ABSTRACT
Siderophore nutrition tests with Caulobacter crescentus strain NA1000 revealed that it utilized a variety of ferric hydroxamate siderophores, including asperchromes, ferrichromes, ferrichrome A, malonichrome, and ferric aerobactin, as well as hemin and hemoglobin. C. crescentus did not transport ferrioxamine B or ferric catecholates. Because it did not use ferric enterobactin, the catecholate aposiderophore was an effective agent for iron deprivation. We determined the kinetics and thermodynamics of [59Fe]apoferrichrome and 59Fe-citrate binding and transport by NA1000. Its affinity and uptake rate for ferrichrome (equilibrium dissociation constant [Kd], 1 nM; Michaelis-Menten constant [KM], 0.1 nM; Vmax, 19 pMol/109 cells/min) were similar to those of Escherichia coli FhuA. Transport properties for 59Fe-citrate were similar to those of E. coli FecA (KM, 5.3 nM; Vmax, 29 pMol/109 cells/min). Bioinformatic analyses implicated Fur-regulated loci 00028, 00138, 02277, and 03023 as TonB-dependent transporters (TBDT) that participate in iron acquisition. We resolved TBDT with elevated expression under high- or low-iron conditions by SDS-PAGE of sodium sarcosinate cell envelope extracts, excised bands of interest, and analyzed them by mass spectrometry. These data identified five TBDT: three were overexpressed during iron deficiency (00028, 02277, and 03023), and 2 were overexpressed during iron repletion (00210 and 01196). CLUSTALW analyses revealed homology of putative TBDT 02277 to Escherichia coli FepA and BtuB. A Δ02277 mutant did not transport hemin or hemoglobin in nutrition tests, leading us to designate the 02277 structural gene as hutA (for heme/hemoglobin utilization).
IMPORTANCE The physiological roles of the 62 putative TBDT of C. crescentus are mostly unknown, as are their evolutionary relationships to TBDT of other bacteria. We biochemically studied the iron uptake systems of C. crescentus, identified potential iron transporters, and clarified the phylogenetic relationships among its numerous TBDT. Our findings identified the first outer membrane protein involved in iron acquisition by C. crescentus, its heme/hemoglobin transporter (HutA).
KEYWORDS: TonB, alphaproteobacteria, heme transport, hemoglobin, iron acquisition, siderophores
INTRODUCTION
Because of its role as a cofactor for metabolic enzymes, oxidoreductases, and electron transport chain components, iron is an essential metal for most organisms. However, in aerobic environments iron oxidizes to Fe3+ and forms insoluble polymers with hydroxide ions. Both commensal and pathogenic microorganisms secrete small organic compounds, called siderophores, that bind and mobilize iron (1, 2) from its hydroxide polymers. Microbes typically produce hydroxamates, alpha-hydroxycarboxylates, or catecholates as ligands for Fe3+ (3, 4). Once acquired from the environment, iron in biological systems is usually complexed in cells by proteins or found as heme groups or iron-sulfur clusters (5).
TonB-dependent transporters (TBDT) (6–8) in the outer membrane (OM) of Gram-negative bacteria use energy from the inner membrane (IM), transmitted by the TonB/ExbBD complex (9), to obtain ferric siderophores from the environment (10). After uptake into the periplasm, iron complexes enter the cytoplasm through ABC transporters (11). Concomitant with IM transport, ferric siderophores undergo reduction, sometimes combined with degradation, that releases Fe2+ to cellular systems (5, 12).
Caulobacter crescentus is a Gram-negative, free-living aquatic alphaproteobacterium that grows in oligotrophic environments. Its asymmetrical cell division generates a sessile, stalked cell and a motile, flagellated cell (13). One atypical aspect of C. crescentus is that its genome encodes 62 TBDT (14) (Fig. 1). By comparison, the similar-size genome of Escherichia coli produces only 8 TBDT, all of which transport metals. The E. coli TBDT acquire iron complexes of hydroxamates (FhuA, ferrichrome [15]; FhuE, coprogen and rhodotorulate [16]), catecholates (FepA, enterobactin [17, 18]; Cir and Fiu, dihydroxybenzoyl serine [19]), mixed-function chelators (IutA, aerobactin [20]), citrate (FecA [21]), and cyanocobalamin (BtuB [22]). Some TBDT of C. crescentus are negatively iron regulated and therefore are expected to function in iron acquisition (23, 24). The genome of NA1000 (14), a prototypic C. crescentus strain, is devoid of the readily recognizable structural genes encoding the large nonribosomal peptide synthetases responsible for siderophore biosynthesis (14, 25). Nevertheless, C. crescentus was reported to utilize ferric rhodotorulic acid and ferrioxamine B (26). Detailed descriptions of its iron acquisition systems are lacking; most information about C. crescentus cell envelope transport physiology originates from genomic and bioinformatic inferences. The numerous other TBDT encoded in the C. crescentus genome may function in utilization of carbohydrates and other nutrients from plants (27). For example, some transport sugars (26, 28, 29), which distinguishes them from the metal-specific TBDT (30–33).
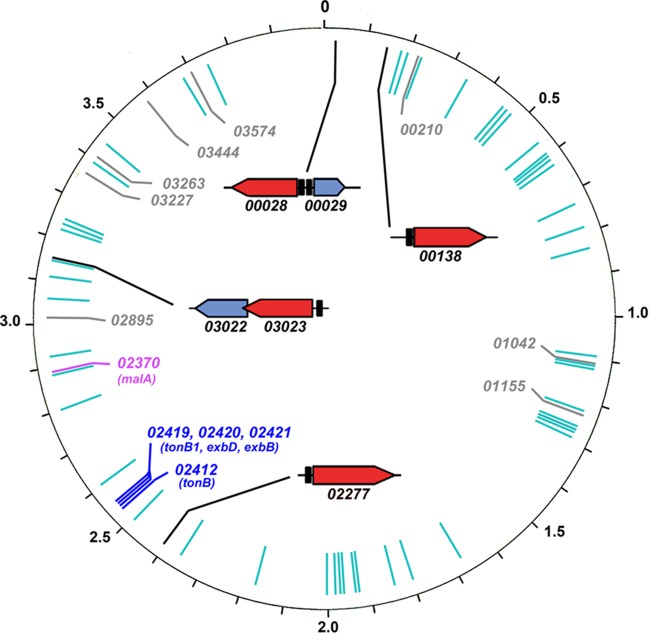
FIG 1.
TBDT in the C. crescentus NA1000 genome. TBDT structural genes that are induced to higher expression levels by iron deprivation (23, 24) or iron repletion are marked with black or gray bars, respectively, at their relative positions in the C. crescentus chromosome (14). The former loci are shown in greater detail (24), with TBDT genes as red arrows denoting the direction of transcription and their fur promoter regions as black boxes. Other TBDT are depicted as cyan bars; Ccr02370 (purple) was identified as a maltose transporter (MalA) (26, 29). We deleted 02277 (see Materials and Methods and Results). The two C. crescentus orthologs of E. coli tonB (29), as well as the orthologs of exbB and exbD (blue), are linked at position ∼2.5 Mb in the genome.
We screened a library of ferric siderophores for uptake by NA1000 and found that it used ferric hydroxamates and ferric aerobactin (an alpha-hydroxycarboxylate chelate) but not ferric catecholates. Enterobactin, a catecholate-based siderophore utilized by many Gram-negative bacterial species (34), did not supply iron to NA1000. Consequently, it was an effective iron chelator that rendered C. crescentus iron deficient in complex media. Results under these conditions showed that C. crescentus possesses membrane transport systems for a variety of hydroxamate iron complexes (except ferrioxamine B) but generally few other types of ferric siderophores. However, NA1000 utilized hemin and hemoglobin. By mass spectrometric (MS) analyses of cell envelope proteins excised from SDS-PAGE gels, we identified five iron-regulated TBDT of C. crescentus. On the basis of their negative regulation and sequence/structure, four proteins (00028, 00138, 02277, and 03023) likely participate in iron uptake. Deletion of locus 02277 revealed it as the OM component of a hemin/hemoglobin utilization system, which we designated HutA.
RESULTS
Siderophore nutrition tests with C. crescentus.
We used siderophore nutrition tests (35) to survey iron utilization by C. crescentus by plating bacteria in nutrient broth (NB) top agar containing a nonutilizable iron chelator and adding paper discs containing iron compounds on the surface of the agar. After overnight incubation at 30°C, iron acquisition from these compounds resulted in growth halos around the filter paper discs (35). We initially used bipyridyl (BP) at 200 μM as the nonutilizable chelator in the media, which revealed utilization of ferrichrome but not ferric enterobactin by C. crescentus (data not shown). We subsequently employed enterobactin (which has the highest known affinity for Fe3+; association equilibrium constant [Ka] of 1052) in media to render C. crescentus iron deficient (Fig. 2). This high affinity creates exclusivity for Fe3+, whereas BP forms complexes with many other cations, potentially including calcium, that C. crescentus requires for optimal growth. Relative to BP, inclusion of enterobactin in agar yielded more well-defined, denser halos (Fig. 2), but we observed the same overall iron uptake profile for C. crescentus on plates with BP, albeit from fainter halos. Besides NB, we tested PYE and minimal medium M2 (36) plates in nutrition tests (data not shown), but NB gave the best and most reproducible results.
