Abstract
Riemerella anatipestifer causes serositis and septicaemia in domestic ducks, geese, and turkeys. Traditionally, the antibiotics were used to treat this disease. Currently, our understanding of R. anatipestifer susceptibility to chloramphenicol and the underlying resistance mechanism is limited. In this study, the cat gene was identified in 69/192 (36%) R. anatipestifer isolated from different regions in China, including R. anatipestifer CH-2 that has been sequenced in previous study. Sequence analysis suggested that there are two copies of cat gene in this strain. Only both two copies of the cat mutant strain showed a significant decrease in resistance to chloramphenicol, exhibiting 4 μg/ml in the minimum inhibitory concentration for this antibiotic, but not for the single cat gene deletion strains. Functional analysis of the cat gene via expression in Escherichia coli BL21 (DE3) cells and in vitro site-directed mutagenesis indicated that His79 is the main catalytic residue of CAT in R. anatipestifer. These results suggested that chloramphenicol resistance of R. anatipestifer CH-2 is mediated by the cat genes. Finally, homology analysis of types A and B CATs indicate that R. anatipestifer comprises type B3 CATs.
Keywords: Riemerella anatipestifer, chloramphenicol acetyltransferase, antibiotics resistance, homologous recombination, site-directed mutagenesis
Introduction
Riemerella anatipestifer is a gram-negative, non-flagellar bacterium belonging to the Flavobacteriaceae family of bacteroidetes that causes serositis and septicaemia in domestic ducks, geese, and turkeys. Currently, the fatality rate of R. anatipestifer-infected ducks has reached up to 75%, thereby resulting in significant economic losses in the duck industry (Ruiz and Sandhu, 2013).
The extensive use of antibiotics for the treatment and prevention of serositis and septicaemia has resulted in multi-drug resistance in R. anatipestifer (Zhong et al., 2009). It was found that 40.5% of R. anatipestifer strains were resistant to chloramphenicol (Chen et al., 2010). Based on the reported whole genome sequence of R. anatipestifer (GenBank accession number: CP004020) (Wang et al., 2014), we searched for resistance genes in R. anatipestifer CH-2 in the Comprehensive Antibiotic Resistance Database (Aakra et al., 2010). We have identified two copies of the cat gene in R. anatipestifer CH-2, namely, G148_1769 and G148_1772.
The cat gene encodes chloramphenicol acetyltransferases (CATs) that inactivate the drugs choramphenicol, thiamphenicol, and azidamfenicol by acetylation, which is the most common mechanism conferring chloramphenicol resistance in bacteria (Schwarz et al., 2004). However, CAT did not inactivate florfenicol because of the replacement of the hydroxyl group at C3 by a fluor residue, and the acceptor site of the acetyl groups was structurally altered in florfenicol (Schwarz et al., 2004). In addition to acetylation inactivation of chloramphenicol, other enzymatic inactivation mechanisms, such as O-phosphorylated (Mosher et al., 1995) and hydrolysis reaction have been identified (Mosher et al., 1990; Tao et al., 2012). Moreover, resistance to chloramphenicol may also be due to mutations/modifications of the target site (Montero et al., 2007), decreased outer membrane permeability (Burns et al., 1989), and the presence of efflux pumps that often act as multidrug extrusion transporters (Daniels and Ramos, 2009), thereby reducing the effective intracellular drug concentration.
In this study, the cat gene was identified in 69/192 (36%) R. anatipestifer isolated from different regions in China by PCR. In order to verify whether cat gene was responsible for chloramphenicol resistance in R. anatipestifer, we constructed the cat gene deletion strains, complement strains and assessed the protein enzyme activity.
Materials and methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. R. anatipestifer strains were grown at 37°C in tryptic soybean broth (TSB, Oxoid) or tryptic soy agar (TSA, Oxoid) in an atmosphere of 5% CO2. Escherichia coli (E. coli) strains were grown on Luria-Bertani (LB, Oxoid) broth or agar at 37°C. When required, antibiotics were added at the following final concentrations (μg/ml): Chloramphenicol (Cm, Sigma), 25; cefoxitin (Cfx, Sigma), 1; kanamycin (Kan, Sigma), 100; ampicillin (Amp, Sigma), 100 or spectinomycin (Spc, Sigma), 70. Diaminopimelic acid (DAP, 50 μg/ml) to E. coli X7213λpir cultures (Edwards et al., 1998).
Table 1.
Strains and plasmids used in this study.
