Summary
The translocation of messenger RNA and transfer RNA through the ribosome is catalyzed by EF-G, a universally conserved GTPase. The mechanism by which the closely related decapeptide antibiotics dityromycin and GE82832 inhibit EF-G-catalyzed translocation is elucidated in this study. Using crystallographic and biochemical experiments we demonstrate that these antibiotics bind to ribosomal protein S12 in solution as well as within the small ribosomal subunit, inducing long-range effects on the ribosomal head. The crystal structure of the antibiotic in complex with the 70S ribosome reveals that the binding involves conserved amino acid residues of S12 whose mutations result in in vitro and in vivo antibiotic resistance and loss of antibiotic binding. The data also suggest that GE82832/dityromycin inhibits EF-G-catalyzed translocation by disrupting a critical contact between EF-G and S12 that is required to stabilize the post-translocational conformation of EF-G, thereby preventing the ribosome-EF-G complex from entering a conformation productive for translocation.
Introduction
The movement of tRNA through the ribosome is catalyzed in bacteria by elongation factor G (EF-G), a universally conserved GTPase that accelerates the translocation of tRNA from the ribosomal A-site to the P-site (Katunin et al., 2002; Rodnina et al., 1997). Although spontaneous translocation occurs, its rate in the absence of EF-G is orders of magnitude slower – too slow to meet the needs of a living cell (Asatryan and Spirin, 1975; Katunin et al., 2002; Peske et al., 2000; Southworth et al., 2002). Several small molecule inhibitors of translocation have been identified (Peske et al., 2004; Walter et al., 2012), providing insight into the workings of EF-G and trapping EF-G-bound conformations of the ribosome for use in structural studies (Gao et al., 2009a; Munro et al., 2010; Ogle et al., 2002; Stark et al., 2000). In addition, inhibitors of translocation can be used as antibiotics, provided they do not also interfere with protein synthesis in higher organisms (Stanley et al., 2010).
Dityromycin is a cyclic decapeptide antibiotic containing several modified amino acids. It was originally discovered in the 1970s (Omura et al., 1977) and structurally characterized a decade later (Teshima et al., 1988) (Fig. 1A). The closely related antibiotic GE82832 was identified as a translation inhibitor by high throughput screening of a library of natural products (Brandi et al., 2006, 2012). Despite the fact that GE82832 and dityromycin are secondary metabolites of different microorganisms, recent studies have shown that both antibiotics target the 30S ribosomal subunit, inhibit translocation and display the same microbiological and functional properties; in addition, their chemical structures are identical, except for a difference of two mass units (Brandi et al., 2012).
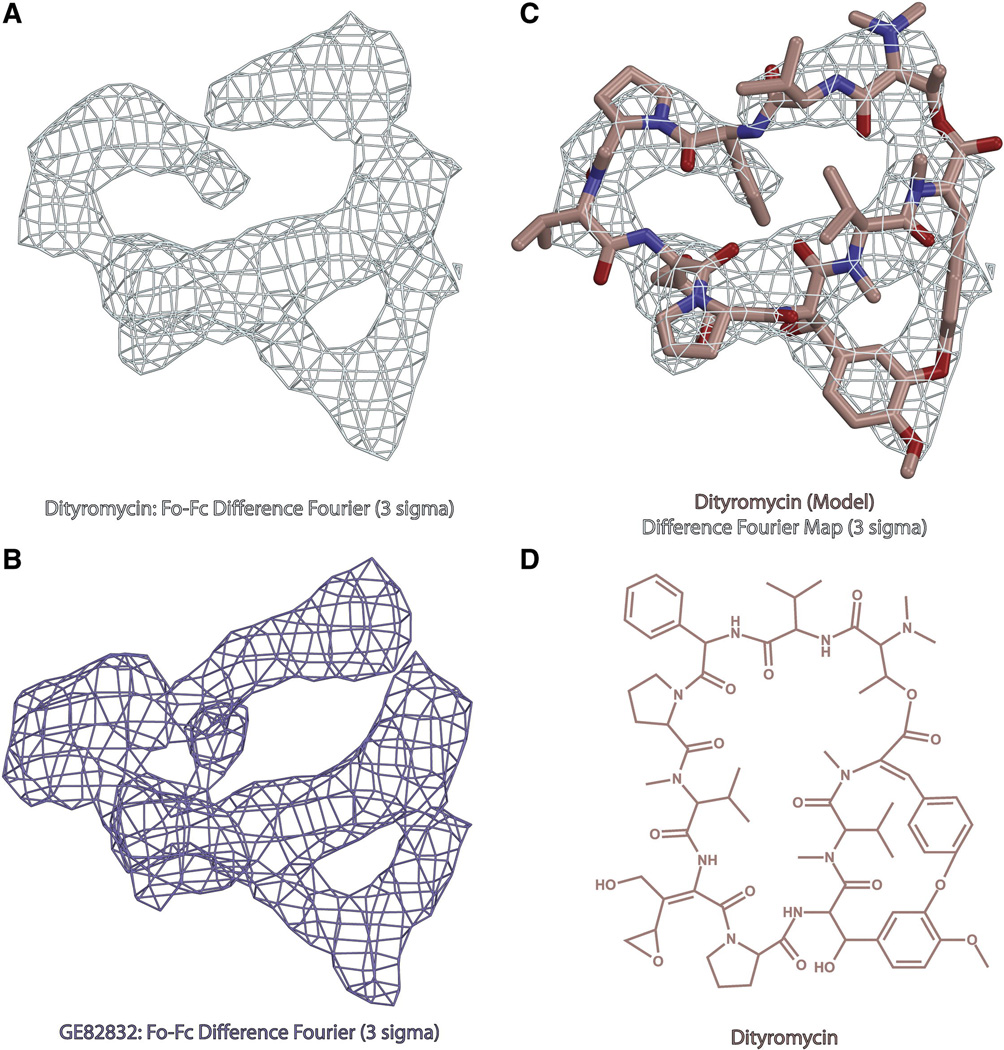
Fig. 1. Structure of dityromycin and comparison with GE82832.
(A) Minimally biased Fo-Fc difference Fourier electron density map contoured at 3σ for dityromycin in complex with the bacterial ribosome from T.thermophilus. (B) Minimally biased Fo-Fc difference Fourier electron density map contoured at 3σ for GE82832 in complex with the bacterial ribosome from T.thermophilus. (C) Model of dityromycin (tan with oxygen colored red and nitrogen blue) docked into the same difference Fourier map shown in panel A. (D) Chemical structure of dityromycin, a secondary metabolite produced by Streptomyces strain AM-2504 (13,14). GE82832 is a secondary metabolite produced by Streptosporangium cinnabarinum (strain GE82832) whose structure is nearly identical to that of dityromycin but for a 2 dalton mass difference likely resulting from an additional point of unsaturation.
Biochemical studies have shown that GE82832/dityromycin blocks the EF-G-catalyzed movement of peptidyl-tRNA and mRNA from the ribosomal A-site to the P-site without preventing the ribosomal binding of the elongation factor, which remains able to stimulate GTP hydrolysis on the ribosome and to dissociate the cleaved γPi, albeit at a somewhat slower rate than in the absence of GE82832/dityromycin (Brandi et al., 2006, 2012).