FIG 2.

Siderophore nutrition tests with C. crescentus. Strain NA1000 was grown in NB to mid-log phase, and 100 μl of the culture was plated on an NB plate in NB top agar containing enterobactin (100 μM). Paper discs were deposited on the surface of the agar, and 10 μl of 50 μM solutions of ferric siderophore complexes were applied to the discs: 1, asperchrome B1; 2, ferrichrome; 3, ferrichrome A; 4, malonichrome; 5, ferrioxamine B; 6, tetraglycyl ferrichrome; 7, rhodotorulate; 8, aerobactin; 9, FeS04; 10, hemin; 11, hemoglobin (15 μM); 12, dimerum acid; 13, mycobactin; 14, schizokinen; 15, coprogen; 16, vibriobactin; 17, corynebactin; and 18, agrobactin. See Table 1 for a compilation of siderophore nutrition results.
C. crescentus NA1000 transported many ferric hydroxamates in siderophore nutrition assays (Fig. 2 and Table 1). These data conflicted with prior negative findings for ferrichrome, ferricrocin, and ferrichrome A (26) that NA1000 efficiently transported and metabolized in our hands. Also contrary to Neugebauer et al. (26), it did not transport ferric rhodotorulic acid or ferrioxamine B (Fig. 2). Our experiments differed in that we used enterobactin as an iron-sequestering agent, but we verified the same results with BP. Both studies utilized wild-type C. crescentus NA1000. Despite different findings for numerous ferric hydroxamates, we confirmed that neither schizokinen nor coprogen supported growth of C. crescentus (26). Among mixed-function siderophores, C. crescentus only used ferric aerobactin; its uptake was best seen at high concentrations (200 μM). NA1000 did not use any iron complexes of catecholates (enterobactin, vibriobactin, corynebactin, and agrobactin), but it utilized both hemin and hemoglobin in nutrition tests.
TABLE 1.
Siderophore nutrition tests with C. crescentus NA1000a
| Siderophore (reference[s]) | Halo size (cm) |
|---|---|
| Hydroxamates | |
| Asperchromesb (39) | 2.5 |
| Ferrichromesc (37) | 2.2 |
| Malonichromed (38) | 2.3 |
| Coprogen (39) | None |
| Ferrioxamine B (39) | 1.4 |
| Rhodotorulic acid (40) | None |
| Dimerum acid (52) | 0.9 |
| Fusarininesd (52) | None |
| Iron porphyrins | |
| Hemin | 1.4 |
| Hemoglobin | 1.5 |
| Catecholates | |
| Enterobactin (2) | None |
| Vibriobactin (42) | None |
| Corynebactin (43, 44) | None |
| Agrobactin (97) | None |
| Mixed chelates | |
| Aerobactine (46) | 0.9 |
| Schizokinen (47) | None |
| Mycobactin (48, 49) | None |
| Pseudobactin (50) | None |
| Albomycin (4, 37) | Rf |
Bacteria were grown in NB, and 108 cells were poured onto NB plates in NB top agar containing 100 μM enterobactin. Purified ferric complexes of the tabulated siderophores were dissolved or prepared in 10 mM NaHPO4, pH 7. Aliquots of 10 μl of 50 μM solutions of the ferric siderophores (except hemoglobin, which was used at 15 μM) were deposited onto paper discs on the surface of media and the plates were incubated at 30°C overnight, after which we measured the growth halos (in centimeters).
Asperchromes B1, B2, C, and D1 (51).
Ferrichrome, ferrichrome A, ferrichrome C, ferrichrysin, ferricrocin, ferrirhodin, ferrirubin, and tetraglycine ferrichrome (37).
Fusarinine and triacetylfusarinine (52).
Growth halos formed around ferric aerobactin at higher concentrations (e.g., 200 μM).
R, resistant.
Growth in iron-deficient media.
We characterized growth of NA1000 in NB containing the two nonutilizable iron chelators. Increasing concentrations of BP progressively reduced the growth rate and final cell density to a maximum effect at 200 μM; enterobactin at 100 μM emphatically inhibited growth of NA1000. BP or enterobactin retarded growth in as little as 4 h, and after 18 h the growth retardation by either compound reduced C. crescentus proliferation by ∼70% (Fig. 3). The addition of ferrichrome after 18 h restored growth to both cultures. In NB plus enterobactin, the addition of 10 μM ferrichrome decreased doubling times from 10 h to 4 h, confirming the specificity of the iron deprivation by the catecholate aposiderophore. The only exception to stimulation by ferrichrome was NB containing 200 μM BP, which did not recover. In this case, the high concentration of chelator blocked growth even when a utilizable ferric siderophore was present to provide iron, underscoring the potentially toxic effects of BP from less specific metal chelation effects.
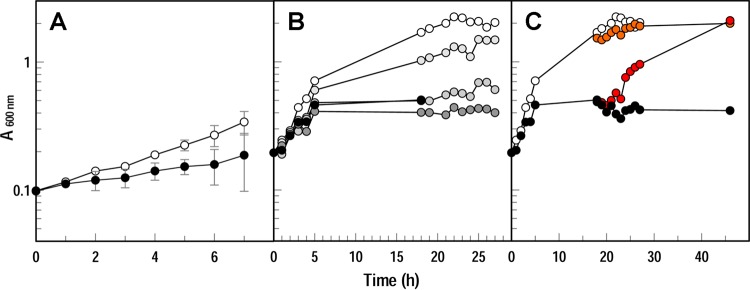
FIG 3.
Growth of C. crescentus NA1000 in iron-deficient media. (A) Growth in NB (○) or NB containing 200 μM BP (•). Data points are means from two experiments; bars show the standard deviations. (B) Concentration dependence of inhibition by BP. White circles show growth in NB; darkening shades of gray circles show the effects of 50, 100, and 200 μM BP, and black circles show the effects of 100 μM enterobactin, added at 5 h. (C) Restoration of growth upon addition of ferrichrome. We added ferrichrome (10 μM) to NA1000 growing in NB (○) or NB plus 100 μM enterobactin (•) (from panel B) at 19 h (orange and red circles), and it restored the original growth rates. We repeated experiments shown in panels B and C several times; the plots represent a single experiment.
Radioisotopic measurements of iron binding and transport.
In light of the predilection of C. crescentus for ferric hydroxamates, we characterized the binding and transport of [59Fe]apoferrichrome ([59Fe]Fc) by NA1000. It showed high-affinity recognition and uptake of [59Fe]Fc, similar to that of E. coli (53) or other enteric bacteria (34), with nanomolar Kd (equilibrium dissociation constant) and KM (Michaelis-Menten constant) values (Fig. 4). The transport rate for [59Fe]Fc (19 pmol/min/109 cells) was also comparable to that of chromosomally encoded ferrichrome uptake by E. coli (53, 54).
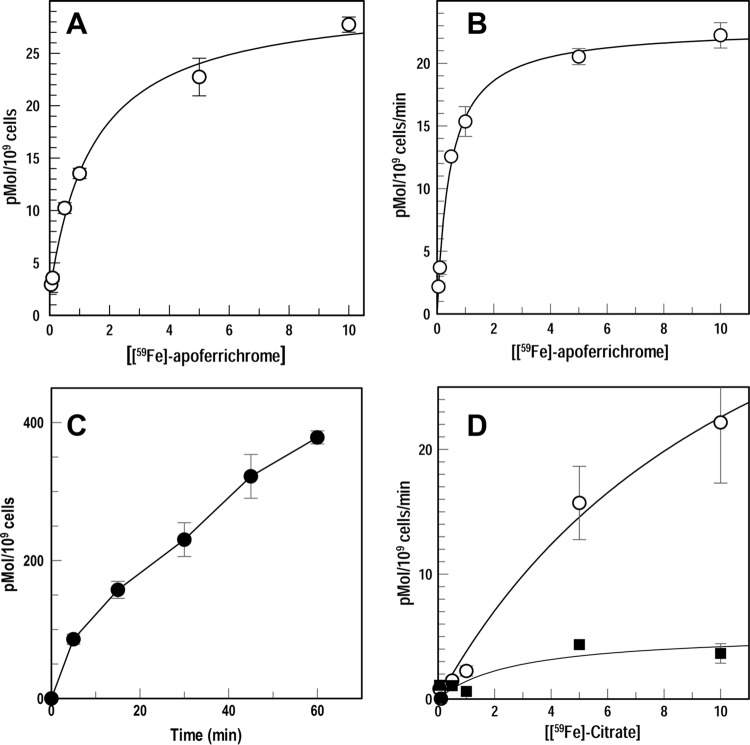
FIG 4.
