secretion (AMS)
A transposon (Tn) insertion mutant library of PAO1 was constructed following the procedures described elsewhere (Lee et al., 2016). In brief, PAO1 was conjugated with Escherichia coli SM10/λpir harboring the pBTK30 plasmid (Table 1). Gentamicin (Gm)-resistant transconjugants were grown on LB agar plates containing 50 μg/ml Gm and 50 μg/ml Irgasan (Sigma). Irgasan was added to eliminate E. coli donor cells. To isolate lysozyme-sensitive mutants, each individual mutant was grown in 0.5 × LB media (50% LB + 50% distilled water) containing 1 mg/mL lysozyme (Sigma) in 96 well plates. Bacterial growth was monitored by measuring the OD600. The Tn insertion site of each defective mutant was determined by arbitrary PCR, followed by DNA sequencing (Lee et al., 2016). Preparation of AMS samples from Normal Healthy Trachea Epithelial (NHTE) cells and bacterial growth in AMS were conducted as previously described (Gi et al., 2015).
Table 1.
Bacterial strains and plasmids used in this study.
| Strains or plasmids | Relevant characteristics | References or source |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 | Standard lab strain | Yoon et al., 2006 |
| ΔPA0420 | PA01, PA0420 gene deleted | This study |
| ΔPA3800 | PAO1, PA3800 gene deleted | This study |
| ΔPA5174 | PAO1, PA5174 gene deleted | This study |
| ΔPA3800/pJN105 | ΔPA3800 harboring pJN105 | This study |
| ΔPA5174/pJN105 | ΔPA5174 harboring pJN105 | This study |
| ΔPA3800/pJN105::PA3800 | ΔPA3800 harboring pJN105::PA3800 | This study |
| ΔPA5174/pJN105::PA5174 | ΔPA5174 harboring pJN105::PA5174 | This study |
| E. coli strains | ||
| SM10/λpir | Kmrthi-1 thr leu tonA lacY supE recA::RP4–2-Tc::Mu pir+, for conjugal transfer | Simon et al., 1983 |
| OTHER BACTERIAL SPECIES | ||
| Staphylococcus aureus | ATCC 29213 | Lab collection |
| Bacillus subtilis | ATCC 6633 | Lab collection |
| Escherichia coli | ATCC 25922 | Lab collection |
| Salmonella typhimurium | LT2 strain | Lab collection |
| Listeria monocytogenes | ATCC 19111 | Lab collection |
| PLASMIDS | ||
| pBTK30 | Transposon vector for construction of a random mutant library, Gmr | Kim et al., 2012 |
| pCVD442 | sacB suicide vector derived from plasmid pUM24 | Lee et al., 2012 |
| pJN105 | araC-PBAD cassette cloned in pBBR1MCS-5, Gmr | Newman and Fuqua, 1999 |
| pJN105::PA3800 | pJN105 with wild-type copy of PA3800 gene under arabinose-inducible promoter | This study |
| pJN105::PA5174 | pJN105 with wild-type copy of PA5174 gene under arabinose-inducible promoter | This study |
Construction of the PA0420, PA3800, and PA5174 clean deletion mutants
The bioA, bamB, and fabY deletion mutants were created by allelic replacement as previously described (Lee et al., 2012). Briefly, flanking sequences (~600 bp) at both ends of each gene were PCR amplified with primers listed in Table 2. Two inner primers (upstream reverse primer and downstream forward primer) are complementary to each other. In this strategy, the 3′-end of the upstream sequence and the 5′-end of the downstream sequence are annealed during PCR amplification without further treatment. The deletion of the PA0420, PA3800, and PA5174 genes was confirmed by PCR and DNA sequencing.
Table 2.
Primers used in this study.
| Gene name | Direction | Primer sequence (5′−3′)a | Restriction enzymes |
|---|---|---|---|
| MUTANT CONSTRUCTION | |||
| PA0420 left | Forward | ACCTTGAGCTCGCCGAGAGATCGTCCAGGGT | SacI |
| PA0420 left | Reverse | TCCGGGTGGAAGTCGCTGACAAGGCCCATGGGCTGTCTCC | |
| PA0420 right | Forward | GGAGACAGCCCATGGGCCTTGTCAGCGACTTCCACCCGGA | |
| PA0420 right | Reverse | TAGAGGAGCTCAGGGCCAGAGGCGGATGGAT | SacI |
| PA3800 left | Forward | ACCTTGCATGCTGGTGGCAGCGCAACGGCAA | SphI |
| PA3800 left | Reverse | ACGAGCTTGCCACCGTTGCCCACCATCTCAGGCCTCTCCC | |
| PA3800 right | Forward | GGGAGAGGCCTGAGATGGTGGGCAACGGTGGCAAGCTCGT | |
| PA3800 right | Reverse | TAGAGGCATGCGATCCCCAGGGCTTCCTGCA | SphI |
| PA5174 left | Forward | ACCTTGAGCTCGTCCAGGCCGCCATCGAGTT | SacI |
| PA5174 left | Reverse | GGCGCTCAGTCGAGCATGTCGCCTGCTGCGTTGTAGCCAC | |
| PA5174 right | Forward | GTGGCTACAACGCAGCAGGCGACATGCTCGACTGAGCGCC | |
| PA5174 right | Reverse | TAGAGGAGCTCCCGGTAGCGAGGAGTTCACC | SacI |
| COMPLEMENTATION | |||
| PA3800 complementation (cloning to pJN105) | Forward | AATTCGAATTCAAGGAGATATACATATGGTGCAATGGAAACACGC | EcoRI |
| Reverse | ATATCTCTAGACTAGCGGATGGTGTAGGCGA | XbaI | |
| PA5174 complementation (cloning to pJN105) | Forward | AATTCGAATTCAAGGAGATATACATATGTCTCGACTACCGGTCATT | EcoRI |
| Reverse | ATATCTCTAGATCAGTCGAGCATGTCGCTGA | XbaI | |
Restriction enzyme recognition sequences are underlined.