| Strains | Description | Source or reference |
|---|---|---|
| Riemerella anatipestifer ATCC 11845 | Serotype 6 | ATCC |
| R. anatipestifer CH-2 | Serotype 2 | Laboratory collection |
| RA-CH2Δ1769 | RA-CH2Δ1769, SpcR | This study |
| RA-CH2Δ1772 | RA-CH2Δ1772, CfxR | This study |
| RA-CH2Δ1769Δ1772 | RA-CH2Δ1769Δ1772, SpcR CfxR | This study |
| RA-ATCC11845 (pLMF02) | R. anatipestifer ATCC11845 carrying pLMF02, AmpR CfxR | This study |
| RA-ATCC11845 (pLMF02:: cat) | R. anatipestifer ATCC11845 carrying pLMF02:: 1769, AmpR CfxR | This study |
| RA-ATCC11845 (pLMF02:: catH79A) | R. anatipestifer ATCC11845 carrying pLMF02:: H79A, AmpR CfxR | This study |
| Escherichia coli strains | Description | Source or reference |
| X7232 | endA1 hsdR17 () glnV44 thi-1 recA1 gyrA relA1Δ(lacZYA-argF)U169λpir deoR (Φ80dlac Δ(lacZ)M15) | Roland et al., 1999 |
| X7232 (pRE112:: 1769USD) | E. coli X7232 pRE112:: 1769USD, SpcR CmR | This study |
| X7232 (pRE112:: 1772UCD) | E. coli X7232 pRE112:: 1772UCD,CfxR CmR | This study |
| X7213 | thi-1 thr-1 leuB6 glnV44 fhuA21 lacY1 recA1 RP4-2-Tc::Mu λpirΔasdA4Δzhf-2:: Tn10 | Roland et al., 1999 |
| X7213 (pRE112:: 1769USD) | E. coli X7213 pRE112::1769USD, DAP, SpcR CmR | This study |
| X7213 (pRE112:: 1772UCD) | E. coli X7213pRE112:: 1772UCD, DAP CfxR CmR | This study |
| BL21(DE3) | E. coli BL21(DE3), expressing host cell | Laboratory collection |
| BL21(DE3) (pET30a) | E. coli BL21(DE3) carrying pET30a, KanR | This study |
| BL21(DE3) (pET30a:: cat-s) | E. coli BL21(DE3) carrying pET30a:: cat-s, KanR | This study |
| BL21(DE3) (pET30a:: catH79A-s) | E. coli BL21(DE3) carrying pET30a:: catH79A-s, KanR | This study |
| S17-1 | Thi-1 thr leu tonA lac Y supE recA::RP4-2-Tc::Mu KanR | Miller and Mekalanos, 1988 |
| S17-1 (pLMF02) | S17-1 carrying pLMF02, AmpR CfxR | This study |
| S17-1 (pLMF02:: cat) | S17-1 carrying pLMF02:: cat, AmpR CfxR | This study |
| S17-1 (pLMF02:: catH79A) | S17-1 carrying pLMF02:: catH79A, AmpR CfxR | This study |
| Plasmids | Description | Source or reference |
| pET30a | pBR322 lacZ, IPTG-inducible promoter, KanR | Laboratory collection |
| pET30a:: cat-s | pET30a carrying cat adding his tag from R. anatipestifer CH-2, KanR | This study |
| pET30a:: catH79A-s | pET30a carrying catH79A adding his tag from R. anatipestifer CH-2, KanR | This study |
| pLMF02 | shuttle vector transferred between E. coli and R. anatipestifer AmpR, KanR | Liu et al., 2016 |
| pLMF02:: cat | pLMF02 carrying cat from R. anatipestifer CH-2, AmpR CfxR | This study |
| pLMF02:: catH79A | pLMF02 carrying catH79A, AmpR CfxR | This study |
| pYES1new | YAC-BAC shuttle plasmid with SpcR | Laboratory collection |
| pRE112 | sacB mobRP4 R6K ori CmR,pRE112-T-vector | Laboratory collection |
| pRE112:: 1769USD | pRE112 carrying 1769USD from R. anatipestifer CH-2and plasmid pYES1new, SpcR CmR | This study |
| pRE112:: 1772UCD | pRE112 carrying 1772UCD from R. anatipestifer CH-2 and plasmid pCP29, CfxR CmR | This study |
ATCC: American Type Culture Collection.
Detection of the cat gene in R. anatipestifer isolates
For this study, 192 R. anatipestifer isolates were collected from different regions of China. All isolates were identified using the Biolog Microbial Identification System (Biolog, Hayward, CA, USA), as well as PCR and biochemical analyses (data not shown). After lysing the bacteria in lysis buffer (0.5% NP-40, Sigma; 200 ng/ml proteinase K, Takara Biotechnology Co., Ltd. Dalian, China), the presence of the cat gene was determined by PCR analysis using primers cat-F1 and cat-R1 (Table 2).
Table 2.
Primers used in this study.
