Abstract
Background
Construction of microbial biocatalysts for the production of biorenewables at economically viable yields and titers is frequently hampered by product toxicity. Membrane damage is often deemed as the principal mechanism of this toxicity, particularly in regards to decreased membrane integrity. Previous studies have attempted to engineer the membrane with the goal of increasing membrane integrity. However, most of these works focused on engineering of phospholipids and efforts to identify membrane proteins that can be targeted to improve fatty acid production have been unsuccessful.
Results
Here we show that deletion of outer membrane protein ompF significantly increased membrane integrity, fatty acid tolerance and fatty acid production, possibly due to prevention of re-entry of short chain fatty acids. In contrast, deletion of fadL resulted in significantly decreased membrane integrity and fatty acid production. Consistently, increased expression of fadL remarkably increased membrane integrity and fatty acid tolerance while also increasing the final fatty acid titer. This 34% increase in the final fatty acid titer was possibly due to increased membrane lipid biosynthesis. Tuning of fadL expression showed that there is a positive relationship between fadL abundance and fatty acid production. Combinatorial deletion of ompF and increased expression of fadL were found to have an additive role in increasing membrane integrity, and was associated with a 53% increase the fatty acid titer, to 2.3 g/L.
Conclusions
These results emphasize the importance of membrane proteins for maintaining membrane integrity and production of biorenewables, such as fatty acids, which expands the targets for membrane engineering.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-017-0650-8) contains supplementary material, which is available to authorized users.
Keywords: Membrane engineering, Membrane integrity, Outer membrane protein, Tolerance, Fatty acid production
Background
Construction of microbial cell factories for production of biorenewable fuels and chemicals is a promising alternative to current petroleum-driven industries [1, 2]. A variety of microorganisms have been engineered for production of bulk chemicals, biofuels and high-value, fine chemicals [3–7]. However, performance of some biocatalysts can be restricted by various detrimental effects, including toxicity of the product or components of the feedstock [8, 9]. A variety of adverse effects could be the cause of this toxicity, e.g. intracellular acidification; DNA, RNA, protein and membrane damage [10]. Among these, membrane damage has been recognized as a common problem [11–15].
Membrane damage can be compared to a reaction vessel that is vulnerable to corrosion by its contents. In this scenario, a typical response would be to change the composition of the reaction vessel in order to increase resistance to corrosion. For microbial biocatalysts, the composition, function and physical properties of the membrane can be altered through targeted, rational genetic manipulation. Such genetic manipulation is consistent with Cameron and Tong’s fifth application of cellular and metabolic engineering, “modification of cell properties” [16]. When enzymes, transporters and regulators are involved in this membrane engineering, it is also consistent with Bailey’s 1991 definition of metabolic engineering as “the improvement of cellular activities by manipulation of enzymatic, transport, and regulatory function of the cell with the use of recombinant DNA technology” [17].
This work focuses on membrane engineering to improve production of fatty acids, an attractive class of biorenewable chemicals which can be catalyzed to a variety of products with a large potential market, e.g. alkanes, olefins, esters, fatty aldehydes, and fatty alcohols [18–22]. Unfortunately, these fatty acids have been reported to cause a decrease in membrane integrity of E. coli during both exogenous challenge and endogenous production [14]. Engineering of membrane phospholipids has proven as a powerful tool in addressing membrane integrity. Decreasing incorporation of medium-chain fatty acids into the membrane increased the average membrane lipid length, decreased the toxicity of fatty acids and increased fatty acid (C12–C14) production in rich medium from 0.60 to 1.36 g/L [23]. Expression of a thioesterase from Geobacillus sp. Y412MC10 that prevents medium-chain unsaturated acyl-ACPs from being incorporated into the phospholipids was shown to increase membrane integrity during fatty acid production, but there was no increase in fatty acid (C8–C14) production after 24 h in rich medium, with titers of 0.65 g/L observed with and without expression of the secondary thioesterase [24]. Both of these works demonstrate the feasibility of engineering the membrane lipid composition in order to increase membrane integrity and possibly enhance fatty acid tolerance and production [23, 24].
As efforts continue to increase the membrane integrity during production of membrane-damaging compounds, it becomes increasingly important to provide a sufficient route of product export. Several studies have shown that increasing the expression of transporters can increase production of inhibitory compounds, such as valine [25] and limonene [8]. With the goal of using this strategy to improve fatty acid production, sixteen possible fatty acid transporters were characterized for their role in fatty acid tolerance and production [26]. This previous study identified several transporters that increased fatty acid tolerance when their expression was increased, but did not identify any such transporters that increased fatty acid production.
The transporters OmpF and FadL were part of the previous study. The OmpF protein exists as a trimer in the outer membrane and participates in the transport of sugars, ions, antibiotics and proteins across the outer membrane [27, 28]. FadL is an outer membrane ligand gated channel that functions in the uptake of exogenous long-chain fatty acids (LCFA), [29, 30], especially palmitic acid (C16:0) and oleic acid (C18:1), yet shows no binding to short-chain fatty acids (SCFA, <C10) [31]. Even though the previous characterization observed that deletion of ompF and fadL had no impact on fatty acid production [26], several other reports related to these two transporters (Table 1) motivated the further exploration of their role in fatty acid tolerance and production described here.
Table 1.
Previous reports of the role of OmpF and FadL in tolerance of membrane-damaging compounds
| Compound | Condition | Result | Reference |
|---|---|---|---|
| OmpF, outer membrane porin F | |||
| C8–C14 fatty acids | Production of ~1 g/L fatty acids during growth in LB with glycerol, 37 °C | Deletion of ompF from a derivative of MG1655 had no impact on cell viability or membrane integrity | [26] |
| Octanoic acid (C8) | Challenge with up to 20 mM C8 in minimal media with glucose, tryptone and yeast extract at pH 7.0 and 37 °C | Deletion of ompF from BW25113 decreased sensitivity to C8, and increased expression of ompF increased sensitivity to C8. Sensitivity was assessed via the maximum OD. Deletion of ompF decreased the magnitude of intracellular acidification | [32] |
| Phenylpropanoids | Challenge with 1 g/L rutin, naringenin or resveratrol in M9 medium with casamino acids and glucose at 30 °C | Increased expression of ompF in BL21 increased the maximum specific growth rate during challenge. Decreased growth rate during challenge was observed when ompF expression was decreased | [33] |
| FadL, long-chain fatty acid outer membrane porin | |||
| C8–C14 fatty acids | Production of ~1 g/L fatty acids during growth in LB with glycerol at 37 °C | Deletion of fadL from a derivative of MG1655 had no impact on cell viability or membrane integrity | [26] |
| Palmitic and ω-hydroxy palmitic acids | Addition of 1 mM palmitic acid in potassium phosphate buffer with glucose or glycerol, 30 °C | Increased expression of fadL increased conversion of palmitic acid to ω-hydroxy palmitic acid. The increase was smaller in the presence of glycerol than glucose | [34] |
| Phenol | Challenge with phenol at 50–75% of the MIC in LB at 37 °C | Deletion of fadL from BW25113 had no impact on survival | [37] |
| Octane | Addition of ~20 vol% octane in LB at 37 °C | Deletion of fadL from a BW25113 derivative abolished conversion of octane to octanol, octanal and octanoic acid | [35] |
| Hexane | Challenge with 10 vol% hexane in LB at 37 °C | Deletion of fadL from BW25113 increased survival, as assessed by OD | [35] |
| Phenylpropanoids | Challenge with 1 g/L rutin, naringenin or resveratrol in minimal medium with casamino acids and glucose at 30 °C | Increased expression of fadL in BL21 increased the maximum specific growth rate during challenge. Decreased growth rate during challenge was observed when fadL expression was decreased | [33] |
Two 2015 publications directly implicated OmpF in tolerance of exogenously supplied inhibitors, though in one case OmpF played a protective role and in the other it played a damaging role. Most relevant to our goal of improving fatty acid production is the demonstration that deletion of ompF dampened octanoic acid toxicity, with evidence that this deletion of ompF reduced SCFA entry into cells [32] (Table 1). This reduced entry of SCFA into cells was assessed by measuring the decrease in intracellular pH during challenge with exogenously supplied octanoic acid. Contrastingly, OmpF was found to be directly related to tolerance of three exogenously provided phenylpropanoids: rutin, naringenin and resveratrol [33]. Specifically, strains with increased expression of OmpF showed increased tolerance to these compounds and strains with decreased expression of OmpF showed decreased tolerance, leading to the proposition that OmpF participates in the removal of phenylpropanoids from the cell interior. Thus, OmpF showed a negative role in SCFA tolerance and a positive role in phenylpropanoid tolerance.