Genetic complementation of the ΔPA3800 and ΔPA5174 mutants
To complement the ΔPA3800 and ΔPA5174, DNA fragments containing the entire PA3800 and PA5174 genes were amplified from the PAO1 genome and ligated into EcoRI/XbaI-treated pJN105. The resultant plasmid and the control empty plasmid (i.e., pJN105) were transferred into the ΔPA3800 and ΔPA5174 mutant by electroporation, respectively. Expression of PA3800 or PA5174 gene is induced by 0.01% L-arabinose (Sigma).
Bacterial growth in airway mucus secretion (AMS)
Bacterial cultures grown in LB for 8 h were diluted in PBS to get bacterial suspensions with the 105 CFU/mL. Ten microliters of each diluent was inoculated into 100 μL AMS to achieve the initial inoculum size of ~103 CFU in AMS. Bacterial cells were then grown for 16 h in a humidified 37°C incubator. Bacterial growth was assessed by measuring the growth index. The values for [CFU after 16 h in AMS / CFU after 0 h in AMS] were calculated and plotted as the growth index.
Mouse infection and Caenorhabditis elegans survival test
Animal experiments were approved by the Committee on the Ethics of Animal Experiments of Yonsei University College of Medicine (IACUC permit number: 2013-0369-5). Animal experiments were conducted following national guidelines provided by the Korean government (Ministry for Food, Agriculture, Forestry and Fisheries) and in strict accordance with the institutional guidelines for animal care and use of laboratory animals. To test the virulence of the ΔbamB and ΔfabY mutants in vivo, 8-week-old C57BL/6N inbred female mice (Orient, Korea) were intranasally infected with 5 × 106 cells of each strain (n = 4). The lungs of the infected mice were removed at 16 h post-infection. The harvested lungs were homogenized, and the number of bacteria in each organ was measured by quantifying CFUs. Caenorhabditis elegans survival tests were performed as described previously (Go et al., 2014).
Electron microscope
Lysozyme-treated bacterial cells were visualized using a scanning electron microscope (SEM), as described previously (Yoon et al., 2011). Briefly, fixed bacterial suspensions were stained with 1% OsO4 (Sigma) and then coated with gold via an ion sputter (IB-3 Eiko, Japan). SEM (FE SEM S-800, Hitachi, Japan) was used at an acceleration voltage of 20 kV. All images were processed using ESCAN 4000 software (Bummi Universe Co., Ltd., Seoul, Korea).
Membrane integrity assays
Bacterial resistance against hypo-osmotic stress was measured by monitoring the decrease of OD600 values. Each bacterial strain, grown to mid-exponential phase was harvested and washed with 1 × PBS. The bacterial cells were resuspended in sterilized deionized water. The OD600 of each bacterial suspension was measured every 10 min for 60 min. The data were normalized with their initial OD600-values. The PAO1, ΔbamB, and ΔfabY mutants were also observed under confocal laser scanning microscope (CLSM) with LIVE/DEAD® BacLight™ Bacterial Viability Kit to detect the difference in membrane integrity among these bacterial strains.
Statistical analysis
The experiments were repeated at least in triplicate, and data are expressed as mean ± standard deviation (SD). Data were analyzed using unpaired Student's t-test, unless otherwise stated and P < 0.05 were considered to be statistically significant. Log-rank test was used to provide statistical significance in the C. elegans lifespan experiments.
Results
P. aeruginosa is highly resistant to lysozyme
Prior literature demonstrated that P. aeruginosa can become lysozyme-sensitive only when co-treated with EDTA at pH 8.0 (Voss, 1964) or pretreated with acetone or heat (Warren et al., 1955). As expected, PAO1 grew unencumbered when grown in LB media supplemented with 1 mg/mL lysozyme (Figure 1). Likewise, two other Gram-negative species, Escherichia coli (Ec) and Salmonella enterica serovar Typhimurium (St), also grew normally in these conditions (Figure 1). These results further support our notion that Gram-negative bacterial cells are intrinsically resistant to lysozyme because their outer membrane prevents lysozyme from accessing the peptidoglycan in the periplasm. On the other hand, two Gram-positive species, Bacillus subtilis (Bs) and Listeria monocytogenes (Lm), lost their viability in response to the same treatment (Figure 1). Consistent with previous reports (Bera et al., 2005, 2006), Staphylococcus aureus (Sa) was completely resistant to the lysozyme treatment (Figure 1). PAO1 growth was not affected, even in the presence of 8 mg/mL lysozyme (data not shown), demonstrating an exceptionally high resistance to lysozyme.
Dentification of lysozyme-sensitive PAO1 mutants
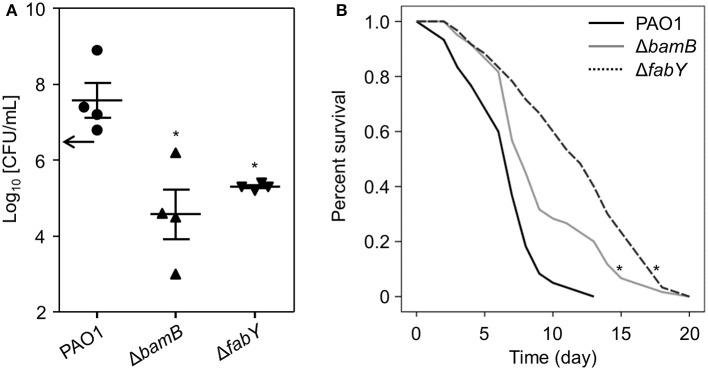
In order to devise a better strategy for P. aeruginosa infection control, it would be desirable to identify lysozyme-sensitive mutants. We therefore constructed a Tn insertion mutant library and screened it for mutants that had lost their resistance to lysozyme treatment. Since lysozyme is more active in environments with reduced ionic strength (Sorrentino et al., 1982; Verhamme et al., 1988), bacterial growth was tested in two-fold diluted LB media (termed 0.5 × LB). Approximately 5,500 mutants were screened, three of which were determined to lack lysozyme resistance. In each mutant, the transposon insertion occurred in PA0420, PA3800, or PA5174 gene. To further verify the effects of these gene disruptions, we generated an in-frame deletion of each gene and examined cell growth in the presence of lysozyme. In 0.5 × LB, ΔPA3800 and ΔPA5174 mutants exhibited significant growth inhibition in the presence of 1.0 mg/mL lysozyme (Figure 2A). After 16 h of growth at 37°C with lysozyme, the CFUs of these two mutants remained similar to those at the time of inoculum (indicated by an arrow on the y-axis), indicating that bacterial growth was prevented by lysozyme at a physiological concentration. PA3800 gene encode β-barrel assembly machinery protein B (BamB) involved in outer membrane protein assembly (Jansen et al., 2012). PA5174 gene encodes probable beta-ketoacyl synthase, which plays a role in fatty acid biosynthesis (Yuan et al., 2012; Figure 2B). The deletion of PA3800 gene did not result in any growth inhibition, whereas the ΔfabY mutant exhibited a slightly affected growth, yielding ~10-fold less viable cell counts after 16 h growth in 0.5 × LB media (Figure 2A). When these mutants were grown with lysozyme, bacterial cells with crumbled rod-shaped morphology were observed (Figure S1), further demonstrating that lysozyme, an enzyme that degrades peptidoglycan polymers, damages the bacterial cell wall structure.