| Primers | Description | Source and reference |
|---|---|---|
| 1769up-F | 5′-ATTCCAGTTTTTCAAATTCAATTCTTCCCTA-3′ | This study |
| 1769up-R | 5′-CTGTCCTGGCTGGTATTTAACATATTTAATTTACA-3′ | This study |
| Spc-F | 5′-ATATGTTAAATACCAGCCAGGACAGAAATGCC-3′ | This study |
| Spc-R | 5′-CTTCTTTTTATTATTTGCCGACTACCTTGGTGA-3′ | This study |
| 1769down-F | 5′-CGGCAAATAATAAAAAGAAGGTTCCGAAAT-3′ | This study |
| 1769down-R | 5′-TTGATGTGGCATTTGCCTGCAGAT-3′ | This study |
| 1769Ident-F | 5′-TTTGCGAAGAAGCGGGCTAA-3′ | This study |
| 1769Ident-R | 5′-CAAAGAGTTCCTCCGCCGCT-3′ | This study |
| 1769Big-F | 5′-TACTTAACCCGCCATTTTGCCA-3′ | This study |
| 1769Big-R | 5′-AACGGTAGCAACCCAAGCAGTG-3′ | This study |
| 1772up-F | 5′-ATTCTCCAGAGTCGGATTCTGTTGAATTTTTTA-3′ | This study |
| 1772up-R | 5′-GCTTCGGGGTCATTATATATTTAACATATTTAATTTACAAT-3′ | This study |
| Cfx-F | 5′-ATATGTTAAATATATAATGACCCCGAAGCAGGGT-3′ | This study |
| Cfx-R | 5′-GAACCTTCTTTTTATTAAGATTTTACTGAAGTTTGCATT-3′ | This study |
| 1772down-F | 5′-TTCAGTAAAATCTTAATAAAAAGAAGGTTCCGAAATTC-3′ | This study |
| 1772down-R | 5′-TCTAATAAACGATTTTTGGTGGGACACAACTTAC-3′ | This study |
| 1772Ident-F | 5′-ATTTTGACGGATTTATTAGTTGTT-3′ | This study |
| 1772Ident-R | 5′-TTCCGTATAAGCTATCTGAAAACT-3′ | This study |
| 1772Big-F | 5′-AATTTTGAACTTAACCCGCC-3′ | This study |
| 1772Big-R | 5′-ACTACGTCGTACAACATCGTATTG-3′ | This study |
| 16SrRNA-F | 5′-CGAAAGTGATAAGTTAGCCACCT-3′ | This study |
| 16SrRNA-R | 5′-GCAGCACCTTGAAAATTGTCC-3′ | This study |
| cat-F1/ MF1 | 5′-GGGAATTCCATATGAAAAATTTCTTCGAAAGTC-3′ | This study |
| cat-R1/MR2 | 5'-CCGCTCGAGTCAGTGGTGGTGGTGGTGGTGTTTCATTTTTCTAAAAAACTT-3′ | This study |
| MR1 | 5′-ATATTTAGCACCTTGATTACCTG -3′ | This study |
| MF2 | 5′-GTAATCAAGGTGCTAAATATGATT-3′ | This study |
| cat-F2 | 5′-CATGCCATGGATGAAAAATTTCTTCGAAAGTC-3′ | This study |
| cat-R2 | 5′-CCGCTCGAGTCATTTCATTTTTCTAAAAAACTT-3′ | This study |
Construction of R. anatipestifer CH-2 cat deletion mutants and generation of catH79A mutant
The cat genes were deleted by homologous recombination using a suicide vector pRE112 (Kong et al., 2011) as described previously (Luo et al., 2015). Briefly, the right flanking sequence (~620 bp) and the left flanking sequence (~620 bp) of the target genes G148_1769 and G148_1772 were amplified using primers 1769up-F and 1769up-R, 1769down-F and 1769down-R, 1772up-F and 1772up-R, and 1772down-F and 1772down-R, respectively (Table 2). The 1,145 bp SpcR cassette and the 1,192 bp CfxR cassette were amplified from plasmid pYES1 (Luo et al., 2015) and pLMF01 (Liu et al., 2016) using primers Spc-F, Spc-R and Cfx-F, Cfx-R, respectively (Table 2). The SpcR cassette and the CfxR cassette were used for deletion of G148_1769 and G148_1772, respectively. The PCR fragments were overlapped using the PCR method (Xiong et al., 2006). The fused PCR fragments were ligated to suicide plasmid pRE112, respectively, to produce pRE112:: 1769USD (SpcR) and pRE112:: 1772UCD (CfxR). Subsequently, the recombinant plasmids were introduced into R. anatipestifer CH-2 by conjugation as described previously (Liao et al., 2015). The transconjugants were selected on TSA plates supplemented with Spc (40 μg/ml) or Cfx (1 μg/ml). The gene-deletion mutant strains, which were designated as RA-CH2Δ1769, RA-CH2Δ1772, and RA-CH2Δ1769Δ1772, were identified by PCR analysis.
The catH79A mutant was constructed by in vitro site-directed mutagenesis. The upstream and downstream mutated regions of the cat gene of R. anatipestifer CH-2 amplified using primers MF1, MR1 and MF2, MR2 his, respectively (Table 2). The fragments were fused by overlap extension PCR to yield the mutant gene catH79A.
Construction of the recombinant vector for complementation and expression
Complete cat and catH79A genes were amplified by PCR from R. anatipestifer CH-2 chromosomal DNA and by in vitro site-directed mutagenesis using primers catF2 and catR2, catF2, MR1 and MF2, catR2 (Table 2), for complementation. Complete cat and catH79A genes were amplified by PCR from R. anatipestifer CH-2 chromosomal DNA and by in vitro site-directed mutagenesis using primers MF1 and MR2 his, MF1, MR1 and MF2, MR2 his, respectively (Table 2), for expression of CAT and CATH79A proteins. The complementation fragments were purified and digested with NcoI and XhoI, and ligated to the pLMF02 plasmid digested with NcoI and XhoI. The expression fragments were purified and digested with NdeI and XhoI, and ligated with the pET30a plasmid digested with corresponding restriction endonucleases. The ligation mixtures were introduced into CaCl2-competent DH5α cells. Transformants were screened by PCR, and positive clones were sequenced.
Construction of R. anatipestifer ATCC 11845 cat and catH79A complementary strains
The plasmids, pLMF02, pLMF02:: cat, and pLMF02:: catH79A, were introduced into R. anatipestifer ATCC 11845, respectively, by the method described previously (Liao et al., 2015). The transconjugants were selected on TSA plates supplemented with Cfx (1 μg/ml) and Kan (40 μg/ml). The complementation strains, RA-ATCC11845 (pLMF02), RA-ATCC11845 (pLMF02:: cat), and RA-ATCC11845 (pLMF02:: catH79A), were identified by PCR analysis.
