ABSTRACT
Transcription of the tryptophan (trp) operon in Bacillus subtilis is regulated by an attenuation mechanism. Attenuation is controlled by the trp RNA-binding attenuation protein (TRAP). TRAP binds to a site in the 5′ leader region of the nascent trp transcript in response to the presence of excess intracellular tryptophan. This binding induces transcription termination upstream of the structural genes of the operon. In prior attenuation models, the role of TRAP was only to alter the secondary structure of the leader region RNA so as to promote formation of the trp attenuator, which was presumed to function as an intrinsic terminator. However, formation of the attenuator alone has been shown to be insufficient to induce efficient termination, indicating that TRAP plays an additional role in this process. To further examine the function of TRAP, we performed a genetic selection for mutant TRAPs that bind tryptophan and RNA but show diminished termination at the trp attenuator. Five such TRAP mutants were obtained. Four of these have substitutions at Glu60, three of which are Lys (E60K) substitutions and the fourth of which is a Val (E60V) substitution. The fifth mutant obtained contains a substitution at Ile63, which is on the same β-strand of TRAP as Glu60. Purified E60K TRAP binds tryptophan and RNA with properties similar to those of the wild type but is defective at inducing termination at the trp attenuator in vitro.
IMPORTANCE Prior models for attenuation control of the B. subtilis trp operon suggested that the only role for TRAP is to bind to the leader region RNA and alter its folding to induce formation of an intrinsic terminator. However, several recent studies suggested that TRAP plays an additional role in the termination mechanism. We hypothesized that this function could involve residues in TRAP other than those required to bind tryptophan and RNA. Here we obtained TRAP mutants with alterations at Glu60 that are deficient at inducing termination in the leader region while maintaining tryptophan and RNA binding properties similar to those of the WT protein. These studies provide additional evidence that TRAP-mediated transcription termination at the trp attenuator is neither intrinsic nor Rho dependent.
KEYWORDS: RNA binding proteins, termination, transcription, tryptophan operon
INTRODUCTION
Expression of the Bacillus subtilis trpEDCFBA operon, which contains six of the seven genes required for tryptophan biosynthesis, is regulated by the trp RNA-binding attenuation protein (TRAP) (1, 2). TRAP is composed of 11 identical 75-amino-acid subunits, each encoded by the mtrB gene (3), which assemble into a symmetric ring complex (4). TRAP is activated to bind RNA by binding up to 11 tryptophan molecules in hydrophobic pockets between adjacent subunits (5). The TRAP binding site in the 203-nucleotide (nt) 5′ leader region of the trp transcript, upstream of trpE, consists of 11 (G/U)AG repeats. TRAP binds to this target by wrapping the RNA around the perimeter of the protein ring (4, 6, 7).
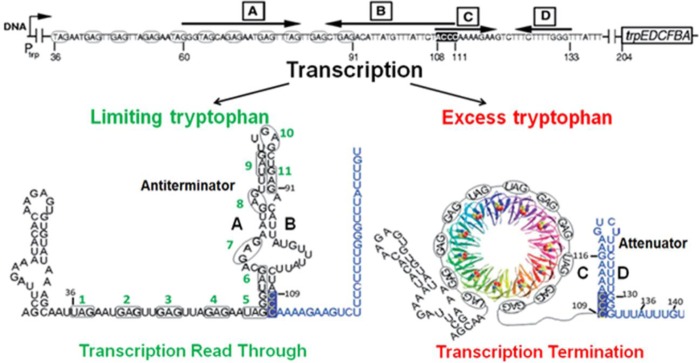
Transcription of the trp operon was originally proposed to be regulated solely by two competing RNA secondary structures, termed the antiterminator and the terminator (attenuator), which form in the 5′ leader region (8, 9). In this model, when intracellular tryptophan levels are in excess, TRAP binding to the trp leader RNA prevents formation of the antiterminator structure, which promotes formation of the attenuator. The attenuator was presumed to function as an intrinsic terminator to halt transcription within the trp leader region and thus prevent expression of the operon (Fig. 1). When tryptophan is limiting, TRAP does not bind RNA and the alternative antiterminator structure forms, allowing transcription to read through into the trp biosynthetic genes (10). The only role of TRAP in this model is to alter the secondary structure of the leader RNA so as to promote formation of the attenuator (8, 11). However, Potter et al. found that formation of the attenuator alone is not sufficient to cause efficient termination but requires TRAP bound to the nascent transcript to do so (12). More recently, we also found that TRAP can induce transcription termination in the trp leader region when the attenuator is mutated or deleted (13). Together these observations suggest that TRAP plays an additional role in attenuation beyond changing the folding pattern of the trp RNA (12).
FIG 1.
Model of transcription attenuation of the B. subtilis trp operon. Bold black letters designate the complementary strands of the attenuator (C and D) (highlighted in blue) and antiterminator (A and B) RNA structures. TRAP is shown as a ribbon diagram, with each subunit in a different color. The 11 (G/U)AG repeats of the TRAP binding site are circled and numbered in green. Small black numbers indicate RNA residues relative to the start of transcription. When tryptophan is limiting, the AB antiterminator RNA structure forms, allowing readthrough of the trp operon. With excess tryptophan, TRAP binds to the nascent RNA and prevents formation of the antiterminator structure, which allows formation of the attenuator, leading to transcription termination.
The additional role of TRAP in the termination mechanism may involve interactions with RNA polymerase (RNAP) or other transcription factors. These interactions may involve specific contact with TRAP residues other than those that interact with tryptophan and RNA. Identifying such residues in TRAP would provide additional evidence for this novel function. Hence, we performed a genetic selection/screen for TRAP mutants that are deficient at inducing termination in the trp leader region but retain the ability to bind tryptophan and RNA. Five TRAP mutants with these properties were obtained. Four of these have substitutions at Glu60, while the fifth has a change at Ile63, which is on the same β-strand as residue 60. Glu60 is located on the side of the TRAP ring opposite the location where tryptophan and RNA bind. Consistent with this location, E60K TRAP binds tryptophan and RNA with properties similar to those of wild-type (WT) TRAP but shows a diminished ability to induce termination at the trp attenuator in vitro. In addition, E60K TRAP shows decreased association with the B. subtilis transcription elongation complex (TEC) in vitro. Together these findings suggest that Glu60 plays a role in TRAP-mediated transcription termination other than binding RNA.
RESULTS
Selection for termination-deficient TRAP mutants yields substitutions at Glu60.
The observation that TRAP can induce transcription termination in mutant trp leader regions with altered or deleted attenuator segments suggests that TRAP plays an active role in transcription termination of B. subtilis RNAP beyond altering the RNA secondary structure in the leader region (13). This additional function may require residues on TRAP other than those involved in binding tryptophan and RNA. We therefore performed a genetic selection/screen for TRAP mutants that are defective at inducing transcription termination at the trp attenuator but retain the ability to bind tryptophan and RNA similarly to WT TRAP.
In B. subtilis NM4909, transcription of the chloramphenicol acetyltransferase (cat) gene is under the control of the native trp promoter and regulatory region (Fig. 2). Since this strain carries a deletion of mtrB, which encodes TRAP, the cat gene is constitutively expressed, yielding resistance to chloramphenicol (CM). However, when WT TRAP is expressed from pHYp59mtrB, NM4909 is resistant to CM in the absence of tryptophan but CM sensitive when grown in the presence of tryptophan due to TRAP-mediated termination in the trp leader region. Strains containing plasmids expressing TRAP mutants that allow transcription to read through the attenuator in the presence of tryptophan were selected as those resistant to both tetracycline (pHYp59mtrB) and CM.
FIG 2.
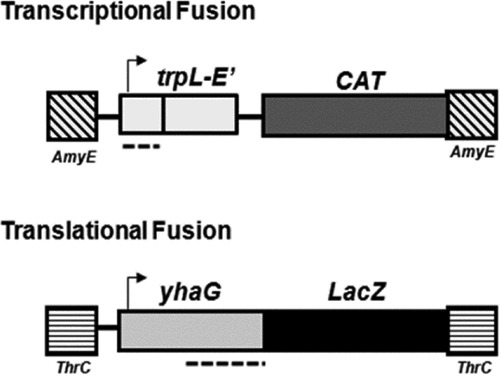
Gene fusions integrated into the genome of B. subtilis strain NM4909 for selection and screening of TRAP mutants are shown. TRAP binding sites are indicated by dotted lines. The diagram is not drawn to scale. (Top) Transcriptional fusion between the trp leader promoter and regulatory region and a portion of trpE and the chloramphenicol acetyltransferase gene (cat), integrated into the genome at the amyE locus. This fusion assesses the ability of TRAP mutants to regulate transcription. (Bottom) Translational fusion between the yhaG leader region and lacZ, integrated at the thrC locus. This fusion tests the ability of TRAP mutants to bind to a site in the RNA that overlaps the ribosome binding site and the start codon of yhaG, which downregulates translation of the lacZ fusion.