Figure 2.
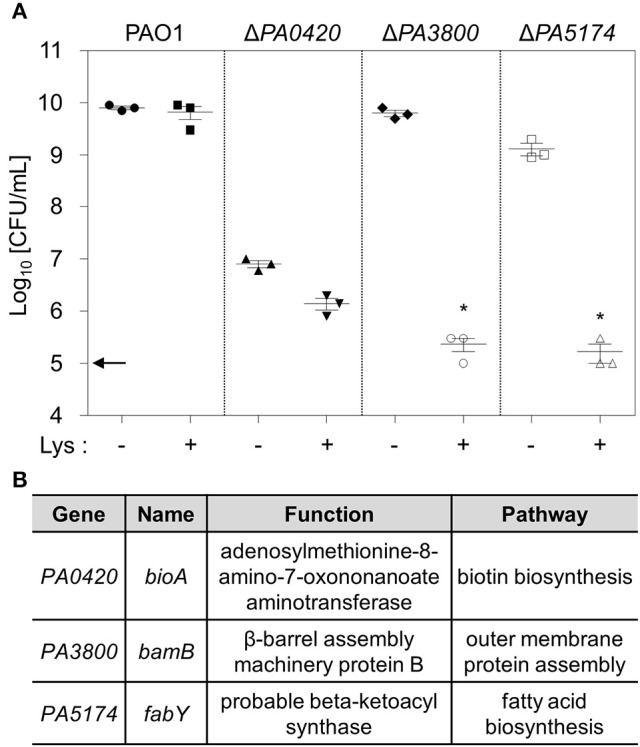
Identification of lysozyme-sensitive PAO1 mutants. (A) Three PAO1 mutant strains were grown in the absence or presence of 1 mg/mL lysozyme in 0.5 × LB. After 16 h growth, aliquots from each culture were diluted and plated for CFU counting. Experimental conditions were identical to those described in the legend of Figure 1. Bacterial cultures were repeated in triplicate, and the mean ± SD values are displayed. *P < 0.05 vs. CFU of untreated cultures. (B) Genetic information for the genes disrupted in each mutant and the functions of the proteins they encoded.
PA0420, bioA gene, produces adenosylmethionine-8-amino-7-oxononanoate aminotransferase, which is responsible for biotin biosynthesis (Beaume et al., 2015). Of note, the ΔbioA mutant exhibited inactive growth, even in the absence of lysozyme treatment, indicating that an interruption in biotin synthesis leads to defective growth in P. aeruginosa. When the mutant was grown in media supplemented with extraneous biotin, its growth and lysozyme susceptibility were completely restored (Figure S2). This result suggests that lysozyme sensitivity of the ΔbioA mutant is likely associated with its faulty growth; therefore, we did not pursue any further investigation on this particular mutant.
When wild type copy of bamB or fabY gene was expressed by the arabinose-inducible promoter, each mutant became fully resistant to lysozyme treatment (Figure 3). These results further confirm that the lysozyme sensitivities observed in the mutants are indeed caused by the deletion of bamB or fabY gene.
Figure 3.
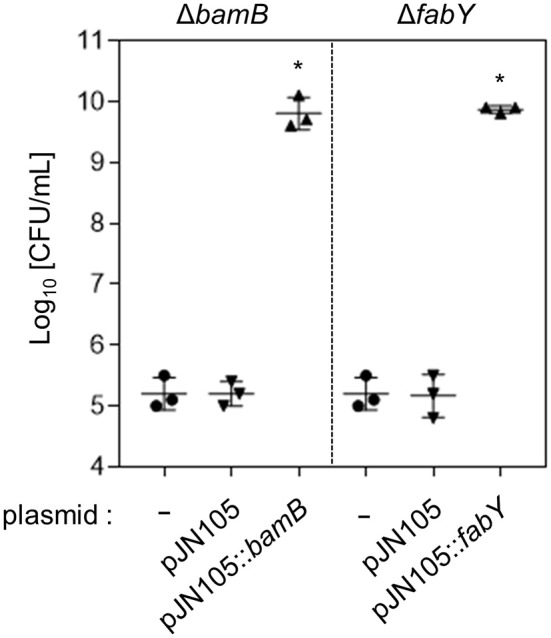
Restored lysozyme resistance of genetically complemented mutants. The ΔbamB and ΔfabY mutants were transformed with pJN105::bamB or pJN105::fabY plasmid, respectively. As a control, each mutant was also transformed with the empty plasmid, pJN105. Bacterial strains were grown in the presence of 1 mg/mL lysozyme for 16 h. To induce the gene expression, 0.01% L-arabinose was also added in each media. Aliquots of each culture were diluted and plated for CFU quantification. Bacterial cultures were performed in triplicate, and the mean ± SD values are displayed. *P < 0.05 vs. CFU of each mutant harboring pJN105.