Expression and purification of CAT and CATH79A his-tagged proteins
Strains E. coli BL21 (DE3) (pET30a:: cat-s) and E. coli BL21 (DE3) (pET30a:: catH79A -s) were grown overnight in LB medium containing Kan (100 μg/ml). Stationary-phase cultures were diluted to an OD600 of 0.05 in 500 ml of LB medium containing Kan (100 μg/ml) and incubated with shaking at 37°C until the culture density reached an OD600 of 0.6. Cells were then induced with 0.4 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG) and reincubated at 37°C. The cells were harvested by centrifugation for 10 min at 8,000 rpm at 4°C, and then the pellet was resuspended in lysis buffer (20 mM Tris-HCl, pH 8.0; 50 ml) and sonicated. The cell lysate was clarified by centrifugation to eliminate cell debris and then applied to a metal affinity resin column that was equilibrated with the same buffer. The column was successively washed with buffers containing 20 mM, 50 mM imidazole, and phosphate buffer (pH 4.4 and 5.0, respectively). Recombinant proteins were ultrafiltered with storage buffer (20 mM Tris-HCl, pH 7.8). The protein purity was assessed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie Blue staining. Protein concentration was determined using the BCA method with bovine serum albumin as the standard.
Minimum inhibitory concentration (MIC) testing
Chloramphenicol MIC tests for deletion mutants and complementary strains were performed in 96-well microtiter plates according to the Clinical and Laboratory Standard Institute criteria (CLSI, 2015). E. coli ATCC 25922 was used as a quality-control strain. The turbidity of the inocula was adjusted to 107 CFU/ml (100 μl/well). An inoculated broth containing no antibiotics was included as positive control, and a tube of uninoculated broth was used as negative control. The experiments were repeated three times.
Determination of mRNA levels of the cat gene by real-time PCR (RT-PCR) analysis
To assess whether the cat gene of R. anatipestifer was regulated by chloramphenicol, the wild-type strain was grown with TSB with or without 1 μg/ml of chloramphenicol. Total RNA was isolated from strains grown to log phase (OD600 ≈ 0.8–1.0) by using the RNAiso Plus kit (TaKaRa). DNA was removed using RNase-Free DNase. cDNA was generated by using the Sensiscript RT kit (TaKaRa), according to the manufacturer's instructions. Real-time quantitative PCR (qPCR) was performed to measure cat mRNA levels using SYBR Premix EX Taq II (TaKaRa). The primers used in real-time qPCR analysis are listed in Table 2. The expression level of the cat gene was normalized to that of the recA gene, which was used as reference. All PCR reactions were performed in triplicate. The efficiency of primer binding was determined by linear regression by plotting the cycle threshold (CT) value vs. the log of the cDNA dilution. Relative quantification of the transcript was determined using the comparative CT method (2−ΔΔCT), calibrated to recA. The experiments were performed multiple times independently and generated comparable results. The findings are presented as fold-change relative to the mRNA expression levels of the control strains.
CAT activity assay
CAT catalyzes the transfer of an acetyl group from acetyl-CoA to Cm, producing acetylated Cm and CoASH. The CATase activity was assayed based on the disappearance of acetyl-CoA during Cm acetylation (Kobayashi et al., 2015). The reaction mixture contained 0.25 ml of 0.2 M Tris-HCl (pH 7.8), 0.05 ml of 1 mM acetyl-CoA, 0.05 ml of 1 mM Cm, 0.05 ml of 10 mM DTNB [5,5′-dithio-bis (2-nitrobenzoic acid)], and 0.1 ml enzyme extract. The reaction was initiated by the addition of Cm. An increase in absorbance at a wavelength of 412 nm, which arises from 5-thio-2-nitrobenzoic acid, was derived from the reaction between free CoASH and DTNB. The concentration of 5-thio-2-nitrobenzoic acid was determined using its molar extinction coefficient at 412 nm (13,600 M−1 cm−1). The value was then used in the determination of the amount of CoASH produced during the reaction. One unit of enzyme activity is defined as the amount of activity catalyzing 1 μmol of acetyl transfer per min under the assay conditions.
Softwares
The changes of mRNAs were expressed as fold expression and calculated using the comparative CT (2−ΔΔCT) method. The results of RT-PCR were performed using GraphPad Prism 6.0 software for Windows (GraphPad Software Inc., La Jolla, USA). Homology analysis of types A and B CATs based on amino acid identity using DNAMAN 8.0 (Lynnon-Biosoft, Ontario, Canada).
Results
Identification and sequence analysis of the cat gene in R. anatipestifer isolates
The cat gene was identified in 69/192 (36%) R. anatipestifer isolates collected from China, thereby suggesting that the cat gene was widely distributed among R. anatipestifer strains. Sequence analysis found that there are two copies of the cat gene (G148_1769: 1854900…1855529 and G148_1772: 1858427…1859056) in R. anatipestifer CH-2. There is no similarity between the cat gene from R. anatipestifer CH-2 and type A cat genes previously reported. However, the cat genes share 99–100% identity in R. anatipestifer strains reported in NCBI.