Strains expressing TRAP mutants defective in tryptophan and/or RNA binding activity also allow transcription through the trp leader region, resulting in CM resistance. However, these mutants are unlikely to contain substitutions in residues that are directly involved in inducing RNAP to terminate. To identify and eliminate TRAP mutants with defects in tryptophan and/or RNA binding, we screened the CM-resistant clones for the ability to downregulate translation of a trpP-lacZ translational fusion in response to tryptophan in B. subtilis NM4909 (Fig. 2). This fusion contains a TRAP binding site overlapping the translational start site of trpP (14). Thus, when NM4909 is grown in the presence of tryptophan, TRAP binding to trpP mRNA inhibits translation initiation of lacZ (14, 15), resulting in white colonies on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Mutations that have a negative impact on the tryptophan and/or RNA binding properties of TRAP allow translation of trpP, yielding blue colonies in the presence of tryptophan and X-Gal. Therefore, NM4909 strains containing TRAP mutants that bind RNA and tryptophan but do not terminate transcription efficiently were identified as CM resistant and white on medium containing CM, X-Gal, and excess tryptophan.
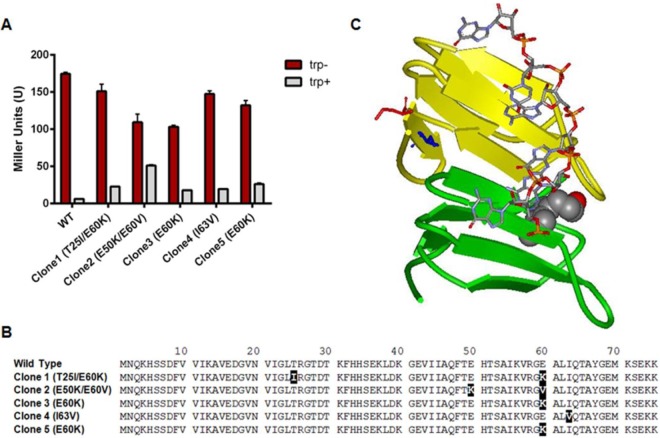
Transforming B. subtilis NM4909 with a pool of approximately 5 × 104 pHYp59 plasmids containing mutant mtrB genes yielded approximately 1 × 103 CM-resistant colonies. The relatively large fraction (∼1/50) of randomly mutated mtrB genes that result in CM resistance is consistent with the presence of only 73 amino acids in TRAP, of which 4 and 9 have been shown to be essential for binding RNA and tryptophan, respectively (5). Thirty-four of the CM-resistant colonies appeared light blue to white on X-Gal. To further assess the RNA binding properties of TRAP in these 34 candidate strains, we assayed β-galactosidase expression from the trpP-lacZ translational fusion. For comparison, WT TRAP expressed from pHYp59mtrB in NM4909 resulted in approximately 175 U and 10 U of β-galactosidase activity when grown in the absence and presence of tryptophan, respectively (Fig. 3A). Twenty-nine of the 34 CM-resistant clones selected based on their appearance on X-Gal yielded more than 100 U of β-galactosidase when grown in the presence of tryptophan, suggesting that TRAP was defective at binding tryptophan and/or RNA in these cells. These clones were not pursued further. Four of the remaining CM-resistant clones (clones 1, 3, 4, and 5) yielded less than 25 U of β-galactosidase, indicating that in these strains, TRAP is capable of downregulating translation of the trpP-lacZ fusion at least 8-fold in response to tryptophan (Fig. 3A). Clone 2 showed intermediate regulation of the fusion, yielding approximately 50 U of β-galactosidase activity in the presence of tryptophan.
FIG 3.
TRAP mutants from genetic selection/screen. (A) Bar graph showing β-galactosidase activities, in Miller units, expressed from the yhaG-lacZ fusion in the absence (trp−) or presence (trp+) of exogenous tryptophan (50 μg/ml) for 5 CM-resistant clones obtained from B. subtilis NM4909 containing plasmids that express mutant mtrB genes. (B) Amino acid sequences of the wild-type and mutant mtrB genes obtained from the genetic selection/screen, with substitutions highlighted in black boxes. (C) Ribbon diagram of two adjacent TRAP subunits, shown in green and yellow. Tryptophan molecules are displayed as van der Waals spheres, and RNA is shown using stick models in CPK colors. Glu60 and Ile63 are shown as red and blue stick models, respectively.
The results of sequencing of the mtrB genes from the plasmids isolated from these 5 clones are shown in Fig. 3B. Three of the four clones (clones 1, 3, and 5) that showed the greatest ability to downregulate the trpP-lacZ fusion in response to tryptophan contained mutations of Glu60 to Lys (E60K). Two of these (clones 3 and 5) had the single E60K mutation, whereas clone 1 was a double mutant with a T25I change in addition to the E60K change. Clone 2, which showed intermediate regulation of trpP-lacZ, was also a double mutant, with mutation of Glu50 to Lys as well as Glu60 to Val. Clone 4 contained a mutation of Ile63 to Val. Since three of the four clones with the greatest ability to regulate the trpP-lacZ fusion (suggesting that TRAP bound RNA efficiently in these cells) contained changes at Glu60, including two with a single change to Lys (clones 3 and 5), we focused our attention on this residue as potentially playing a specific role in TRAP-mediated transcription termination other than altering RNA folding.
Charge alterations at residue 60 of TRAP inhibit transcription termination without affecting RNA and tryptophan binding.
Selection for CM resistance in NM4909 is based on impaired TRAP-mediated transcription termination in the regulatory trp leader region of the trpE′-′cat fusion. To quantify this effect, we transformed pHYp59 plasmids expressing WT, E60K, E60R, and E60D TRAP into B. subtilis NM421. This strain contains a transcriptional fusion of lacZ under the control of the trp promoter and regulatory region; thus, β-galactosidase expression reflects transcriptional readthrough of the trp attenuator. The native mtrB gene is deleted in NM421, and approximately 300 U of β-galactosidase activity was expressed from this fusion in the absence or presence of tryptophan (Table 1). When WT TRAP was expressed from pHYp59mtrB in NM421, β-galactosidase expression was reduced 45-fold, to 7 U, when the cells were grown in the presence of tryptophan (Table 1). Changing Glu60 to either Lys (E60K) or Arg (E60R) impaired the ability of TRAP to induce transcription termination in the trp leader region approximately 3-fold, such that ∼20 U (compared to 7 U) of β-galactosidase was produced in the presence of tryptophan (Table 1). Replacing Glu60 with another acidic residue (E60D) did not alter the ability of TRAP to regulate transcription of this fusion in vivo (Table 1). These results show that an acidic side chain at residue 60 of TRAP is needed to fully induce transcription termination of B. subtilis RNAP at the trp attenuator in vivo.
TABLE 1.
Regulation of trp leader transcription in vivo
| TRAP | Trpa | β-Galactosidase activity (U)b | Fold regulationc |
|---|---|---|---|
| No pHYp59mtrB | − | 312 ± 56 | |
| + | 286 ± 33 | 1.1 | |
| WT | + | 7 ± 1 | 45 |
| E60K mutant | + | 23 ± 2 | 14 |
| E60R mutant | + | 19 ± 1 | 16 |
| E60D mutant | + | 7 ± 1 | 45 |
−, absence of exogenous l-tryptophan; +, presence of 50 μg/ml l-tryptophan.
Values are the averages ± standard deviations for two or three independent experiments, each performed in triplicate.
Fold regulation of TRAP in response to tryptophan-activated TRAP, calculated by dividing the value for no pHYp59mtrB plasmid with no added Trp (312 U) by each value for TRAP with added Trp.
The goal of our selection/screen was to identify residues on TRAP that are required to induce efficient transcription termination at the trp attenuator independent of tryptophan and RNA binding. Our results show that changing Glu60 to Lys impairs the ability of TRAP to induce termination. Moreover, the observation that E60K TRAP downregulated translation of the trpP-lacZ fusion in NM4909 to nearly the same extent as that with WT TRAP (Fig. 3A) suggests that the mutant protein binds RNA similarly to WT TRAP. To confirm that the impaired ability of E60K TRAP to induce transcription termination was not due to changes in the tryptophan or RNA binding properties of the protein, we examined these activities with purified E60K TRAP compared to WT TRAP. Consistent with prior studies (4, 5, 16, 17), WT B. subtilis TRAP bound tryptophan, with an S0.5 of 7.4 μM, and (GAGAU)11 RNA, with a dissociation constant (Kd) of 3.8 nM. E60K TRAP showed a somewhat greater affinity for tryptophan (S0.5 = 1.4 μM) and a similar affinity for RNA (Kd = 3.9 nM). Hence, the reduced ability of E60K TRAP to induce termination at the trp attenuator in vivo does not appear to result from altered tryptophan or RNA binding activity. These findings are consistent with the location of Glu60, which is on the side of TRAP opposite the location where tryptophan and RNA bind (Fig. 3C) (5).