Binding and uptake of [59Fe]Fc and [59Fe]Cit by C. crescentus. We grew NA1000 in NB at 30°C until mid-log phase, added enterobactin to 100 μM, and allowed the culture to shake for 3 h. (A and B) We measured binding (A) and uptake (B) of [59Fe]Fc by filtration assays (see Materials and Methods). From experiments performed in triplicate, we calculated the mean values and used Grafit 6.011 (Erathicus Ltd., West Sussex, United Kingdom) to determine Kd (1.4 nM), capacity (26 pMol/109 cells), KM (0.03 nM), and Vmax (19 pMol/109 cells/min) values. Error bars represent the standard deviations of the means from triplicate trials. The estimates of Kd and capacity using a nonlinear fit to the bound-versus-total equation of Grafit 6.011 gave standard errors of 11% and 3%, respectively; estimates of KM and Vmax, by nonlinear fit to the Michaelis-Menten kinetic model of Grafit 6.011, gave standard errors of 13% and 4%, respectively. (C and D) We also measured uptake of [59Fe]Cit. We grew NA1000 in NB at 30°C until mid-log phase, added enterobactin and citrate to 100 μM, and allowed the culture to shake for 3 h. (C) We exposed NA1000 to 1 μM [59Fe]Cit and measured accumulation of the radionuclide for 1 h. (D) Lastly, we measured the uptake of increasing concentrations of [59Fe]Cit by NA1000 in the absence (circles) or presence (squares) of 0.5 mM CCCP. Analysis of the NA1000 uptake data (D) by Grafit 6.011 produced KM (5.3 nM) and Vmax (29 pMol/109 cells/min) values. Error bars represent the standard deviations of the means from triplicate trials. Estimates of KM and Vmax by nonlinear fits of the Michaelis-Menten kinetic model, using Grafit 6.011, gave standard errors of 19% and 6%, respectively.
E. coli FecA contains an extended N-terminal domain that simultaneously upregulates fecA transcription in response to ferric citrate (FeCit) transport (55). The chromosome of C. crescentus NA1000 encodes four potential TBDT with N-terminal homology to E. coli FecA (14, 25), intimating the strain's potential for ferric citrate uptake. Siderophore nutrition tests are ineffective for FeCit, because both enterobactin and BP remove Fe3+ from its complexes with citrate. Consequently, we formed 59Fe-citrate ([59Fe]Cit) and characterized its uptake by NA1000 in filter binding assays. Biosynthesis of E. coli FecA is both repressed by Fur and induced by external FeCit (56). After growth in the presence of enterobactin and/or 100 μM citrate, C. crescentus transported [59Fe]Cit much like its uptake of [59Fe]Fc (KM = 5 nM; Vmax = 29 pmol/min/109 cells) (Fig. 4). The reaction was inhibited by 0.5 mM carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (Fig. 4), as expected for a TonB-dependent process.
Identification of potential iron transporters.
We used CLUSTALW2 alignments to analyze the TBDT in the C. crescentus genome (14, 25) with regard to their sequence relatedness. Using the 8 ferric siderophore and porphyrin receptors of E. coli as a basis of comparison to the 62 annotated TBDT of C. crescentus, we found 4 different phylogenetic branches (colored magenta, purple, green, and blue in Fig. S2 in the supplemental material). The full-length TBDT of E. coli, which are all metal transporters, subdivided into two branches, with the catecholate transporters FepA and Cir, the porphyrin transporter BtuB, and the ferric aerobactin transporter IutA in one branch (magenta) and the ferric hydroxamate transporters FhuA and FhuE, the ferric citrate transporter FecA, and the ferric catecholate transporter Fiu in another branch (blue). Similarly, alignment of their N domain sequences bifurcated the E. coli TBDT into the same two groups: FepA, Cir, BtuB, and IutA together, distinct from FhuA, FhuE, FecA, and Fiu. However, when we aligned the C domain sequences (β-barrels) of all of the proteins, the ferric siderophore receptors grouped together and the E. coli BtuB (EcoBtuB) β-barrel became an outlier. Finally, CLUSTALW2 alignment of the TonB box regions clustered all of the E. coli TBDT together and distinct from those of the C. crescentus TBDT.
The iron-regulated TBDT of C. crescentus (24) fell in two groups that showed higher expression during either iron deprivation or iron repletion. The first group (Fig. S2, black boxes) contained proteins 00028, 00138, 02277, and 03023; their Fur-like negative regulation (24) suggested that they function in iron acquisition. These proteins are related to the TonB-dependent OM proteins of E. coli, generally at a level of 20 to 25% identity. They all also contain a recognizable TonB box sequence near their N termini. TBDT Ccr02277 (Fig. S2, magenta branch) has the closest overall homology to E. coli ferric catecholate (Cir and FepA) and porphyrin (vitamin B12 and BtuB) receptors. The other three, 00028, 00138, and 03023, segregated in the blue branch, with the closest relationship to E. coli ferric hydroxamate receptors. These identities, consistent with ferric siderophore transporters, combined with their Fur-regulated, enhanced expression under iron-deficient conditions, suggested that proteins 00028, 00138, 02277, and 03023 are iron transporters in C. crescentus. The other group of C. crescentus TBDT (Fig. S3), which were overexpressed by high levels of extracellular iron (24), are typified by 00210 in the green branch. At present, their physiological functions are unknown.
Iron-regulated cell envelope proteins.
To identify the iron-regulated OM proteins of C. crescentus and the extent of their derepression by iron sufficiency or iron deficiency, we inoculated NA1000 in NB and added either FeSO4 (50 μM) or enterobactin (100 nM) when the culture entered exponential growth phase. Following growth for an additional 5 to 6 h at 30°C, we prepared the cell envelopes and extracted them with sodium sarcosinate (Sarkosyl). Differential extraction with 0.5% Sarkosyl usually separates IM and OM fractions of Gram-negative bacteria (45). However, the OM proteins of C. crescentus were more susceptible to solubilization (Fig. 5). This conclusion came from the facile solubilization of its TBDT, which are known OM proteins from their β-barrel architecture (63), by exposure to 0.5% Sarkosyl for 30 min at 25°C. Decreasing the temperature to 4°C and the time to 20 min improved the differential extraction of IM and OM, but reducing the Sarkosyl concentration to 0.1% resulted in a typical separation of inner and outer membranes (Fig. 5B) in which the bulk of TBDT remained in the insoluble fraction. Sarkosyl solubilizations also improved SDS-PAGE resolution of the protein components, and SDS-PAGE gels of Sarkosyl-solubilized and Sarkosyl-resistant fractions revealed numerous iron-regulated cell envelope proteins (Fig. 5) that illustrated the similar response of C. crescentus to that of other Gram-negative bacteria (34) during iron deprivation. We also performed isopycnic sucrose gradients in an attempt to separate the IM and OM of C. crescentus (58, 59), but despite apparent separation of the two fractions (Fig. S1A), SDS-PAGE of their component proteins did not reveal many differences (Fig. S1B), suggesting that the membranes did not genuinely separate from each other.
FIG 5.
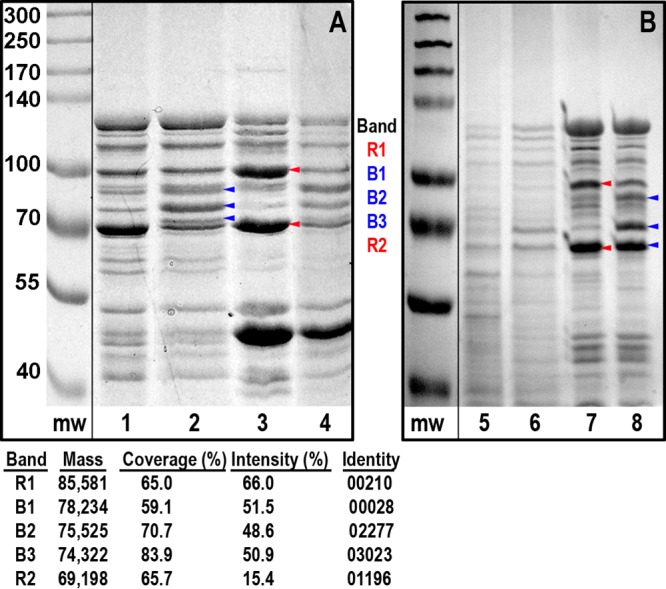
Iron-regulated cell envelope proteins of C. crescentus. Cell envelopes from NA1000 grown under iron-replete (odd lanes) or iron-deficient (even lanes) conditions (see Materials and Methods) were analyzed by SDS-PAGE. In both panels the molecular weight markers (mw) were from another portion of the same gel and juxtaposed to the lanes of interest. (A) Solubilization with 0.5% Sarkosyl. Cell envelope fractions were extracted with 0.5% Sarkosyl at 4°C for 20 min, and insoluble material was pelleted by centrifugation. We collected the supernatants (lanes 1 and 2) and resuspended the pellets (lanes 3 and 4) in distilled water, added SDS-PAGE sample buffer, analyzed the proteins (∼30 μg/lane) by SDS-PAGE, and stained them with Coomassie blue. After destaining, we excised the bands marked with arrowheads, which were overexpressed during iron deprivation (100 μM enterobactin; blue) or iron repletion (50 μM FeSO4; red), determined the masses of their tryptic peptides, and identified them from their correspondence with the predicted masses of TBDT in the C. crescentus genome. The tryptic peptides and noted proteins had 49 to 66% identity and 59 to 83% coverage of the mature sequences. The predicted masses of mature 00210, 00028, 02277, 03023, and O1196 were 85.6, 78.2, 75.5, 74.3, and 69.2 kDa, respectively, which agreed with their relative SDS-PAGE mobilities. (B) Solubilization with 0.1% Sarkosyl. Because of the tendency of TBDT to equally partition between the 0.5% Sarkosyl-soluble and -insoluble fractions (A), we conducted a series of extractions with reducing concentrations of the detergent. SDS-PAGE of the supernatants (lanes 5 and 6) and the pellets (lanes 7 and 8) from extractions with 0.1% Sarkosyl for 20 min at 4°C showed a more typical fractionation in which known OM proteins (e.g., TBDT) remained in the insoluble fraction.