While GE82832/dityromycin arrests the translocation of tRNA through the ribosome, the compound has almost no effect on the accommodation of tRNA into either the P- or A-site, either in the context of the 30S subunit or the full 70S ribosome (Brandi et al., 2006). Additional experiments showed that pre-incubation of 30S subunits with GE82832/dityromycin inactivates protein synthesis while the same treatment of 50S subunits has little effect, thereby suggesting that GE82832/dityromycin interacts almost exclusively with the 30S subunit. Furthermore, experiments of chemical and hydroxyl radical cleavage protection of 16S rRNA suggest that GE82832/dityromycin modulates the reactivity/accessibility of nucleotides located in the head of the small subunit, raising the possibility that the compound may alter the conformational dynamics of the ribosome (Brandi et al., 2006, 2012). However, despite these data and the suggestion that GE82832/dityromycin might interfere with intersubunit rotation and ratcheting (Brandi et al., 2006), the exact mechanism whereby this antibiotic inhibits translocation remains unknown.
Thus, to better understand the mechanism of action of GE82832/dityromycin, we determined the structure of its complex with the bacterial ribosome from Thermus thermophilus. We find that GE82832/dityromycin interacts with a region of the ribosome that has not been previously identified as an antibiotic binding site (Wilson, 2009), but has been shown to be functionally important in controlling the translocation of tRNA through the ribosome (Cukras et al., 2003; Mcmahont and Landau, 1982; Vila-Sanjurjo et al., 2007). Based on this observation, we suggest that the antibiotic acts through a previously-unknown mechanism and further outline a new region of the ribosome that can be the target of future structure-based antibiotic design efforts.
Results
After demonstrating that GE82832 is active in inhibiting translation in a T. thermophilus system at both 50° and 70° C (Fig. S1), the crystal structures of GE82832 and dityromycin in complex with the bacterial ribosome from T. thermophilus were determined and statistics for the data are shown in Table 1. The structures were solved by molecular replacement using an existing model of the 70S ribosome (Polikanov et al., 2012) in which P-site tRNA and mRNA were bound, but no other ligands were present. Coordinates for the antibiotics were withheld during refinement in PHENIX (Adams et al., 2010), and minimally biased Fobs-Fcalc difference Fourier electron density maps were used to localize the antibiotics with respect to the ribosome. An atomic model of dityromycin was generated from the chemical structure of the antibiotic (Teshima et al., 1988) using the PRODRG server (Schüttelkopf and Van Aalten, 2004) and restraints based on the idealized three-dimensional geometry of dityromycin were used to refine dityromycin into our electron density maps. The final model for dityromycin, along with difference Fourier maps for GE82832 and dityromycin are shown in Figure 1.
Table 1.
Data collection and refinement statistics
| Crystal | 70S-tRNA-Dityromycin | 70S-tRNA-GE82832 | |
|---|---|---|---|
| Diffraction Data | |||
| Space Group | P212121 | P212121 | |
| Unit Cell Dimensions, Å (a × b × c) | 210.08 × 449.83 × 619.65 | 209.68 × 450.64 × 622.54 | |
| Wavelength, Å | 1.100 | 1.100 | |
| Resolution range (outer shell), Å | 49.7–3.00 (3.08–3.00) | 49.7–3.10 (3.18–3.10) | |
| I/σI (outer shell with I/σI=1) | 6.19 (1.00) | 5.19 (0.96) | |
| Resolution at which I/σI=2, Å | 3.27 | 3.45 | |
| Wilson B-factor, Å2 | 66.79 | 70.99 | |
| Completeness (outer shell), % | 98.8 (99.1) | 98.2 (99.2) | |
| Rmerge, % | 19.9 (140.4) | 20.5 (121.9) | |
| CC(1/2) at which I/σI=1, % | 24.0 | 27.5 | |
| No. of crystals used | 1 | 1 | |
| No. of Reflections Used: | Observed | 3,948,062 | 3,118,277 |
| Unique | 1,143,138 | 1,034,964 | |
| Redundancy | 3.45 | 3.01 | |