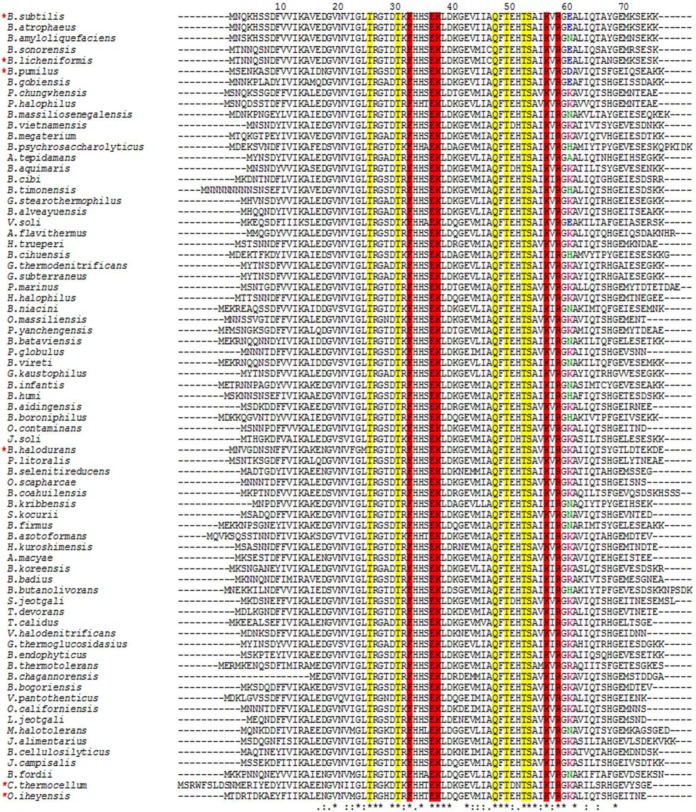
Residue 60 in TRAPs from other species.
An amino acid sequence alignment of TRAPs from 73 bacterial species shows that the sequence is highly conserved between residues 8 and 69 (Fig. 4). Residues directly involved in binding tryptophan (highlighted in yellow) and RNA (highlighted in red) (4, 5) are 100% identical among these proteins. Residue 60 shows an unusual pattern among the TRAP sequences. Only seven TRAPs, including that of B. subtilis, have acidic residues (Glu or Asp) at position 60 (blue), 17 TRAPs have a neutral polar Asn or His residue at position 60, and one contains Ala (green). The remaining TRAPs all contain a basic Lys or Arg residue at position 60 (fuchsia).
FIG 4.
Sequence alignment of TRAPs from different bacterial species. The amino acid sequence alignment was generated using Clustal Omega for 73 TRAP sequences from bacterial species (39). The residues involved in tryptophan binding (yellow) and RNA binding (red) are highlighted. Acidic residues at position 60 are shown in blue, basic residues are displayed in fuchsia, and uncharged residues are shown in green. Asterisks show positions that are 100% conserved, colons show positions with conservation of strongly similar amino acids, and dots indicate conservation between weakly similar amino acids. Numbers correspond to the B. subtilis TRAP sequence. Red asterisks at the left indicate the TRAPs examined for in vitro transcription attenuation with B. subtilis RNA polymerase (see Fig. 5).
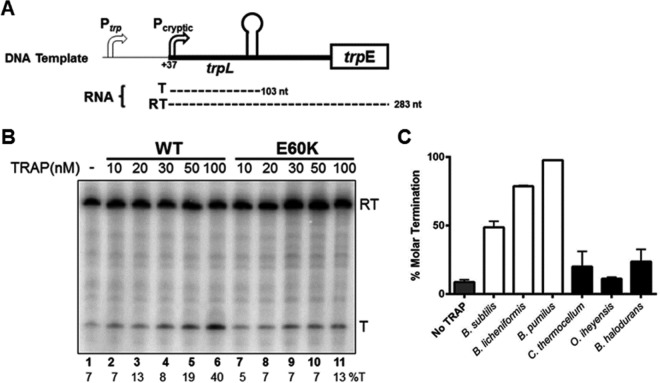
To further examine the importance of residue 60 in the attenuation mechanism, we tested the ability of TRAPs isolated from several different bacterial species to induce termination at the B. subtilis trp attenuator in vitro by using B. subtilis RNAP. Figure 5B shows a representative denaturing polyacrylamide gel with the results of in vitro transcription of a double-stranded DNA (dsDNA) fragment containing the trp promoter, the regulatory leader region, and 116 bp of trpE (the first structural gene of the trp operon) (Fig. 5A). As seen previously (13), two major RNA products were obtained. Both transcripts initiate from a modified consensus promoter that initiates at position +37 relative to the start of native transcripts (18) (see Materials and Methods). Readthrough (RT) transcripts (283 nt) are obtained when RNAP reads through the trp attenuator in the leader region and continues to the end of the template. In addition, when transcription terminates at the trp attenuator in the leader region, ∼103-nt terminated (T) transcripts are obtained. Adding increasing amounts of WT B. subtilis TRAP increased the fraction of terminated transcripts produced while decreasing the amount of RT transcripts, which is indicative of TRAP-mediated termination at the attenuator (Fig. 5B, lanes 1 to 6). Consistent with the in vivo observations described above, similar amounts of E60K TRAP yielded less termination than that with WT TRAP (Fig. 5B, lanes 7 to 11). With 100 nM E60K TRAP, only 13% of the transcripts terminated at the attenuator, compared to 40% for WT TRAP (Fig. 5B, lanes 6 and 11).
FIG 5.
In vitro attenuation assays. (A) Schematic diagram of the DNA template used for in vitro transcription attenuation assays. Transcription initiates at position +37 (relative to the WT trp promoter), from a cryptic promoter modified to match the consensus −10 and −35 sequences. The two major transcripts produced from this template are shown below the diagram and include a 103-nt transcript that terminates at the trp attenuator (T) and a 283-nt transcript that reads through the attenuator and continues to the end of the template (RT). (B) Representative 6% polyacrylamide–8 M urea gel electrophoresis analysis of the products of in vitro transcription of the trp leader region by use of B. subtilis RNAP. Reactions were performed in the absence or presence of various concentrations of WT or E60K TRAP. Positions of readthrough (RT; 283 nt) and terminated (T; 103 nt) transcripts are indicated on the right. The percentage of transcripts terminating at the attenuator (%T) for each reaction is shown at the bottom of each lane. (C) Bar graph representation of average percent termination from three independent analyses of in vitro transcription attenuation in the absence or presence of 100 nM TRAPs from several bacterial species. The TRAPs examined contained either an acidic (open bars) or basic (closed bars) residue at position 60.
We then compared the abilities of TRAPs from 6 different bacterial species to induce B. subtilis RNAP to terminate at the trp attenuator in vitro. Three of these have an acidic residue at position 60, including those of B. subtilis (Glu60), B. licheniformis (Glu60), and B. pumilus (Asp60). The other three TRAPs tested contain a basic lysine at residue 60. TRAPs with acidic residues at position 60 yielded 50 to 100% termination when present at 100 nM in the assay mixture (Fig. 5C, open bars). In contrast, all of the proteins with Lys at residue 60 displayed only 15 to 20% termination under these conditions (Fig. 5C, closed bars). All of the TRAPs tested in this assay bound (GAGAU)11 RNA similarly (apparent Kd of 0.4 to 4 nM), with the exception of Oceanobacillus iheyensis TRAP, which had a Kd of 22 nM (data not shown). Hence, the presence of a 100 nM concentration of each TRAP in these attenuation assays should ensure that the differences in termination observed are not due to differences in the ability of these proteins to bind the nascent transcript. Moreover, there was no correlation between the affinity for RNA and the presence of an acidic or basic amino acid at residue 60 (data not shown).
Residue 60 is involved in association with the TEC.