There are also reports of FadL being involved in fatty acid production and tolerance to some inhibitors (Table 1). Increased expression of fadL resulted in increased conversion of exogenously supplied palmitic acid to ω-hydroxy palmitic acid [34]. This improved organism performance was attributed to increased uptake of palmitic acid, as data indicated that FadL was not involved in export of the hydroxylated product. Similarly, FadL seemed to play a crucial role in the import of octane for production of octanol, octanal and octanoic acid [35]. Specifically, production of these compounds from exogenously supplied octane was abolished when fadL was deleted. However, it was noted that this deletion of fadL increased survival during challenge with hexane, with the conclusion that FadL was the main route of hexane entry into the cell [35]. The phenylpropanoid studies described above also noted that FadL abundance was directly related to tolerance of exogenously supplied rutin, naringenin and resveratrol, the same trend was observed for OmpF, with the interpretation that FadL was involved in repairing membrane damage caused by these compounds [33]. However, even though phenol toxicity is often attributed to membrane damage [36], deletion of fadL had no impact on survival during phenol challenge [37]. Thus, FadL appears to be important to the uptake of some fatty acids and alkanes, provides protection from the inhibitory effects of phenylpropanoids, provides entry to some harmful alkanes and yet possibly plays no role in repairing the membrane damage caused by phenol.
Here we have taken another look at the role of OmpF and FadL in fatty acid tolerance and production, with the conclusion that OmpF and FadL have opposite effects. Specifically, fatty acid tolerance, fatty acid production and membrane integrity were all increased when ompF was deleted or when expression of fadL was increased. Concurrent utilization of these two engineering strategies enabled a roughly 50% increase in production of fatty acids (primarily C14, C16:1 and C16), resulting in a final titer of 2.3 g/L. Although we employed a thioesterase specific for LCFA (C14–C16), some SCFAs (e.g. C8 and C10) were also produced. We propose that deletion of ompF prevents re-entry of the SCFA and their corresponding toxic effects. Contrastingly, it seems that FadL may enable the recapture of some of the LCFA for use in membrane biosynthesis and repair.
Methods
Strains and plasmids
All plasmids and strains used in this study are listed in Table 2. One-step recombination method (FLP-FRT) was employed to perform genetic modifications [38]. E. coli K-12 MG1655 was employed as the host strain. For modulating expression of fadL, the FRT-cat-FRT selection marker linked with four different promoters (M1-12, M1-37, M1-46, M1-93) [6, 39, 40] with varying strengths was employed to regulate expression of the original fadL gene, yielding engineered strains M1-12-fadL, M1-37-fadL, M1-46-fadL and M1-93-fadL, respectively.
Table 2.
Strains and plasmids used in this study
| Strains/plasmids | Genetic characteristics | Source |
|---|---|---|
| Strains | ||
| MG1655 | Wild type E. coli K-12 strain | Lab collection |
| ΔompF | MG1655, ΔompF | This study |
| ΔfadD | MG1655, ΔfadD | This study |
| ΔfadL | MG1655, ΔfadL | This study |
| Pla-empty | MG1655, pACYC184-Kan | This study |
| Pla-fadL | MG1655, pACYC184-Kan-fadL | This study |
| Gen-empty | MG1655, ldhA::FRT-cat-FRT | This study |
| Gen-fadL | MG1655, ldhA::FRT-cat-FRT, fadL | This study |
| M1-12-fadL | MG1655, FRT-cat-FRT, M1-12-fadL | This study |
| M1-37-fadL | MG1655, FRT-cat-FRT, M1-37-fadL | This study |
| M1-46-fadL | MG1655, FRT-cat-FRT, M1-46-fadL | This study |
| M1-93-fadL | MG1655, FRT-cat-FRT, M1-93-fadL | This study |
| ΔompF + Pla-empty | MG1655, ΔompF, pACYC184-Kan | This study |
| ΔompF + Pla-fadL | MG1655, ΔompF, pACYC184-Kan-fadL | This study |
| Plasmids | ||
| pACYC184-Kan | p15A, pACYC184, Kanr | This study |
| pACYC184-Kan-fadL | pACYC184-Kan harboring fadL, Kanr | This study |
| pXZ18Z (TE) | pTrc99a-Ricinus communis thioesterase-fabZ, Ampr | [42] |
For increasing expression of fadL, two different strategies were employed. First, the low-copy plasmid pACYC184-Kan-fadL, which harbors the native promoter, open reading frame (ORF), and terminator of fadL was transformed to MG1655, resulting in Pla-fadL. MG1655 with empty pACYC184-Kan served as the corresponding control (Pla-empty). Second, for increased expression of fadL from the chromosome, a second copy of the fadL gene was inserted into the MG1655 genome at the ldhA site, resulting in Gen-fadL. The ldhA gene was also deleted from MG1655 to generate strain Gen-empty, which serves as a control for strain Gen-fadL. Selection of ldhA as the integration site was motivated by previous reports [41].
The pXZ18Z plasmid [42] harboring a thioesterase from Ricinus communis and the E. coli 3-hydroxyacyl-ACP dehydratase (fabZ) was used for long-chain fatty acid (LCFA) production. When necessary, ampicillin, kanamycin and chloramphenicol were used at final concentrations of 100, 50 and 34 mg/L, respectively.
Strain tolerance characterization
Octanoic acid tolerance was characterized in 50 mL MOPS defined minimal medium with 2.0% (wt/v) dextrose and 10 mM octanoic acid (1.44 g/L) in 250 mL baffled flasks at 220 rpm and initial pH at 7.0, 30 °C. MOPS media contains the following: 8.37 g/L 3-(N-morpholino)propanesulfonic acid (MOPS), 0.72 g/L tricine, 2.92 g/L NaCl, 0.51 g/L NH4Cl, 1.6 g/L KOH, 50 mg/L MgCl2, 48 mg/L K2SO4, 348 mg/L K2HPO4, 0.215 mg/L Na2SeO3, 0.303 mg/L Na2MoO4·2H2O, 0.17 mg/L ZnCl2, 2.5 μg/L FeCl2·4H2O, 0.092 μg/L CaCl2·2H2O, 0.031 μg/L H3BO3, 0.020 μg/L MnCl2·4H2O, 0.0090 μg/L CoCl2·4H2O, and 0.0020 μg/L CuCl2·4H2O [43, 44]. Specific growth rate μ (h−1) was calculated by fitting the equation OD = OD0eμt over the duration of the exponential growth phase. OD was measured at 550 nm and all estimated μ values had an R2 of at least 0.95 [45]. Dry cell weight (DCW) was calculated from the optical density at 550 nm (1 OD550 = 0.333 g DCW/L).
Membrane integrity characterization
Cells were centrifuged, washed twice, and then resuspended in PBS buffer (pH 7.0) at a final OD550 of ~1. One hundred microliter (100 μL) of this suspension was mixed with 900 μL of PBS buffer and SYTOX Green (Invitrogen) was added to a final concentration of 5.0 μM. After resting at room temperature for 15 min, cells were analyzed by a BD Biosciences FACSCanto II flow cytometer equipped with standard factory-installed 488 nm excitation laser, signal collection optics, and fluorescence emission filter configuration. Instrument sheath fluid was filtered (0.22 μm) PBS buffer. Green fluorescence from stained cells was collected in the FL1 channel (525/50 nm). Forward scatter (FSC), side scatter (SSC), and FL1 (Green) parameters were collected as logarithmic signals. All data collections were performed at low flow rate setting (~12 μL/min) and cell concentrations were such that the event rate was below 5000 events/s. All samples were analyzed immediately after staining. Background noise and small debris was eliminated from data collection via a side scatter signal threshold that was established by examining samples containing only SYTOX Green staining buffer. Bacteria in SYTOX Green-stained samples were readily identified on the basis of FSC and SSC signals and an appropriate “Cell” gate was drawn to limit FL1 analysis to bacteria and exclude non-cell events. A minimum of 20,000 cell-gated events were collected for each sample. Green fluorescence data for these “cell” events were plotted as histograms showing the signal distribution of bacteria in the sample [14]. Flow cytometry data for this work is available via Flow Repository (https://flowrepository.org) FR-FCM-ZY2B.
Membrane lipid composition characterization
The membrane lipids were extracted by using the Bligh and Dyer method with minor modifications [14, 46]. Cells were centrifuged, washed twice with cold double-distilled water (ddH2O), resuspended in 1.4 mL methanol and transferred to a new glass tube. Ten μicroliter of 1 μg/μL pentadecanoic acid (C15:0) dissolved in ethanol was added as internal standard. Then, samples were sonicated, incubated at 70 °C for 15 min and centrifuged at 5000×g for 5 min. The supernatant was transferred to a new glass tube and the cell pellet was resuspended in 0.75 mL of chloroform, shaken at 37 °C, 150 rpm for 5 min. Transferred supernatant and pellet suspension were combined, vortexed for 1 min and centrifuged at 5000×g for 2 min. The bottom phase was transferred to a new glass tube and dried under nitrogen gas. Two milliliter of methanol:sulfuric acid (98:2 v/v) mixture was added and the mixture was vortexed and incubated at 80 °C for 30 min. Finally, 2 mL of 0.9% (wt/v) sodium chloride (NaCl) and 1 mL of hexane were added, vortexed and centrifuged at 2000×g for 2 min. The top hexane layer was then analyzed by gas chromatography–mass spectrometry (GC–MS). The temperature for GC–MS analysis was initially held at 50 °C for 2 min, ramped to 200 °C at 25 °C/min, held for 1 min, then raised to 315 °C at 25 °C/min, held for 2 min. Helium was used as a carrier gas and the flow rate was 1 mL/min through a DB-5MS separation column (30 m, 0.25 mm ID, 0.25 μm, Agilent). Methods for calculating average membrane lipid length and lipid saturated:unsaturated ratio can be found in [14].