| Refinement | |||
| Rwork/Rfree, % | 20.3/25.9 | 20.3/25.9 | |
| No. of Non-Hydrogen Atoms | |||
| RNA | 190,374 | 190,375 | |
| Protein | 91,181 | 91,173 | |
| Ions (Mg, Zn, Fe) | 2,003 | 2,003 | |
| Waters | 2,520 | 2,520 | |
| Ramachandran Plot | |||
| Favored regions, % | 93.43 | 92.25 | |
| Allowed regions, % | 5.65 | 6.54 | |
| Outliers, % | 0.92 | 1.21 | |
| Deviations from ideal values (RMSD) | |||
| Bond, Å | 0.010 | 0.009 | |
| Angle, degrees | 1.515 | 1.466 | |
| Chirality | 0.062 | 0.060 | |
| Planrity | 0.007 | 0.007 | |
| Dihedral, degrees | 17.113 | 16.907 | |
| Average B-factor (overall), Å2 | 62.3 | 62.6 | |
Rmerge = Σ |I − <I>| / Σ I, where I is the observed intensity and <I> is the average intensity from multiple measurements.
Rwork = Σ|Fobs − Fcalc| / Σ Fobs. For recalculation of Rfree, 5% of the truncated data set was excluded from the refinement.
GE82832/dityromycin interacts with ribosomal protein S12
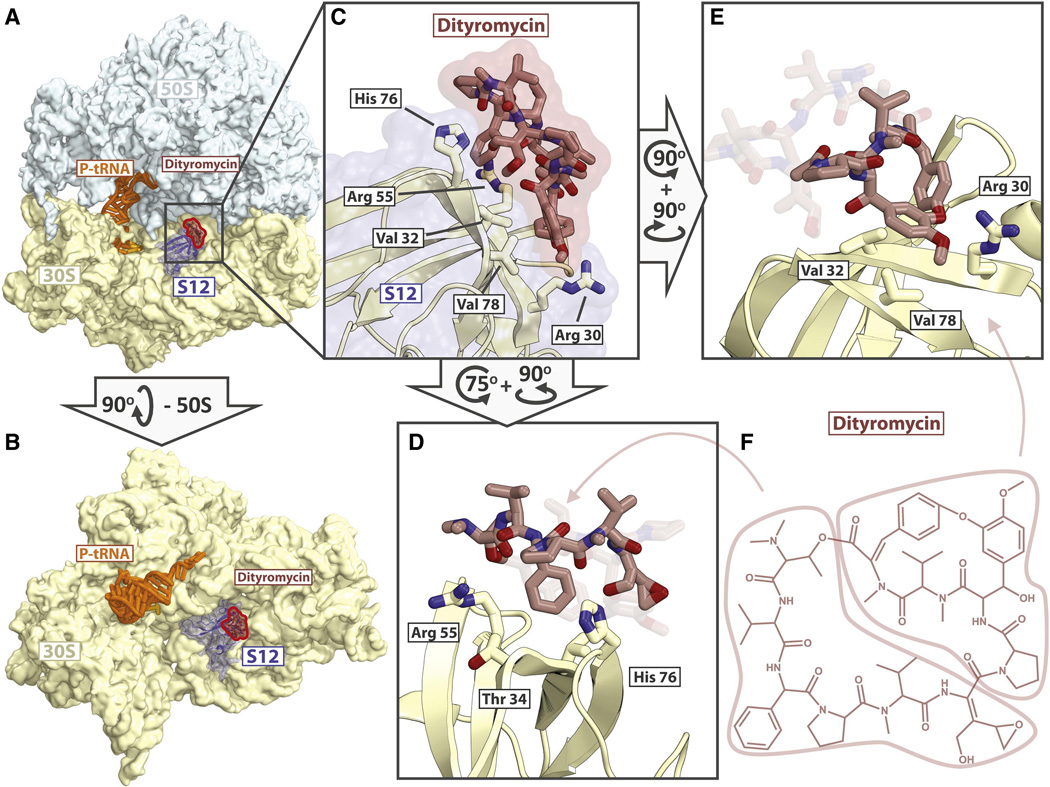
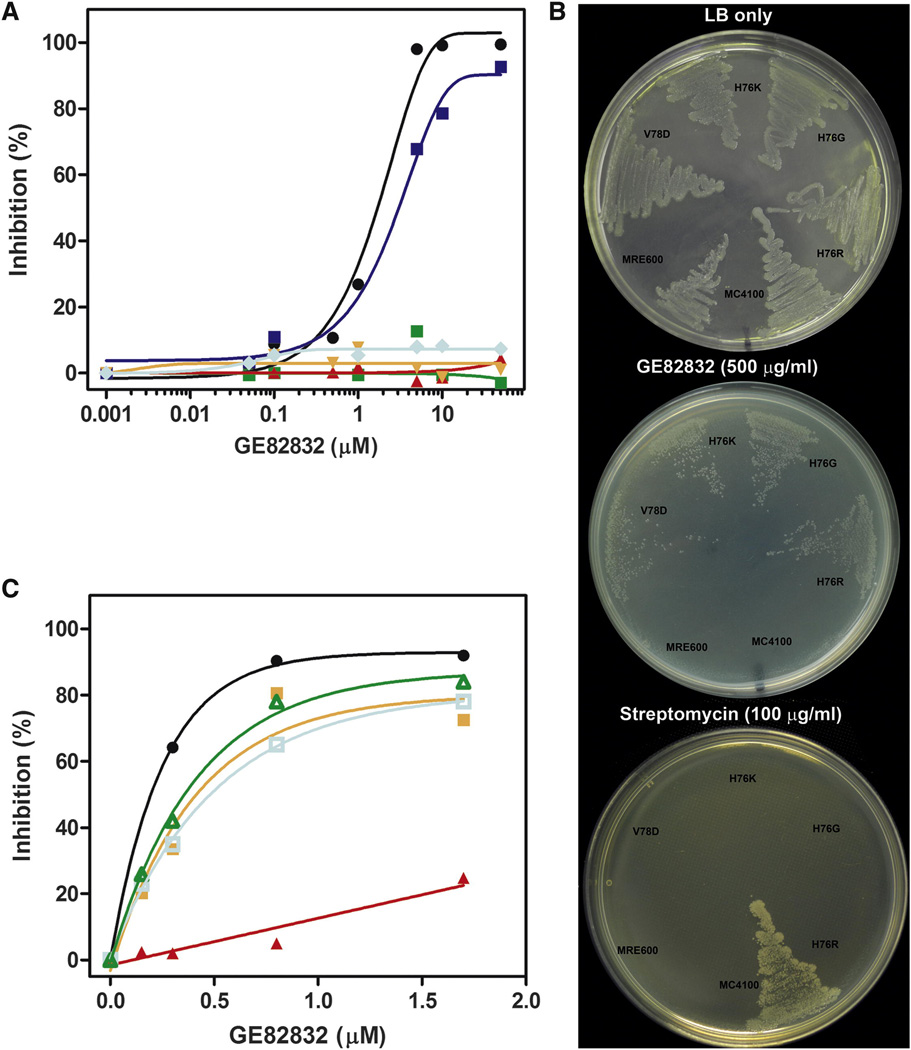
GE82832/dityromycin binds to the shoulder of the 30S subunit and interacts exclusively with ribosomal protein S12 on the small subunit (Fig. 2A, B). No direct interaction between dityromycin and the ribosomal RNA is observed; the antibiotic is 5.5Å away from the phosphate of residue G361 and 7Å away from the phosphate of G362 (16S rRNA, E. coli numbering). On the ribosome, GE82832/dityromycin contacts histidine 76 of S12, probably forming a hydrogen bond between the delta nitrogen of His76 and the hydroxyl group of the epoxy hydroxy dehydroisoleucine residue of GE82832/dityromycin (Fig. 2C, D). The phenyl glycine residue of GE82832/dityromycin also appears to stack against the aromatic surface of His76, further stabilizing this contact. The second major contact is formed by the cyclic di-tyrosine moiety of GE82832/dityromycin, which stacks against the guanidinium group of arginine 30 and is supported by hydrophobic interactions with the side chains of valine 32 and valine 78 of S12 (Fig. 2E). The N,N-dimethyl valine residue of GE82832/dityromycin also comes within 3.5 Å of Arg55 of S12 (Fig 2D) The interaction of the antibiotic with His76 and Val78 is confirmed by the finding that E. coli ribosomes bearing mutations of residues His76 (R,G or K) or Val78D of protein S12 are resistant to inhibition of in vitro mRNA translation by GE82832 (Fig. 3 A). Furthermore, the same mutations confer in vivo resistance to GE82832, whereas both wild type E. coli MRE600, as well as a standard laboratory strain, MC4100, which carries an S12 substitution (K42R) conferring streptomycin resistance (Casadaban, 1976) (see Experimental Procedures) are sensitive to the antibiotic (Fig. 3 B, middle plate). However, the same S12 GE82832-resistant mutants display a streptomycin-sensitive phenotype, unlike E. coli MC4100 (Fig. 3 B, lower plate).
Fig. 2. Binding site of GE82832/dityromycin with the ribosome and their interactions with S12.
(A) Overview of the 70S ribosome (50S light blue, 30S yellow) in complex with dityromycin (red highlight) with S12 (blue) shown. (B) Another view of the binding site of GE82832/dityromycin in which the large subunit has been removed for clarity. (C) Close up of dityromycin (tan, red highlight) bound to S12 (white, blue highlight). (D) Another view of dityromycin (tan) interacting with S12 (white), highlighting the interaction between His76 of S12 and the phenyl glycine and epoxy hydroxy dehydroisoleucine residues of dityromycin. Arg55 packs against the N,N-dimethyl valine of dityromycin, and Thr34 comes within 4 Å of the phenyl glycine residue. (E) The di-tyrosine moiety of dityromycin stacks with Arg30 and rests on a hydrophobic surface formed by Val32 and Val78. (F) Chemical structure of dityromycin, indicating the regions of the antibiotic highlighted in figures D and E.
Fig. 3. Involvement of ribosomal protein S12 in GE82832/dityromycin binding.