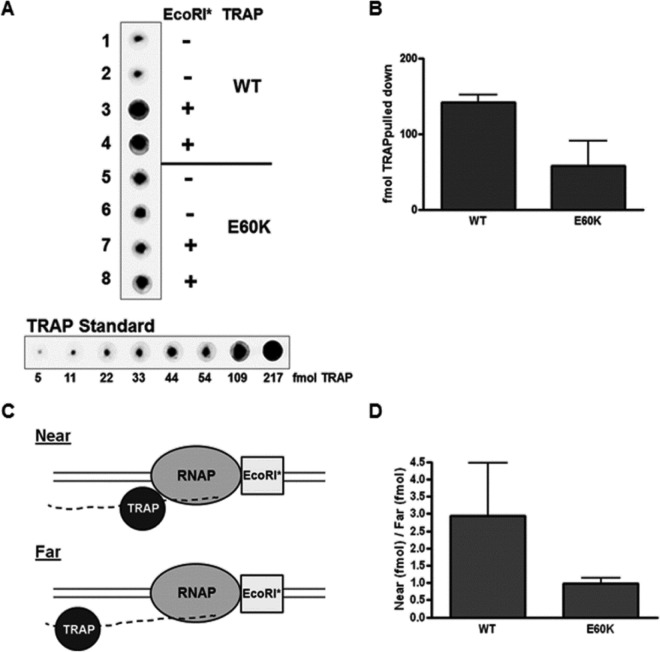
The results presented here indicate that Glu60 is specifically involved in TRAP-mediated transcription termination. Residue 60 is on the opposite side of the TRAP ring from where tryptophan and RNA bind to the protein (Fig. 3C). Consistent with this location, changes in residue 60 do not affect the tryptophan or RNA binding properties of TRAP. Together these observations suggest that Glu60 may interact with some component of the transcription elongation complex (TEC) other than the nascent RNA to induce termination. If so, then this interaction may contribute to the stability of TRAP-TEC. To test this possibility, we used pulldown assays to compare the association of WT and E60K TRAP with the TEC when the first 10 (G/U)AG repeats of the TRAP binding site are exposed on the nascent RNA. To do so, transcription elongation was blocked with a cleavage-defective E111Q mutant EcoRI (EcoRI*) protein on a template that contains an EcoRI (GAATTC) site starting at position +116 of the trp leader region (12). EcoRI* bound to the DNA template blocks TECs such that 90 residues have exited from RNAP and are available for TRAP binding (19–21). After allowing TRAP to bind the nascent transcript, TECs were isolated and washed, and the amount of associated TRAP was quantified by immunoblotting (Fig. 6A). Reactions in the absence of the EcoRI* block were used to assess the amount of nonspecifically bound TRAP, which was subtracted from the amount of TRAP pulled down in the presence of the EcoRI* block to determine the amount specifically pulled down with the TEC. Using this approach, we found that approximately three times more WT than E60K TRAP was specifically pulled down with the TEC (Fig. 6A and B). Since WT and E60K TRAP bind RNA with similar affinities, these results suggest that Glu60 participates in an interaction with some other component of the TEC and that this interaction is disrupted by substitution with Lys.
FIG 6.
In vitro transcription pulldown assays. (A) Western blots to estimate the amount of WT or E60K TRAP interacting with the TEC. Transcription was performed with EcoRI* bound to the DNA at a GAATTC recognition site starting at position +116, which stalls the elongation complex such that the nascent trp RNA is exposed from RNAP up to approximately position +90 after the start of transcription. Reactions in the absence of EcoRI* were performed to measure nonspecific interaction of TRAP, which was subtracted from the amount of TRAP pulled down in the presence of EcoRI* to determine the amount of specifically bound TRAP. The blots at the bottom show known amounts of TRAP, which served as a standard for quantification. (B) Bar graph showing the average for three repeats determining the amount (in femtomoles) of WT or E60K TRAP specifically pulled down with the TEC. (C) Schematic diagram of the Near and Far transcription templates. (Top) The TEC stalls on the Near template such that the last triplet repeat of the RNA binding site is directly adjacent to RNAP. (Bottom) The Far template includes 16 additional RNA residues after the last triplet repeat and the stalled RNAP. (D) Bar graph displaying the averages for 3 repeats determining the ratio of the amounts of TRAP pulled down with the Near template and the Far template.
The association of TRAP with the TEC described above may be stabilized by interactions that are facilitated by close proximity of TRAP bound to the nascent RNA when RNAP is immediately adjacent to the RNA exit channel. To test this possibility, we created two different transcription templates that expose 8 or 9 (G/U)AG repeats on the nascent RNA when RNAP is blocked with EcoRI*. Since the affinities of TRAP are similar for RNAs containing 8 to 11 (G/U)AG repeats (22), differences in association of TRAP with these templates should reflect interactions with some other component of the TEC. The first template (Near) consists of the trp leader region with an EcoRI recognition site inserted starting at position +106. When transcription is blocked by EcoRI* on this template, 8 (G/U)AG repeats are exposed on the nascent transcript, with the last repeat being immediately adjacent to the exit channel on RNAP. The second template (Far) contains 16 bp inserted between the 3′ end of the TRAP binding site and the EcoRI site at position +106. Hence, when the Far template is transcribed up to the EcoRI* block, 9 (G/U)AG repeats are exposed, and there are ∼16 nt of RNA between the 3′-most repeat and RNAP (Fig. 6C). The associations of WT and E60K TRAP with the TEC blocked at EcoRI* for each of these templates were compared by using a pulldown assay. We found that approximately three times more WT TRAP associated with TECs stalled on the Near template than on the Far template (Fig. 6D). In contrast, the same amount of E60K TRAP was associated with TECs stalled on both the Near and Far templates. Together these results suggest that Glu60 interacts with some component of the TEC and that this interaction is stabilized by tethering TRAP close to the TEC when it is bound to the nascent RNA.
DISCUSSION
In the original model proposed for attenuation control of the B. subtilis trp operon, transcription is regulated by two alternative RNA secondary structures in the 5′ leader region (untranslated region [UTR]) (Fig. 1) (11, 23, 24). These structures include an attenuator (terminator) and an upstream antiterminator. The last three (9) or four (11) residues of the antiterminator are shared with the attenuator, and hence, formation of the two structures is mutually exclusive. If the antiterminator forms, transcription continues through the structural trp genes. TRAP binding to the leader region RNA induces formation of the terminator and halts transcription prior to trpE. In this model, the attenuator RNA structure was proposed to function as an intrinsic terminator. Hence, the only role of TRAP in the attenuation mechanism was to alter the secondary structure of the nascent trp leader RNA so as to promote formation of the attenuator. However, recent studies have shown that (i) the trp attenuator is not an efficient intrinsic terminator (12) and (ii) TRAP can induce termination in the trp leader region in vivo when the attenuator is mutated or even deleted (13). Together these observations indicate that TRAP plays an additional, as yet uncharacterized role in the attenuation mechanism beyond altering the RNA structure in the leader region.
The tryptophan and RNA binding properties of TRAP have been studied extensively (4, 5, 16, 17), and amino acid residues involved in both activities have been identified (5). The crystal structure has shown that up to 11 tryptophan molecules bind in pockets between adjacent subunits and that the RNA wraps around the perimeter of the same side of the TRAP ring that binds tryptophan (Fig. 3C) (4). No amino acid residues have previously been identified in TRAP that are specifically required for transcription termination independent of tryptophan and RNA binding. Here we used a genetic selection/screen to obtain TRAP mutants with a diminished ability to induce transcription termination in the trp leader region while retaining the ability to bind tryptophan and RNA. The expectation was that such mutants would contain changes in amino acid residues that are specifically involved in the additional role that TRAP plays in the termination mechanism.
Five TRAP mutants were obtained that show reduced transcription termination at the trp attenuator while maintaining RNA and tryptophan binding properties similar to those of WT TRAP. Four of these mutants contain changes at Glu60, including two isolates with a single substitution of Lys at residue 60 (E60K); the other two contain secondary mutations in addition to the change at Glu60. A prior study (5) also found that changing Glu60 to Ala reduced the ability of TRAP to regulate a trpE′-′lacZ fusion in vivo 14-fold compared to that of WT TRAP, without altering the tryptophan and RNA binding properties of the protein. Together the above observations suggest that Glu60 may play a specific role in TRAP-mediated termination distinct from binding trp leader RNA. Glu60 is located at the start of β-strand F (5), on the opposite side of the TRAP ring from the location where tryptophan and RNA bind (Fig. 3C). The side chain of Glu60 is solvent exposed on the surface of TRAP such that it can interact with another factor when TRAP is in complex with the nascent RNA. The only clone obtained in our selection/screen that did not contain a substitution at Glu60 was clone 4, in which Ile63 is replaced with Val (I63V). Ile63 is located on the same β-strand as Glu60, and its hydrophobic side chain faces the interior of TRAP (Fig. 3C). The Ile63 side chain contacts several residues on adjacent β-strands, including Ile12, Val57, and Met70. Hence, the subtle change of removing one methyl group when Ile63 is replaced with Val may perturb the arrangement of this region of TRAP, possibly including Glu60.