Lysozyme-sensitive mutants are also susceptible to treatment with airway mucus secretion
Airway mucus secretion (AMS) plays a role in the host innate defense and contains various antimicrobial components including lysozyme (Gi et al., 2015). To examine bacterial response to AMS, we treated the bacterial strains with AMS collected from primary cultures of three different human tracheal tissues. In each treatment, 103 bacterial cells were inoculated and incubated for 16 h. After the treatments, the number of PAO1 cells increased to ~5 × 106 cells (growth index of ~5,000), demonstrating the capability of wild type P. aeruginosa to propagate in AMS (Figure 4). The growth of the ΔbamB mutant was not as robust as that of PAO1. The average growth index of the mutant in AMS was ~68 (Figure 4). Of note, the growth of the ΔfabY mutant was more severely affected, yielding the growth index of ~1.0. Together, these results suggest that two lysozyme-sensitive mutants are less capable of proliferating in AMS, a frontline substance that P. aeruginosa encounters during the early stage of airway infection.
Figure 4.

Bacterial growth in airway mucus secretion (AMS) prepared from primary culture of human tracheal tissues. The wild type PAO1 and its two mutants, ΔbamB and ΔfabY, were grown in three separate AMS preparations for 16 h. Aliquots of each culture were diluted and plated for CFU quantification. The values for CFU in16h / CFU inoculum were calculated and plotted as the growth index. Bacterial cultures were performed in triplicate, and the mean ± SD values are displayed. *P < 0.05 vs. CFU of PAO1 cells.
In vivo infectivity of the lysozyme-sensitive mutants was ameliorated
We next examined whether these mutants were also defective in establishing infections in vivo. To address this question, we utilized two different animal infection models: mouse and nematode. First, mice were intranasally infected with bacterial cells. At 16 h post-infection, mouse lung homogenates were prepared, and the bacterial cells were enumerated. Increased numbers of PAO1 cells were recovered following the 16 h infection (Figure 5A), demonstrating that wild type P. aeruginosa can replicate inside the mouse airway. By comparison, decreased numbers of ΔbamB and ΔfabY mutant cells were recovered, indicating a compromise in their ability to multiply inside a host airway (Figure 5A).
Figure 5.
Survival fitness and virulence of ΔbamB and ΔfabY mutants in vivo. (A) Bacterial counts recovered from the left lung lobes. The infection dose was 5.0 × 106 cells per mouse (black arrow on the y-axis). Mice were infected with the indicated bacterial strains for 16 h. *P < 0.05 vs. CFU of PAO1 cells. (B) Survival curves of the C. elegans N2 strain fed with PAO1 (blue), ΔbamB (green), and ΔfabY (black). *P < 0.05 vs. the survival rate of the PAO1-fed worms. In each group, 20 worms were used. Statistical significance was determined by log-rank analysis.
C. elegans is a nematode that has been widely used to study host-microbe interactions (Go et al., 2014). Furthermore, C. elegans is known to express 15 homologs of lysozyme that act as digestive enzymes for bacterial prey and, therefore, as key players for innate immunity (Mallo et al., 2002; Boehnisch et al., 2011). Given these characteristics, we postulated that C. elegans could serve as an appropriate model to study in vivo infectivity of lysozyme-sensitive mutants. As shown in Figure 5B, C. elegans lived significantly longer when fed ΔbamB and, to a greater extent, ΔfabY mutants; the average lifespans were 9.2 ± 0.5 (days) and 11.6 ± 0.6 (days), respectively. As expected, a considerably shorter lifespan (6.6 ± 0.3 days) was observed in worms grown by feeding on PAO1 cells (Figure 5B). Together, these results demonstrate that lysozyme-sensitive mutants are also less capable of establishing in vivo virulence.
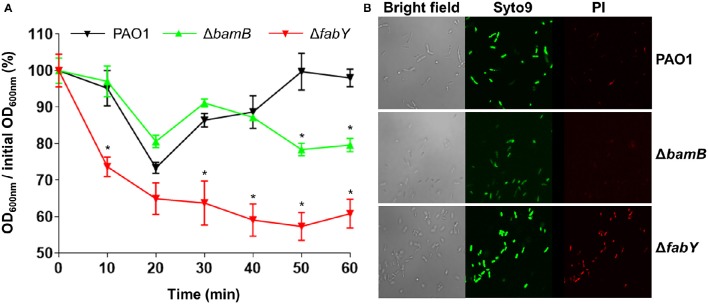
Bacterial cell membrane integrity was mildly and severely affected in ΔbamB and ΔfabY mutants, respectively
Because lysozyme targets bacterial peptidoglycan, we postulated that the increased lysozyme sensitivity of the mutants may be associated with altered bacterial cell membrane integrity. To address this issue, we analyzed how the lysozyme-sensitive mutants responded to hypo-osmotic stress. We also assessed the penetration of propidium iodide, a fluorescent dye that normally impermeable of healthy bacterial membrane, into bacterial cells of each strain. When PBS-washed bacterial cells were resuspended in distilled water, a marked and persistent drop in OD600-values were observed in the ΔfabY mutant over 1 h time period (Figure 6A). In contrast, bacterial cell density monitored by measuring OD600-values was decreased only in the later stage of the experiment for the case of the ΔbamB mutant (Figure 6A). Consistent with the hypo-osmotic resistance result, ΔfabY mutant cells were more permeable to propidium iodide, a red fluorescent dye that only stains bacterial nucleic acids when the membrane integrity was compromised. Together, these results demonstrate that bacterial membrane integrity is affected, especially in the ΔfabY mutant and such a defect likely accounts for the enhanced sensitivity (i) to the AMS treatment (Figure 4) and (ii) inside the C. elegans (Figure 5B).
Figure 6.
Bacterial membrane integrity tests of two lysozyme-sensitive mutants. (A) Bacterial cells resuspended in distilled water were monitored for their changes in OD600 values at 10 min intervals for 1 hr. Experiments were performed in triplicate, and the mean ± SD values are displayed. *P < 0.05 vs. OD600 of PAO1 at each time point. (B) Each bacterial culture was stained with Syto9 and propidium iodide and visualized by confocal laser scanning microscope (CLSM) as described in Section Materials and Methods.