To identify the proteins of interest, we excised bands from the gels, digested them with trypsin, and subjected the samples to matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometry. When matched with predicted tryptic degradation patterns of C. crescentus TBDT, the procedure discovered several of the iron-regulated proteins in the samples. The results reiterated the derepression of TBDT proteins 00028, 02277, and 03023 by iron deficiency (24). The predicted masses of the three mature proteins were 78.2, 75.5, and 74.3 kDa, respectively, which agreed with their mobility on SDS-PAGE gels (Fig. 5). We also identified two proteins that were overexpressed by iron repletion: 00210 and 01196. The former protein was previously identified as an iron-regulated TBDT (24); the latter was denoted as an iron-regulated outer membrane virulence protein (25) but was not previously recognized as a TBDT or known to be iron regulated (24). However, 01196 was overexpressed during growth of C. crescentus with 50 μM FeSO4 (Fig. 5), and its C domain has 18% identity to the β-barrel of E. coli FepA (EcoFepA) (see below), confirming its relationship to the TBDT family. The predicted masses of the mature 00210 and 01196 proteins, 85.6 and 69.2 kDa, concurred with their mobility in SDS-PAGE.
Ccr02277 (HutA) is a receptor for heme/hemoglobin.
The 25% identity between Ccr02277 and EcoCir indicates a comparable structural fold (60–62). In that sense, Ccr02277 is a typical TBDT (7, 32, 63), with only slightly less identity to EcoFepA (23%) and EcoBtuB (22%). The extent of Ccr02277 relatedness to these E. coli TBDT is higher than the overall identity among all E. coli TBDT (18.2%). These data, and its well-defined TonB box (DKVTVTAT), implied the TonB dependence of Ccr02277. We engineered a precise deletion of ccr02277 and characterized the resulting strain's iron acquisition properties. The mutant was unable to acquire hemin or hemoglobin in nutrition tests and, as expected, lacked the 75.5-kDa iron-regulated OM protein in Sarkosyl extracts of C. crescentus cell envelopes (Fig. 6). Consequently, we named the locus hutA, for its role in hemin/hemoglobin utilization.
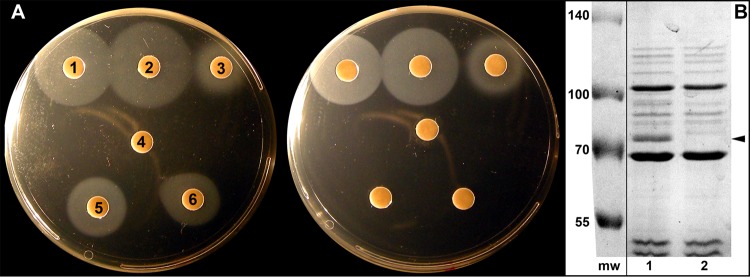
FIG 6.
Analysis of the ccrΔ02277 (ΔhutA) strain. (A) Siderophore nutrition tests were conducted, as described in the legend to Fig. 2, with C. crescentus NA1000 (left) and its Δ02277 derivative (right). Paper discs on the agar surface contained 10 μl of 50 μM solutions of the following: 1, ferrichrome; 2, ferrichrome A; 3, ferric aerobactin (200 μM); 4, ferrioxamine B (200 μM); 5, hemin; and 6, hemoglobin (15 μM). (B) SDS-PAGE of C. crescentus using 0.1% sodium sarcosinate-insoluble cell envelope (OM) fractions prepared as described for Fig. 5. The Coomassie blue-stained gels confirmed that relative to NA1000 (lane 1), the Δ02277 strain (lane 2) lacked a 75.5-kDa band (arrow), as expected. The molecular weight markers (mw) were from another portion of the same gel and juxtaposed to the lanes of interest.
DISCUSSION
Our findings demonstrate that C. crescentus NA1000, which does not elaborate siderophores, utilizes a variety of ferric hydroxamates and a mixed alpha-hydroxycarboxylate ferric chelate. This strategy of xenosiderophore piracy combats the low availability of iron in aquatic environments, where cyanobacteria of Anabaena spp., for example, produce both hydroxamate (64) and catecholate (65) siderophores. Like C. crescentus, blue-green algae obtain iron through TonB-dependent uptake systems (66, 67), so this overall mechanism of Gram-negative bacterial iron acquisition is common in aquatic milieu. On the other hand, the inability of NA1000 to utilize ferric enterobactin or any other ferric catecholates we tested distinguishes it from many free-living, commensal, and pathogenic Gram-negative species. Ninety-four percent of tested Gram-negative bacteria transport ferric enterobactin (34), the native catecholate siderophore of Enterobacteriaceae (1, 2), including strains from the genera Escherichia, Shigella, Salmonella, Klebsiella, Citrobacter, Serratia, Enterobacter, Edwardsiella, Hafnia, Yersinia, Providencia, Morganella, Proteus, Haemophilus, and Neisseria. C. crescentus likely encounters ferric catecholates in its natural environment, given that soil and surface water are also habitats for Klebsiella, Enterobacter, and Erwinia spp., all of which produce enterobactin (68). It is curious that C. crescentus does not take advantage of the presence of enterobactin. Instead, the apocatecholate that has the highest affinity for iron among natural or synthetic chelators is a potent inhibitor of C. crescentus growth.
Hemin/hemoglobin uptake, conferred by the newly discovered TBDT HutA, was a second noteworthy attribute of C. crescentus iron metabolism. This ability may pertain to the aquatic environment it inhabits, where iron or other metal porphyrins may be present in decaying biomatter. The utilization of iron from hemin/hemoglobin by the TonB-dependent OM protein HutA is the first demonstration of this ability for this protein, its cognate transporter, or that of any iron transporter in C. crescentus. However, other well-studied soil alphaproteobacteria, including the two nitrogen-fixing symbionts Sinorhizobium meliloti (69) and Bradyrhizobium japonicum (70), are known to use hemin. In the latter case heme uptake is not essential for nitrogen fixation, but it gives B. japonicum an alternative iron source in its nonsymbiotic state. Although little is known about heme utilization by freshwater bacteria, marine alphaproteobacteria from the Roseobacter clade also use it as an iron source (71).
Fur, the ferric uptake regulator, senses iron levels in bacterial cells and controls gene expression according to iron availability (72, 73): iron limitation triggers expression of Fur-regulated genes. Low iron stress of C. crescentus induces four TBDT to higher expression levels, as shown by global transcriptomic analyses (23, 24) and our studies. Transcription from 00028, 00138, 03023, and hutA (02277) is repressed by Fur when iron is abundant (23, 24). We observed and identified the latter protein in our experiments. As expected, addition of 100 μM enterobactin increased its synthesis and 50 μM FeSO4 decreased it. Its regulation concurs with the fact that HutA acquires iron as hemin/hemoglobin. We expect that other negatively regulated TBDT transport the ferric hydroxamates that conferred growth in nutrition tests, but this remains for further study. The biochemical functions of TBDT that are overexpressed during iron repletion are more difficult to rationalize. Expression of 00210 and 01196, for example, increased as iron became more readily available. 00210 is one of six positively iron-regulated proteins (23, 24) in the same branch of the cladogram of full-length proteins. Although we cannot yet postulate their function or transport substrates, the results suggest that they are functionally or physiologically related to cellular iron metabolism. 01196 was not previously implicated as a TBDT, but our studies identified it as one.