The timing of TRAP binding to the leader segment of trp RNA also plays a role in the attenuation mechanism. TRAP must bind to trp leader RNA before RNAP has transcribed beyond the trp attenuator for efficient transcription termination to occur (25). Several features of the system have been identified that influence this timing. Previous studies have shown that NusA and NusG stimulate B. subtilis RNAP to pause at positions within the trp leader region (9, 18, 26). Pausing at position 107 has been suggested to provide time for TRAP to bind the nascent RNA before RNAP progresses beyond the attenuator region (18, 26). In addition, a stem-loop structure forms at the 5′ end of the trp transcript (5′SL) (Fig. 1) (27). This structure has been shown to increase the rate of TRAP binding to the leader RNA, and thus the efficiency of termination. One role for Glu60 may involve interaction with the 5′SL. The E60K TRAP mutant was selected as having a diminished ability to induce transcription termination in the trp leader region in vivo by use of a gene fusion containing the 5′SL (Fig. 2), and then the purified mutant protein was shown to be defective at inducing termination in vitro with a template lacking the 5′SL. Moreover, photochemical cross-linking studies indicate that the 5′SL interacts with TRAP in close proximity to His34 and His51 (28). These two residues are located near the bound tryptophan, which is on the opposite side of TRAP from Glu60 (Fig. 3C). Based on this and other observations, McGraw et al. (28) proposed a model for the interaction of TRAP with the 5′SL. In this model, the RNA is distant from Glu60. Together these observations suggest that it is unlikely that the role of Glu60 involves interacting with the 5′SL.
Transcriptional pausing has also been shown to play a role in TRAP-mediated control of the trp operon.
Canonical intrinsic terminators are composed of a GC-rich hairpin stem followed by a run of 7 to 9 U residues (29, 30). Transcription of the U tract induces RNAP to pause, after which formation of the stem-loop induces termination. Several models have been proposed for the mechanism by which formation of the stem-loop induces release of the nascent transcript and dissociation of RNAP from the DNA (31). In the allosteric model, the stem-loop is proposed to interact specifically with the polymerase to induce termination (32). In the forward translocation model, formation of the hairpin physically drives the polymerase forward on the DNA in the absence of nucleotide addition (33). This movement shears the DNA-RNA hybrid and displaces the 3′ end of the nascent RNA from the active site of RNAP, leading to termination (34, 35).
The GC content of the trp attenuator hairpin stem is low (40%), and the U stretch is interrupted twice within the first 7 residues (Fig. 1). Alleviating either of these anomalies converts the attenuator into an intrinsic terminator such that, in the absence of the competing antiterminator, TRAP is no longer required to induce efficient termination (12). Both of these features of intrinsic terminators have been shown to be required for forward translocation (31). Prior studies have shown that the B. subtilis trp attenuator requires the presence of TRAP bound to the nascent trp mRNA to induce efficient termination (12). It was proposed that the additional function provided by TRAP induces forward translocation of RNAP. To do so, TRAP would interact either directly or indirectly with the TEC, and this interaction may involve Glu60. Consistent with this suggestion, we found that almost three times more WT than E60K TRAP associated with the TEC in a pulldown assay with RNAP blocked such that 10 (G/U)AG repeats of the TRAP binding site were exposed (Fig. 6). Since both proteins bind RNA with similar affinities, this observation suggests that Glu60 contacts some component of the TEC. Moreover, this effect depended on TRAP being bound to the nascent RNA immediately adjacent to the exit channel on RNAP (Fig. 6D). A simple explanation for these observations is that TRAP (in particular Glu60) interacts directly with RNAP. However, several attempts to demonstrate a direct interaction between TRAP and RNAP in a stalled TEC failed (C. Szyjka and P. Gollnick, unpublished observations). Moreover, the observation that WT TRAP can induce significant transcription termination in a trp leader region in which the attenuator segment is severely altered or deleted in vivo but not in vitro suggests that TRAP may interact with an additional factor that is missing in the purified in vitro transcription system (13).
With the exception of 5 to 7 residues at both the amino and carboxyl ends, the amino acid sequence of TRAP is highly conserved (Fig. 4). This conservation is particularly true of residues that are directly involved in binding tryptophan or RNA, which are 100% conserved. However, residue 60 shows an unusual pattern among the TRAPs from 73 species aligned in Fig. 4. Only 7 proteins, including that from B. subtilis, contain an acidic residue at position 60, 17 species contain an uncharged polar residue (Asn or His), 1 species contains an Ala residue, and the remaining TRAPs (from 48 species) all contain basic residues. No other residue within the central 60 (from positions 10 to 70) residues of TRAP shows such wide variation among TRAPs from these species (Fig. 4). Moreover, we found that purified TRAPs from 3 bacterial species that contain an acidic residue (Glu or Asp) at position 60 were more active at inducing termination at the B. subtilis trp attenuator than TRAPs from three species that contain Lys at position 60 when examined using an in vitro attenuation assay with B. subtilis RNAP (Fig. 5A and B). These assays were performed at low nucleoside triphosphate (NTP) concentrations to reduce the elongation rate and to minimize any potential effects of differences in the rate of TRAP binding to the nascent RNA (25). This, together with the high concentration (100 μM) of TRAP used in these assays, suggests that the reduced ability of Lys60-containing TRAPs to induce termination at the B. subtilis trp attenuator is unlikely to be due to differences in the RNA binding properties of these proteins.
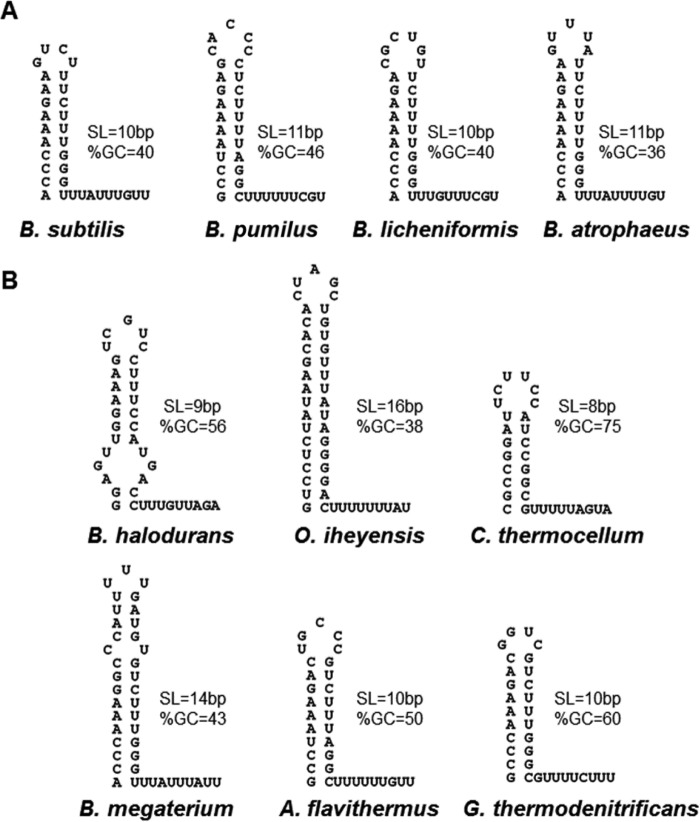
There are differences in the predicted attenuators between bacteria with TRAPs that contain a Glu/Asp at residue 60 and those with Lys60 (Fig. 7). In general, attenuators from species with an acidic residue 60 are similar to that from B. subtilis (Fig. 7A). All of these contain 10- or 11-bp stems with <50% G+C content. In all cases, there are 3 G·C pairs near the base of the stem, which are usually preceded by a single A·U pair (although it should be noted that this A·U pair does not appear to form in the B. subtilis attenuator). In all cases except that of B. pumilus, the U stretch following the stem is interrupted after the first 2 or 3 U's.
FIG 7.
Predicted trp attenuator structures from various bacteria. RNA attenuator structures were predicted by Mfold (59) for the trp leader regions of several TRAP-containing bacterial species that contain either an acidic (A) or basic (B) amino acid at residue 60. SL and %GC indicate the number of base pairs and the percentage of G·C base pairs in the predicted stems, respectively.
The attenuators from species with Lys at residue 60 of TRAP show more variation than those described above. The base-paired stems range from 8 to 16 bp, with GC contents of 37 to 75% (Fig. 7B). Several of these predicted attenuators, such as that from Clostridium thermocellum, closely resemble canonical intrinsic terminators with high-GC-content stem-loops followed by at least 5 uninterrupted U's. Others, such as that from B. halodurans, bear little to no resemblance to intrinsic terminators (24). However, we have shown that B. halodurans TRAP (Lys60) mediates transcription termination at or near this attenuator both in vivo and in vitro (36). Together the above observations suggest that residue 60 of TRAP may have evolved to be acidic (such as in B. subtilis) so as to perform a specific function in the transcription termination mechanism in conjunction with a particular type of trp attenuator. This mechanism may involve interacting with another protein that is not involved in the mechanism in other species. In contrast, the termination mechanism with Lys60-containing TRAP, such as that in B. halodurans, may be different. Interestingly, recent studies have identified numerous NusA-dependent terminators in B. subtilis with weak hairpins and U-tract interruptions (37). We previously found that adding NusA to our in vitro attenuation assays increased TRAP-mediated termination from mutant attenuators with U-tract interruptions but did not affect termination from terminators with alterations in the stem-loop. Thus, while the trp attenuator may have characteristics similar to those of NusA-dependent terminators, TRAP is still needed for efficient transcription termination. Therefore, NusA and TRAP may provide similar yet distinct functions in the transcription termination mechanism at the trp attenuator.