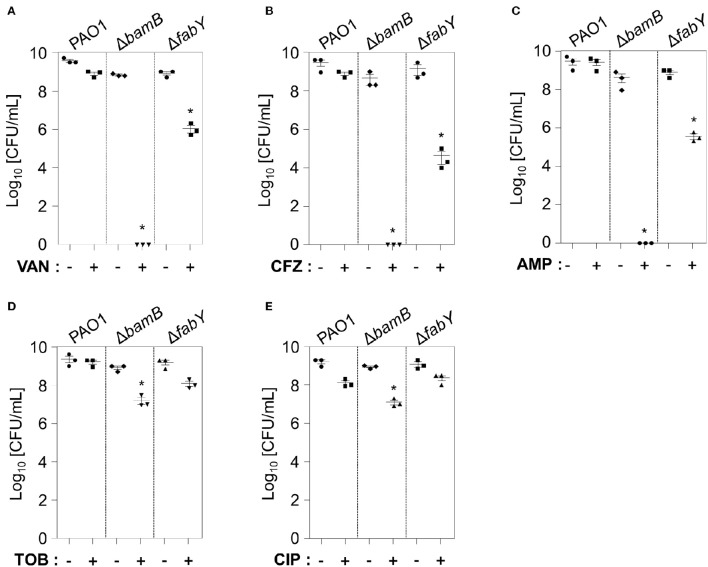
ΔbamB mutant is extremely sensitive to antibiotics that target cell wall synthesis
We then explored whether lysozyme-sensitive mutants also exhibited elevated sensitivity to antibiotic treatments. To examine bacterial responsiveness to antibiotics of diverse classes, we tested vancomycin (Day et al., 1993), ceftazidime (O'Callaghan, 1986), ampicillin (Ghobashy and Chiori, 1984), tobramycin (Hoff et al., 1974), and ciprofloxacin (Roy et al., 1983), the first three of which target cell wall synthesis. Tobramycin and ciprofloxacin inhibit protein synthesis and DNA replication, respectively. At the concentrations tested, PAO1 was either unaffected or only mildly affected (Figures 7A–E). Importantly, the ΔbamB mutant completely lost its viability in response to treatment with vancomycin, ceftazidime, and ampicillin (Figures 7A–C). The viability of the ΔfabY mutant was also decreased about 100~1,000 fold in response to the same treatment (Figures 7A–C). The degree of growth inhibition in these two mutants was not as great when treated with tobramycin and ciprofloxacin (Figures 7D,E). Collectively, these results suggest that (i) the ΔbamB mutant is exceptionally sensitive to cell wall-targeting antibiotics and, therefore, (ii) BamB may be an effective target in the elimination of P. aeruginosa.
Figure 7.
Antibiotic susceptibility of two lysozyme-sensitive PAO1 mutants. PAO1, ΔbamB, and ΔfabY mutants were treated in LB for 16 h with 100 μg/mL Vancomycin (A), 0.5 μg/mL Ceftazidime (B), 25 μg/mL ampicillin (C), 0.2 μg/mL tobramycin (D), or 0.1 μg/mL ciprofloxacin (E). After a16-h incubation, aliquots from each culture were diluted and plated for CFU enumeration. *P < 0.05 vs. CFU of untreated cultures. Antibiotic concentrations that do not affect PAO1 growth were chosen for analysis.
Discussion
Recalcitrant P. aeruginosa infection remains a huge threat to public healthcare. Increasing numbers of metallo-β-lactamase-producing clinical isolates have been identified around the world (Corvec et al., 2006; Chin et al., 2011; Jovcic et al., 2011; Piyakul et al., 2012; Van der Bij et al., 2012), and strains exhibiting resistance to colistin, a “last hope” antibiotic, have also been isolated (Lee et al., 2014). Therefore, novel anti-Pseudomonas strategies that do not generate escape mutants are required. We postulate that one such approach is to exploit lysozyme, an anti-bacterial innate immunoprotein that is abundantly present in the human airway. In this study, we identified PAO1 mutants that became sensitive to lysozyme treatment and provided the clinical perspectives to control P. aeruginosa infections.
The outer membrane of Gram-negative bacterial cells is a physical barrier that prevents lysozyme from gaining access to the peptidoglycan layer. EDTA-treated P. aeruginosa was found to be sensitive to lysozyme; when grown with Mg2+, the effect of EDTA-mediated increased susceptibility to lysozyme was reversed (Witholt et al., 1976; Ayres et al., 1998). These results suggest that outer membrane integrity is important for lysozyme resistance. In this respect, it is not surprising that lysozyme-sensitive mutants were also susceptible to antibiotics that target peptidoglycan synthesis. Of particular interest, however, is the extreme sensitivity of the ΔbamB mutant against cell wall-targeting antibiotics (Figure 7). Complete loss of viability was observed in response to treatment with vancomycin, ceftazidime, or ampicillin. These results demonstrate the potential of BamB as a drug target, the inhibition of which could result in efficient elimination of P. aeruginosa via a low dose of cell wall-targeting antibiotics. The crystal structure of the BamB indicates that it contains (i) a β-propeller fold with a central pore region and (ii) protruding loops that mediate its association with BamA (Jansen et al., 2012). BamA homologs in diverse bacterial species are considered essential for viability (Voulhoux et al., 2003; Gentle et al., 2004; Jansen et al., 2012). Given that BamB is an outer membrane protein, we can seek to identify a potential inhibitor that binds to the central pore region of BamB, thereby interfering with its function. Alternatively, the association of BamB with BamA could be targeted for inhibition. Mori and colleagues showed that peptides homologous to a portion of BamA-binding region of BamD can increase the efficacy of antibiotic-mediated killing of P. aeruginosa (Mori et al., 2012). It will be important to ask whether these peptides can also potentiate bactericidal action of lysozyme in vitro and inside the patient's airway as well.
The ΔfabY mutant produced hypoacylated lipid A and this change was proposed to be responsible for the increased antibiotic sensitivity (Six et al., 2014). Our result in Figure 6 shows that the ΔfabY mutant is more vulnerable to hypo-osmotic stress than ΔbamB. On the other hand, ΔbamB mutant cells are far more sensitive than ΔfabY to cell wall-targeting antibiotics (Figure 7). These findings suggest that alterations in fatty acid synthesis or outer membrane protein assembly may lead to unique cell surface changes, which result in distinct consequences. Importantly, both ΔbamB and ΔfabY mutant are equally susceptible to the treatment with 1.0 mg/ml lysozyme (Figure 2). We therefore hypothesize that lysozyme can be widely used in combination with any intervention that targets bacterial cell surface either at the lipid or protein level. We anticipate that the results provided here will stimulate future investigations.