(A) Effect of S12 mutations on the in vitro ribosome susceptibility to GE82832/ dityromycin. 027IF2Cp(A) mRNA translation was tested in the presence of the concentrations of GE82832 indicated in abscissa in an E. coli MRE600 translational system reconstituted with post-ribosomal supernatant and purified wt ribosomes (●) or ribosomes lacking ribosomal protein S13 ( ) or carrying the following amino acid substitutions in S12: H76R (
) or carrying the following amino acid substitutions in S12: H76R ( ); H76G (
); H76G ( ); H76K (
); H76K ( ); V78D (
); V78D ( ). The % inhibition reported in the ordinate was calculated by comparison with mRNA translation in the absence of antibiotic. (B) Effect of S12 mutations on the in vivo susceptibility to GE82832 and streptomycin; Wild type E. coli MRE600, E. coli MC4100 (carrying a K42R substitution in S12 conferring streptomycin resistance) and E. coli MC41 carrying H76R, H76 G, H76K and Val78D mutations in S12 were plated as indicated in each case on LB-agar plates containing no antibiotics (upper plate), 500 µg/ml GE82832 (middle plate) or 100 µg/ml streptomycin (lower plate). (C) Binding of GE82832 to isolated S12. 7 µM of GE82832 were incubated for 10 min at 20 °C in 100 µl of buffer (10mM Tris-HCl pH7.1; 10 mM MgAcetate; 60mM NH4Cl; 1mM DTT) without further additions (
). The % inhibition reported in the ordinate was calculated by comparison with mRNA translation in the absence of antibiotic. (B) Effect of S12 mutations on the in vivo susceptibility to GE82832 and streptomycin; Wild type E. coli MRE600, E. coli MC4100 (carrying a K42R substitution in S12 conferring streptomycin resistance) and E. coli MC41 carrying H76R, H76 G, H76K and Val78D mutations in S12 were plated as indicated in each case on LB-agar plates containing no antibiotics (upper plate), 500 µg/ml GE82832 (middle plate) or 100 µg/ml streptomycin (lower plate). (C) Binding of GE82832 to isolated S12. 7 µM of GE82832 were incubated for 10 min at 20 °C in 100 µl of buffer (10mM Tris-HCl pH7.1; 10 mM MgAcetate; 60mM NH4Cl; 1mM DTT) without further additions ( ) or containing 20 µM of purified ribosomal protein S12 (
) or containing 20 µM of purified ribosomal protein S12 ( ) or 20 µM of initiation factor IF3 (
) or 20 µM of initiation factor IF3 ( ) or 20 µM of bovine serum albumin (
) or 20 µM of bovine serum albumin ( ). The samples were the loaded on a Amicon spin filter (MWCO 3 KDa). After 10’ of centrifugation at 12K rpm, the eluate was collected and increasing aliquots tested for their capacity to inhibit 027IF2Cp(A) mRNA translation in a standard E. coli-based system. A sample of non-centrifuged antibiotic was also tested as a control (●).
). The samples were the loaded on a Amicon spin filter (MWCO 3 KDa). After 10’ of centrifugation at 12K rpm, the eluate was collected and increasing aliquots tested for their capacity to inhibit 027IF2Cp(A) mRNA translation in a standard E. coli-based system. A sample of non-centrifuged antibiotic was also tested as a control (●).
It has long been known that decoding fidelity is influenced by ribosomal protein S12 (Gorini, 1971; Strigini and Brickman,1973); thus, the evidence that GE82832/dityromycin binds to this protein prompted us to investigate a possible effect of this antibiotic on the accuracy of mRNA translation. To do so, the level of translation measured from the incorporation of a radioactive precursor in the acid-insoluble product was compared to the level of correctly synthesized protein, as immunologically quantified by ELISA (Fabbretti et al., 2012). By contrast to what occurs in the presence of streptomycin, both tests show the same extent of inhibition in the presence of GE82832/dityromycin, indicating that this antibiotic does not induce codon misreading (Fig. S2).
The interactions observed between GE82832/dityromycin and S12 suggest that the thermodynamic stability of the protein-drug complex could be quite high. In light of this, the possibility that the antibiotic might establish an interaction with the isolated ribosomal protein was also investigated. The results obtained in this experiment clearly show that, unlike two control proteins (serum albumin and initiation factor IF3), GE82832/dityromycin co-purifies with protein S12, indicating that this isolated ribosomal protein can indeed establish a stable interaction with the antibiotic (Fig. 3C).
Previous chemical (Brandi et al., 2006) and hydroxyl radical (Brandi et al., 2012) probing experiments had shown that GE82832/dityromycin affects the accessibility of 16S rRNA bases located in the head of the 30S subunit, near protein S13, but no effect was detected near S12. Whereas the latter result would be fully compatible with the crystallographic data showing the lack of contacts between the antibiotic and 16S rRNA, whose closest phosphate group is >5 Å away (see above), the altered RNA accessibility near S13 deserved renewed attention, also in light of the role played by this protein and by the 30S head rotation in translocation (Cukras et al., 2003; Pulk and Cate, 2013; Tourigny et al., 2013; Zhou et al., 2013). Therefore, a possible role of protein S13 in the mechanism of translation inhibition by this antibiotic was investigated using ribosomes from cells bearing an rpsM (the gene encoding S13) null mutation (Cukras and Green, 2005). However, the in vitro mRNA translation test carried out with these ribosomes demonstrates that the lack of S13 does not alter their sensitivity to GE82832/dityromycin inhibition (Fig. 3A blue tracing).
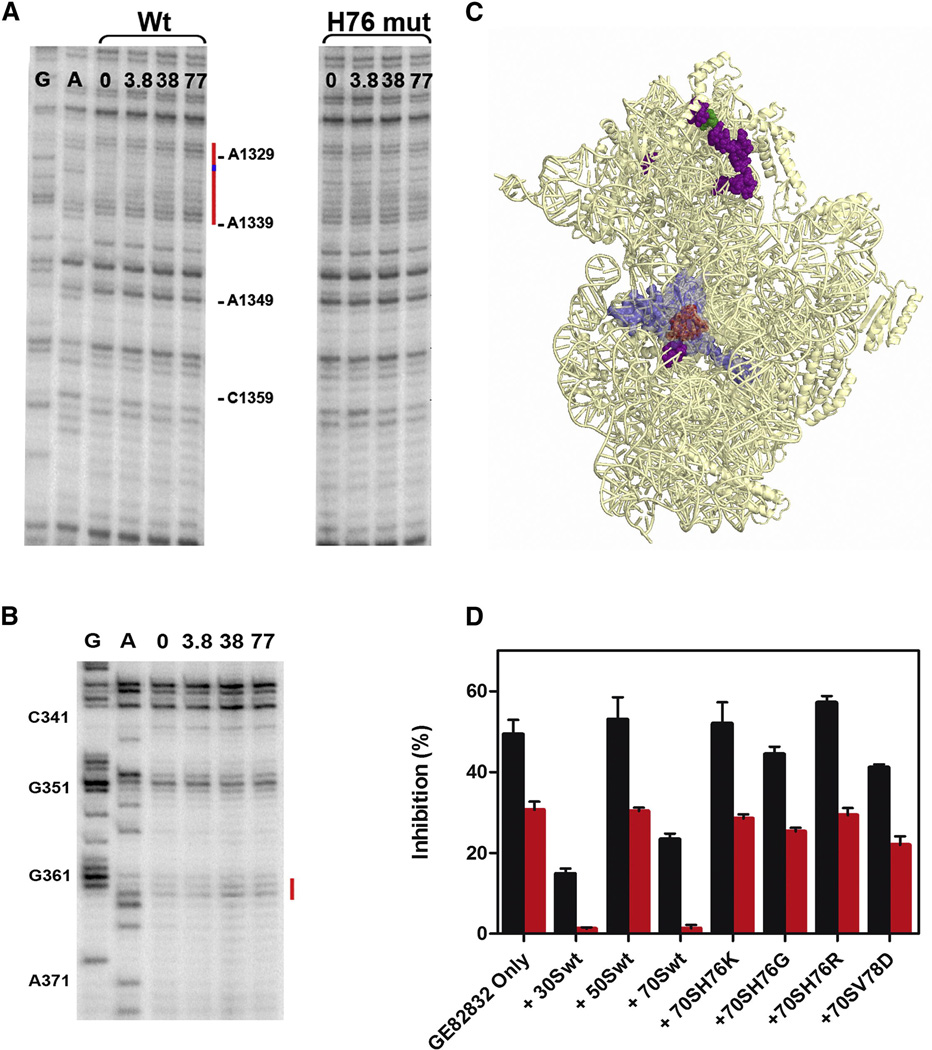
Finally, in situ probing of the rRNA was analyzed (Fig. 4A, B) by comparing the effects of GE82832/dityromycin on the accessibility of 16S rRNA to hydroxyl radical cleavage in wt 30S subunits with its accessibility in mutated 30S subunits containing an H76 substitution that made them GE82832/dityromycin-resistant. As seen in Figure 4 A, the altered accessibility to hydroxyl radical cleavage of 16S rRNA bases in the 30S subunit head near S13 was fully confirmed in wt 30S subunits, with a clear enhancement of exposure for bases A1329-2330, A1332-1339, 1344 and C1359 and a slight protection of G1331; however, no comparable effects could be detected in the 30S subunits bearing the S12 H76 mutation. Remarkably, all the bases whose accessibilities are affected by the antibiotic are very far from S12 (Fig.4 B). Concerning the S12- proximal region of the 30S subunit, it can be seen that the antibiotic has no effect on the accessibility of bases in this region, aside from a slight increased exposure of bases 362–363 (Fig. 4 B).