Recent in vitro studies with B. subtilis anti-TRAP (AT) also showed a difference in function between E60K and WT TRAP (38). AT is a protein antagonist of TRAP that is produced in response to an accumulation of uncharged tRNATrp (39, 40). AT binds as a trimer to TRAP and prevents it from binding to RNA, thereby increasing trp gene expression (41). AT can prevent TRAP-mediated transcription termination by inducing dissociation of TRAP from the nascent RNA when the protein has bound to fewer than all 11 (G/U)AG repeats (38). For AT to induce dissociation of TRAP from the nascent RNA, TRAP must be in close proximity to a stalled RNAP (38). Therefore, an interaction between TRAP and the TEC appears to be important for AT-mediated dissociation of TRAP from the nascent transcript. It was found that AT was three times less efficient at inducing dissociation of E60K TRAP from nascent RNA than at inducing dissociation of the WT protein (38), which further supports the hypothesis that this mutant TRAP is deficient in some interaction with the TEC.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli K802 was used as a host for plasmid construction and propagation. B. subtilis strains NM421 (CMr ΔmtrB amyE::[Ptrp-trpL-trpE-lacZ]), NM4409 (CMr ΔmtrB argC amyE::[Ptrp-trpL′-cat]), and NM4909 (CMr Spectr ΔmtrB argC amyE::[Ptrp-trpL′-cat] thrC::[PtrpP′-′lacZ]) were created as described below and used for selection and screening of TRAP mutants. B. subtilis was transformed by natural competence (42), and transformants were selected on plates containing Vogel and Bonner (VB) minimal salts (43), 0.2% acid-hydrolyzed casein (ACH), 0.2% (wt/vol) glucose, 50 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 10 μg/ml l-arginine, and 20 μg/ml tetracycline.
TRAP mutants were selected from a pool of mutants with PCR-induced mutations (44) in the mtrB gene (1). These genes were expressed from the p59 promoter, which was taken from pHB201 (residues 520 to 892) (45) and inserted between the EcoRI and BamHI sites of pHY300PLK (46). The 3′ end of the WT mtrB gene contains a run of 8 adenine residues followed by a TAA stop codon (3). This region is highly susceptible to spontaneous mutations that change the TAA stop codon into an AAA Lys codon, which extends the open reading frame by 6 codons (3). The mutant TRAP produced from this mtrB264 allele is defective in regulating the trp operon and acts as a dominant negative mutant (3). To prevent a high background level of this mtrB allele in our selection, we created an alternative version of the WT mtrB gene, termed mtrBΔ264. In this mtrB allele, the sequence at the 3′ end was altered from GAA AAA AAA TAA to GAG AAG AAA TGA TAG by QuikChange site-directed mutagenesis (Agilent). These changes do not alter the Glu-Lys-Lys-Lys sequence at the C terminus of TRAP, but they effectively reduce spontaneous mtrB264 mutations. This optimized mtrB sequence was cloned into pHYp59 between BglII and HindIII sites, creating the plasmid pHYp59mtrBΔ264. Mutagenic PCR (44) was performed using pHYp59mtrBΔ264 as the template, with the primers MtrB Mut PCR For BglII (5′-AGC TAA CAG ATC TCG CCA GGA CTA ATA AAG ATA GAG G-3′ [BglII site underlined]) and MtrB Mut PCR Rev HindIII (5′-TAC TGA AAG CTT CAG CGG GGA CAG CC-3′ [HindIII site underlined]). The resulting PCR products were digested with BglII and HindIII, ligated into similarly digested pHYp59 (as described above), and transformed into E. coli K802. Approximately 5 × 104 transformants were selected on LB agar with 20 μg/ml tetracycline, combined into one pool, and grown for 5 h at 37°C in 100 ml of LB, and plasmids were isolated from the pool of cells.
Mutant TRAPs with a decreased ability to induce termination at the trp attenuator were selected in B. subtilis NM4409, which contains a transcriptional fusion between the trp promoter and 5′ regulatory leader region and the chloramphenicol acetyltransferase (cat) gene. To make this fusion, the native promoter was first removed from the cat gene. To do so, an EcoRI site was created at position 4094 in pDH32 (47) by QuikChange site-directed mutagenesis (Agilent). This placed the cat promoter between two EcoRI sites, at positions 3821 and 4094 of pDH32. This 273-bp EcoRI fragment was removed by digestion with EcoRI and religation of the plasmid to yield pDH32-ΔPcat. B. subtilis transformed with pDH32-ΔPcat is chloramphenicol sensitive. The DNA segment between positions −98 and +170 (relative to the start of transcription) of the trp operon was amplified by PCR, with EcoRI and BamHI recognition sites introduced into the upstream and downstream ends of the DNA fragment. The PCR product was digested with EcoRI and BamHI and ligated into the multiple-cloning site of similarly cut pDH32-ΔPcat. The resulting plasmid, pDH32-PtrpL-cat, has the trp promoter and leader region controlling transcription of the cat gene. This plasmid was linearized with PstI and integrated as a single copy at the amyE locus (23) of B. subtilis BG4233 (ΔmtrB argC4) to create NM4409 (CMr ΔmtrB argC amyE::[Ptrp-trpL′-cat]).
A translational fusion between trpP and lacZ was also inserted into NM4409 to allow screening of the TRAP mutants for the ability to bind tryptophan and RNA. trpP mRNA contains a TRAP binding site overlapping its ribosome binding site (rbs) and start codon (14). TRAP binding to this site blocks access to the rbs and inhibits translation of trpP (14, 48). To create a translational fusion between trpP and lacZ, QuikChange site-directed mutagenesis was used to remove a ClaI site at position 7273 of pDG1729 (49), making the ClaI site at position 3043 unique (49). The region between the EcoRI and ClaI sites was then replaced with the EcoRI-ClaI fragment of pRS552 (50) to generate pPDG1729, creating a translational fusion with lacZ. The DNA region from positions −80 to +50 (relative to the start of transcription) of trpP was inserted into pPDG1729 between the EcoRI and BamHI sites, resulting in pPDG1729trpP, containing a translational fusion between trpP and lacZ. pPDG1729trpP was linearized with SmaI, transformed into NM4409, and integrated at the thrC locus (49) to create NM4909 (CMr Spectr ΔmtrB argC amyE::[Ptrp-trpL′-cat] thrC::[PtrpP′-′lacZ]).
Genetic selection/screen.
B. subtilis NM4909 (ΔmtrB) is CM resistant due to unregulated transcriptional readthrough of the trp leader region in the absence of TRAP. When this strain is transformed with the plasmid pHYp59mtrBΔ264, which expresses WT TRAP, the resulting strain remains CM resistant in the absence of tryptophan. However, when this strain is grown in the presence of excess tryptophan, it is sensitive to CM due to TRAP-mediated transcription termination in the trp leader region upstream of the cat gene. We determined the lowest concentration of CM that prevents growth of this strain in the presence of tryptophan but permits growth in its absence to be 10 μg/ml. Hence, we transformed NM4909 with our pool of plasmids containing mutant mtrB genes (described above) and selected for CM-resistant colonies on LB agar containing 10 μg/ml CM, 20 μg/ml tetracycline, and 50 μg/ml X-Gal. CM-resistant colonies were screened for the ability of TRAP to downregulate translation of the trpP-lacZ fusion, which yields white colonies on X-Gal, whereas colonies with TRAP mutants defective in tryptophan/RNA binding are blue.
In vivo TRAP-mediated regulation of transcription of the trp operon.
The ability of mutant TRAPs to regulate transcription of the trp operon was examined quantitatively in vivo by using B. subtilis NM421 (Cmr ΔmtrB amyE::[Ptrp-trpL-trpE-lacZ]), in which the genomic copy of mtrB is deleted. This strain contains a transcriptional lacZ fusion containing the trp promoter and leader region and the first 40 codons of trpE followed by a stop codon and then the entire lacZ coding segment (13). β-Galactosidase assays were performed as described previously (12, 13).
Amino acid sequence alignment of TRAPs from 73 bacterial species.
The amino acid sequence of B. subtilis TRAP was used to perform a search against a representative microbial genome database by using a protein query (TBLASTN) (51). The taxonomy report from the search was analyzed, and 73 mtrB sequences that were annotated as being from identified bacterial species were aligned using Clustal Omega (39).
TRAP expression and purification.
TRAPs were expressed from pET9a plasmids (Novagen) containing a WT or mutant mtrB gene in E. coli BL21(DE3). TRAP was purified by immunoaffinity chromatography (52) or with phenyl-Sepharose as described previously (53).
In vitro transcription.
