ABSTRACT
Rickettsia rickettsii, the causative agent of Rocky Mountain spotted fever, contains two immunodominant proteins, rOmpA and rOmpB, in the outer membrane. Both rOmpA and rOmpB are conserved throughout spotted fever group rickettsiae as members of a family of autotransporter proteins. Previously, it was demonstrated that rOmpB is proteolytically processed, with the cleavage site residing near the autotransporter domain at the carboxy-terminal end of the protein, cleaving the 168-kDa precursor into apparent 120-kDa and 32-kDa fragments. The 120- and 32-kDa fragments remain noncovalently associated on the surface of the bacterium, with implications that the 32-kDa fragment functions as the membrane anchor domain. Here we present evidence for a similar posttranslational processing of rOmpA. rOmpA is expressed as a predicted 224-kDa precursor yet is observed on SDS-PAGE as a 190-kDa protein. A small rOmpA fragment of ∼32 kDa was discovered during surface proteome analysis and identified as the carboxy-terminal end of the protein. A rabbit polyclonal antibody was generated to the autotransporter region of rOmpA and confirmed a 32-kDa fragment corresponding to the calculated mass of a proteolytically cleaved rOmpA autotransporter region. N-terminal amino acid sequencing revealed a cleavage site on the carboxy-terminal side of Ser-1958 in rOmpA. An avirulent strain of R. rickettsii Iowa deficient in rOmpB processing was also defective in the processing of rOmpA. The similarities of the cleavage sites and the failure of R. rickettsii Iowa to process either rOmpA or rOmpB suggest that a single enzyme may be responsible for both processing events.
IMPORTANCE Members of the spotted fever group of rickettsiae, including R. rickettsii, the etiologic agent of Rocky Mountain spotted fever, express at least four autotransporter proteins that are protective antigens or putative virulence determinants. One member of this class of proteins, rOmpB, is proteolytically processed to a passenger domain and an autotransporter domain that remain associated on the rickettsial outer membrane. The protease responsible for this posttranslation processing remains unknown. Here we show that another autotransporter, rOmpA, is similarly processed by R. rickettsii. Similarities in sequence at the cleavage site and predicted secondary protein structure suggest that all four R. rickettsii autotransporters may be processed by the same outer membrane protease.
KEYWORDS: Rickettsia, autotransporter proteins
INTRODUCTION
Rickettsia rickettsii, the tick-borne causative agent of Rocky Mountain spotted fever (RMSF), is an obligate intracellular Gram-negative bacterium which grows and replicates in the cytosol of host cells. R. rickettsii is a member of the spotted fever group (SFG) of rickettsiae, which includes some species that cause disease in humans but also species that do not cause apparent disease and appear to be commensals in ticks (1, 2). R. rickettsii is the most virulent of the SFG rickettsiae but even within the species shows dramatic variation in virulence. The factors contributing to virulence are poorly understood.
Two immunodominant outer membrane proteins, rOmpA and rOmpB, comprise a significant portion of the outer membrane proteome and S-layer (3–5) of SFG rickettsiae. Both rOmpA and rOmpB are members of the Sca (surface cell antigen) family of autotransporters (6, 7), which includes 17 orthologous proteins that are variably present throughout the genus Rickettsia (8). R. rickettsii specifically contains intact forms of sca0 (rOmpA), sca1, sca2, sca4, and sca5 (rOmpB). Sca2 plays a role in actin polymerization and cytoplasmic escape (9, 10), and Sca4 binds to vinculin (11) but lacks the autotransporter domain. rOmpA, rOmpB, Sca1, and Sca2 have all been implicated in rickettsial adherence and invasion of host cells (12–17).
Autotransporter proteins, also known as the type V secretion pathway, are highly diverse and widely distributed across Gram-negative bacteria (18). In the classical autotransporter model, an N-terminal Sec signal is followed by a hypervariable passenger domain and a C-terminal translocator, also known as the transmembrane or β domain. Once in the periplasm, the transmembrane domain forms a pore in the outer membrane through which the passenger domain is translocated. The passenger domain then becomes surface exposed or secreted into the extracellular space (18–20). The transmembrane domain “anchor” may remain as a single peptide linked to the passenger domain, or it may be cleaved. This cleavage can occur by accessory proteolytic enzymes or autocatalysis, after which the transmembrane domain may stay noncovalently linked to the passenger domain or completely disassociate (20–23).