Cerulenin, an anti-fungal agent, inhibits fatty acid biosynthesis by inhibiting β-ketoacyl-acyl carrier protein synthetase in E. coli (Buttke and Ingram, 1978). PAO1 FabY protein, however, exhibits low homology to the E. coli counterpart. Our experiments demonstrated that cerulenin at up to 200 μg/ml does not inhibit P. aeruginosa growth (data not shown). To date, a wide range of fatty acid synthesis inhibitors have been identified. These include isoniazid (Chan and Vogel, 2010), triclosan (Wright and Reynolds, 2007), Irgasan (Nishi et al., 2016), Kaempferol (Thors et al., 2008), Quercetin (Zhao et al., 2014a), Apigenin (Brusselmans et al., 2005), 1,2,3,4,6-Penta-O-galloyl-β-D-glucose (Zhao et al., 2014b), and Epigallocatechin gallage (Brusselmans et al., 2005). Although many of these inhibitors are proved to be effective in eukaryotic cells, these have not been actively tested in bacterial cells. It is worthwhile to examine whether any of these inhibitors can exert synergistic effects on inhibiting P. aeruginosa growth, when used together with lysozyme.
In conclusion, this study uncovered a common aspect of two different P. aeruginosa mutants with pleiotropic phenotypes. Our results show that interventions that affect fatty acid synthesis or outer membrane protein assembly may facilitate the eradication of P. aeruginosa with the aid of physiological concentration of lysozyme. Insights gleaned from this study will be useful in devising new strategies to combat one of the most resistant infections.
Author contributions
KML and SY conceived, designed, and coordinated the study. KML, KL, JG, IP, JS, JC, and HK performed the experiment and interpreted the data. KML, KL, and SY wrote the manuscript. All the authors participated in discussions of the results and reviewed the final draft.
Funding
This work was supported by grants from the National Research Foundation (NRF) of Korea, funded by the Korean government (2014R1A2A2A01002861, 2014R1A4A1008625, and 2014R1A1A2059520). This work was also made possible by a grant from the Korea Healthcare Technology R&D Project of the Ministry for Health, Welfare, and Family Affairs (HI15C0694).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00059/full#supplementary-material
References
- Ayres H. M., Furr J. R., Russell A. D. (1998). Effect of divalent cations on permeabilizer-induced lysozyme lysis of Pseudomonas aeruginosa. Lett. Appl. Microbiol. 27, 372–374. 10.1046/j.1472-765X.1998.00455.x [DOI] [PubMed] [Google Scholar]
- Beaume M., Kohler T., Fontana T., Tognon M., Renzoni A., van Delden C. (2015). Metabolic pathways of Pseudomonas aeruginosa involved in competition with respiratory bacterial pathogens. Front. Microbiol. 6:321. 10.3389/fmicb.2015.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera A., Biswas R., Herbert S., Gotz F. (2006). The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect. Immun. 74, 4598–4604. 10.1128/IAI.00301-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berra L., Sampson J., Wiener-Kronish J. (2010). Pseudomonas aeruginosa: acute lung injury or ventilator-associated pneumonia? Minerva Anestesiol. 76, 824–832. 10.2147/IDR.S50669 [DOI] [PubMed] [Google Scholar]
- Bera A., Herbert S., Jakob A., Vollmer W., Gotz F. (2005). Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55, 778–787. 10.1111/j.1365-2958.2004.04446.x [DOI] [PubMed] [Google Scholar]
- Boehnisch C., Wong D., Habig M., Isermann K., Michiels N. K., Roeder T., et al. (2011). Protist-type lysozymes of the nematode Caenorhabditis elegans contribute to resistance against pathogenic Bacillus thuringiensis. PLoS ONE 6:e24619. 10.1371/journal.pone.0024619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusselmans K., Vrolix R., Verhoeven G., Swinnen J. V. (2005). Induction of cancer cell apoptosis by flavonoids is associated with their ability to inhibit fatty acid synthase activity. J. Biol. Chem. 280, 5636–5645. 10.1074/jbc.M408177200 [DOI] [PubMed] [Google Scholar]
- Buttke T. M., Ingram L. O. (1978). Inhibition of unsaturated fatty acid synthesis in escherichia coli by the antibiotic cerulenin. Biochemistry 17, 5282–5286. 10.1021/bi00617a031 [DOI] [PubMed] [Google Scholar]
- Chan D. I., Vogel H. J. (2010). Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem. J. 430, 1–19. 10.1042/BJ20100462 [DOI] [PubMed] [Google Scholar]
- Chin B. S., Han S. H., Choi S. H., Lee H. S., Jeong S. J., Choi H. K., et al. (2011). The characteristics of metallo-beta-lactamase-producing gram-negative bacilli isolated from sputum and urine: a single center experience in Korea. Yonsei Med. J. 52, 351–357. 10.3349/ymj.2011.52.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvec S., Poirel L., Decousser J. W., Allouch P. Y., Drugeon H., Nordmann P. (2006). Emergence of carbapenem-hydrolysing metallo-beta-lactamase VIM-1 in Pseudomonas aeruginosa isolates in France. Clin. Microbiol. Infect. 12, 941–942. 10.1111/j.1469-0691.2006.1532_1.x [DOI] [PubMed] [Google Scholar]
- Dajani R., Zhang Y., Taft P. J., Travis S. M., Starner T. D., Olsen A., et al. (2005). Lysozyme secretion by submucosal glands protects the airway from bacterial infection. Am. J. Respir. Cell Mol. Biol. 32, 548–552. 10.1165/rcmb.2005-0059OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day C. A., Marceau-Day M. L., Day D. F. (1993). Increased susceptibility of Pseudomonas aeruginosa to ciprofloxacin in the presence of vancomycin. Antimicrob. Agents Chemother. 37, 2506–2508. 10.1128/AAC.37.11.2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin R. F., Robinson S. K., Widdicombe J. H. (2004). Secretion of lactoferrin and lysozyme by cultures of human airway epithelium. Am. J. Physiol. Lung Cell. Mol. Physiol. 286, L750–L755. 10.1152/ajplung.00326.2003 [DOI] [PubMed] [Google Scholar]