Fig. 4. Long-distance effects of GE82832 on the in situ 16S rRNA cleavage by hydroxyl radicals.
Primer extension analysis of the in situ cleavage pattern of 16S rRNA (A) in the 1340 region (near S13) in wt 30S (left lanes) and S12 H76K mutant 30S ribosomal subunits (right lanes) and (B) in the 360 region (near S12) in wt 30S subunits in the absence (0) and in the presence of 3.8, 38 and 77 µM GE82832, as indicated above each lane. The lanes marked G and A are sequencing gels. (C) Image of the 30S ribosomal subunit in which the bases whose accessibility to cleavage is increased (purple) or decreased (green) by GE82832 are highlighted; the position of protein S12 (light blue) and of GE82832/dityromycin binding site (light red) are also indicated. (D) Lack of GE82832 binding by ribosomes carrying His76 and Val78 substitutions. The extent (%) of inhibition of mRNA translation by eluates of Amicon spin filter (MWCO 50 KDa) on which 3 µM (red bars) or 9 µM (black bars) of GE82832 were loaded alone or together with 30 µM of wt 30S, 50S and 70S particles or 70S ribosomes carrying H76K, H76G, H76R and V78D substitutions in S12, as indicated under each histogram bar. The other experimental details are described in the legend of Fig.3.
Taken together, these findings could be explained either by the existence of multiple antibiotic binding sites on the 30S subunit and/or by long-range conformational changes of the subunit induced by GE82832/dityromycin binding. However, the crystallographic data are consistent with a single ribosomal binding site for the antibiotic; furthermore, although our “pull down” experiments do not allow us to rule out the existence of additional, low affinity antibiotic binding site(s) (Fig. 4D), the nature of most of the effects (increased exposure) observed near S13 in wt 30S and the fact that no alterations of the cleavage patterns are seen with the 30S subunit bearing the H76 mutation in S12 give a clear indication that GE82832 causes a long range conformational change in the head of the subunit. The fact that no conformational rearrangements resulting from GE82832/dityromycin binding were detectable in the crystal structure can be explained by the fact that the crystal packing has probably stabilized a single conformation of the subunit.
Discussion
Protein S12 plays a critical role in bacterial translation (Cukras et al., 2003; Gregory et al., 2009; Mcmahont and Landau, 1982; Sharma et al., 2007; Vila-Sanjurjo et al., 2007; Yates, 1979; Zengel et al., 1977). Protein S12 is positioned on the shoulder of the 30S subunit, where it reaches into the decoding center and acts as a control element in tRNA selection (Yates, 1979) and the translocation of tRNA-mRNA through the ribosome (Cukras et al., 2003). S12 has also been shown to influence the inhibition of translation by several antibiotics, including streptomycin (Vila-Sanjurjo et al., 2007) and paromomycin (Sharma et al., 2007). The structural and biochemical data presented here show that S12 is the target of GE82832/dityromycin inhibition through an interaction which is distinct from that of any other ribosomal inhibitor and also pivotal in a GE82832/dityromycin-mediated conformational change of the head of the 30S subunit that might be related to the mechanism of translation inhibition.
The long distance S12-mediated effects of GE82832 on the accessibility of 16S rRNA bases of the 30S subunit head underlie the well-established dynamics of this region of the subunit (Pulk and Cate, 2013; Tourigny et al., 2013; Wang et al., 2012; Zhou et al., 2013), as well as the ability of protein S12 to act as a conformational relay for the subunit (Gregory et al., 2009; Vila-Sanjurjo et al., 2007; Zengel et al., 1977) and the functional interrelationship between S12 and S13 (Cukras et al., 2003). However, it is not possible to conclude whether these effects, which do not occur in the GE82832-resistant 30S subunits, have any causal relationship with the mechanism of translocation inhibition.
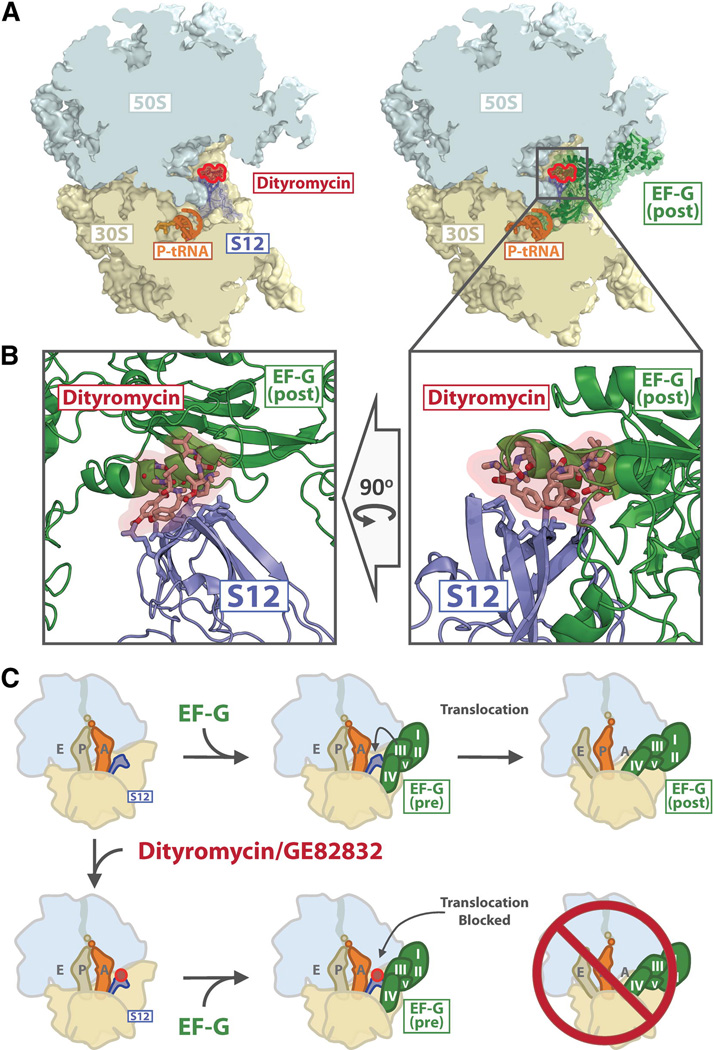
EF-G-catalyzed translocation has been shown to involve a number of conformational rearrangements of EF-G, the ribosome, and ribosome-bound tRNA-mRNA (Ermolenko and Noller, 2011; Frank et al., 2007; Munro et al., 2010; Peske et al., 2000, 2004; Ratje et al., 2010; Savelsbergh et al., 2003). In addition to biochemical evidence indicating that interactions between S12 and EF-G control translocation (Cukras et al., 2003), crystal structures of EF-G bound to the ribosome in post-translocational and ratcheted pre-translocational conformations show that EF-G interacts extensively with S12 (Feng et al., 2013; Gao et al., 2009b; Pulk and Cate, 2013; Tourigny et al., 2013; Zhou et al., 2013). Critically, domain 3 of EF-G binds to S12 in the region of His 76 (E. coli), and comparison of the ribosome bound to EF-G versus GE82832/dityromycin shows that both molecules interact with a similar region of S12 (Fig. 5). Thus, our structure indicates that the binding of GE82832/dityromycin sterically occludes EF-G from entering a post-translocational conformation on the ribosome by blocking the interaction between domain 3 of EF-G and protein S12 on the small ribosomal subunit.