Membrane lipid content measurement
Thirty milliliters of mid-log E. coli cells were centrifuged, washed by ddH2O and adjusted to OD550 ~10. Then, 1.8 mL of cell suspension was centrifuged at 14,000×g for 5 min and the resulting cell pellets were resuspended in 1.4 mL methanol. As described in “Membrane lipid composition characterization” section, the total membrane bound fatty acid was measured. Given that membrane-bound fatty acids account for 71% (w/w) of lipid mass [47], we use the following formula to calculate the membrane lipid content: total membrane lipid (mg/g DCW) = membrane fatty acids (mg)/0.71 × g DCW.
Real-time quantitative PCR
Bacterial cultures were grown and collected by centrifugation at 10,000×g for 2 min. Total RNA was extracted by using the RNeasy Mini Kit (Qiagen), and the residual DNA was removed by TURBO DNA-free™ Kit (Life Technology). Superscript III First-Strand Synthesis SuperMix (Invitrogen) was employed for the cDNA synthesis, then the cDNA was diluted 100-fold and used as template for quantitative real-time PCR (qRT-PCR) analysis with SYBR Green ER™ qPCR SuperMix (Invitrogen). The E. coli 16S rrsA gene was employed as the housekeeping gene for fadL mRNA abundance analysis. Sequences of fadL primers for qRT-PCR are CTGAAATGTGGGAAGTGTC/GAAGGTCCAGTTATCATCGT, Primers for rrsA are TGGCTCAGATTGAACGC/ATCCGATGGCAAGAGGC. The qRT-PCR was performed with the StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific). The PCR mixture was held at 95 °C for 10 min and then subjected to 40 cycles of incubation at 95 °C for 15 s, then 60 °C for 1 min.
Fermentation for fatty acid production
Individual colonies were selected from Luria Broth (LB) plates with ampicillin and inoculated into 3 mL of LB liquid medium with ampicillin for 4 h. Then, 0.5 mL of culture was added to 20 mL LB with ampicillin at 30 °C, 220 rpm overnight for seed culture preparation. Seed culture was collected, resuspended in MOPS 2.0% (wt/v) dextrose medium, and transferred into 50 mL MOPS 2.0% (wt/v) dextrose containing ampicillin and 1 mM of isopropyl-β-d-thiogalactopyranoside (IPTG) in 250 mL baffled flasks. The target initial cell density was OD550 ~0.1. Cultures were grown in 250 mL baffled flasks with initial pH 7.0 at 30 °C, 220 rpm for 72 h.
Determination of carboxylic acid titers
Carboxylic acid production was quantified by an Agilent 7890 gas chromatograph equipped with an Agilent 5975 mass spectroscope using flame ionization detector and mass spectrometer (GC–MS) after carboxylic acid extraction. Briefly, 100 μL of whole liquid media sample was taken and 10 μL of 1 μg/μL C7:0/C11:0/C17:0 was added as internal standards. Two milliliter of ethanol: sulfuric acid (98:2 v/v) mixture was added, mixed and incubated at 65 °C for 30 min. Then, 2 mL of 0.9% (wt/v) NaCl solution and 1 mL of hexane were added, vortexed and centrifuged at 2000×g for 2 min. The top hexane layer was then analyzed by GC–MS, as described in “Strain tolerance characterization” section.
Statistical analysis
The two-tailed t test method was employed to analyze the statistical significance of all data in this study and P value <0.05 is deemed statistically significant.
Results
Effects of ompF or fadL deletion on tolerance and production of fatty acids
It was previously reported that OmpF facilitates transport of SCFA, such as octanoic acid (C8), into E. coli, and that deletion of ompF in E. coli BW25113 decreased the impact of C8 on biomass production [32]. To evaluate the effect of OmpF on C8 tolerance in MG1655, we also constructed an ompF deletion strain (ΔompF) and confirmed that this engineering strategy improved tolerance to C8. In the absence of C8, the specific growth rates (µ) of both strains were approximately 0.39 h−1. During C8 challenge, the specific growth rate of the ΔompF mutant was 0.33 h−1, which is 7% higher than that of MG1655 (0.31 h−1) (Fig. 1a), which is consistent with the previous report [32].
Fig. 1.
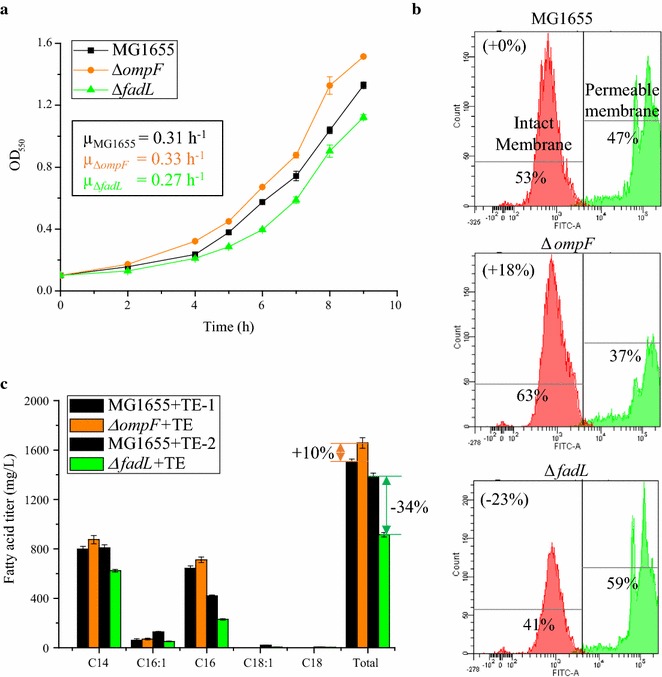
Effects of ompF or fadL deletion on membrane integrity during short-chain fatty acid challenge, short-chain fatty acid tolerance and production of C12 and C14 fatty acids. a Deletion of ompF or fadL impact the specific growth rate relative to the wild type MG1655 during challenge with 10 mM C8. Inset values are the specific growth rate, h−1. b Deletion of ompF or fadL alters the percentage of cells with intact membranes (membrane integrity), assessed using SYTOX Green, during challenge with 10 mM C8. c Deletion of ompF increased fatty acid production and deletion of fadL decreased fatty acid production. MG1655 + TE-1 and MG1655 + TE-2 indicates experiments performed with the same strain, but on different days. For a and b, experiments were performed in MOPS + 2% (wt/v) dextrose shake flasks at 220 rpm 30 °C with an initial pH of 7.0, 10 mM octanoic acid (C8). For c, strains carry the pXZ18Z plasmid (TE) for LCFA (C14–C16) production. Fermentations were performed in MOPS + 2% (wt/v) dextrose shake flasks at 220 rpm 30 °C with an initial pH of 7.0, 1.0 mM IPTG. Values are the average of at least three biological replicates with error bars indicating one standard deviation. Percent increase values are shown only for differences that were deemed statistically significant (P < 0.05)
Decreased membrane integrity has been previously described as a primary cause of C8 toxicity, where decreased membrane integrity is evidenced by leakage of metabolites and ions, such as Mg2+, out of the cell or the entry of membrane-impermeable molecules, such as SYTOX, into the cell [14, 24, 48]. We next characterized the membrane integrity changes after disruption of ompF. Consistent with the growth results, deletion of ompF dampened the impact of C8 on membrane integrity. Specifically, the percentage of cells with intact membranes, i.e. SYTOX impermeable, during challenge with exogenously provided 10 mM C8, increased by 18% compared with the wild-type control strain (P < 0.05) (Fig. 1b).
Given that increased tolerance might lead to increased production of bio-products, we next applied the ompF deletion strategy to fatty acid production. The plasmid pXZ18Z (TE) harboring the heterologous thioesterase from R. communis [42], which primarily releases tetradecanoic acid (C14:0), palmitoleic acid (C16:1) and hexadecanoic acid (C16:0), was transformed into the ΔompF strain and the corresponding control for fatty acid production in minimal MOPS 2.0% (wt/v) dextrose medium. We observed that deletion of ompF increased fatty acid production (Fig. 1c): in the ΔompF + TE mutant, the titer of C14:0 was increased by 10% (P = 0.03) to 875 mg/L, C16:1 was increased by 17% (P = 0.24) to 71 mg/L and C16:0 was increased by 11% (P = 0.01) to 711 mg/L. All of these increases led to a 10% improvement of total fatty acids produced by the ΔompF + TE mutant compared to MG1655 + TE strain, with titers of 1500 ± 20 and 1660 ± 40 mg/L, respectively (P = 0.005). It should be noted that previous studies concluded that deletion of ompF from E. coli strain TY05 did not significantly increase fatty acid (C8–C14) production [26]. The difference from this previous report and the findings presented here may be due to the use of different thioesterases (from U. californica vs. from R. communis), growth media (nutrient-rich LB + 0.4% (v/v) glycerol vs. minimal MOPS + 2% (wt/v) glucose) and temperature (37 vs. 30 °C).