- Duszyk M. (2001). CFTR and lysozyme secretion in human airway epithelial cells. Pflugers Arch. 443(Suppl. 1), S45–S49. 10.1007/s004240100643 [DOI] [PubMed] [Google Scholar]
- Gentle I., Gabriel K., Beech P., Waller R., Lithgow T. (2004). The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164, 19–24. 10.1083/jcb.200310092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghobashy A. A., Chiori C. O. (1984). The combined activity of ampicillin with streptomycin or chloramphenicol against Pseudomonas aeruginosa. Arzneimittelforschung 34, 255–257. [PubMed] [Google Scholar]
- Gi M., Lee K. M., Kim S. C., Yoon J. H., Yoon S. S., Choi J. Y. (2015). A novel siderophore system is essential for the growth of Pseudomonas aeruginosa in airway mucus. Sci. Rep. 5:14644. 10.1038/srep14644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go J., Lee K. M., Park Y., Yoon S. S. (2014). Extended longevity and robust early-stage development of Caenorhabditis elegans by a soil microbe, Lysinibacillus sphaericus. Environ. Microbiol. Rep. 6, 730–737. 10.1111/1758-2229.12196 [DOI] [PubMed] [Google Scholar]
- Hoff G. E., Schiotz P. O., Paulsen J. (1974). Tobramycin treatment of pseudomonas aeruginosa infections in cystic fibrosis. Scand. J. Infect. Dis. 6, 333–337. 10.3109/inf.1974.6.issue-4.07 [DOI] [PubMed] [Google Scholar]
- Hughes W. T., Koblin B. A., Rosenstein B. J. (1982). Lysozyme activity in cystic fibrosis. Pediatr. Res. 16, 874–876. 10.1203/00006450-198210000-00014 [DOI] [PubMed] [Google Scholar]
- Jacquot J., Tournier J. M., Puchelle E. (1985). In vitro evidence that human airway lysozyme is cleaved and inactivated by Pseudomonas aeruginosa elastase and not by human leukocyte elastase. Infect. Immun. 47, 555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen K. B., Baker S. L., Sousa M. C. (2012). Crystal structure of BamB from Pseudomonas aeruginosa and functional evaluation of its conserved structural features. PLoS ONE 7:e49749. 10.1371/journal.pone.0049749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovcic B., Lepsanovic Z., Suljagic V., Rackov G., Begovic J., Topisirovic L., et al. (2011). Emergence of NDM-1 metallo-beta-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob. Agents Chemother. 55, 3929–3931. 10.1128/AAC.00226-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. R. M., Seol S. Y., Yoon S. S., Kim J. (2012). Pseudomonas aeruginosa bacteriophage PA1O requires type IV pili for infection and shows broad bactericidal and biofilm removal activities. Appl. Environ. Microbiol. 78, 6380–6385. 10.1128/A.E.M.00648-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Na I. Y., Park Y. K., Ko K. S. (2014). Genomic variations between colistin-susceptible and -resistant Pseudomonas aeruginosa clinical isolates and their effects on colistin resistance. J. Antimicrob. Chemother. 69, 1248–1256. 10.1093/jac/dkt531 [DOI] [PubMed] [Google Scholar]
- Lee K., Lee K. M., Go J., Ryu J. C., Ryu J. H., Yoon S. S. (2016). The ferrichrome receptor A as a new target for Pseudomonas aeruginosa virulence attenuation. FEMS Microbiol. Lett. 363. 10.1093/femsle/fnw104 [DOI] [PubMed] [Google Scholar]
- Lee K. M., Park Y., Bari W., Yoon M. Y., Go J., Kim S. C., et al. (2012). Activation of cholera toxin production by anaerobic respiration of trimethylamine N-oxide in Vibrio cholerae. J. Biol. Chem. 287, 39742–39752. 10.1074/jbc.M112.394932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo G. V., Kurz C. L., Couillault C., Pujol N., Granjeaud S., Kohara Y., et al. (2002). Inducible antibacterial defense system in C. elegans. Curr. Biol. 12, 1209–1214. 10.1016/S0960-9822(02)00928-4 [DOI] [PubMed] [Google Scholar]
- Marx J., Jacquot J., Berjot M., Puchelle E., Alix A. J. (1986). Characterization and conformational analysis by Raman spectroscopy of human airway lysozyme. Biochim. Biophys. Acta 870, 488–494. 10.1016/0167-4838(86)90257-8 [DOI] [PubMed] [Google Scholar]
- Merlo C. A., Boyle M. P., Diener-West M., Marshall B. C., Goss C. H., Lechtzin N. (2007). Incidence and risk factors for multiple antibiotic-resistant Pseudomonas aeruginosa in cystic fibrosis. Chest 132, 562–568. 10.1378/chest.06-2888 [DOI] [PubMed] [Google Scholar]
- Mori N., Ishii Y., Tateda K., Kimura S., Kouyama Y., Inoko H., et al. (2012). A peptide based on homologous sequences of the beta-barrel assembly machinery component BamD potentiates antibiotic susceptibility of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 67, 2173–2181. 10.1093/jac/dks174 [DOI] [PubMed] [Google Scholar]
- Newman J. R., Fuqua C. (1999). Broad-host-range expression vectors that carry the-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227, 197–203. 10.1016/S0378-1119(98)00601-5 [DOI] [PubMed] [Google Scholar]
- Nishi K. S. K., Sawamoto J., Tokizawa Y., Iwase Y., Yumita N., Ikeda T. (2016). Inhibition of fatty acid synthesis induces apoptosis of human pancreatic cancer cells. Anticancer Res. 36, 4655–4660. 10.21873/anticanres.11016 [DOI] [PubMed] [Google Scholar]
- O'Callaghan H. (1986). Ceftazidime, a broad spectrum cephalosporin with activity against Ps. aeruginosa. J. Hyg. Epidemiol. Microbiol. Immunol. 30, 449–453. [PubMed] [Google Scholar]