MIC of chloramphenicol for R. anatipestifer CH-2 and other strains
To verify whether the cat genes of R. anatipestifer CH-2 were responsible for chloramphenicol resistance, the deletion strains and complementation strains were constructed. Table 3 showed that the chloramphenicol MICs of R. anatipestifer CH-2 and RA-CH2Δ1769 were 32 and 64 μg/ml, respectively. Compared to the MIC of the wild-type strain, the MIC of RA-CH2Δ1769 increased (Table 3). Similarly, another signal cat gene deletion strain RA-CH2Δ1772 had no obviously decreased in resistance to chloramphenicol, exhibiting 32 μg/ml in the minimum inhibitory concentration for chloramphenicol (Table 3). Thus, we supposed that the two copies of cat gene in R. anatipestifer CH-2 were involved in chloramphenicol resistance. The two copies of the cat gene deletion strain RA-CH2Δ1769Δ1772 was constructed. The level of chloramphenicol resistance was determined to be significantly reduced, 4 μg/ml.
Table 3.
Minimal inhibitory concentration (MIC) of chloramphenicol on R. anatipestifer and other strains.
| Strain | MIC (μg/ml) |
|---|---|
| RA-CH-2 | 32 |
| RA-CH2Δ1769 | 64 |
| RA-CH2Δ1772 | 32 |
| RA-CH2Δ1769Δ1772 | 4 |
| RA-ATCC11845(pLMF02) | <2 |
| RA-ATCC11845(pLMF02:: cat) | 32 |
| RA-ATCC11845(pLMF02:: catH79A) | <2 |
To further verify that the cat genes are related to chloramphenicol resistance in R. anatipestifer, shuttle plasmid pLMF02 with the cat gene was introduced into R. anatipestifer ATCC 11845, which is sensitive to chloramphenicol. It was restored the level of chloramphenicol resistance (Table 3). These results strongly suggested that the cat gene was responsible for chloramphenicol resistance in R. anatipestifer.
The transcription of cat gene was increased in RA-CH2Δ1769
According to the study described above, the minimum inhibitory concentrations for chloramphenicol between RA-CH2Δ1769 and RA-CH2Δ1772 are not same (Table 3). To explore whether the transcription of cat gene is affected by single deletion strain, RT-PCR analysis was performed. The result revealed that G148-1772 was upregulated 3.82-fold in the RA-CH2Δ1769 mutant (Figure 1). However, the mRNA level of G148-1769 in RA-CH2Δ1772 did not increased significantly. This information could explain why the resistance level of RA-CH2Δ1769 is greater than RA-CH2Δ1772 and wild-type strain. The result showed that the cat genes do mediate the production of chloramphenicol resistance and the relationship of the two cat copies is complementary and cooperative in R. anatipestifer CH-2.
Figure 1.

Real-time RT-PCR analysis of cat gene expression in the RA-CH2, RA-CH2Δ1769, RA-CH2Δ1772, and RA-CH2Δ1769Δ1772 strains. The changes of mRNAs were expressed as fold expression and calculated using the comparative CT (2−ΔΔCT) method. The error bars represent the standard deviation of three independent experiments.
The transcription of cat gene was induced by chloramphenicol
In order to study the cellular strategies used by R. anatipestifer CH-2 and mutant strains in the presence of chloramphenicol, we decided to analyze transcriptional changes of cat gene in these strains growing in the presence of 1 μg/ml of this antibiotic. Chloramphenicol treatment had no bactericidal effect when sub-inhibitory concentration of chloramphenicol were applied (data not shown). We found that the mRNA level of cat genes was increased 11-fold, 13.94- and 18.31-fold in wild strain and mutant strains RA-CH2Δ1769, RA-CH2Δ1772, respectively (Figure 2). These results suggested that cat genes were regulated by chloramphenicol.
Figure 2.
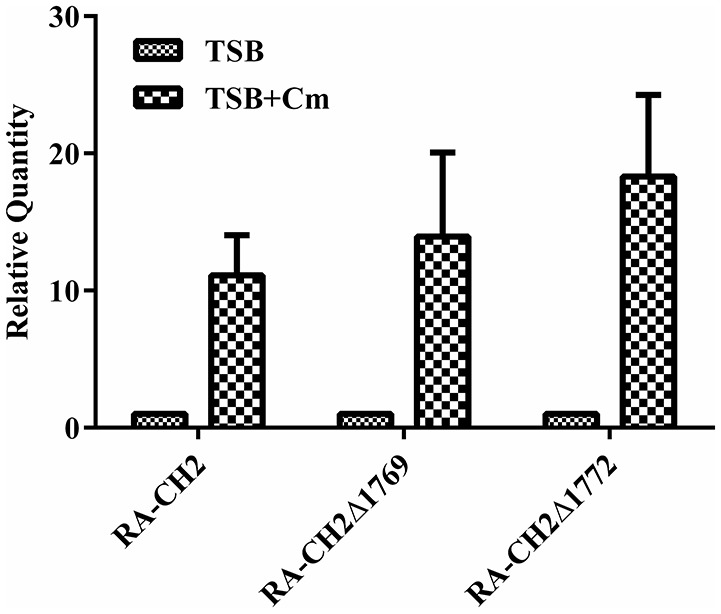
mRNA level of cat genes in chloramphenicol-induced RA-CH2 and mutant strains RA-CH2Δ1769, RA-CH2Δ1772. The changes of mRNAs were expressed as fold expression and calculated using the comparative CT (2−ΔΔCT) method. The error bars represent the standard deviation of three independent experiments.