The trp leader region was previously found to contain a cryptic promoter that directs transcription to initiate at position +37 in vitro and, when altered to the consensus −10 and −35 sequences, yields more efficient transcription at low NTP concentrations than the upstream native promoter (18). We used PCR amplification with the pUCtrpL plasmid (25) as the template and the M13 reverse primer together with a +37trpL primer (5′-CAGCTTGACAAATACACAAGAGTGTGTTATAATGCAATTAGAATG 3′) that binds from positions +31 to +42 in the trp leader region and converts the cryptic promoter into a consensus promoter. PCR products were purified on 1% agarose gels by use of a Qiagen MinElute gel extraction kit.
Single-round in vitro transcription reactions were initiated in the absence of CTP with 50 μg/ml B. subtilis σA RNA polymerase (RNAP), 20 nM DNA template, 20 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 0.1 mM EDTA, 4 mM spermidine, 5 mM dithiothreitol (DTT), 8 μM (each) ATP and GTP, 2 μM UTP, and 1 μCi [α-32P]UTP (3,000 Ci/mmol) in 20 μl (12), and reaction mixtures were incubated at 37°C for 10 min. The B. subtilis RNAP holoenzyme was purified from B. subtilis MH5636 as described previously (54). Transcription of this template in the absence of CTP allows initiation and elongation from positions +37 to +65, yielding a stable transcription elongation complex (TEC) with a 29-nt nascent transcript. A single round of transcription elongation was induced by addition of all 4 NTPs at 50 μM (each) in the presence of 0.1 mg/ml heparin, TRAP (0 to 100 nM), and 1 mM tryptophan. Reaction mixtures were incubated at 37°C for 10 min and then stopped by adding an equal volume of stop solution (95% formamide, 20 mM EDTA, 0.02% bromophenol blue, and 0.02% xylene cyanol). The samples were heated at 95°C for 2 min, and the resulting RNAs were separated in 6% denaturing polyacrylamide gels. The gels were dried, exposed to a phosphorstorage screen, and quantified using a Storm phosphorimager and ImageQuant software (GE Healthcare). The numbers of U residues in the terminated and readthrough transcripts were used to calculate the molar percentage of termination (12).
RNA binding assay.
32P-labeled RNA composed of 11 tandem GAGUU repeats was prepared by in vitro transcription of HindIII-linearized pTZGAGUU-11 (55) by use of T7 RNA polymerase and [α-32P]UTP as described previously (52). The RNA was gel purified and extracted from 6% (wt/vol) polyacrylamide–8 M urea gels by crushing and soaking (56). Nitrocellulose filter binding using the (GAGUU)11 RNA in the presence of WT and mutant TRAPs was performed and analyzed as described previously (57).
Tryptophan binding assay.
Fluorescence spectroscopy using competition with 1-anilinonaphthalene-8-sulfonic acid (ANS) was used to measure tryptophan binding to TRAP (16). Fluorescence intensity was measured using an LS-50B fluorometer (PerkinElmer) with excitation at 372 nm and emission at 465 nm. Data were analyzed as described previously (16).
TRAP pulldown assay.
Pulldown assays were performed to examine the association of TRAP with the nascent RNA on stalled B. subtilis TECs. The E111Q mutant EcoRI (EcoRI*) protein binds to its GAATTC recognition site but does not cleave the DNA (58). EcoRI* bound to the DNA template blocks the TEC such that the active site of RNAP is stalled approximately 12 to 13 bp upstream of the G of the GAATTC recognition site (19). EcoRI* was purified as described previously (12). QuikChange site-directed mutagenesis was used to introduce an EcoRI recognition site starting at position +116 (relative to the start of transcription) of the trp leader DNA in pUCtrpL, creating the plasmid pUCtrpLEco116. Bead-bound transcription templates were created by PCR amplification of pUCtrpLEco116 (25) with a 5′-biotinylated +37trpL primer (see “In vitro transcription”) and the M13 reverse primer (18). PCR products were purified in 1% agarose gels, followed by extraction using a Qiagen MiniElute gel extraction kit. Purified DNA was coupled to streptavidin MagneSphere beads (Promega) per the manufacturer's instructions. Site-directed mutagenesis was also used to modify the pUC119trpLEco116 plasmid to delete the 10th and 11th repeats of the TRAP binding site, from positions +82 to +91 (relative to the start of transcription), leaving 9 repeats in plasmid pUCtrpLΔ10-11Eco116 (Near template) (38). When the TEC is blocked by EcoRI* on this template, 8 (G/U)AG repeats of the TRAP binding site are exposed on the nascent transcript, with the 3′ end of the TRAP binding site immediately adjacent to the stalled polymerase. The pUCtrpLΔ10-11Eco116EX plasmid contains 16 bp of random sequence between the last (9th) repeat and the EcoRI site. This results in 16 nucleotides of RNA between the 3′ end of the TRAP binding site and the stalled polymerase (Far template), with 9 (G/U)AG repeats of the TRAP binding site exposed on the nascent transcript (38).
For TRAP pulldown assays, single-round in vitro transcription of bead-bound templates with EcoRI bound to the DNA was performed with B. subtilis RNAP. To provide enough blocked TECs for TRAP to be detected by Western blotting, reaction mixtures were scaled up 8-fold, to 160 μl. Transcription was initiated in the presence of TRAP as described above and then elongated to the EcoRI* roadblock bound to the DNA, and the TECs stalled on the bead-bound templates were collected by use of a magnet. Unbound TRAP was removed by washing the beads twice in transcription buffer containing 1 mM l-tryptophan. Transcription complexes were freed from the beads with 0.15 mg/ml DNase for 15 min at 37°C. The supernatant was filtered through a nitrocellulose membrane. TRAP was detected on the membrane by using rabbit polyclonal antibodies against B. subtilis TRAP and Amersham ECL Plus Western blotting detection reagents (GE Healthcare) as described previously (12). Known concentrations of TRAP filtered onto the same membrane served as standards to quantify the amount of TRAP pulled down with each template.
ACKNOWLEDGMENTS
We thank Sally May Enriquez for technical help with preparing the proteins used for this study. We also thank Kristine Potter for useful discussions and advice.
N.M.M. performed the bulk of the experiments described here and participated in writing. A.P. performed some of the work presented, and P.G. oversaw the project, its design, and the writing of the manuscript.
This work was funded by grant MCB 1019969 from the National Science Foundation.
REFERENCES
- 1.Gollnick P. 1994. Regulation of the Bacillus subtilis trp operon by an RNA-binding protein. Mol Microbiol 11:991–997. doi: 10.1111/j.1365-2958.1994.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 2.Carlton BC, Whitt DD. 1969. The isolation and genetic characterization of mutants of the tryptophan system of Bacillus subtilis. Genetics 62:445–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gollnick P, Ishino S, Kuroda MI, Henner DJ, Yanofsky C. 1990. The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis. Proc Natl Acad Sci U S A 87:8726–8730. doi: 10.1073/pnas.87.22.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antson AA, Otridge J, Brzozowski AM, Dodson EJ, Dodson GG, Wilson KS, Smith TM, Yang M, Kurecki T, Gollnick P. 1995. The structure of trp RNA-binding attenuation protein. Nature 374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- 5.Yang M, Chen X, Militello K, Hoffman R, Fernandez B, Baumann C, Gollnick P. 1997. Alanine-scanning mutagenesis of Bacillus subtilis trp RNA-binding attenuation protein (TRAP) reveals residues involved in tryptophan binding and RNA binding. J Mol Biol 270:696–710. doi: 10.1006/jmbi.1997.1149. [DOI] [PubMed] [Google Scholar]
- 6.Babitzke P, Bear DG, Yanofsky C. 1995. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides. Proc Natl Acad Sci U S A 92:7916–7920. doi: 10.1073/pnas.92.17.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gollnick P, Baumann C, Yang M, Otridge J, Antson A. 1995. Interaction of the 11-subunit trp RNA-binding attenuation protein (TRAP) with its RNA target. Nucleic Acids Symp Ser 1995:43–45. [PubMed] [Google Scholar]