Many autotransporters have a functional role in pathogenesis (24). The classical example of autotransporters, Neisseria gonorrhoeae's IgA protease, has immunomodulatory properties and cleaves human antibodies (25). Other autotransporters function as adhesins, toxins, and esterases; however, the mechanisms by which several autotransporters contribute to disease are poorly understood (24).
A previous study revealed that rOmpB, the most abundant outer membrane protein in R. rickettsii, was posttranslationally processed by an unknown protease from a predicted 168-kDa full-length precursor to mature 120-kDa and 32-kDa fragments (21). The 32-kDa fragment made up of the transmembrane domain β-barrel remained noncovalently associated with the 120-kDa passenger region. N-terminal sequencing of the 32-kDa peptide revealed a cleavage site in rOmpB between Ala-1361 and Gly-1362. This cleavage site is highly conserved among the virulent and avirulent strains of R. rickettsii. In this study, we present evidence for a similar posttranslational processing of the autotransporter rOmpA.
RESULTS
Identification of rOmpA β fragment.
The outer membrane proteome of R. rickettsii Sheila Smith was examined by standard outer membrane fractionation, followed by separation on a sucrose gradient (Fig. 1A). The outer membrane fraction was recovered and run on an SDS-PAGE gel for mass spectrometry (MS) analysis. A prominent band at ∼32 kDa was excised and analyzed (Fig. 1B). The majority of peptides analyzed from the excised band originated from the rOmpB β fragment; however, two peptides specific to the rOmpA transmembrane domain were identified. In order to confirm the findings of a possible rOmpA-derived β peptide, the outer membrane fractionation and MS analysis were repeated. An additional two rOmpA-specific peptides were identified (Fig. 1C). These four peptides were the only non-rOmpB peptides recovered by MS analysis in the 32-kDa band. The size of the excised band and identification of these peptides suggest that a β fragment of rOmpA exists in the rickettsial outer membrane, most likely originating from a posttranslational processing event similar to that of rOmpB.
FIG 1.
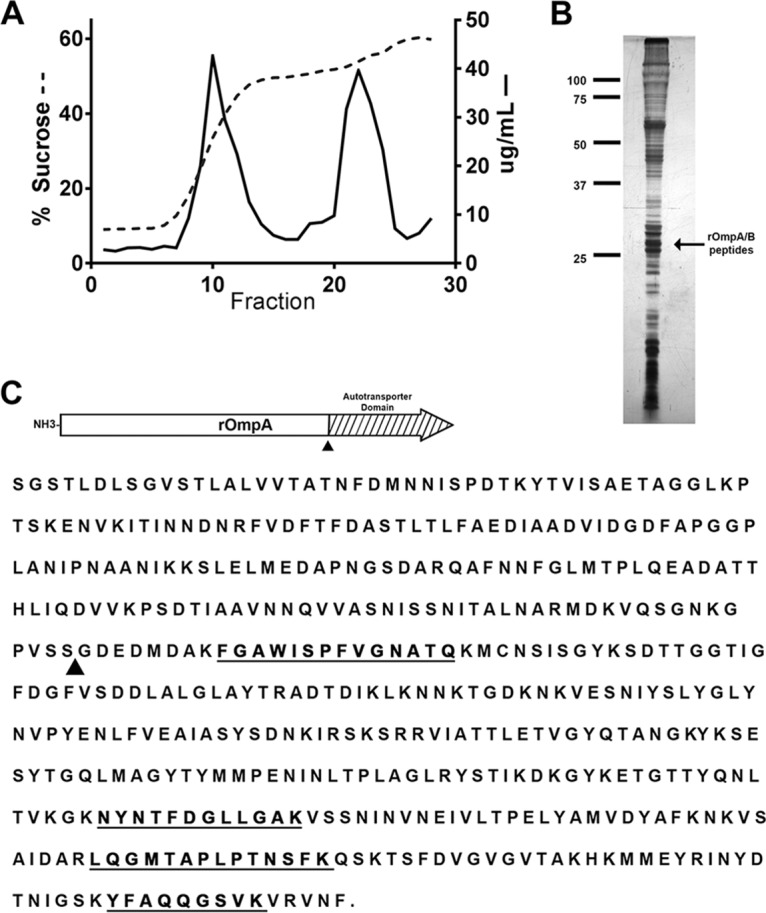
Identification of rOmpA autotransporter β fragment. (A) Sucrose gradient fractionation of the inner and outer membranes of R. rickettsia Sheila Smith. The percent sucrose is shown on the left y axis, and the protein concentration (μg/ml) is shown on the right y axis. The lower band from the sucrose gradient was collected for analysis. (B) Silver-stained polyacrylamide gel of the outer membrane fraction of R. rickettsii Sheila Smith. An arrow indicates the band from which rOmpA and rOmpB autotransporter peptides were identified. (C) Alignment of sequences of peptides identified by mass spectrometry with the amino acid sequence of rOmpA. An arrowhead indicates the position of the predicated cleavage site. Peptides identified by mass spectrometry are in bold and underlined.