Catalytic activity of the CAT and CATH79A proteins
In a previous study involving Pseudomonas aeruginosa, His79 served as a major catalytic residue (Beaman et al., 1998). The two amino acid sequences of cat from P. aeruginosa and R. anatipestifer, respectively, showed 86.95% identity (Figure 3). To examine the main catalytic site of CAT from R. anatipestifer CH-2, the cat gene and the catH79A gene were expressed in E. coli cells. The gene products, which contained C-terminal His6-tag, were purified by Ni-agarose affinity chromatography, yielding a distinct protein band in the SDS-PAGE gel, with an approximately molecular weight of ~25 kDa. The catalytic activities of CAT and CATH79A were analyzed at 37°C. The detailed information of the reaction mixtures is described in the Materials and Methods. The specific activities of CAT and CATH79A were 8.33 ± 0.38 and 0 U·mg−1, respectively (Table 4). Meanwhile, the MIC of ATCC 11845 harbored catH79A was significantly lower than that of ATCC 11845 carried the cat gene (Table 3). Thus, the H79A substitution had a significant effect on CAT activity.
Figure 3.
Sequence alignments of CAT from R. anatipestifer (RA) and P. aeruginosa (PA) using CLUSTAL X.
Table 4.
The CAT and CATH79A activity.
| Protein | Specific activity (U/mg) | |||
|---|---|---|---|---|
| I | II | III | Avg | |
| CAT | 8.75 | 8.25 | 8.0 | 8.33 ± 0.38 |
| CATH79A | 0 | 0 | 0 | 0 |
Discussion
CATs inactivate chloramphenicol via acetylation, which is the most prevalent mechanism of resistance to chloramphenicol in bacteria (Shaw, 1983; Murray and Shaw, 1997; Schwarz et al., 2004). CATs have been described in both gram-positive and gram-negative bacteria. There are two defined types of CATs that distinctly differ in their structure: The classical CATs, which are referred to as type A CATs, and the novel CATs, which are also known as type B CATs (Schwarz et al., 2004). There are at least 16 distinct groups of catA genes (A1–A16) and at least 5 different groups of type B cat genes (B1–B5) (Schwarz et al., 2004). Types A and B CATs are both capable of acetylating the hydroxyl group at C3 of chloramphenicol.
In our case, there was two copies of the cat gene in R. anatipestifer CH-2. Not surprisingly, the phenomenon of having 2 copies of the cat gene was found in other bacteria, for example Clostridium sporogenes (CP009225) (Zhang et al., 2015), Chryseobacterium sp. (AP014624) (Morohoshi et al., 2014), M. odoratimimus (CP013690) (Hu et al., 2016), and Aliivibrio wodanis (LN554847). Mutant strains were constructed. Only both two copies of the cat mutant strain showed a significant reduction in resistance to chloramphenicol, but not for the single cat gene deletion strains. ATCC 11845 is a R. anatipestifer strain that was isolated from ducklings in 1932, and genome analysis indicated that it does not harbor the cat gene and is sensitive to chloramphenicol. Complementation ATCC 11845 with the cat gene from R. anatipestifer CH-2 restored the level of chloramphenicol resistance. These results showed that the cat genes do mediate the production of chloramphenicol resistance and the relationship of the two cat copies is complementary and cooperative in R. anatipestifer.
To explore the function and the active site of the cat gene in R. anatipestifer, CAT and CATH79A were expressed and purified. Enzymatic activity analysis of CAT and CATH79A produced by in vitro site-directed mutagenesis indicated that CATH79A had no catalytic activity, thereby suggesting that His79 is the main catalytic residue of CAT. In addition, the present study further demonstrated that the cat gene is involved in chloramphenicol resistance, thus supporting our hypothesis that the cat genes are chloramphenicol resistance determinant factors in R. anatipestifer.
Type B CATs can be further classified into at least five groups. We constructed a homology tree of types A and B CATs (Figure 4) based on their reported amino acid sequence (van Hoek et al., 2011; Roberts et al., 2012). Types A and B CATs showed 10% similarity. We also determined that the R. anatipestifer CAT forms a separate branch from the type B CATs. In addition, types A4 and A7 CATs were observed to be 100% similarity. Thus, the classification of CATs should be revisited. Two types of genes that encode CATs could be based on their structure, namely, types A and B, by using the criterion of ≥80% amino acid identity to define a subgroup (Roberts and Schwarz, 2009). The sequence information of types A and B CATs is listed in the Supplementary Table 1. A total of 15 distinct groups were identified, A1–A15 for type A CATs and five different groups for type B CATs, B1–B5. Types A4 and A7 share 100% identity and belong to a subclass that we designated as A4. Groups A8–A16 were renamed as A7–A15. Groups B2, B3, and B6 showed >80% homology. These three categories are classified as a subclass, namely, B2. The CAT of R. anatipestifer was designated as B3. The rest of the type B classifications remained the same.
Figure 4.

Homology analysis of types A and B CATs based on amino acid identity using DNAMAN 8.0 (Lynnon-Biosoft, Ontario, Canada). The sequence information of types A and B CATs are listed in the Supplementary Table 1.
It was reported that the cat genes identified in gram-positive bacteria Bacillus spp. and Straphylococcus were inducibly expressed by chloramphenicol (Mongkolsuk et al., 1984; Bruckner and Matzura, 1985; Duvall et al., 1985). To verify whether the cat genes were induced by chloramphenicol in R. anatipestifer CH-2, RT-PCR was performed to determine the cat transcript level of the wild-type strain and mutant strains in the presence or absence of chloramphenicol at a concentration of 1 μg/ml. The results exhibited that the level of transcription of the cat gene increased in the presence of chloramphenicol. However, the inducing mechanism is not understood at this time in R. anatipestifer.