While OmpF has been previously characterized in terms of SCFA transport, FadL predominantly functions in the uptake of LCFA [29, 30]. To investigate the effect of FadL on fatty acid tolerance and production, a fadL deletion mutant (ΔfadL) was constructed. Interestingly, the ΔfadL mutant showed decreased tolerance to C8. For example, the specific growth rate of the ΔfadL strain was 12% lower than that of MG1655 (0.27 vs. 0.31 h−1) (P < 0.05) (Fig. 1a). Further membrane characterization showed that the percentage of cells with intact membranes was 23% lower for the ΔfadL strain than MG1655 (P < 0.05) (Fig. 1b). When this fadL deletion strategy was applied to fatty acid production (+TE), titers of C14:0 decreased by 23% to 623 mg/L, C16:1 decreased by 60% to 51 mg/L and C16:0 decreased by 45% to 230 mg/L. Each of these changes had a P value less than 0.05. Together, these changes led to a 34% reduction of total fatty acids in the ΔfadL + TE mutant compared with MG1655 + TE strain (from 1390 ± 30 to 920 ± 20 mg/L) (P < 0.05) (Fig. 1c). It should be noted that the fatty acid titer of MG1655 + TE here (1390 ± 30 mg/L) is slightly lower than the 1500 ± 20 mg/L described above for the ompF results, due to differences between batches, similar to the results described elsewhere [26]. As with deletion of ompF, our results differ from previous reports of the effect of fadL deletion on fatty acid production. This previous characterization employed E. coli strain TY05 in rich medium with glycerol and found no significant change in production of C8–C14 fatty acids upon deletion of fadL [26]. However, our observation that deletion of fadL can increase sensitivity to membrane-damaging short-chain fatty acids is consistent with observations made for phenylpropanoid tolerance [33].
Increased expression of fadL increased fatty acid tolerance and production
Given that the deletion of fadL decreased fatty acid tolerance and production, it is reasonable to expect that increased expression of fadL might improve fatty acid tolerance and production. To this end, two different strategies were employed in E. coli MG1655 for increased expression of fadL: plasmid expression (Pla-fadL) and genomic integration of a second copy of fadL (Gen-fadL). Consistent with our hypothesis, both of these increased expression strategies significantly improved C8 tolerance. Specifically, the specific growth rate of Pla-fadL (0.33 h−1) and Gen-fadL (0.33 h−1) were 8 and 7% higher than Pla-empty (0.31 h−1) and Gen-empty (0.31 h−1) (P < 0.05) (Fig. 2a). Membrane damage, as evidenced by entry of the SYTOX nucleic acid dye into the cell, was decreased in the two strains engineered for increased fadL expression. Specifically, Pla-fadL showed a 25% increase in membrane integrity and Gen-fadL had a 14% increase in membrane integrity (P < 0.05) (Fig. 2b).
Fig. 2.
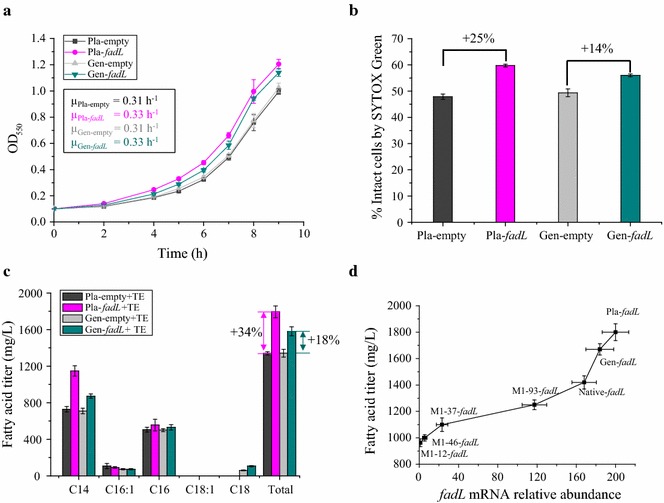
Increased expression of fadL increases membrane integrity, fatty acid tolerance and production. a Increased expression of fadL from a plasmid (Pla-fadL) or a genomic insertion (Gen-fadL) both increase the specific growth rate relative to the corresponding controls (Pla-empty, Gen-empty) during challenge with 10 mM C8. Inset values are the specific growth rate, h−1. b Percentage of cells with intact membrane (membrane integrity), assessed using SYTOX Green. Strains with increased expression of fadL, Pla-fadL and Gen-fadL, have improved membrane integrity relative to their corresponding controls, Pla-empty and Gen-empty, during challenge with 10 mM C8. c Strains with increased expression of fadL, Pla-fadL and Gen-fadL, produce more fatty acid than the corresponding controls, Pla-empty and Gen-empty. d The fadL mRNA relative abundance at 48 h has a positive relationship with the fatty acids titer after 72 h. Four different promoters (M1-12, M1-37, M1-46 and M1-93) were used to replace the native promoter of fadL. The mRNA abundance of fadL in M1-12-fadL strain was set as 1. The 16S rrsA gene was used as normalizing factor. For a and b, experiments were performed in shake flasks containing MOPS + 2% (wt/v) dextrose with 10 mM octanoic acid (C8) at an initial pH of 7.0, shaken at 220 rpm, and maintained at 30 °C. For c and d, all strains carry the pXZ18Z plasmid (TE, fabZ) for LCFA (C14–C16) production. Fermentations were performed in MOPS + 2% (wt/v) dextrose shake flasks at 220 rpm 30 °C with an initial pH of 7.0, 1.0 mM IPTG. Values are the average of at least three biological replicates with error bars indicating one standard deviation. Percent increase values are shown only for differences that were deemed statistically significant (P < 0.05). Pla-empty: MG1655 + pACYC184-Kan; Pla-fadL: MG1655 + pACYC184-Kan-fadL; Gen-empty: MG1655 ldhA::FRT-cat-FRT; Gen-fadL: MG1655 ldhA::FRT-cat-FRT, fadL. TE: pXZ18Z plasmid
Further characterization showed that both of the strains with increased fadL expression also had increased fatty acid production capability. This significantly (P < 0.05) increased fatty acid titer was observed for C14:0 and the total fatty acid pool, though the increase was slightly higher for C16:1 and C16:0 in both cases (Fig. 2c). Specifically, the plasmid-based strain produced 1150 mg/L of C14:0, 556 mg/L of C16:0 and 1800 mg/L of total fatty acid, which was 57, 10 and 34% higher than the corresponding control encoding the thioesterase and an empty plasmid. This control strain produced 728 mg/L C14:0, 505 mg/L C16:0 and 1340 mg/L total fatty acids. A similar trend was also observed for genome-based fadL expression tuning: 872 mg/L of C14:0, 531 mg/L of C16:0 and 1580 mg/L of total fatty acids were produced by the engineered Gen-fadL + TE strain, which was 23, 6 and 18% higher than in the 710, 500 and 1340 mg/L produced by the corresponding Gen-empty + TE control. These results demonstrate the effectiveness of increasing fadL expression for increasing fatty acid production.
In order to further characterize the relationship between the expression level of fadL and fatty acid production, additional strains were constructed (+TE) and characterized. Specifically, different promoters (M1-12, M1-37, M1-46, M-93) with varied strengths [6, 39, 40] were employed to regulate the expression of the native fadL (Fig. 2d). A positive relationship between mRNA relative abundance of fadL and fatty acid titers was observed (Fig. 2d). For instance, mRNA relative abundance of fadL increased nearly 120-fold in M1-93-fadL strain relative to M1-12-fadL (of which fadL expression level was deemed as 1), and it also produced 1250 mg/L of fatty acid, which is 37% higher than the 915 mg/L produced by M1-12-fadL. It should be noted that expression level of fadL under all artificial promoters used here is lower than the native promoter, which suggests that expression of fadL is held at a relatively high level in E. coli MG1655.
Deletion of ompF and increased expression of fadL have an additive effect in increasing fatty acid production
Given that deletion of ompF and increased expression of fadL were each found to increase tolerance and production of fatty acids, we proposed that combinatorial utilization of both engineering strategies would further increase performance. To this end, the plasmid-based expression of fadL was selected as the strategy for increasing expression of fadL, due to its substantial increase in tolerance and production of fatty acid.
Consistent with our hypothesis, combinatorial utilization of the ompF deletion and increased expression of fadL was found to have an additive effect for improving tolerance to C8 (Fig. 3a). The specific growth rate of ΔompF + Pla-fadL strain reached up to 0.36 h−1 in the presence of 10 mM C8, which exceeds that of Pla-empty (μ = 0.31 h−1) by 18%, and is also 10% higher than individual deletion of ompF (ΔompF + Pla-empty, μ = 0.33 h−1) and 12% higher than individual increased expression of fadL (Pla-fadL, μ = 0.32 h−1) (P < 0.05) (Fig. 3a). Besides increased tolerance, membrane integrity was significantly increased in the ΔompF + Pla-fadL strain during challenge with C8. Compared with Pla-empty, the percentage of ΔompF + Pla-fadL cells with intact membranes increased by 37% (P < 0.05) (Fig. 3b).