Bioinformatic analysis of rOmpA and rOmpB autotransporter domains.
Type Va autotransporter proteins, of which rOmpA and rOmpB are members, follow a classic structure of N-terminal Sec signal, hypervariable passenger domain, and C-terminal transmembrane domain. rOmpA is encoded by a 6,750-bp open reading frame (ORF) in the R. rickettsii genome, with an estimated molecular mass of 224 kDa (26). Within its predicted passager domain, rOmpA contains 13 direct repeat units of approximately 75 amino acids each (26). Empirical evidence has shown that rOmpA migrates to an apparent molecular mass of 190 kDa (or 155 kDa) on SDS-PAGE (26–29). The discovery of an rOmpA β fragment has revealed that the discrepancy between the calculated and observed sizes is likely due to the posttranslational processing of rOmpA.
rOmpB is encoded by a 4,965-bp ORF with a calculated molecular mass of 168 kDa and processed into mature bands with apparent molecular masses of 120 and 32 kDa for the passenger domain and β fragment, respectively (30). Although both are members of the Sca autotransporter family (8), the passenger domains of these proteins are highly dissimilar. Alignment of the transmembrane domains of the two proteins displayed a high level of similarity and identity (45% and 28%, respectively). While the general features of the transmembrane domain are well conserved, the relatively low levels of specific identity allowed peptides identified by MS to be conclusively assigned to either rOmpA or rOmpB.
The cleavage site of rOmpB was previously known and compared to the sequence for rOmpA. A 40-amino-acid sequence beginning at the known cleavage site of rOmpB was globally aligned with the rOmpA sequence. A single alignment having 42% identify and 57% similarity beginning at Gly-1959 was seen in rOmpA. The alignment further showed that four out of the first six rOmpB β fragment amino acids (GDEXXD) were conserved in rOmpA as well. This site, which would cleave rOmpA into 190- and 32-kDa fragments, was considered a putative cleavage site for proteolytic processing. All additional Sca proteins in the R. rickettsii strain Sheila Smith genome containing intact transmembrane domains were aligned against this consensus sequence. Alignment revealed that all intact Sca proteins in strain Sheila Smith contain a conserved GDE sequence (Table 1).
TABLE 1.
Cleavage sites of the rOmpA and rOmpB transporters compared to the predicted cleavage sites of Sca1 and Sca2a
| Protein | Passenger domain | Autotransporter domainb |
|---|---|---|
| rOmpB | VAA | 1362GDEAIDNVAYG |
| rOmpA | VSS | 1959GDEDMDAKFGA |
| Sca1 | VGA | 1568GDEEESHIKRG |
| Sca2 | IGA | 1561GDEETSINRGV |
The last 3 amino acids of the passenger domain and the first 10 N-terminal amino acids of the autotransporter domain are shown.
The conserved GDE sequence is shown in bold.