It has been demonstrated that catA86 and catA112 were regulated by a mechanism known as translation attenuation in the previous studies (Lovett, 1996). Later, translational attenuation has been proposed as the regulatory mechanism for the chloramphenicol-inducible catB1 gene of Agrobacterium tumefaciens (Rogers et al., 2002). Sequence analysis found that CAT from R. anatipestifer shared 65% identity with that of A. tumefaciens. It is unclear whether they have the same inducing mechanism. Further studies determining the regulatory mechanism underlying the cat gene in R. anatipestifer are warranted.
Author contributions
DZ and AC conceived and designed the project; LH and HY constructed the cat deletion mutant of R. anatipestifer and detected resistance; ML and XZ detected the mRNA levels of the cat gene by RT-PCR; LH and HY constructed ATCC 11845 cat and catH79A complementary strains. LH, RJ, and SC performed expression and purification of CAT and CATH79A His-tagged proteins; LH, QY, and YW performed CAT activity assay; MW, KS, and XC detected the cat gene in R. anatipestifer isolates; LH and DZ drafted and revised the manuscript. All authors have read and approved the final version manuscript.
Funding
This work was supported by the National Natural Science Foundation of China under Grant No. 31372468; National Science and Technology Support Program under Grant No. 2015BAD12B05; China Agricultural Research System under Grant No. CARS-43-8; Youth Science and Technology Innovation Research Team of Sichuan Province for Waterfowl Diseases Prevention and Control under Grant No. 2013TD0015; Integration and Demonstration of Key Technologies for Duck Industrial in Sichuan Province under Grant No. 2014NZ0030.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank professor Francis Biville, Département Infection et Epidémiologie, Institut Pasteur, for his helpful suggestions which have improved the quality of this paper.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00297/full#supplementary-material
References
- Aakra A., Vebo H., Indahl U., Snipen L., Gjerstad O., Lunde M., et al. (2010). The response of Enterococcus faecalis V583 to chloramphenicol treatment. Int. J. Microbiol. 2010:483048. 10.1155/2010/483048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaman T. W., Sugantino M., Roderick S. L. (1998). Structure of the hexapeptide xenobiotic acetyltransferase from Pseudomonas aeruginosa. Biochemistry 37, 6689–6696. 10.1021/bi980106v [DOI] [PubMed] [Google Scholar]
- Bruckner R., Matzura H. (1985). Regulation of the inducible chloramphenicol acetyltransferase gene of the Staphylococcus aureus plasmid pUB112. EMBO J. 4, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J. L., Hedin L. A., Lien D. M. (1989). Chloramphenicol resistance in Pseudomonas cepacia because of decreased permeability. Antimicrob. Agents Chemother. 33, 136–141. 10.1128/AAC.33.2.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. P., Tsao M. Y., Lee S. H., Chou C. H., Tsai H. J. (2010). Prevalence and molecular characterization of chloramphenicol resistance in Riemerella anatipestifer isolated from ducks and geese in Taiwan. Avian Pathol. 39, 333–338. 10.1080/03079457.2010.507761 [DOI] [PubMed] [Google Scholar]
- CLSI (2015). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fifth Informational Supplement. CLSI document M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Daniels C., Ramos J. L. (2009). Adaptive drug resistance mediated by root-nodulation-cell division efflux pumps. Clin. Microbiol. Infect. 15(Suppl. 1), 32–36. 10.1111/j.1469-0691.2008.02693.x [DOI] [PubMed] [Google Scholar]
- Duvall E. J., Mongkolsuk S., Kim U. J., Lovett P. S., Henkin T. M., Chambliss G. H. (1985). Induction of the chloramphenicol acetyltransferase gene cat-86 through the action of the ribosomal antibiotic amicetin: involvement of a Bacillus subtilis ribosomal component in cat induction. J. Bacteriol. 161, 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R. A., Keller L. H., Schifferli D. M. (1998). Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157. 10.1016/S0378-1119(97)00619-7 [DOI] [PubMed] [Google Scholar]
- Hu S. H., Yuan S. X., Qu H., Jiang T., Zhou Y. J., Wang M. X., et al. (2016). Antibiotic resistance mechanisms of Myroides sp. J. Zhejiang Univ. Sci. B 17, 188–199. 10.1631/jzus.B1500068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi J., Furukawa M., Ohshiro T., Suzuki H. (2015). Thermoadaptation-directed evolution of chloramphenicol acetyltransferase in an error-prone thermophile using improved procedures. Appl. Microbiol. Biotechnol. 99, 5563–5572. 10.1007/s00253-015-6522-4 [DOI] [PubMed] [Google Scholar]
- Kong Q., Yang J., Liu Q., Alamuri P., Roland K. L., Curtiss R., III. (2011). Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect. Immun. 79, 4227–4239. 10.1128/IAI.05398-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Cheng X., Zhu D., Wang M., Jia R., Chen S., et al. (2015). TonB energy transduction systems of Riemerella anatipestifer are required for iron and hemin utilization. PLoS ONE 10:e0127506. 10.1371/journal.pone.0127506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Wang M., Zhu D., Wang M., Jia R., Chen S., et al. (2016). Investigation of TbfA in Riemerella anatipestifer using plasmid-based methods for gene over-expression and knockdown. Sci. Rep. 6:37159. 10.1038/srep37159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S. (1996). Translation attenuation regulation of chloramphenicol resistance in bacteria. Gene 179, 157–162. 10.1016/S0378-1119(96)00420-9 [DOI] [PubMed] [Google Scholar]