Caulobacters thrive in natural aquatic environments of low organic content, in part by producing an arsenal of enzymes that allows them to obtain nutrients from sources like the plant polysaccharides cellulose, xylan, lignin, glucan, and pectin (14). Their attachment to surfaces via the holdfast present at the tip of the stalk (74, 75), including the surfaces of other microrganisms, such as green and blue-green algae and diatoms, distinguishes the caulobacters from most other aquatic, chemoheterotrophic bacteria. This capacity potentially allows them to scavenge nutrients, including iron, from other organisms. C. crescentus may rely on TBDTs to perform most of its nutrient uptake, as suggested by the 62 TBDT structural genes in its genome (14). Besides transporting iron, its TBDTs act to accumulate maltose (26, 29) and potentially other sugars (27, 28). The localization of TBDT 02370, which transports maltodextrins (MalA) (26, 29), in the purple branch of the full-length TBDT cladogram suggests that the 18 other TBDTs in the same branch also act in sugar or solute uptake. Unlike the gut microenvironment of enteric bacteria, in natural aqueous surroundings carbon sources such as maltose likely do not reach high enough concentrations to traverse the OM by passive diffusion through general porins or even facilitated diffusion through specific porins (76). TBDTs solve this dilemma with sufficiently high affinity to absorb and transport ligands from solution. This function must be reconciled with the bioenergetics of TonB-dependent transport. FepA-mediated OM transport of ferric enterobactin requires expenditure of ∼4 ATPs per iron atom (77). However, aerobic bacterial catabolism of glucose yields as much as 38 ATPs/molecule, so TonB-dependent transport of di- or oligosaccharides like maltose or maltodextrin is potentially energetically affordable despite extra energetic cost at the OM stage. Second, like Vibrio spp. (78), Pseudomonas spp. (79–81), and Acinetobacter spp. (82) that encode 2, 3, and 3 TonB proteins, respectively, annotations of the C. crescentus genome (14) predict that it contains two TonB proteins (29). It is of interest to determine why multiple TonB proteins are needed in these organisms, whether they have specific partners among the multitude of TBDTs in the C. crescentus OM, and if so, how they achieve their specificity.
In summary, the experiments reported here revealed numerous attributes of C. crescentus iron acquisition. (i) It utilizes many ferric hydroxamates, some mixed ferric siderophores and hemin, but not ferric catecholates. In this sense it resembles the saprophytic bacterium and human pathogen Listeria monocytogenes, which likewise does not produce siderophores but utilizes ferric hydroxamates in wild environments and heme/hemoglobin in the host (83–86). (ii) Inability to transport ferric enterobactin makes (apo)enterobactin an effective agent for iron deprivation of C. crescentus, which will aid the study of its cell envelope iron acquisition biochemistry. (iii) The affinities and transport rates of C. crescentus ferric siderophore uptake systems match those of Enterobacteriaceae, indicating equivalent selectivity and discrimination of iron complexes. (iv) The 62 TBDT of C. crescentus fall into 4 bioinformatic groups or branches in their cladogram of sequence homology: two contain putative iron transporters, a third contains TBDT that are overexpressed in iron repletion, and the fourth appears specialized in uptake of other solutes. (v) Under iron-limiting conditions, C. crescentus overexpresses four OM TBDT that are closely structurally related to iron-specific TBDT of Enterobacteriaceae. (vi) Conversely, unlike enteric bacteria, the 62 TBDTs of C. crescentus include OM proteins that are overexpressed by iron repletion. (vii) The identities of five TBDT are now known: three were overexpressed in iron-deficient conditions (00028, 02277, and 03023), and 2 were overexpressed in iron-replete conditions (00210 and 01196). (viii) ccr02277 (which we designated hutA) encodes an OM TBDT for hemin/hemoglobin that is Fur regulated and derepressed by growth under low-iron conditions. Additional biochemical characterizations of these newly discovered iron-regulated transporters will allow us to understand each one's role in Caulobacter iron homeostasis.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
C. crescentus strain NA1000 (87) and its Δ02277 derivative, strain MM90, were grown at 30°C in PYE (36) or nutrient broth (NB) (88) medium containing 0.5 mM CaCl2. We supplemented media with nalidixic acid (20 μg/ml) or kanamycin (Km; 5 μg/ml for C. crescentus and 50 μg/ml for E. coli) as necessary. Escherichia coli strain S17-1 (89) was grown in Luria-Bertani medium containing tetracycline (12.5 μg/ml). Conjugation with S17-1 allowed introduction of plasmids into C. crescentus NA1000 (89). Solid medium contained 1.5% agar, and for siderophore nutrition tests we used 0.75% agar in NB. After culturing NA1000 in NB with 10 mM MgCl2 or in brain heart infusion (BHI) broth, we added a nonutilizable iron chelator at mid-log phase to deprive the bacteria of adventitious iron. We initially used α,α-bipyridyl (BP; 0.2 mM) to elicit low-iron stress but later switched to enterobactin, because the potent E. coli siderophore was not utilized by C. crescentus (see Results).
Bioinformatic analysis of C. crescentus TBDT.
We accessed information on the genome of C. crescentus NA1000 from the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg-bin/show_organism?org=ccs), which is searchable for chromosomal loci according to 5-digit identifiers that convey their relative positions on the genomic map. Here, we designate loci of interest according to this numerical convention. For sequence analyses and ensuing comparisons we used the complete protein sequences of C. crescentus putative TBDTs (14, 25), except locus 00779 (which encoded a protein only 133 residues in length). We initially aligned them using CLUSTALW2 (90), which identified their TonB box and N domain-C domain junction regions from homology to E. coli FepA (EcoFepA). We then analyzed sequence relationships among the proteins by separate CLUSTALW2 comparisons of their TonB boxes, N domains, C domains (β-barrel), and full-length mature proteins. We used SignalP 4.1 to predict the mature protein sequences (http://www.cbs.dtu.dk/services/SignalP/).
Construction of strain MM90 (Δ02277).
We generated a deletion of the gene encoding putative TBDT 02277 by amplifying its flanking regions from NA1000 genomic DNA with the oligonucleotide pair 2277-1F (AAAAGCTTCGACTGGATCTGGTAGACGC)/2277-2R (AAGGATCCCCCCTCTAAACACTAAAACATG) or 2277-3F (AAGGATCCCTGACGGACGAGAAATACTGG)/2277-4R (GCCGTACTTGCCATACCAGC) (restriction sites are underlined). The PCR mixtures contained 12.5 pmol oligonucleotide primers, 0.3 mM deoxynucleoside triphosphates (dNTPs) (Invitrogen), 5% dimethyl sulfoxide, and 0.625 U Pfu DNA polymerase (Fermentas) in enzyme reaction buffer. The PCR conditions were 95°C for 6 min, followed by 35 cycles at 95°C for 1 min, 55°C or 64°C for 30 s, and 72°C for 2 min. At completion we incubated the reaction mixtures at 72°C for an additional 7 min and stored them at 4°C. We individually cloned the products into pGEM T-Easy (Promega) and confirmed them by DNA sequencing. After digesting the plasmids with HindIII/BamHI (for the product of 2277-1 and 2277-2) or BamHI/EcoRI (for the product of 2277-3 and 2277-4, using an internal EcoRI site), we cloned the amplified regions in tandem into suicide vector pNPTS138 (a gift from D. Alley). pNPTS138 is a derivative of pNPTS129 (91), which contains a Km resistance marker. It cannot replicate in C. crescentus, so growth in the presence of Km selects for its insertion into the chromosome by homologous recombination mediated by the cloned fragments. Subsequent growth in the presence of sucrose selects for loss of the vector sequences by a second recombination event, producing clones that contained the desired deletion of the target gene. We confirmed the deletion that precisely removed the entire structural gene by DNA sequencing of PCR products from the chromosome.
Siderophores.
For preparation of catecholate ferric siderophores, a micromole of purified, crystalline aposiderophore (agrobactin, corynebactin, enterobactin, and vibriobactin) was dissolved in 0.5 ml of methanol and mixed with 0.5 ml of equimolar aqueous FeSO4, and after a few minutes we added NaHPO4, pH 6.9, to 50 μM. After an hour we purified the iron complexes by chromatography on Sephadex LH20 (35). For preparation of ferric hydroxamate siderophores, we dissolved the crystalline compounds in sterile distilled water to a stock concentration of 1 mM; for ferrichrome A, we neutralized the solution with 30 mM NaOH. We performed wavelength scans from 350 to 700 nm to determine the concentrations of the ferric siderophores and to verify their purity.
For [59Fe]Fc, we mixed 10 μl of 4.2 mM 59FeCl3 (NEN-037; Perkin-Elmer Corp.) with excess apoferrichrome (approximately 10 μl of a 10 mM solution) in 450 μl of 0.01 N HCl, added 50 μl of 0.5 M NaHPO4, pH 7, incubated the mixture for 1 h at room temperature, and chromatographed the solution on an anion exchange (DE52) column in 50 mM Tris-Cl, pH 7. Ferrichrome is uncharged, so [59Fe]Fc passed through the resin. We spectrophotometrically determined its concentration at 425 nM (εmM = 2.9). For [59Fe]Cit, we mixed a 50-fold excess of sodium citrate at pH 7 with 59FeCl3 and used it without further purification.
Siderophore nutrition assays (35).
After overnight growth of C. crescentus in NB, we diluted the culture to an optical density at 600 nm (OD600) of 0.1 in NB and allowed the cells to shake at 30°C until they grew to an OD600 of 0.3. For standard petri dishes, we plated 200 μl of the culture in 20 ml of NB top agar containing BP (200 μM) or enterobactin (100 μM). For 6-well plates, we added 100 μl of bacterial culture to 3 ml of NB top agar and poured the seeded agar into the individual wells. After the agar solidified, in standard petri plates we deposited 6 or more paper discs (6-mm diameter) on the surface; for 6-well plates, we placed a single filter paper disc in the center of each well. We then added 10 μl of a 50 μM solution of ferric siderophore to each disc (35). After incubation at 30°C for 24 h, we observed and measured bacterial growth halos around the paper disks.