Fig. 5. Dityromycin blocks the transition from pre- to post-translocational conformations of EF-G.
(A) The left panel shows a cross section of the ribosome (30S yellow, 50S light blue) with dityromycin (red) and S12 (blue) highlighted. The right panel shows the same view, but with EF-G (green) bound in the post translocational conformation (Gao et al., 2009a), PDB accession code 2WRI. (B) Close-up view of the clash between dityromycin and EF-G bound in the post translocational conformation. (C) Schematic of translocation and its inhibition by dityromycin/GE82832. The 50S (light blue) and 30S (yellow) subunits, deacylated tRNA (light brown), peptidyl tRNA (light grey) and S12 (blue) are shown, along with EF-G (green with domains I–V indicated) and dityromycin (red).
While our crystal structure indicates that GE82832/dityromycin blocks the binding of EF-G in the post-translocational conformation, EF-G can still catalyze the hydrolysis of GTP on the ribosome, though the kinetics of phosphate release are altered (Brandi et al., 2012). Because GTP hydrolysis and phosphate release occur in steps separate from the engagement of domain 4 of EF-G with the A-site (Munro et al., 2010), the possibility that GTP hydrolysis and translocation are decoupled by GE82832/dityromycin is not unreasonable; viomycin and spectinomycin also inhibit translocation, but do not block GTP hydrolysis (Pan et al., 2007), while mutations in EF-G have also been shown to decouple translocation and GTP hydrolysis (Savelsbergh et al., 2000).
Both biochemical and structural experiments have established that S12 plays a critical role in translocation (Cukras et al., 2003; Gao et al., 2009a; Pulk and Cate, 2013; Tourigny et al., 2013; Zhou et al., 2013). Entering the post-translocational state in the presence of EF-G requires that domain 3 of EF-G engages S12. S12 also acts as a control element in translocation; removal of S12 along with proteins S11 or S13 stimulates factor-independent translocation, while removal of S12 and S5, S7, S8 or S14 blocks factor-catalyzed translocation (Cukras et al., 2003). Interestingly, removal of protein S13 does not reduce the sensitivity of E. coli ribosomes to GE82832/dityromycin (Fig. 3A). Overall, the results indicate that GE82832/dityromycin may inhibit translation by preventing the complex between EF-G and the ribosome from adopting a conformation that is productive for translocation. This premise is consistent with the finding that even an excess of EF-G does not relieve the inhibition (Fig. S3), a result which is consistent with GE82832/dityromycin affecting the position of EF-G on the ribosome and not its binding.
While the structure of GE82832/dityromycin in complex with the bacterial ribosome explains its activity as a translocation inhibitor, our structure would also be consistent with GE82832/dityromycin affecting the ability of EF-Tu to deliver aminoacyl-tRNA to the ribosomal A-site. The mutation of several residues of protein S12 that are distant from the decoding center have been shown to increase miscoding errors; two of these, Thr57 and Val78 (E. coli) (Agarwal et al., 2011), form part of the binding pocket for GE82832/dityromycin. In addition, His76 (E. coli), the same residue that we show to be critical for GE82832/dityromycin binding (Fig. 3 and 4), is involved in signaling EF-Tu when codon recognition has taken place (Gregory et al., 2009). However, only at high concentrations (~10 µM) does GE82832/dityromycin inhibit (~50%) the delivery of tRNA to the A-site in the absence of EF-Tu, whereas at the same concentration it has virtually no effect when EF-Tu is present (Brandi et al., 2006). While this is probably due to the fact that aa-tRNA and EF-Tu outcompete the antibiotic from its binding site, it should be noted that overall protein synthesis and translocation are inhibited at the same rate by GE82832/dityromycin (Brandi et al., 2006).
Unlike EF-G, the affinity of the ternary complex of EF-Tu with tRNA and GTP for the ribosome depends heavily on the interaction between the anticodon of the EF-TU-bound tRNA and the codon of the mRNA: the correct pairing of tRNA and mRNA in the decoding center can increase the affinity of the ternary complex for the ribosome by as much as 1000-fold as compared to a single mismatch (Gromadski and Rodnina, 2004; Pape et al., 1999; Thompson et al., 1986) and the equilibrium dissociation constant of the correct EF-Tu/tRNA/GTP ternary complex for the ribosome is on the order of 1 nM (Harrington et al., 1993; Louie and Jurnak, 1985). In contrast, the binding of domain IV of EF-G in the decoding center does not appear to be as critical to the overall affinity of the factor for the ribosome, as evidenced by the fact that a conformationally-restrictied EF-G which is unable to translocate tRNA (and therefore likely does not bind with domain IV in the A-site) has only a slightly reduced affinity for the ribosome and remains capable of catalyzing GTP hydrolysis on the ribosome (Peske et al., 2000). Therefore, we speculate that the affinity of the EF-Tu/tRNA/GTP ternary complex for the decoding center is sufficient to outcompete GE82832/dityromycin bound to the ribosome. Since the MIC for GE82832/dityromycin is only in the low µM, it is possible that an antibiotic which bound to the same region of the ribosome with increased affinity would be capable of inhibiting the delivery of aminoacyl-tRNA by EF-Tu.
The binding site of GE82832/dityromycin also overlaps with that of ribosome recycling factor (RRF) (Gao et al., 2007). Because RRF and EF-G work together in recycling, it is unclear whether the effects of GE82832/dityromycin on RRF could be disentangled from its effects on EF-G alone, but a superposition of RRF bound to both the E. coli and T. thermophilus ribosomes shows that RRF and GE82832/dityromycin share a contact point with S12 (Borovinskaya et al., 2007; Weixlbaumer et al., 2007; Yokoyama et al., 2012). Superposition with the ratcheted E. coli ribosome bound to RRF shows that GE82832/dityromycin would sterically clash with domain 2 of RRF, while only a slight clash would exist between GE82832/dityromycin and RRF bound to a classical-state ribosome (Fig. S4).