- Luo H., Liu M., Wang L., Zhou W., Wang M., Cheng A., et al. (2015). Identification of ribosomal RNA methyltransferase gene ermF in Riemerella anatipestifer. Avian Pathol. 44, 162–168. 10.1080/03079457.2015.1019828 [DOI] [PubMed] [Google Scholar]
- Miller V. L., Mekalanos J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583. 10.1128/jb.170.6.2575-2583.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Ambulos N. P., Jr., Lovett P. S. (1984). Chloramphenicol-inducible gene expression in Bacillus subtilis is independent of the chloramphenicol acetyltransferase structural gene and its promoter. J. Bacteriol. 160, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero C. I., Johnson M. R., Chou C. J., Conners S. B., Geouge S. G., Tachdjian S., et al. (2007). Responses of wild-type and resistant strains of the hyperthermophilic bacterium Thermotoga maritima to chloramphenicol challenge. Appl. Environ. Microbiol. 73, 5058–5065. 10.1128/AEM.00453-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi T., Wang W. Z., Someya N., Ikeda T. (2014). Complete genome sequence of Chryseobacterium sp. Strain StRB126, an N-Acylhomoserine lactone-degrading bacterium isolated from potato root. Genome Announc. 2:e00952-14. 10.1128/genomea.00952-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R. H., Camp D. J., Yang K., Brown M. P., Shaw W. V., Vining L. C. (1995). Inactivation of chloramphenicol by o-phosphorylation: a novel resistance mechanism in Streptomyces venezuelae isp5230, a chloramphenicol producer. J. Biol. Chem. 270, 27000–27006. 10.1074/jbc.270.45.27000 [DOI] [PubMed] [Google Scholar]
- Mosher R. H., Ranade N. P., Schrempf H., Vining L. C. (1990). Chloramphenicol resistance in Streptomyces: cloning and characterization of a chloramphenicol hydrolase gene from Streptomyces venezuelae. J. Gen. Microbiol. 36, 293–301. 10.1099/00221287-136-2-293 [DOI] [PubMed] [Google Scholar]
- Murray I. A., Shaw W. V. (1997). O-Acetyltransferases for chloramphenicol and other natural products. Antimicrob. Agents Chemother. 41, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. C., Schwarz S. (2009). Tetracycline and chloramphenicol resistance mechanisms, in Mechanisms of Drug Resistance, ed Mayers D. L. (New York, NY: Humana Press; ), 183–193. [Google Scholar]
- Roberts M. C., Schwarz S., Aarts H. J. (2012). Erratum: acquired antibiotic resistance genes: an overview. Front. Microbiol. 3:384. 10.3389/fmicb.2012.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E. J., Rahman M. S., Hill R. T., Lovett P. S. (2002). The chloramphenicol-inducible catB gene in Agrobacterium tumefaciens is regulated by translation attenuation. J. Bacteriol. 184, 4296–4300. 10.1128/JB.184.15.4296-4300.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland K., Curtiss R., III., Sizemore D. (1999). Construction and evaluation of a delta cya delta crp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 43, 429–441. 10.2307/1592640 [DOI] [PubMed] [Google Scholar]
- Ruiz J., Sandhu T. S. (2013). Riemerella anatipestifer infection, in Diseases of Poultry, 13th Edn. eds Swayne D. E., Glisson J. R., McDougald L. R., Nolan L. K., Suarez D. L., Nair V. L. (Hoboken, NJ: John Wiley & Sons, Inc.), 823–828. [Google Scholar]
- Schwarz S., Kehrenberg C., Doublet B., Cloeckaert A. (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28, 519–542. 10.1016/j.femsre.2004.04.001 [DOI] [PubMed] [Google Scholar]
- Shaw W. V. (1983). Chloramphenicol acetyltransferase: enzymology and molecular biology. CRC Crit. Rev. Biochem. 14, 1–46. 10.3109/10409238309102789 [DOI] [PubMed] [Google Scholar]
- Tao W., Lee M. H., Wu J., Kim N. H., Kim J. C., Chung E., et al. (2012). Inactivation of chloramphenicol and florfenicol by a novel chloramphenicol hydrolase. Appl. Environ. Microbiol. 78, 6295–6301. 10.1128/AEM.01154-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoek A. H., Mevius D., Guerra B., Mullany P., Roberts A. P., Aarts H. J. (2011). Acquired antibiotic resistance genes: an overview. Front. Microbiol. 2:203. 10.3389/fmicb.2011.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Liu W., Zhu D., Yang L., Liu M., Yin S., et al. (2014). Comparative genomics of Riemerella anatipestifer reveals genetic diversity. BMC Genomics 15:479. 10.1186/1471-2164-15-479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong A. S., Yao Q. H., Peng R. H., Duan H., Li X., Fan H. Q., et al. (2006). PCR-based accurate synthesis of long DNA sequences. Nat. Protoc. 1, 791–797. 10.1038/nprot.2006.103 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Grosse-Honebrink A., Minton N. P. (2015). A universal mariner transposon system for forward genetic studies in the genus Clostridium. PLoS ONE 10:e0122411. 10.1371/journal.pone.0122411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C. Y., Cheng A. C., Wang M. S., Zhu de K., Luo Q. H., Zhong C. D., et al. (2009). Antibiotic susceptibility of Riemerella anatipestifer field isolates. Avian Dis. 53, 601–607. 10.1637/8552-120408-ResNote.1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