- 8.Shimotsu H, Kuroda MI, Yanofsky C, Henner DJ. 1986. Novel form of transcription attenuation regulates expression of the Bacillus subtilis tryptophan operon. J Bacteriol 166:461–471. doi: 10.1128/jb.166.2.461-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yakhnin AV, Babitzke P. 2010. Mechanism of NusG-stimulated pausing, hairpin-dependent pause site selection and intrinsic termination at overlapping pause and termination sites in the Bacillus subtilis trp leader. Mol Microbiol 76:690–705. doi: 10.1111/j.1365-2958.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babitzke P, Yanofsky C. 1993. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc Natl Acad Sci U S A 90:133–137. doi: 10.1073/pnas.90.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuroda MI, Henner D, Yanofsky C. 1988. cis-acting sites in the transcript of the Bacillus subtilis trp operon regulate expression of the operon. J Bacteriol 170:3080–3088. doi: 10.1128/jb.170.7.3080-3088.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potter KD, Merlino NM, Jacobs T, Gollnick P. 2011. TRAP binding to the Bacillus subtilis trp leader region RNA causes efficient transcription termination at a weak intrinsic terminator. Nucleic Acids Res 39:2092–2102. doi: 10.1093/nar/gkq965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAdams NM, Gollnick P. 2014. The Bacillus subtilis TRAP protein can induce transcription termination in the leader region of the tryptophan biosynthetic (trp) operon independent of the trp attenuator RNA. PLoS One 9:e88097. doi: 10.1371/journal.pone.0088097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarsero JP, Merino E, Yanofsky C. 2000. A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport. J Bacteriol 182:2329–2331. doi: 10.1128/JB.182.8.2329-2331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yakhnin H, Yakhnin AV, Babitzke P. 2007. Translation control of trpG from transcripts originating from the folate operon promoter of Bacillus subtilis is influenced by translation-mediated displacement of bound TRAP, while translation control of transcripts originating from a newly identified trpG promoter is not. J Bacteriol 189:872–879. doi: 10.1128/JB.01398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li PT, Gollnick P. 2004. Characterization of a trp RNA-binding attenuation protein (TRAP) mutant with tryptophan independent RNA binding activity. J Mol Biol 335:707–722. doi: 10.1016/j.jmb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Li PT, Gollnick P. 2002. Using hetero-11-mers composed of wild type and mutant subunits to study tryptophan binding to TRAP and its role in activating RNA binding. J Biol Chem 277:35567–35573. doi: 10.1074/jbc.M205910200. [DOI] [PubMed] [Google Scholar]
- 18.Yakhnin AV, Yakhnin H, Babitzke P. 2006. RNA polymerase pausing regulates translation initiation by providing additional time for TRAP-RNA interaction. Mol Cell 24:547–557. doi: 10.1016/j.molcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Grundy FJ, Yousef MR, Henkin TM. 2005. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J Mol Biol 346:73–81. doi: 10.1016/j.jmb.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 20.Pavco PA, Steege DA. 1990. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem 265:9960–9969. [PubMed] [Google Scholar]
- 21.Komissarova N, Kashlev M. 1998. Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc Natl Acad Sci U S A 95:14699–14704. doi: 10.1073/pnas.95.25.14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott MB, Gottlieb PA, Gollnick P. 2001. The mechanism of RNA binding to TRAP: initiation and cooperative interactions. RNA 7:85–93. doi: 10.1017/S135583820100173X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimotsu H, Henner DJ. 1986. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene 43:85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- 24.Gollnick P, Babitzke P, Antson A, Yanofsky C. 2005. Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis. Annu Rev Genet 39:47–68. doi: 10.1146/annurev.genet.39.073003.093745. [DOI] [PubMed] [Google Scholar]
- 25.Barbolina MV, Kristoforov R, Manfredo A, Chen Y, Gollnick P. 2007. The rate of TRAP binding to RNA is crucial for transcription attenuation control of the B. subtilis trp operon. J Mol Biol 370:925–938. doi: 10.1016/j.jmb.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yakhnin AV, Babitzke P. 2002. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. Proc Natl Acad Sci U S A 99:11067–11072. doi: 10.1073/pnas.162373299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGraw AP, Bevilacqua PC, Babitzke P. 2007. TRAP-5′ stem loop interaction increases the efficiency of transcription termination in the Bacillus subtilis trpEDCFBA operon leader region. RNA 13:2020–2033. doi: 10.1261/rna.719507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGraw AP, Mokdad A, Major F, Bevilacqua PC, Babitzke P. 2009. Molecular basis of TRAP-5′SL RNA interaction in the Bacillus subtilis trp operon transcription attenuation mechanism. RNA 15:55–66. doi: 10.1261/rna.1314409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.d'Aubenton Carafa Y, Brody E, Thermes C. 1990. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol 216:835–858. [DOI] [PubMed] [Google Scholar]
- 30.Wilson KS, von Hippel PH. 1995. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc Natl Acad Sci U S A 92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters JM, Vangeloff AD, Landick R. 2011. Bacterial transcription terminators: the RNA 3′-end chronicles. J Mol Biol 412:793–813. doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. 2007. An allosteric path to transcription termination. Mol Cell 28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Larson MH, Greenleaf WJ, Landick R, Block SM. 2008. Applied force reveals mechanistic and energetic details of transcription termination. Cell 132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarnell WS, Roberts JW. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 35.Santangelo TJ, Roberts JW. 2004. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol Cell 14:117–126. doi: 10.1016/S1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- 36.Szigeti R, Milescu M, Gollnick P. 2004. Regulation of the tryptophan biosynthetic genes in Bacillus halodurans: common elements but different strategies than those used by Bacillus subtilis. J Bacteriol 186:818–828. doi: 10.1128/JB.186.3.818-828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mondal S, Yakhnin AV, Sebastian A, Albert I, Babitzke P. 2016. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat Microbiol 1:15007. doi: 10.1038/nmicrobiol.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma S, Gollnick P. 2014. Modulating TRAP-mediated transcription termination by AT during transcription of the leader region of the Bacillus subtilis trp operon. Nucleic Acids Res 42:5543–5555. doi: 10.1093/nar/gku211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarsero JP, Merino E, Yanofsky C. 2000. A Bacillus subtilis operon containing genes of unknown function senses tRNATrp charging and regulates expression of the genes of tryptophan biosynthesis. Proc Natl Acad Sci U S A 97:2656–2661. doi: 10.1073/pnas.050578997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valbuzzi A, Gollnick P, Babitzke P, Yanofsky C. 2002. The anti-trp RNA-binding attenuation protein (anti-TRAP), AT, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein. J Biol Chem 277:10608–10613. doi: 10.1074/jbc.M111813200. [DOI] [PubMed] [Google Scholar]
- 42.Anagnostopoulos C, Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J Bacteriol 81:741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogel HJ, Bonner DM. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218:97–106. [PubMed] [Google Scholar]
- 44.Cadwell RC, Joyce GF. 1994. Mutagenic PCR. PCR Methods Appl 3:S136–S140. doi: 10.1101/gr.3.6.S136. [DOI] [PubMed] [Google Scholar]
- 45.Bron S, Bolhuis A, Tjalsma H, Holsappel S, Venema G, van Dijl JM. 1998. Protein secretion and possible roles for multiple signal peptidases for precursor processing in bacilli. J Biotechnol 64:3–13. doi: 10.1016/S0168-1656(98)00099-6. [DOI] [PubMed] [Google Scholar]
- 46.Shimizu-Kadota M, Shibahara-Sone H, Ishiwa H. 1991. Shuttle plasmid vectors for Lactobacillus casei and Escherichia coli with a minus origin. Appl Environ Microbiol 57:3292–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henner DJ. 1990. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol 185:223–228. doi: 10.1016/0076-6879(90)85022-G. [DOI] [PubMed] [Google Scholar]
- 48.Yakhnin H, Zhang H, Yakhnin AV, Babitzke P. 2004. The trp RNA-binding attenuation protein of Bacillus subtilis regulates translation of the tryptophan transport gene trpP (yhaG) by blocking ribosome binding. J Bacteriol 186:278–286. doi: 10.1128/JB.186.2.278-286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guerout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61. doi: 10.1016/S0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 50.Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 51.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otridge J, Gollnick P. 1993. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner. Proc Natl Acad Sci U S A 90:128–132. doi: 10.1073/pnas.90.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antson AA, Brzozowski AM, Dodson EJ, Dauter Z, Wilson KS, Kurecki T, Otridge J, Gollnick P. 1994. 11-fold symmetry of the trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis determined by X-ray analysis. J Mol Biol 244:1–5. doi: 10.1006/jmbi.1994.1698. [DOI] [PubMed] [Google Scholar]
- 54.Qi Y, Hulett FM. 1998. PhoP-P and RNA polymerase sigmaA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Mol Microbiol 28:1187–1197. doi: 10.1046/j.1365-2958.1998.00882.x. [DOI] [PubMed] [Google Scholar]
- 55.Hopcroft NH, Manfredo A, Wendt AL, Brzozowski AM, Gollnick P, Antson AA. 2004. The interaction of RNA with TRAP: the role of triplet repeats and separating spacer nucleotides. J Mol Biol 338:43–53. doi: 10.1016/j.jmb.2004.02.038. [DOI] [PubMed] [Google Scholar]
- 56.Milligan JF, Uhlenbeck OC. 1989. Determination of RNA-protein contacts using thiophosphate substitutions. Biochemistry 28:2849–2855. doi: 10.1021/bi00433a016. [DOI] [PubMed] [Google Scholar]
- 57.Baumann C, Otridge J, Gollnick P. 1996. Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J Biol Chem 271:12269–12274. doi: 10.1074/jbc.271.21.12269. [DOI] [PubMed] [Google Scholar]
- 58.Wright DJ, King K, Modrich P. 1989. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem 264:11816–11821. [PubMed] [Google Scholar]
- 59.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]