Growth in liquid media.
Following overnight growth of C. crescentus in NB, we diluted the culture to an OD600 of 0.05 and continued shaking at 30°C. At an OD600 of 0.2 we divided the culture into five aliquots: three contained BP (50, 100, or 200 μM final concentrations), one had 100 μM enterobactin, and one remained as an unaltered, NB control. We measured growth by OD600 determinations at regular intervals. After 18 h, we split the cultures and added ferrichrome to half of the samples to a final concentration of 10 μM. The procedure created iron-deficient and iron-replete cultures; we monitored their growth rates by OD600 determinations at regular intervals.
59Fe-siderophore binding and transport measurements.
For [59Fe]Fc binding and transport determinations, we cultured C. crescentus in NB and rendered it iron deficient by the addition of enterobactin to 100 μM, followed by shaking for 3 h. For [59Fe]Fc adsorption measurements, we pelleted the cells by centrifugation, chilled them on ice for an hour, and measured their affinity (Kd) and capacity for [59Fe]Fc (specific activity of 25 to 100 cpm/pMol) by filter binding assays (92). For [59Fe]Fc uptake determinations, we directly used the cells in NB at 30°C and measured the affinity (KM) and rate (Vmax) of ferric siderophore transport (92).
In E. coli, FeCit uptake is negatively regulated by Fur and positively regulated by extracellular citrate (56). For measurement of 59Fe-citrate uptake by C. crescentus, we grew NA1000 in NB to mid-log phase, added both enterobactin and sodium citrate to 100 μM, and incubated the cultures for 3 h at 30°C with shaking. After washing twice with phosphate-buffered saline (PBS), we resuspended the cells in PBS plus 0.2% glucose at 30°C and incubated them with increasing concentrations of [59Fe]Cit (specific activity of 25 to 100 cpm/pMol) for 3 min, with the organic ligand in 50-fold molar excess. At the end of the incubation, we collected the cells by filtration on 0.45-μm HAWP filters (Millipore), washed the filters with 0.9% LiCl, and recorded radioactivity in a Packard Cobra gamma counter. We also performed uptake measurements in the presence of the proton ionophore CCCP after preincubating the cells with 0.5 mM CCCP for 15 min at 37°C before initiation of the 59Fe-citrate uptake assays.
For both [59Fe]Fc and [59Fe]Cit, we measured the concentration dependence of adsorption and transport by adding appropriate amounts of 59Fe complexes to two aliquots of 2 × 107 cells of NA1000 and incubating the aliquots for 5 s and 185 s, respectively, before collecting and washing the cells on 0.45-μm HAWP filters. The 5-s time point measured the amount initially bound to the cells, which, when subtracted from the second time point, gave the amount transported during a 3-min period. At each concentration we collected data in triplicate, averaged the values, and determined the KD and capacity of 59Fe complex binding with the “bound-versus-total” equation of Grafit 6.09 (Erithacus, Ltd., Middlesex, United Kingdom) or the KM and Vmax of transport with the “enzyme kinetics” equation.
Detergent extractions of cell envelope proteins.
Detergent solubilization facilitates the electrophoretic resolution of C. crescentus cell envelope proteins (57, 93–95); we utilized 0.1 to 0.5% sodium sarcosinate (Sarkosyl) to solubilize TBDT for mass spectrometry. After growth in NB containing either 50 μM FeSO4 (iron-replete conditions) or 100 μM enterobactin (iron-deficient conditions [see Results]), we pelleted the cells by centrifugation, resuspended them in ice-cold 50 mM Tris-Cl, pH 7.4, containing trace amounts of DNase and RNase, and lysed them by passage through a French press at 14,000 lb/in2. After a low-speed centrifugation to remove unbroken cells and debris, we spun the lysate for 1 h in a microcentrifuge at 13,000 rpm to pellet the cell envelopes, resuspended the pellets in PBS, and added Sarkosyl to 0.1 to 0.5% at 4°C for 20 min, followed by centrifugation for 1 h in a microcentrifuge at 13,000 rpm to separate Sarkosyl-soluble (supernatant) and Sarkosyl-insoluble (pellet) proteins (34). We also subjected C. crescentus cell envelopes to isopycnic sucrose gradients (45, 58) and differential extractions with Triton X-100 (41) to fractionate inner and outer membranes, but these approaches were less effective (see the supplemental material).
SDS-PAGE.
We analyzed the proteins in bacterial cell lysates or cell envelope fractions by SDS-PAGE (92, 96). Equal amounts of protein extracts were suspended in SDS sample buffer plus 3% β-mercaptoethanol, boiled for 5 min, and electrophoresed at room temperature, and the gels were stained with a solution of Coomassie blue.
Mass spectrometry.
We excised protein bands of interest from Coomassie blue-stained polyacrylamide gels, destained them in a 1:1 mix of 200 mM ammonium bicarbonate (NH4HCO3) and acetonitrile (ACN), denatured the proteins with 50 mM dithiothreitol, and carbamidomethylated them with 100 mM iodoacetamide. The bands were dehydrated in 100% ACN and digested with 100 ng of Trypsin Gold (Promega) in 50 mM NH4HCO3. After 18 h, we extracted the peptides from the gel with a 1:2 mix of 1% trifluoroacetic acid (TFA) in ACN, concentrated them in a speed vacuum, and applied them to a MALDI-TOF mass spectrometer (Bruker Ultraflex III) using 50 mg/ml 2,5-dihydroxybenzoic acid (DHB) matrix in 1:1 ACN and 0.1% TFA. The data were processed with the open-source software mMass (http://www.mmass.org), and the peak list was searched against the NCBI MASCOT database. Protein candidates were further analyzed by using BLASTP to match similar peptides to C. crescentus genomic data from KEGG strain NA1000 (25) (http://www.genome.jp/kegg-bin/show_organism?org=ccs) and UCSC strain CB15 (95) (http://archaea.ucsc.edu/cgi-bin/hgTracks?hgs,id=1974688). The matched proteins from strain NA1000 were in situ digested in mMass, and the MALDI data were matched to determine sequence and peptide intensity coverage, both of which were typically >50%.
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by National Science Foundation grant MCB0952299 and National Institutes of Health grants GM53836 and AI115187 (to P.E.K. and S.M.N.), as well as by Fundação de Amparo à Pesquisa do Estado de São Paulo-FAPESP grant 2014/04046-8 (to M.V.M.), doctoral fellowship grants 2009/52883-8 and 2013/06873-6 (to H.B.), and postdoctoral grant 2015/07386-7 (to L.L.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00723-16.