While questions remain regarding the impact of GE82832/dityromycin on RRF, EF-Tu and the accommodation of tRNA, our structure provides a clear mechanism for how this antibiotic is able to inhibit the EF-G-catalyzed translocation of tRNA. When bound in the post-translocational conformation, domain 3 of EF-G makes extensive contacts with S12 in the region of His 76 (E. coli) (Gao et al., 2009b; Pulk and Cate, 2013; Tourigny et al., 2013; Zhou et al., 2013). A comparison of S12 bound to EF-G versus GE82832/dityromycin shows that both molecules engage a similar region of S12 and that both cannot simultaneously bind S12 (Fig. 5). Therefore, the binding of GE82832/dityromycin sterically occludes EF-G from entering a post-translocational conformation on the ribosome.
Experimental Procedures
GE82832 was originally identified, isolated and purified from Streptosporangium cinnabarinum strain GE82832 at Biosearch Italia spa (Gerenzano, Italy) within the European Commission project ‘‘Ribosome Inhibitors’’ (contract QLRT-2001-00892EC). Dityromycin, purified from Streptomyces sp. strain AM-2504 was a kind gift of Prof. O. Omura (Tokyo, Japan). 70S ribosomes from T. thermophilus were purified and crystallized according to previously published protocols (Bulkley et al., 2010). Prior to crystallization, dityromycin or GE82832 was added at a concentration of 100 µM and incubated at 37°C for 5 min. 15 µM mRNA with sequence GGCAAGGAGGUAAAAAUGUUC was then added and further incubated for 5 min at 37° C after which fMet-tRNAfMet was added at a concentration of 8 µM, and ribosomes were further incubated for 5 min at 37°C. Ribosomes were then diluted to a final concentration of 5 µM in buffer composed of 5 mM Hepes pH 7.5, 50 mM KCl, 10 mM NH4Cl and 10 mM MgAc2 and then equilibrated via sitting drop vapor diffusion against a reservoir solution made up of 2.9% (w/v) PEG 20K, 9% (v/v) MPD, 175 mM L-arginine and 100 mM Tris-HCl pH 7.6. Immediately prior to equilibration, 3 to 4 µL of reservoir solution were added to 3 µL of ribosome-containing solution.
Crystals appeared after approximately 4 days and were harvested after 8 days. Following stabilization by gradually increasing the concentration of 2-methyl-2,4-pentanediol to 40% (v/v), crystals were equilibrated for 12 hours and flash frozen in a nitrogen cryostream at 80 K. Diffraction data were collected at the National Synchrotron Light Source on beamline X-25 using a Pilatus 6M detector. Data reduction was performed with the program XDS (Kabsch, 2010) and an initial solution was generated by molecular replacement with PHASER (McCoy et al., 2007) using the T. thermophilus 70S ribosome from which all ligands were removed as a search model (Polikanov et al., 2012). The solution was then refined with the PHENIX package (Adams et al., 2010).
The model for dityromycin was generated using the PRODGR server (Schüttelkopf and Van Aalten, 2004) based on the chemical structure of dityromycin (Teshima et al., 1988). Restraints from PRODRG were originally used, but modified restraints with idealized geometry were generated and used for the final refinement. The antibiotic model was placed into Fo-Fc difference Fourier maps using ribosome models that had been refined in the absence of antibiotic coordinates. Final coordinates for the antibiotic ribosome complexes are deposited in the PDB and the accession codes are listed below.
In vitro translation and miscoding tests were carried out in E. coli or T. thermophilus systems programmed with 027IF2Cp(A) mRNA as described (Brandi et al., 2007; Fabbretti et al., 2012). In situ probing the 16S rRNA by hydroxyl radical cleavage was carried out as described (Fabbretti et al., 2007).
E coli strains carrying mutations in S12 were constructed by recombineering, as previously described (Agarwal et al., 2011). Briefly, chromosomal DNA from a strain carrying a chloramphenicol acetyl transferase (cat) cassette integrated upstream of the rpsL gene was used as a template in a two-step, extension overlap PCR, with oligonucleotides randomized at codon positions 76 or 78. The amplified fragments carrying the mutated rpsL gene and cat cassette were electroporated into MC323 (Agarwal et al., 2011), selecting for chloramphenicol resistance. The rpsL mutations were identified by sequencing and the desired alleles were then transferred into strain MC41 (F− Δ(lac-pro) thi−). The cat cassette was removed by transient expression of plasmid-borne Flp recombinase (Datsenko and Wanner, 2000).
Supplementary Material
Acknowledgments
LB, AF and COG are grateful to all their colleagues and friends in Europe and the USA who have made their work possible, providing the necessary chemicals and biochemicals, thereby compensating for the lack of financial support from Italian public and private agencies to this type of research. DB and YSP were supported by HHMI and NIH grant GM022778 awarded to TAS. DB and YSP would like to thank Jinzhong Lin and Matthieu Gagnon for helpful discussions, the staff of the Yale Center for Structural Biology, particularly Michael Strickler and David Keller, and the beamlines at Brookhaven National Laboratory (X-25 and X29) and Argonne National Laboratory (NECAT, 24-ID). Conflict of interest statement: Thomas A. Steitz is a scientific advisor for Rib-X Pharmaceuticals, a product-driven small molecule drug discovery and development company focused on the structure-based design of unique classes of antibiotics.
Footnotes
Accession Numbers
Coordinates for the complex of dityromycin with the 70S ribosome have been deposited in the PDB under accession codes 4NVU, 4NVV, 4NVW, 4NVX (dityromycin) and 4NVY, 4NVZ, 4NW0, 4NW1 (GE82832).