- Pankhaniya R. R., Tamura M., Allmond L. R., Moriyama K., Ajayi T., Wiener-Kronish J. P., et al. (2004). Pseudomonas aeruginosa causes acute lung injury via the catalytic activity of the patatin-like phospholipase domain of ExoU. Crit. Care Med. 32, 2293–2299. 10.1097/01.CCM.0000145588.79063.07 [DOI] [PubMed] [Google Scholar]
- Pendleton J. N., Gorman S. P., Gilmore B. F. (2013). Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 11, 297–308. 10.1586/eri.13.12 [DOI] [PubMed] [Google Scholar]
- Piyakul C., Tiyawisutsri R., Boonbumrung K. (2012). Emergence of metallo-beta-lactamase IMP-14 and VIM-2 in Pseudomonas aeruginosa clinical isolates from a tertiary-level hospital in Thailand. Epidemiol. Infect. 140, 539–541. 10.1017/S0950268811001294 [DOI] [PubMed] [Google Scholar]
- Roy C., Foz A., Segura C., Tirado M., Teixell M., Teruel D. (1983). Activity of ciprofloxacin (BAYo 9867) against Pseudomonas aeruginosa and ampicillin-resistant Enterobacteriaceae. Infection 11, 326–328. 10.1007/BF01641358 [DOI] [PubMed] [Google Scholar]
- Sagel S. D., Sontag M. K., Accurso F. J. (2009). Relationship between antimicrobial proteins and airway inflammation and infection in cystic fibrosis. Pediatr. Pulmonol. 44, 402–409. 10.1002/ppul.21028 [DOI] [PubMed] [Google Scholar]
- Sawa T. (2014). The molecular mechanism of acute lung injury caused by Pseudomonas aeruginosa: from bacterial pathogenesis to host response. J. Intensive Care 2:10. 10.1186/2052-0492-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1, 784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Six D. A., Yuan Y., Leeds J. A., Meredith T. C. (2014). Deletion of the beta-acetoacetyl synthase FabY in Pseudomonas aeruginosa induces hypoacylation of lipopolysaccharide and increases antimicrobial susceptibility. Antimicrob. Agents Chemother. 58, 153–161. 10.1128/AAC.01804-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino S., Yakovlev G. I., Libonati M. (1982). Dimerization of deoxyribonuclease I, lysozyme and papain. Effects of ionic strength on enzymic activity. Eur. J. Biochem. 124, 183–189. 10.1111/j.1432-1033.1982.tb05923.x [DOI] [PubMed] [Google Scholar]
- Thors L., Belghiti M., Fowler C. J. (2008). Inhibition of fatty acid amide hydrolase by kaempferol and related naturally occurring flavonoids. Br. J. Pharmacol. 155, 244–252. 10.1038/bjp.2008.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Bij A. K., Van der Zwan D., Peirano G., Severin J. A., Pitout J. D., Van Westreenen M., et al. (2012). Metallo-beta-lactamase-producing Pseudomonas aeruginosa in the Netherlands: the nationwide emergence of a single sequence type. Clin. Microbiol. Infect. 18, E369–E372. 10.1111/j.1469-0691.2012.03969.x [DOI] [PubMed] [Google Scholar]
- Vanderwinkel E., de Pauw P., Philipp D., Ten Have J. P., Bainter K. (1995). The human and mammalian N-acetylmuramyl-L-alanine amidase: distribution, action on different bacterial peptidoglycans, and comparison with the human lysozyme activities. Biochem. Mol. Med. 54, 26–32. 10.1006/bmme.1995.1004 [DOI] [PubMed] [Google Scholar]
- Verhamme I. M., Van Dedem G. W., Lauwers A. R. (1988). Ionic-strength-dependent substrate inhibition of the lysis of Micrococcus luteus by hen egg-white lysozyme. Eur. J. Biochem. 172, 615–620. 10.1111/j.1432-1033.1988.tb13933.x [DOI] [PubMed] [Google Scholar]
- Voss J. G. (1964). Lysozyme lysis of gram-negative bacteria without production of spheroplasts. J. Gen. Microbiol. 35, 313–317. 10.1099/00221287-35-2-313 [DOI] [PubMed] [Google Scholar]
- Voulhoux R., Bos M. P., Geurtsen J., Mols M., Tommassen J. (2003). Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265. 10.1126/science.1078973 [DOI] [PubMed] [Google Scholar]
- Warren G. H., Gray J., Bartell P. (1955). The lysis of Pseudomonas aeruginosa by lysozyme. J. Bacteriol. 70, 614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witholt B., Heerikhuizen H. V., De Leij L. (1976). How does lysozyme penetrate through the bacterial outer membrane? Biochim. Biophys. Acta 443, 534–544. [DOI] [PubMed] [Google Scholar]
- Wright H. T., Reynolds K. A. (2007). Antibacterial targets in fatty acid biosynthesis. Curr. Opin. Microbiol. 10, 447–453. 10.1016/j.mib.2007.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M. Y., Lee K. M., Park Y., Yoon S. S. (2011). Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS ONE 6:e16105. 10.1371/journal.pone.0016105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S. S., Coakley R., Lau G. W., Lymar S. V., Gaston B., Karabulut A. C., et al. (2006). Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J. Clin. Invest. 116, 436–446. 10.1172/JCI24684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Sachdeva M., Leeds J. A., Meredith T. C. (2012). Fatty acid biosynthesis in Pseudomonas aeruginosa is initiated by the FabY class of beta-ketoacyl acyl carrier protein synthases. J. Bacteriol. 194, 5171–5184. 10.1128/JB.00792-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Mao J. M., Zhang S. Y., Zhou Z. Q., Tan Y., Zhang Y. (2014a). Quercetin induces HepG2 cell apoptosis by inhibiting fatty acid biosynthesis. Oncol. Lett. 8, 765–769. 10.3892/ol.2014.2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Wang Y., Hao W., Zhao M., Peng S. (2014b). In vitro inhibition of fatty acid synthase by 1,2,3,4,6-penta-O-galloyl-beta-D-glucose plays a vital role in anti-tumour activity. Biochem. Biophys. Res. Commun. 445, 346–351. 10.1016/j.bbrc.2014.01.191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.