REFERENCES
- 1.Neilands JB. 1976. Siderophores: diverse roles in microbial and human physiology. Ciba Found Symp 51:107–124. [DOI] [PubMed] [Google Scholar]
- 2.Neilands JB. 1995. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem 270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 3.Neilands JB, Peterson T, Leong SA. 1980. High affinity iron transport in microorganisms, p 263–278. In Martell AE. (ed), Inorganic chemistry in biology and medicine, vol 140 ACS symposium series. ACS Publications, Washington, DC. [Google Scholar]
- 4.Neilands JB. 1981. Microbial iron compounds. Annu Rev Biochem 50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 5.Braun V, Hantke K. 2011. Recent insights into iron import by bacteria. Curr Opin Chem Biol 15:328–334. doi: 10.1016/j.cbpa.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saier MH., Jr 2000. Families of proteins forming transmembrane channels. J Membr Biol 175:165–180. doi: 10.1007/s00232001065. [DOI] [PubMed] [Google Scholar]
- 8.Schauer K, Rodionov DA, de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the “tip of the iceberg”? Trends Biochem Sci 6:6. [DOI] [PubMed] [Google Scholar]
- 9.Jordan LD, Zhou Y, Smallwood CR, Lill Y, Ritchie K, Yip WT, Newton SM, Klebba PE. 2013. Energy-dependent motion of TonB in the Gram-negative bacterial inner membrane. Proc Natl Acad Sci U S A 110:11553–11558. doi: 10.1073/pnas.1304243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klebba PE. 2016. ROSET model of TonB action in Gram-negative bacterial iron acquisition. J Bacteriol 198:1013–1021. doi: 10.1128/JB.00823-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui J, Davidson AL. 2011. ABC solute importers in bacteria. Essays Biochem 50:85–99. doi: 10.1042/bse0500085. [DOI] [PubMed] [Google Scholar]
- 12.Runyen-Janecky LJ. 2013. Role and regulation of heme iron acquisition in gram-negative pathogens. Front Cell Infect Microbiol 3:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laub MT, Shapiro L, McAdams HH. 2007. Systems biology of Caulobacter. Annu Rev Genet 41:429–441. doi: 10.1146/annurev.genet.41.110306.130346. [DOI] [PubMed] [Google Scholar]
- 14.Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen JA, Heidelberg JF, Alley MR, Ohta N, Maddock JR, Potocka I, Nelson WC, Newton A, Stephens C, Phadke ND, Ely B, DeBoy RT, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Kolonay JF, Smit J, Craven MB, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg SL, Venter JC, Shapiro L, Fraser CM. 2001. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci U S A 98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wayne R, Neilands JB. 1975. Evidence for common binding sites for ferrichrome compounds and bacteriophage phi 80 in the cell envelope of Escherichia coli. J Bacteriol 121:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hantke K. 1983. Identification of an iron uptake system specific for coprogen and rhodotorulic acid in Escherichia coli K12. Mol Gen Genet 191:301–306. doi: 10.1007/BF00334830. [DOI] [PubMed] [Google Scholar]
- 17.Hollifield WC Jr, Fiss EH, Neilands JB. 1978. Modification of a ferric enterobactin receptor protein from the outer membrane of Escherichia coli. Biochem Biophys Res Commun 83:739–746. doi: 10.1016/0006-291X(78)91051-3. [DOI] [PubMed] [Google Scholar]
- 18.Wookey P, Rosenberg H. 1978. Involvement of inner and outer membrane components in the transport of iron and in colicin B action in Escherichia coli. J Bacteriol 133:661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikaido H, Rosenberg EY. 1990. Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: study with beta-lactam antibiotics containing catechol and analogous groups. J Bacteriol 172:1361–1367. doi: 10.1128/jb.172.3.1361-1367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Lorenzo V, Bindereif A, Paw BH, Neilands JB. 1986. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J Bacteriol 165:570–578. doi: 10.1128/jb.165.2.570-578.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagegg W, Braun V. 1981. Ferric citrate transport in Escherichia coli requires outer membrane receptor protein fecA. J Bacteriol 145:156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassford PJ Jr, Kadner RJ. 1977. Genetic analysis of components involved in vitamin B12 uptake in Escherichia coli. J Bacteriol 132:796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.da Silva Neto JF, Braz VS, Italiani VC, Marques MV. 2009. Fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Res 37:4812–4825. doi: 10.1093/nar/gkp509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.da Silva Neto JF, Lourenco RF, Marques MV. 2013. Global transcriptional response of Caulobacter crescentus to iron availability. BMC Genomics 14:549. doi: 10.1186/1471-2164-14-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marks ME, Castro-Rojas CM, Teiling C, Du L, Kapatral V, Walunas TL, Crosson S. 2010. The genetic basis of laboratory adaptation in Caulobacter crescentus. J Bacteriol 192:3678–3688. doi: 10.1128/JB.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neugebauer H, Herrmann C, Kammer W, Schwarz G, Nordheim A, Braun V. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J Bacteriol 187:8300–8311. doi: 10.1128/JB.187.24.8300-8311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denance N, Vasse J, Lauber E, Arlat M. 2007. Plant carbohydrate scavenging through tonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisenbeis S, Lohmiller S, Valdebenito M, Leicht S, Braun V. 2008. NagA-dependent uptake of N-acetyl-glucosamine and N-acetyl-chitin oligosaccharides across the outer membrane of Caulobacter crescentus. J Bacteriol 190:5230–5238. doi: 10.1128/JB.00194-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmiller S, Hantke K, Patzer SI, Braun V. 2008. TonB-dependent maltose transport by Caulobacter crescentus. Microbiology 154:1748–1754. doi: 10.1099/mic.0.2008/017350-0. [DOI] [PubMed] [Google Scholar]
- 30.Cornelis P. 2010. Iron uptake and metabolism in pseudomonads. Appl Microbiol Biotechnol 86:1637–1645. doi: 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 31.Kuehl CJ, Crosa JH. 2010. The TonB energy transduction systems in Vibrio species. Future Microbiol 5:1403–1412. doi: 10.2217/fmb.10.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyckoff EE, Mey AR, Payne SM. 2007. Iron acquisition in Vibrio cholerae. Biometals 20:405–416. doi: 10.1007/s10534-006-9073-4. [DOI] [PubMed] [Google Scholar]
- 34.Rutz JM, Abdullah T, Singh SP, Kalve VI, Klebba PE. 1991. Evolution of the ferric enterobactin receptor in gram-negative bacteria. J Bacteriol 173:5964–5974. doi: 10.1128/jb.173.19.5964-5974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wayne R, Frick K, Neilands JB. 1976. Siderophore protection against colicins M, B, V, and Ia in Escherichia coli. J Bacteriol 126:7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol 204:372–384. doi: 10.1016/0076-6879(91)04019-K. [DOI] [PubMed] [Google Scholar]
- 37.Llinas M, Klein MP, Neilands JB. 1972. The solution conformation of the ferrichromes. II. Proton magnetic resonance of metal-free ferricrocin and ferrichrysin, conformational implications. Int J Pept Protein Res 4:157–166. [PubMed] [Google Scholar]
- 38.Emery T. 1980. Malonichrome, a new iron chelate from Fusarium roseum. Biochim Biophys Acta 629:382–390. doi: 10.1016/0304-4165(80)90110-5. [DOI] [PubMed] [Google Scholar]
- 39.Wiebe C, Winkelmann G. 1975. Kinetic studies on the specificity of chelate-iron uptake in Aspergillus. J Bacteriol 123:837–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atkin CL, Neilands JB. 1968. Rhodotorulic acid, a diketopiperazine dihydroxamic acid with growth-factor activity. I. Isolation and characterization. Biochemistry 7:3734–3739. [DOI] [PubMed] [Google Scholar]
- 41.Schnaitman CA. 1973. Outer membrane proteins of Escherichia coli. I. Effect of preparative conditions on the migration of protein in polyacrylamide gels. Arch Biochem Biophys 157:541–552. [DOI] [PubMed] [Google Scholar]
- 42.Griffiths GL, Sigel SP, Payne SM, Neilands JB. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem 259:383–385. [PubMed] [Google Scholar]
- 43.Budzikiewicz H, Bössenkamp A, Taraz K, Pandey A, Meyer JM. 1997. Corynebactin, a cyclic catecholate siderophore from Corynebacterium glutamicum ATCC14067 (Brevibacterium sp. DSM 20411). Z Naturforsch C 52:551–554. [Google Scholar]
- 44.May JJ, Wendrich TM, Marahiel MA. 2001. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J Biol Chem 276:7209–7217. doi: 10.1074/jbc.M009140200. [DOI] [PubMed] [Google Scholar]
- 45.Smit J, Kamio Y, Nikaido H. 1975. Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studies with lipopolysaccharide mutants. J Bacteriol 124:942–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson F, Magrath DI. 1969. The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-I. Biochim Biophys Acta 192:175–184. doi: 10.1016/0304-4165(69)90353-5. [DOI] [PubMed] [Google Scholar]
- 47.Mullis KB, Pollack JR, Neilands JB. 1971. Structure of schizokinen, an iron-transport compound from Bacillus megaterium. Biochemistry 10:4894–4898. doi: 10.1021/bi00802a010. [DOI] [PubMed] [Google Scholar]
- 48.Snow GA. 1965. The structure of mycobactin P, a growth factor for Mycobacterium johnei, and the significance of its iron complex. Biochem J 94:160–165. doi: 10.1042/bj0940160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barclay R, Ewing DF, Ratledge C. 1985. Isolation, identification, and structural analysis of the mycobactins of Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium scrofulaceum, and Mycobacterium paratuberculosis. J Bacteriol 164:896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teintze M, Hossain MB, Barnes CL, Leong J, van der Helm D. 1981. Structure of ferric pseudobactin, a siderophore from a plant growth promoting Pseudomonas. Biochemistry 20:6446–6457. doi: 10.1021/bi00525a025. [DOI] [PubMed] [Google Scholar]