Fig. 3.
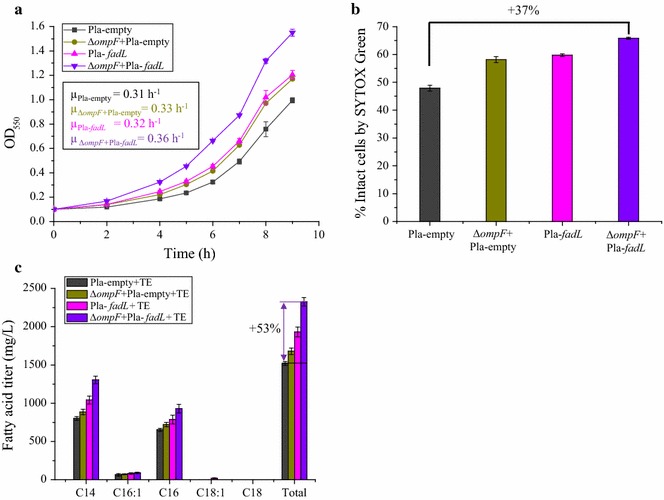
Deletion of ompF and increased expression of fadL have an additive effect on increasing membrane integrity, fatty acid tolerance and production. a Combinatorial deletion of ompF (ΔompF) and increased expression of fadL (Pla-fadL) increases the specific growth rate during challenge with 10 mM C8 relative to the starting strain (Pla-empty), individual ompF deletion strain (ΔompF + Pla-empty), and individual overexpression of fadL (Pla-fadL). Inset values are the specific growth rate, h−1 b Percentage of cells with intact membrane (membrane integrity), assessed using SYTOX Green. Combinatorial deletion of ompF and increased expression of fadL improves membrane integrity during challenge with 10 mM C8 relative to Pla-empty, ΔompF + Pla-empty and Pla-fadL strains. c The combined implementation of ompF deletion and increased expression of fadL supports increased fatty acid titers relative to each engineering strategy implemented individually. For a and b, experiments were performed in MOPS + 2% (wt/v) dextrose shake flasks at 220 rpm 30 °C with an initial pH of 7.0, 10 mM octanoic acid (C8). For c, all strains carry the pXZ18Z plasmid (TE, fabZ) for LCFA (C14–C16) production. Fermentations were performed in MOPS + 2% (wt/v) dextrose shake flasks at 220 rpm 30 °C with an initial pH of 7.0, 1.0 mM IPTG. Values are the average of at least three biological replicates with error bars indicating one standard deviation. Percent increase values are shown only for differences that were deemed statistically significant (P < 0.05). Pla-empty: MG1655 + pACYC184-Kan; ΔompF + Pla-empty: MG1655, ΔompF + pACYC184-Kan; Pla-fadL: MG1655 + pACYC184-Kan-fadL; ΔompF + Pla-fadL: MG1655, ΔompF + pACYC184-Kan-fadL
Combination of ompF deletion and increased expression of fadL also increased the specific growth rate during fatty acid production (data not shown), and final fatty acid titers (Fig. 3c). Specifically, the combination of these engineering strategies in the ΔompF + Pla-fadL + TE strain resulted in a specific growth rate of 0.25 h−1 in the first 12 h of fermentation, where this exceeds that of Pla-empty (μ = 0.16 h−1) by 53% (P < 0.05). Correspondingly, the ΔompF + Pla-fadL + TE strain produced 1310 mg/L of C14:0, 90 mg/L of C16:1, 930 mg/L of C16:0 and 2330 mg/L of total fatty acids after 72 h fermentation. These titers are 47, 25, 29 and 38% higher than the strain in which only the ompF deletion was implemented (ΔompF + Pla-empty + TE, 885 mg/L of C14:0, 72 mg/L of C16:1, 722 mg/L of C16:0 and 1680 mg/L of total fatty acid) and 25, 10, 18 and 20% higher than the strain in which only the fadL overexpression was implemented (Pla-fadL + TE, 1040 mg/L of C14:0, 83 mg/L of C16:1, 786 mg/L of C16:0 and 1930 mg/L of total fatty acid). Note that all of these comparisons have P < 0.05, except for C16:1. The combined strain has an approximately 50% improvement in fatty acid titers relative to the corresponding un-engineered control, Pla-empty + TE, which produced 801 mg/L of C14:0, 65 mg/L of C16:1, 653 mg/L of C16:0 and 1520 mg/L of total fatty acid (Fig. 3c). These results again demonstrate the effectiveness of concurrent utilization of ompF deletion and increased expression of fadL for increasing fatty acid production.
Functional mechanism of OmpF and FadL on increased membrane integrity
In this study, engineering the abundance of the membrane proteins OmpF and FadL increased membrane integrity, fatty acid tolerance and fatty acid production. Prior studies showed that increasing the average length or the saturated:unsaturated (S/U) ratio of E. coli membrane lipids can alleviate the decreased membrane integrity caused by fatty acids [23, 24]. In order to determine whether the increased membrane integrity here could be attributed to such changes in the phospholipid tail distribution, we measured the membrane lipid composition in the wild-type MG1655, ΔompF, ΔfadL and Pla-fadL strains (Table 3). However, no significant changes in membrane composition were observed. Similarly, the average lipid length in wild-type MG1655 was 16.4 ± 0.2, which is comparable to the value observed for the ΔompF, ΔfadL and Pla-fadL strains (Table 3). Additionally, the membrane lipid S/U ratio in the wild-type MG1655 was 1.06 ± 0.02, which is similar to the ratios for the ΔompF, ΔfadL and Pla-fadL strains (Table 3). These results indicate that the previously-described membrane engineering mechanisms of increasing the membrane lipid and S/U ratio are not the underlying reason for increased membrane integrity here.
Table 3.
Membrane lipid content and composition changes in the wild type MG1655, ΔompF, ΔfadL, Pla-fadL strains
| Strain | Membrane lipid content (mg/g DCW) | Membrane lipid composition (mol %) | Membrane lipid length | Membrane lipid S/U ratio | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C14:0 | C16:1 | C16:0 | C17cyc | C18:1 | C18:0 | C19cyc | ||||
| MG1655 | 69.4 ± 0.3 | 1.3 ± 0.1 | 13.6 ± 0.2 | 48.5 ± 0.2 | 14.1 ± 0.1 | 19.1 ± 0.4 | 1.70 ± 0.03 | 1.8 ± 0.1 | 16.4 ± 0.2 | 1.06 ± 0.02 |
| ΔompF | 71.3 ± 0.5 (+2.7%) | 1.1 ± 0.1 | 13.6 ± 0.1 | 48.7 ± 0.1 | 13.6 ± 0.3 | 19.4 ± 0.1 | 1.9 ± 0.1 | 1.7 ± 0.1 | 16.4 ± 0.1 | 1.07 ± 0.01 |
| ΔfadL | 62 ± 3 (−10%) | 1.2 ± 0.1 | 12.7 ± 0.1 | 48.3 ± 0.4 | 14.5 ± 0.1 | 19.4 ± 0.3 | 1.9 ± 0.1 | 1.9 ± 0.1 | 16.4 ± 0.2 | 1.06 ± 0.01 |
| Pla-fadL | 78 ± 1 (+13%) | 1.3 ± 0.1 | 13.2 ± 0.2 | 48.2 ± 0.1 | 13.4 ± 0.1 | 20.7 ± 0.1 | 1.9 ± 0.2 | 1.2 ± 0.1 | 16.4 ± 0.1 | 1.00 ± 0.02 |
Each value is an average and standard deviation of three biological replicates
All experiments were performed in MOPS + 2% (wt/v) dextrose shake flasks at 220 rpm 30 °C with an initial pH of 7.0, 10 mM octanoic acid (C8). All values are the average of at least three biological replicates with the associated standard deviation indicated. Percent increase values are only shown for differences that were deemed statistically significant (P < 0.05)
DCW dry cell weight, S/U ratio membrane saturated: unsaturated lipid ratio
Since the membrane consists of lipids and proteins, altering the abundance of FadL and OmpF might affect the total membrane lipid content. The ΔompF strain had a comparable membrane lipid content to MG1655 (Table 3), which indicates that ompF deletion did not significantly impact membrane lipid production. However, unlike ompF, altering the abundance of fadL remarkably affected membrane lipid content. For example, the membrane lipid content of ΔfadL is only 62 ± 3 mg/g DCW, which is an 11% decrease compared to MG1655 (P < 0.05). Consistently, Pla-fadL had a 13% increase in membrane lipid content relative to MG1655 (P < 0.05) (Table 3). This result indicates that, unlike OmpF, FadL might be involved in membrane lipid synthesis, and therefore altering the abundance of fadL affects the membrane lipid content and thus membrane integrity. It should be noted that the relative distribution of the lipid tails is not changed in the Pla-fadL strain (Table 3).