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal D, Gregory ST, O’Connor M. Error-prone and error-restrictive mutations affecting ribosomal protein S12. Journal of Molecular Biology. 2011;410:1–9. doi: 10.1016/j.jmb.2011.04.068. [DOI] [PubMed] [Google Scholar]
- Asatryan LS, Spirin aS. Non-enzymatic translocation in ribosomes from streptomycin-resistant mutants of Escherichia coli. Molecular & General Genetics : MGG. 1975;138:315–321. doi: 10.1007/BF00264801. [DOI] [PubMed] [Google Scholar]
- Borovinskaya Ma, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JHD. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nature Structural & Molecular Biology. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- Brandi L, Fabbretti A, Di Stefano M, Lazzarini A, Abbondi M, Gualerzi CO. Characterization of GE82832, a peptide inhibitor of translocation interacting with bacterial 30S ribosomal subunits. RNA (New York, N.Y.) 2006;12:1262–1270. doi: 10.1261/rna.61206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi L, Fabbretti A, Milon P, Carotti M, Pon CL, Gualerzi CO. Methods for identifying compounds that specifically target translation. Methods in Enzymology. 2007;431:229–267. doi: 10.1016/S0076-6879(07)31012-4. [DOI] [PubMed] [Google Scholar]
- Brandi L, Maffioli S, Donadio S, Quaglia F, Sette M, Milón P, Gualerzi CO, Fabbretti A. Structural and functional characterization of the bacterial translocation inhibitor GE82832. FEBS Letters. 2012;586:3373–3378. doi: 10.1016/j.febslet.2012.07.040. [DOI] [PubMed] [Google Scholar]
- Bulkley D, Innis CA, Blaha G, Steitz TA. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. Journal of Molecular Biology. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Cukras AR, Green R. Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. Journal of Molecular Biology. 2005;349:47–59. doi: 10.1016/j.jmb.2005.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R. Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Molecular Cell. 2003;12:321–328. doi: 10.1016/s1097-2765(03)00275-2. [DOI] [PubMed] [Google Scholar]
- Datsenko Ka, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nature Structural & Molecular Biology. 2011;18:457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbretti A, Pon CL, Hennelly SP, Hill WE, Lodmell JS, Gualerzi CO. The real-time path of translation factor IF3 onto and off the ribosome. Molecular Cell. 2007;25:285–296. doi: 10.1016/j.molcel.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Fabbretti A, Brandi L, Petrelli D, Pon CL, Castañedo NR, Medina R, Gualerzi CO. The antibiotic Furvina® targets the P-site of 30S ribosomal subunits and inhibits translation initiation displaying start codon bias. Nucleic Acids Research. 2012;40:10366–10374. doi: 10.1093/nar/gks822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Chen Y, Gao Y-G. Crystal Structure of 70S Ribosome with Both Cognate tRNAs in the E and P Sites Representing an Authentic Elongation Complex. PloS One. 2013;8:e58829. doi: 10.1371/journal.pone.0058829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Zavialov AV, Ehrenberg M, Frank J. Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits. Journal of Molecular Biology. 2007;374:1345–1358. doi: 10.1016/j.jmb.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The Structure of the Ribosome with Elongation Factor G trapped in the Posttranslocational state. Science. 2009a;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y-G, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science (New York, N.Y.) 2009b;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini L. Ribosomal Discrimination of tRNAs. Nature New Biology. 1971;234:261–264. doi: 10.1038/newbio234261a0. [DOI] [PubMed] [Google Scholar]
- Gregory ST, Carr JF, Dahlberg AE. A signal relay between ribosomal protein S12 and elongation factor EF-Tu during decoding of mRNA. RNA (New York, N.Y.) 2009;15:208–214. doi: 10.1261/rna.1355709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Molecular Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- Harrington KM, Nazarenko Ia, Dix DB, Thompson RC, Uhlenbeck OC. In vitro analysis of translational rate and accuracy with an unmodified tRNA. Biochemistry. 1993;32:7617–7622. doi: 10.1021/bi00081a003. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallographica. Section D, Biological Crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry. 2002;41:12806–12812. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- Louie a, Jurnak F. Kinetic studies of Escherichia coli elongation factor Tu-guanosine 5’-triphosphate-aminoacyl-tRNA complexes. Biochemistry. 1985;24:6433–6439. doi: 10.1021/bi00344a019. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcmahont G, Landau JV. Effect of S12 ribosomal mutations on peptide chain elongation in Escherichia coli : a hydrostatic pressure Effect of S12 Ribosomal Mutations on Peptide Chain Elongation in Escherichia coli : a Hydrostatic Pressure Study. 1982 doi: 10.1128/jb.151.1.516-520.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nature Structural & Molecular Biology. 2010;17:1470–1477. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Omura S, Iwai Y, Hirano A, Awaya J, Suzuki Y, Matsumoto K. A New Antibiotic, AM-2504. Agricultural and Biological Chemistry. 1977;41:1827–1828. [Google Scholar]
- Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Molecular Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. The EMBO Journal. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Matassova NB, Savelsbergh a, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Molecular Cell. 2000;6:501–505. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. Journal of Molecular Biology. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- Polikanov YS, Blaha GM, Steitz Ta. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science (New York, N.Y.) 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulk a, Cate JHD. Control of Ribosomal Subunit Rotation by Elongation Factor G. Science. 2013;340:1235970–1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brünner M, Hildebrand PW, Starosta AL, Dönhöfer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP be elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Savelsbergh a, Matassova NB, Rodnina MV, Wintermeyer W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. Journal of Molecular Biology. 2000;300:951–961. doi: 10.1006/jmbi.2000.3886. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Molecular Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Schüttelkopf AW, Van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica. Section D, Biological Crystallography. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Sharma D, Cukras AR, Rogers EJ, Southworth DR, Green R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. Journal of Molecular Biology. 2007;374:1065–1076. doi: 10.1016/j.jmb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth DR, Brunelle JL, Green R. EFG-independent Translocation of the mRNA:tRNA Complex is Promoted by Modification of the Ribosome with Thiol-specific Reagents. Journal of Molecular Biology. 2002;324:611–623. doi: 10.1016/s0022-2836(02)01196-8. [DOI] [PubMed] [Google Scholar]
- Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz Ta. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nature Structural & Molecular Biology. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, Van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- Strigini P, Brickman E. Analysis of specific misreading in Escherichia coli. Journal of Molecular Biology. 1973;75:659–672. doi: 10.1016/0022-2836(73)90299-4. [DOI] [PubMed] [Google Scholar]
- Teshima T, Nishikawa M, Kuota I, Shiba T, Iwai Y, Omura S. The structure of an antibiotic, dityromycin. Tetrahedron Letters. 1988;29:1963–1966. [Google Scholar]
- Thompson RC, Dix DB, Karim AM. The Reaction of Ribosomes with Elongation Factor Tu-GTP Complexes. The Journal of Biological Chemistry. 1986;261:4868–4874. [PubMed] [Google Scholar]
- Tourigny DS, Fernandez IS, Kelley aC, Ramakrishnan V. Elongation Factor G Bound to the Ribosome in an Intermediate State of Translocation. Science. 2013;340:1235490–1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Vila-Sanjurjo A, Lu Y, Aragonez JL, Starkweather RE, Sasikumar M, O’Connor M. Modulation of 16S rRNA function by ribosomal protein S12. Biochimica Et Biophysica Acta. 2007;1769:462–471. doi: 10.1016/j.bbaexp.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Walter JD, Hunter M, Cobb M, Traeger G, Spiegel PC. Thiostrepton inhibits stable 70S ribosome binding and ribosome-dependent GTPase activation of elongation factor G and elongation factor 4. Nucleic Acids Research. 2012;40:360–370. doi: 10.1093/nar/gkr623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Pulk A, Wasserman MR, Feldman MB, Altman RB, Cate JHD, Blanchard SC. Allosteric control of the ribosome by small-molecule antibiotics. Nature Structural & Molecular Biology. 2012;19:957–963. doi: 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Petry S, Dunham CM, Selmer M, Kelley AC, Ramakrishnan V. Crystal structure of the ribosome recycling factor bound to the ribosome. Nature Structural & Molecular Biology. 2007;14:733–737. doi: 10.1038/nsmb1282. [DOI] [PubMed] [Google Scholar]
- Wilson DN. The A-Z of bacterial translation inhibitors. Critical Reviews in Biochemistry and Molecular Biology. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- Yates JL. Role of ribosomal protein S12 in discrimination of aminoacyl-tRNA. The Journal of Biological Chemistry. 1979;254:11550–11554. [PubMed] [Google Scholar]
- Yokoyama T, Shaikh TR, Iwakura N, Kaji H, Kaji A, Agrawal RK. Structural insights into initial and intermediate steps of the ribosome-recycling process. The EMBO Journal. 2012;31:1836–1846. doi: 10.1038/emboj.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel JM, Young R, Dennis PP, Nomura M. Role of ribosomal protein S12 in peptide chain elongation: analysis of pleiotropic, streptomycin-resistant mutants of Escherichia coli. Journal of Bacteriology. 1977;129:1320–1329. doi: 10.1128/jb.129.3.1320-1329.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal Structures of EF-G-Ribosome Complexes Trapped in Intermediate States of Translocation. Science. 2013;340:1236086–1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.