- 51.Jalal MA, Mocharla R, Barnes CL, Hossain MB, Powell DR, Eng-Wilmot DL, Grayson SL, Benson BA, van der Helm D. 1984. Extracellular siderophores from Aspergillus ochraceous. J Bacteriol 158:683–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jalal MA, Love SK, van der Helm D. 1986. Siderophore mediated iron(III) uptake in Gliocladium virens. 1. Properties of cis-fusarinine, trans-fusarinine, dimerum acid, and their ferric complexes. J Inorg Biochem 28:417–430. [DOI] [PubMed] [Google Scholar]
- 53.Scott DC, Cao Z, Qi Z, Bauler M, Igo JD, Newton SM, Klebba PE. 2001. Exchangeability of N termini in the ligand-gated porins of Escherichia coli. J Biol Chem 276:13025–13033. doi: 10.1074/jbc.M011282200. [DOI] [PubMed] [Google Scholar]
- 54.Thulasiraman P, Newton SM, Xu J, Raymond KN, Mai C, Hall A, Montague MA, Klebba PE. 1998. Selectivity of ferric enterobactin binding and cooperativity of transport in gram-negative bacteria. J Bacteriol 180:6689–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Angerer A, Enz S, Ochs M, Braun V. 1995. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. Fecl belongs to a new subfamily of sigma 70-type factors that respond to extracytoplasmic stimuli. Mol Microbiol 18:163–174. [DOI] [PubMed] [Google Scholar]
- 56.Hantke K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet 182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 57.Phadke ND, Molloy MP, Steinhoff SA, Ulintz PJ, Andrews PC, Maddock JR. 2001. Analysis of the outer membrane proteome of Caulobacter crescentus by two-dimensional electrophoresis and mass spectrometry. Proteomics 1:705–720. doi:. [DOI] [PubMed] [Google Scholar]
- 58.Clancy MJ, Newton A. 1982. Localization of proteins in the inner and outer membranes of Caulobacter crescentus. Biochim Biophys Acta 686:160–169. doi: 10.1016/0005-2736(82)90108-0. [DOI] [PubMed] [Google Scholar]
- 59.Ryan KR, Taylor JA, Bowers LM. 2010. The BAM complex subunit BamE (SmpA) is required for membrane integrity, stalk growth and normal levels of outer membrane β-barrel proteins in Caulobacter crescentus. Microbiology 156:742–756. doi: 10.1099/mic.0.035055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ginalski K. 2006. Comparative modeling for protein structure prediction. Curr Opin Struct Biol 16:172–177. doi: 10.1016/j.sbi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 61.Kryshtafovych A, Venclovas C, Fidelis K, Moult J. 2005. Progress over the first decade of CASP experiments. Proteins 61(Suppl 7):S225–S236. [DOI] [PubMed] [Google Scholar]
- 62.Stokes-Rees I, Sliz P. 2010. Protein structure determination by exhaustive search of Protein Data Bank derived databases. Proc Natl Acad Sci U S A 107:21476–21481. doi: 10.1073/pnas.1012095107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buchanan SK, Lukacik P, Grizot S, Ghirlando R, Ali MM, Barnard TJ, Jakes KS, Kienker PK, Esser L. 2007. Structure of colicin I receptor bound to the R-domain of colicin Ia: implications for protein import. EMBO J 26:2594–2604. doi: 10.1038/sj.emboj.7601693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goldman SJ, Lammers PJ, Berman MS, Sanders-Loehr J. 1983. Siderophore-mediated iron uptake in different strains of Anabaena sp. J Bacteriol 156:1144–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beiderbeck H, Taraz K, Budzikiewicz H, Walsby AE. 2000. Anachelin, the siderophore of the cyanobacterium Anabaena cylindrica CCAP 1403/2A. Z Naturforsch C 55:681–687. [DOI] [PubMed] [Google Scholar]
- 66.Mirus O, Strauss S, Nicolaisen K, von Haeseler A, Schleiff E. 2009. TonB-dependent transporters and their occurrence in cyanobacteria. BMC Biol 7:68. doi: 10.1186/1741-7007-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rudolf M, Stevanovic M, Kranzler C, Pernil R, Keren N, Schleiff E. 2016. Multiplicity and specificity of siderophore uptake in the cyanobacterium Anabaena sp. PCC 7120. Plant Mol Biol 92:57–69. [DOI] [PubMed] [Google Scholar]
- 68.Hider RC, Kong X. 2010. Chemistry and biology of siderophores. Nat Prod Rep 27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 69.Battistoni F, Platero R, Duran R, Cervenansky C, Battistoni J, Arias A, Fabiano E. 2002. Identification of an iron-regulated, hemin-binding outer membrane protein in Sinorhizobium meliloti. Appl Environ Microbiol 68:5877–5881. doi: 10.1128/AEM.68.12.5877-5881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nienaber A, Hennecke H, Fischer HM. 2001. Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol Microbiol 41:787–800. [DOI] [PubMed] [Google Scholar]
- 71.Roe KL, Hogle SL, Barbeau KA. 2013. Utilization of heme as an iron source by marine Alphaproteobacteria in the Roseobacter clade. Appl Environ Microbiol 79:5753–5762. doi: 10.1128/AEM.01562-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Escolar L, Perez-Martin J, de Lorenzo V. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol 181:6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 74.Hardy GG, Allen RC, Toh E, Long M, Brown PJ, Cole-Tobian JL, Brun YV. 2010. A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol 76:409–427. doi: 10.1111/j.1365-2958.2010.07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ong CJ, Wong ML, Smit J. 1990. Attachment of the adhesive holdfast organelle to the cellular stalk of Caulobacter crescentus. J Bacteriol 172:1448–1456. doi: 10.1128/jb.172.3.1448-1456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klebba PE, Newton SM. 1998. Mechanisms of solute transport through outer membrane porins: burning down the house. Curr Opin Microbiol 1:238–247. doi: 10.1016/S1369-5274(98)80017-9. [DOI] [PubMed] [Google Scholar]
- 77.Newton SM, Trinh V, Pi H, Klebba PE. 2010. Direct measurements of the outer membrane stage of ferric enterobactin transport: postuptake binding. J Biol Chem 285:17488–17497. doi: 10.1074/jbc.M109.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Occhino DA, Wyckoff EE, Henderson DP, Wrona TJ, Payne SM. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol Microbiol 29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 79.Huang B, Ru K, Yuan Z, Whitchurch CB, Mattick JS. 2004. tonB3 is required for normal twitching motility and extracellular assembly of type IV pili. J Bacteriol 186:4387–4389. doi: 10.1128/JB.186.13.4387-4389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Poole K, Zhao Q, Neshat S, Heinrichs DE, Dean CR. 1996. The Pseudomonas aeruginosa tonB gene encodes a novel TonB protein. Microbiology 142:1449–1458. doi: 10.1099/13500872-142-6-1449. [DOI] [PubMed] [Google Scholar]
- 81.Zhao Q, Poole K. 2000. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol Lett 184:127–132. doi: 10.1111/j.1574-6968.2000.tb09002.x. [DOI] [PubMed] [Google Scholar]
- 82.Beddek AJ, Sheehan BJ, Bosse JT, Rycroft AN, Kroll JS, Langford PR. 2004. Two TonB systems in Actinobacillus pleuropneumoniae: their roles in iron acquisition and virulence. Infect Immun 72:701–708. doi: 10.1128/IAI.72.2.701-708.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin B, Newton SM, Shao Y, Jiang X, Charbit A, Klebba PE. 2005. Iron acquisition systems for ferric hydroxamates, haemin and haemoglobin in Listeria monocytogenes. Mol Microbiol 59:1185–1198. [DOI] [PubMed] [Google Scholar]
- 84.Klebba PE, Charbit A, Xiao Q, Jiang X, Newton SM. 2012. Mechanisms of iron and haem transport by Listeria monocytogenes. Mol Membr Biol 29:69–86. doi: 10.3109/09687688.2012.694485. [DOI] [PubMed] [Google Scholar]
- 85.Newton SM, Klebba PE, Raynaud C, Shao Y, Jiang X, Dubail I, Archer C, Frehel C, Charbit A. 2005. The svpA-srtB locus of Listeria monocytogenes: Fur-mediated iron regulation and effect on virulence. Mol Microbiol 55:927–940. [DOI] [PubMed] [Google Scholar]
- 86.Xiao Q, Jiang X, Moore KJ, Shao Y, Pi H, Dubail I, Charbit A, Newton SM, Klebba PE. 2011. Sortase independent and dependent systems for acquisition of haem and haemoglobin in Listeria monocytogenes. Mol Microbiol 80:1581–1597. doi: 10.1111/j.1365-2958.2011.07667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Evinger M, Agabian N. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol 132:294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 89.Simon R, O'Connell M, Labes M, Puhler A. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol 118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 90.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 91.Tsai JW, Alley MR. 2000. Proteolysis of the McpA chemoreceptor does not require the Caulobacter major chemotaxis operon. J Bacteriol 182:504–507. doi: 10.1128/JB.182.2.504-507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Newton SM, Igo JD, Scott DC, Klebba PE. 1999. Effect of loop deletions on the binding and transport of ferric enterobactin by FepA. Mol Microbiol 32:1153–1165. doi: 10.1046/j.1365-2958.1999.01424.x. [DOI] [PubMed] [Google Scholar]
- 93.Cao Y, Bazemore-Walker CR. 2014. Proteomic profiling of the surface-exposed cell envelope proteins of Caulobacter crescentus. J Proteomics 97:187–194. doi: 10.1016/j.jprot.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 94.Cao Y, Johnson HM, Bazemore-Walker CR. 2012. Improved enrichment and proteomic identification of outer membrane proteins from a Gram-negative bacterium: focus on Caulobacter crescentus. Proteomics 12:251–262. doi: 10.1002/pmic.201100288. [DOI] [PubMed] [Google Scholar]
- 95.Molloy MP, Phadke ND, Chen H, Tyldesley R, Garfin DE, Maddock JR, Andrews PC. 2002. Profiling the alkaline membrane proteome of Caulobacter crescentus with two-dimensional electrophoresis and mass spectrometry. Proteomics 2:899–910. doi:. [DOI] [PubMed] [Google Scholar]
- 96.Ames GF, Spudich EN, Nikaido H. 1974. Protein composition of the outer membrane of Salmonella typhimurium: effect of lipopolysaccharide mutations. J Bacteriol 117:406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ong SA, Peterson T, Neilands JB. 1979. Agrobactin, a siderophore from Agrobacterium tumefaciens. J Biol Chem 254:1860–1865. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.