Discussion
Product toxicity is often an obstacle for cost-effective production of biofuels and chemicals [9, 10]. Therefore, construction of robust production organisms tolerant to these biorenewables is critical for industrial applications and has attracted increasing attention in recent years [12, 45, 49, 50]. Given its importance to overall cell function, membrane integrity has become an attractive engineering target for enhancing robustness [13, 24]. In the case of fatty acids, a variety of engineering efforts have been applied to increasing membrane integrity, with mixed results. Most of these engineering strategies focused on altering the distribution of the membrane lipids of E. coli, such as by altering the average lipids length or degree of saturation [23, 24], though there have also been efforts to identify an efflux system that can improve fatty acid production [26].
Here we focused on two membrane proteins, OmpF and FadL, and found that they have distinct effects on maintaining membrane integrity during fatty acid challenge and production. OmpF has been reported to function as the general diffusion porin of E. coli, through which a variety of inhibitory molecules, e.g. antibiotics, colicin and SCFA, can enter the cell [32, 51, 52]. Rodriguez-Moya et al. showed that OmpF facilitates transport of C8 into E. coli, disrupting intracellular pH and oxidative balance [32]. It has also been suggested that OmpF is involved in the removal of phenylpropanoids from the cell interior [33]. In this study, we further characterized the role of OmpF in maintaining membrane integrity and used the ompF deletion strategy to increase fatty acid production. Although we employed the thioesterase specific for release of LCFA (C14–C16), some SCFAs were produced (e.g. C8 and C10) (Additional file 1: Figure S1). These endogenously produced SCFAs can be exported, i.e. by AcrAB-TolC [26], to the extracellular environment. Conversely, they can also re-enter across the outer membrane through E. coli porins (e.g. OmpF) (Fig. 4), which can cause severe membrane damage to E. coli even at low concentrations [14].
Fig. 4.
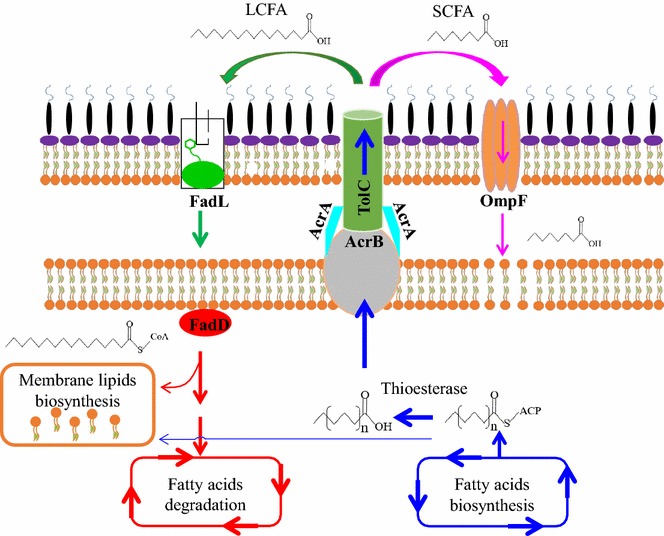
Schematic of the proposed role of ompF and fadL in maintenance of membrane integrity during fatty acid production in E. coli. The elongated acyl-ACP formed during the fatty acids biosynthesis will have two major destinations. Partial acyl-ACPs are hydrolyzed by thioesterase to release free fatty acids. Residual acyl-ACPs serve as precursor for membrane lipids biosynthesis. Among the produced free fatty acids, LCFA (C14–C16) predominates while there is still some SCFA (<C10). It is proposed that LCFA and SCFA are both transported from the cytoplasm directly to the extracellular medium with the AcrAB-TolC complex [26]. However, the low abundance of these compounds in the periplasmic space relative to the extracellular medium results in a driving force for SCFA entry via OmpF and LCFA entry via FadL. LCFAs imported by FadL can be catalyzed by FadD to acyl-CoA, which then serve as fatty acyl precursors for synthesis of phospholipids or enter the β-oxidation cycle for degradation. SCFAs that enter the cell through OmpF, can damage the inner membrane. Increased expression of fadL contributes to import of exogenous LCFA, providing precursors for membrane lipids biosynthesis, thereby increasing membrane integrity and supporting fatty acids production, while deletion of OmpF prevents re-entry of the harmful SCFA. LCFA, long chain fatty acids; SCFA, short-chain fatty acids
One possible explanation for our observations is that after the endogenously produced fatty acids exit the cell, presumably via AcrAB-TolC [26], some of the SCFA re-enter the cell via OmpF. Deletion of ompF blocks this re-entry and thereby increases membrane integrity, which in turn reduces the leakage of important cellular molecules such as Mg2+ [14, 53], thereby elevating fatty acid tolerance and production (Fig. 4). The unexpected driving force for such transport may be due to the nature of the AcrAB-TolC transporter. Specifically, this transporter spans the periplasmic space [54–56] and thus the periplasm should be relatively depleted in fatty acids.
Our results demonstrate that, in addition to membrane engineering strategies that alter the distribution of the membrane lipid tails, altering the abundance of membrane protein OmpF can also affect membrane integrity and production of fatty acids, which provides another strategy for future membrane engineering. Increasing the expression of an efflux pump has been shown to improve the production of inhibitory products, such as valine [25] and limonene [8] and these efflux pumps are also an important part of antibiotic resistance [57]. To the best of our knowledge, this is the first demonstration that deletion of a transporter is associated with increased production of a membrane-damaging compound.
In contrast to the ompF deletion strategy, deletion of fadL was found to decrease membrane integrity, tolerance and production of fatty acid. FadL is the only known outer membrane protein capable of importing exogenous hydrophobic LCFA compounds in E. coli [32, 34, 58, 59]. Imported LCFA can be degraded through the β-oxidation pathway as sources of carbon and energy, or serve as precursors for membrane phospholipid biosynthesis [30, 59–61]. Since there was still residual glucose at the end of our experiments (data not shown), it is not likely that the decreased fatty acid tolerance and decreased fatty acid production of the ΔfadL mutant was caused by carbon or energy limitations. Membrane lipid biosynthesis in E. coli requires acyl chains (C16:0, C16:1 and C18:1), of which there are two sources: (1) endogenous long chain acyl-ACP produced by the fatty acid biosynthesis pathway; and (2) long chain acyl-CoA derived from exogenous LCFA [62, 63]. Upon inactivation of FadL, uptake of exogenous LCFA will be decreased and thus membrane lipid biosynthesis will be impaired (Fig. 4). Our experimental results verify this hypothesis, as membrane lipid content was decreased in the ΔfadL strain and increased in the Pla-fadL strain. Since lipids are the primary structural component of the membrane, changing the membrane lipid content is likely to alter the membrane integrity. This altered membrane lipid content by ΔfadL or Pla-fadL does not change the distribution of the different membrane lipid types (Table 3), which suggests that FadL is only responsible for supplying LCFA precursors instead of directly participating in the biosynthesis of phospholipids.
As with OmpF, a driving force for fatty acid uptake via FadL is not expected to exist during fatty acid production. Here, we again refer to the nature of the AcrAB-TolC efflux pump as a possible reason for the existence of this driving force. Since the AcrAB-TolC system spans the periplasmic space [54–56], the periplasm may be depleted of fatty acids relative to the extracellular medium. This direct relationship between fadL expression and tolerance of membrane-damaging compounds has been noted elsewhere, specifically in regards to phenylpropanoids [33]. This protective effect of FadL against rutin, naringenin and resveratrol was attributed to FadL’s role in repairing membrane damage, though there is no apparent exogenous source of the fatty acids used for this membrane repair [33].
Current membrane engineering strategies focus on altering membrane lipids composition, such as with the goal of increasing membrane lipid length or S/U ratio, to increase membrane integrity. Our results show that increasing the whole membrane lipid content possibly also contributes to increased membrane integrity, tolerance and production of fatty acids, which may serve as a novel strategy for membrane engineering in the future.
Our qRT-PCR results showed that there is a positive relationship between fadL mRNA abundance and fatty acid titer, and they also show that the native fadL gene is maintained at a high expression level, which indicates the importance of FadL in maintaining normal phospholipids biosynthesis. Concurrent deletion of ompF and increased expression of fadL synergistically increased fatty acid tolerance and production, accompanied by increased membrane integrity, possibly due to an increase in membrane lipid content and prevention of re-entry of the SCFA.
Bae et al. [34] found that deletion of fadD and overexpression of fadL in E. coli increased hydroxy long-chain fatty acid production. In that study, it was concluded that overexpression of fadL contributes to the improvement in the production of ω-hydroxy palmitic acid, primarily due to increased ability to transport exogenously fed palmitic acid (C16). The present work mainly focuses on the effect of fadL overexpression on the import of exogenous LCFA for membrane lipid synthesis and thus maintaining membrane integrity during the production of or challenge with membrane-damaging fatty acids. Prior research showed that deletion of ompF or fadL in E. coli did not affect fatty acid production [26], which is different from our results. There are two possible reasons for this difference: (A) the use of different thioesterases; and (B) the use of different growth conditions. The previous studies used a C8–C14-producing thioesterase enzyme from U. californica, while here we used a C14–C16-producing thioesterase from R. communis. This previous study also used nutrient-rich LB with 0.4% (v/v) glycerol at 37 °C, while we used the nutrient-poor minimal MOPS with 2% (wt/v) dextrose at 30 °C. It is interesting to note that the studies that identified a positive relationship between OmpF abundance, FadL abundance and phenylpropanoid tolerance were also performed at 30 °C [33]. The use of glycerol in the previous fatty acid production studies may also be a complicating factor. The increase in hydroxy-palmitic acid production upon overexpression of FadL was smaller in the presence of glycerol relative to glucose [34] and the presence of glycerol has previously been reported to alter the phospholipid composition of microbial cell membranes [64–66]. Under different growth conditions, the membrane composition and associated amount of membrane damage caused by the fatty acids is expected to vary, and therefore the roles of OmpF and FadL may differ.
This engineering method appears to increase fatty acid production as a direct function of increased abundance of the microbial biocatalyst. Thus, it differs from a previously described membrane engineering method that increased fatty acid titers by 50% without impacting the final culture OD [23] and evolutionary strain development that improved fatty acid production fivefold while only increasing growth during fatty acid production threefold [50]. The strategy described here also differs from provision of valine-producing E. coli with a valine exporter, which increased valine titers by 50% without changing the final OD [25]. Thus, additional strain engineering would be needed in order for this strategy to be effective in improving fatty acid production in fed-batch or continuous culture systems. However, this work clearly demonstrates that these two membrane proteins are two viable engineering targets for improving fatty acid production.
Conclusions
Membrane damage of the microbial biocatalyst is a widespread problem in the problem of biorenewable fuels and chemicals. Here we have demonstrated two strategies for dealing with membrane damage in our condition. The first is to increase the abundance of FadL, which we propose increases the ability of the organism to repair the membrane damage incurred by fatty acids. The second method is to delete OmpF, which we propose prevents re-entry of the inhibitory product.
Authors’ contributions
ZT and LRJ designed research. ZT, WB, and JMY performed research. ZT, JVS and LRJ analyzed data, and LRJ wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We thank ISU Flow Cytometry Facility for help with SYTOX Green cells analysis, ISU DNA Facility for help with real-time quantitative PCR analysis and ISU W.M. Keck Metabolomics Research Laboratory for help with membrane fluidity analysis and GC–MS analysis. We would also like to thank Edward Yu and Thomas Mansell for helpful discussion of these results.
Competing interests
This work will be included in patent applications by Iowa State University.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article.
Funding
This work was supported by the NSF Engineering Research Center for Biorenewable Chemicals (CBiRC), NSF Award number EEC-0813570. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- OmpF
outer membrane porin F
- FadL
long-chain fatty acid outer membrane porin
- SCFA
short-chain fatty acids
- LCFA
long chain fatty acids
- C7
heptanoic acid
- C8
octanoic acid
- C11
undecanoic acid
- C14
tetradecanoic acid
- C15
pentadecanoic acid
- C16:1
palmitoleic acid
- C16:0
hexadecanoic acid
- C17
heptadecanoic acid
- TE
pXZ18Z
- DCW
dry cell weight
- IPTG
isopropyl-β-d-thiogalactopyranoside
- ORF
open reading frame
- ldhA
lactate dehydrogenase gene
Additional file
Additional file 1: Figure S1. Fatty acids profile of E. coli MG1655 harboring pXZ18Z plasmid which carries thioesterase gene from Ricinus communis and fabZ gene from E. coli. Some short chain fatty acids (e.g. butanedioic acid, octanoic acid and decanoic acid) were found in the fermentation broth.
Contributor Information
Zaigao Tan, Email: tcltzg1987@gmail.com.
William Black, Email: wblack@uci.edu.
Jong Moon Yoon, Email: jmyoon99@gmail.com.
Jacqueline V. Shanks, Email: jshanks@iastate.edu
Laura R. Jarboe, Phone: +1-515-294-2319, Email: ljarboe@iastate.edu
References
- 1.Gallezot P. Process options for converting renewable feedstocks to bioproducts. Green Chem. 2007;9:295–302. doi: 10.1039/b615413a. [DOI] [Google Scholar]
- 2.Larson ED. A review of life-cycle analysis studies on liquid biofuel systems for the transport sector. Energy Sustain Dev. 2006;10:109–126. doi: 10.1016/S0973-0826(08)60536-0. [DOI] [Google Scholar]
- 3.Thakker C, Martinez I, San KY, Bennett GN. Succinate production in Escherichia coli. Biotechnol J. 2012;7:213–224. doi: 10.1002/biot.201100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park J, Rodriguez-Moya M, Li M, Pichersky E, San KY, Gonzalez R. Synthesis of methyl ketones by metabolically engineered Escherichia coli. J Ind Microbiol Biotechnol. 2012;39:1703–1712. doi: 10.1007/s10295-012-1178-x. [DOI] [PubMed] [Google Scholar]
- 5.McKenna R, Nielsen DR. Styrene biosynthesis from glucose by engineered E. coli. Metab Eng. 2011;13:544–554. doi: 10.1016/j.ymben.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Zhu X, Tan Z, Xu H, Chen J, Tang J, Zhang X. Metabolic evolution of two reducing equivalent-conserving pathways for high-yield succinate production in Escherichia coli. Metab Eng. 2014;24C:87–96. doi: 10.1016/j.ymben.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJ, Hanai T, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng. 2008;10:305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A. Engineering microbial biofuel tolerance and export using efflux pumps. Mol Syst Biol. 2011;7:487. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarboe LR, Liu P, Royce LA. Engineering inhibitor tolerance for the production of biorenewable fuels and chemicals. Curr Opin Chem Eng. 2011;1:38–42. doi: 10.1016/j.coche.2011.08.003. [DOI] [Google Scholar]
- 10.Nicolaou SA, Gaida SM, Papoutsakis ET. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng. 2010;12:307–331. doi: 10.1016/j.ymben.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Huffer S, Clark ME, Ning JC, Blanch HW, Clark DS. Role of alcohols in growth, lipid composition, and membrane fluidity of yeasts, bacteria, and archaea. Appl Environ Microbiol. 2011;77:6400–6408. doi: 10.1128/AEM.00694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lennen RM, Kruziki MA, Kumar K, Zinkel RA, Burnum KE, Lipton MS, Hoover SW, Ranatunga DR, Wittkopp TM, Marner WD, 2nd, Pfleger BF. Membrane stresses induced by overproduction of free fatty acids in Escherichia coli. Appl Environ Microbiol. 2011;77:8114–8128. doi: 10.1128/AEM.05421-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu P, Chernyshov A, Najdi T, Fu Y, Dickerson J, Sandmeyer S, Jarboe L. Membrane stress caused by octanoic acid in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2013;97:3239–3251. doi: 10.1007/s00253-013-4773-5. [DOI] [PubMed] [Google Scholar]
- 14.Royce LA, Liu P, Stebbins MJ, Hanson BC, Jarboe LR. The damaging effects of short chain fatty acids on Escherichia coli membranes. Appl Microbiol Biotechnol. 2013;97:8317–8327. doi: 10.1007/s00253-013-5113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaldivar J, Ingram LO. Effect of organic acids on the growth and fermentation of ethanologenic Escherichia coli LY01. Biotechnol Bioeng. 1999;66:203–210. doi: 10.1002/(SICI)1097-0290(1999)66:4<203::AID-BIT1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Cameron DC, Tong IT. Cellular and metabolic engineering—an overview. Appl Biochem Biotechnol. 1993;38:105–140. doi: 10.1007/BF02916416. [DOI] [PubMed] [Google Scholar]
- 17.Bailey JE. Toward a science of metabolic engineering. Science. 1991;252:1668–1675. doi: 10.1126/science.2047876. [DOI] [PubMed] [Google Scholar]
- 18.Korstanje TJ, van der Vlugt JI, Elsevier CJ, de Bruin B. Hydrogenation of carboxylic acids with a homogeneous cobalt catalyst. Science. 2015;350:298–302. doi: 10.1126/science.aaa8938. [DOI] [PubMed] [Google Scholar]
- 19.Lennen RM, Braden DJ, West RM, Dumesic JA, Pfleger BF. A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnol Bioeng. 2010;106:193–202. doi: 10.1002/bit.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Ruiz JA, Davis RJ. Decarbonylation of heptanoic acid over carbon-supported platinum nanoparticles. Green Chem. 2014;16:683–694. doi: 10.1039/C3GC41287C. [DOI] [Google Scholar]
- 21.Kim S, Cheong S, Chou A, Gonzalez R. Engineered fatty acid catabolism for fuel and chemical production. Curr Opin Biotechnol. 2016;42:206–215. doi: 10.1016/j.copbio.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez MA, Torres GC, Mazzieri VA, Pieck CL. Selective hydrogenation of fatty acids and methyl esters of fatty acids to obtain fatty alcohols—a review. J Chem Technol Biotechnol. 2017;92:27–42. doi: 10.1002/jctb.5039. [DOI] [Google Scholar]
- 23.Sherkhanov S, Korman TP, Bowie JU. Improving the tolerance of Escherichia coli to medium-chain fatty acid production. Metab Eng. 2014;25:1–7. doi: 10.1016/j.ymben.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Lennen RM, Pfleger BF. Modulating membrane composition alters free fatty acid tolerance in Escherichia coli. PLoS ONE. 2013;8:54031. doi: 10.1371/journal.pone.0054031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JH, Lee KH, Kim TY, Lee SY. Metabolic engineering of Escherichia coli for the production of l-valine based on transcriptome analysis and in silico gene knockout simulation. Proc Natl Acad Sci USA. 2007;104:7797–7802. doi: 10.1073/pnas.0702609104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lennen RM, Politz MG, Kruziki MA, Pfleger BF. Identification of transport proteins involved in free fatty acid efflux in Escherichia coli. J Bacteriol. 2013;195:135–144. doi: 10.1128/JB.01477-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 28.Nikaido H. Outer-membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33:1831–1836. doi: 10.1128/AAC.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lepore BW, Indic M, Pham H, Hearn EM, Patel DR, van den Berg B. Ligand-gated diffusion across the bacterial outer membrane. Proc Natl Acad Sci USA. 2011;108:10121–10126. doi: 10.1073/pnas.1018532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Berg B, Black PN, Clemons WM, Jr, Rapoport TA. Crystal structure of the long-chain fatty acid transporter FadL. Science. 2004;304:1506–1509. doi: 10.1126/science.1097524. [DOI] [PubMed] [Google Scholar]
- 31.Black PN. Characterization of FadL-specific fatty-acid binding in Escherichia coli. Biochim Biophys Acta. 1990;1046:97–105. doi: 10.1016/0005-2760(90)90099-J. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Moya M, Gonzalez R. Proteomic analysis of the response of Escherichia coli to short-chain fatty acids. J Proteom. 2015;122:86–99. doi: 10.1016/j.jprot.2015.03.033. [DOI] [PubMed] [Google Scholar]
- 33.Zhou JW, Wang K, Xu S, Wu JJ, Liu PR, Du GC, Li JH, Chen J. Identification of membrane proteins associated with phenylpropanoid tolerance and transport in Escherichia coli BL21. J Proteom. 2015;113:15–28. doi: 10.1016/j.jprot.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Bae JH, Park BG, Jung E, Lee PG, Kim BG. fadD deletion and fadL overexpression in Escherichia coli increase hydroxy long-chain fatty acid productivity. Appl Microbiol Biotechnol. 2014;98:8917–8925. doi: 10.1007/s00253-014-5974-2. [DOI] [PubMed] [Google Scholar]
- 35.Call TP, Akhtar MK, Baganz F, Grant C. Modulating the import of medium-chain alkanes in E. coli through tuned expression of FadL. J Biol Eng. 2016;10:5. doi: 10.1186/s13036-016-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heipieper HJ, Keweloh H, Rehm HJ. Influence of phenols on growth and membrane-permeability of free and immobilized Escherichia coli. Appl Environ Microbiol. 1991;57:1213–1217. doi: 10.1128/aem.57.4.1213-1217.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang DF, Li H, Lin XM, Wang SY, Peng XX. Characterization of outer membrane proteins of Escherichia coli in response to phenol stress. Curr Microbiol. 2011;62:777–783. doi: 10.1007/s00284-010-9786-z. [DOI] [PubMed] [Google Scholar]
- 38.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang J, Zhu X, Lu J, Liu P, Xu H, Tan Z, Zhang X. Recruiting alternative glucose utilization pathways for improving succinate production. Appl Microbiol Biotechnol. 2013;97:2513–2520. doi: 10.1007/s00253-012-4344-1. [DOI] [PubMed] [Google Scholar]
- 40.Tan Z, Zhu X, Chen J, Li Q, Zhang X. Activating phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in combination for improvement of succinate production. Appl Environ Microbiol. 2013;79:4838–4844. doi: 10.1128/AEM.00826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Jantama K, Moore JC, Shanmugam KT, Ingram LO. Production of l-alanine by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol. 2007;77:355–366. doi: 10.1007/s00253-007-1170-y. [DOI] [PubMed] [Google Scholar]
- 42.San K-Y, Li M, Zhang X. Bacteria and method for synthesizing fatty acids. Google Patents; 2011.
- 43.Neidhard FC, Bloch PL, Smith DF. Culture medium for Enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wanner BL. Methods in molecular genetics. New York: Academic; 1994. [Google Scholar]
- 45.Tan Z, Yoon JM, Nielsen DR, Shanks JV, Jarboe LR. Membrane engineering via trans unsaturated fatty acids production improves Escherichia coli robustness and production of biorenewables. Metab Eng. 2016;35:105–113. doi: 10.1016/j.ymben.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 47.Torella JP, Ford TJ, Kim SN, Chen AM, Way JC, Silver PA. Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proc Natl Acad Sci USA. 2013;110:11290–11295. doi: 10.1073/pnas.1307129110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lian J, McKenna R, Rover MR, Nielsen DR, Wen Z, Jarboe LR. Production of biorenewable styrene: utilization of biomass-derived sugars and insights into toxicity. J Ind Microbiol Biotechnol. 2016;43:595–604. doi: 10.1007/s10295-016-1734-x. [DOI] [PubMed] [Google Scholar]
- 49.Chubukov V, Mingardon F, Schackwitz W, Baidoo EEK, Alonso-Gutierrez J, Hu QJ, Lee TS, Keasling JD, Mukhopadhyay A. Acute limonene toxicity in Escherichia coli is caused by limonene hydroperoxide and alleviated by a point mutation in alkyl hydroperoxidase AhpC. Appl Environ Microbiol. 2015;81:4690–4696. doi: 10.1128/AEM.01102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Royce LA, Yoon JM, Chen Y, Rickenbach E, Shanks JV, Jarboe LR. Evolution for exogenous octanoic acid tolerance improves carboxylic acid production and membrane integrity. Metab Eng. 2015;29:180–188. doi: 10.1016/j.ymben.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 51.Kim YC, Tarr AW, Penfold CN. Colicin import into E. coli cells: a model system for insights into the import mechanisms of bacteriocins. Biochim Biophys Acta. 2014;1843:1717–1731. doi: 10.1016/j.bbamcr.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 52.Ziervogel BK, Roux B. The binding of antibiotics in OmpF porin. Structure. 2013;21:76–87. doi: 10.1016/j.str.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jarboe LR, Royce LA, Liu P. Understanding biocatalyst inhibition by carboxylic acids. Front Microbiol. 2013;4:272. doi: 10.3389/fmicb.2013.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du DJ, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tikhonova EB, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J Biol Chem. 2004;279:32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- 56.Touze T, Eswaran J, Bokma E, Koronakis E, Hughes C, Koronakis V. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol Microbiol. 2004;53:697–706. doi: 10.1111/j.1365-2958.2004.04158.x. [DOI] [PubMed] [Google Scholar]
- 57.Blair JMA, Richmond GE, Piddock LJV. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014;9:1165–1177. doi: 10.2217/fmb.14.66. [DOI] [PubMed] [Google Scholar]
- 58.Black PN, DiRusso CC. Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification. Microbiol Mol Biol Rev. 2003;67:454–472. doi: 10.1128/MMBR.67.3.454-472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hearn EM, Patel DR, Lepore BW, Indic M, van den Berg B. Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature. 2009;458:367–370. doi: 10.1038/nature07678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hearn EM, Patel DR, van den Berg B. Outer-membrane transport of aromatic hydrocarbons as a first step in biodegradation. Proc Natl Acad Sci USA. 2008;105:8601–8606. doi: 10.1073/pnas.0801264105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fujita Y, Matsuoka H, Hirooka K. Regulation of fatty acid metabolism in bacteria. Mol Microbiol. 2007;66:829–839. doi: 10.1111/j.1365-2958.2007.05947.x. [DOI] [PubMed] [Google Scholar]
- 62.Rock CO. Fatty acid and phospholipid metabolism in prokaryotes. In: Biochemistry of lipids, lipoproteins and membranes. 5th ed. 2008; p. 59–96.
- 63.Rock CO, Jackowski S. Pathways for the incorporation of exogenous fatty-acids into phosphatidylethanolamine in Escherichia coli. J Biol Chem. 1985;260:2720–2724. [PubMed] [Google Scholar]
- 64.Du GC, Yang G, Qu YB, Chen J, Lun SY. Effects of glycerol on the production of poly(gamma-glutamic acid) by Bacillus licheniformis. Process Biochem. 2005;40:2143–2147. doi: 10.1016/j.procbio.2004.08.005. [DOI] [Google Scholar]
- 65.Kautharapu KB, Rathmacher J, Jarboe LR. Growth condition optimization for docosahexaenoic acid (DHA) production by Moritella marina MP-1. Appl Microbiol Biotechnol. 2013;97:2859–2866. doi: 10.1007/s00253-012-4529-7. [DOI] [PubMed] [Google Scholar]
- 66.Pramanik J, Keasling JD. Effect of Escherichia coli biomass composition on central metabolic fluxes predicted by a stoichiometric model. Biotechnol Bioeng. 1998;60:230–238. doi: 10.1002/(SICI)1097-0290(19981020)60:2<230::AID-BIT10>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article.