The rOmpB and rOmpA β fragments maintain a high degree of biochemical similarity and are virtually inseparable on a standard one- or two-dimensional polyacrylamide gel. The rOmpA and rOmpB β fragments had calculated masses of 32.1 and 31.8 kDa and pIs of 9.03 and 9.09, respectively. Hydrophobicity plots and structural modeling of the β fragments revealed that the two fragments had a high degree of conformational similarity, with comparable compositions of hydrophobic and polar amino acids (Fig. 2). In order to confirm the cleavage site of rOmpA, recovery of the β fragment and N-terminal amino acid sequencing were performed. The recovery of the rOmpA β fragment was not possible due to the nearly identical biophysical properties between the rOmpA and rOmpB β fragments. Furthermore, the rOmpA β fragment is much less abundant, as it has been reported that the molar ratio of rOmpB to rOmpA is 9:1 (3). Because of the nearly identical properties of the β fragments of rOmpA and rOmpB, the band contained a mixture of both rOmpA and rOmpB. N-terminal Edman degradation revealed predominantly the rOmpB β fragment; however, secondary signals corresponding to 5 of the 6 unique amino acids in the first 10 amino acids of the rOmpA β fragment were also observed (Table 2). The last two amino acids of this sequence overlapped the first two amino acids of the first tryptic fragment of rOmpA observed by mass spectroscopy (Fig. 1C). The sequence obtained by these peptides confirmed the predicted cleavage site of rOmpA between Ser-1958 and Gly-1959.
FIG 2.
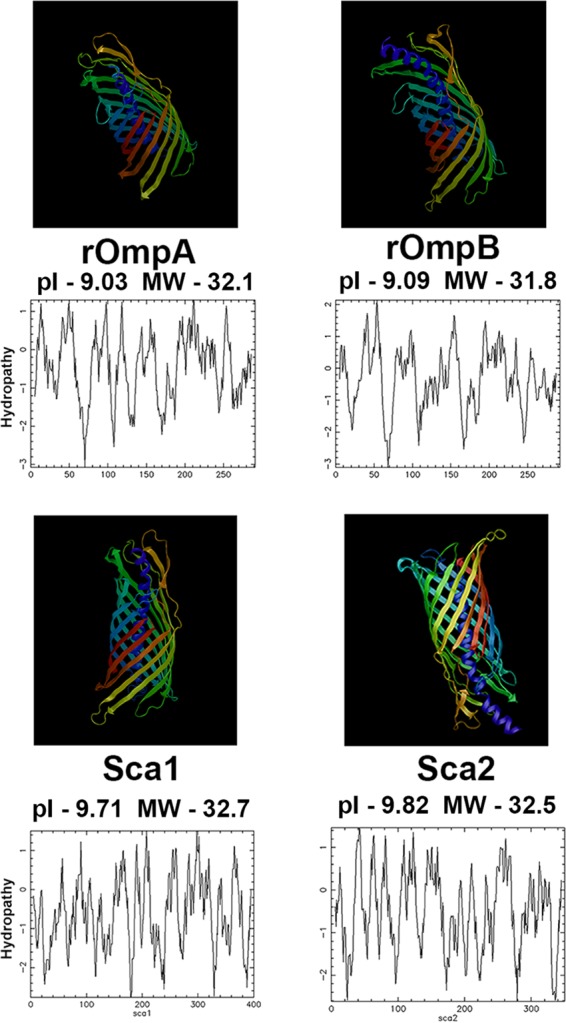
Structural prediction and hydrophobicity plots of rOmpA, rOmpB, Sca1, and Sca2 autotransporter regions. Amino acid sequences of the autotransporter region of rOmpA, rOmpB, Sca1, and Sca2 were analyzed by the Phyre2 protein modeling and prediction program. The model is colored based on sequence position from the N (blue) to the C (red) terminus. Ribbons represent β pleated sheets, coils represent α helices, and thin strands represent amino acid chains not predicted to affect secondary structure. Hydropathy plots of the autotransporter regions were created using EMBOSS Pepwindow. MW, molecular weight (in thousands).
TABLE 2.
N-terminal sequence identification of the rOmpA and rOmpB β fragments
| Fragment | Sequencea | Protein |
|---|---|---|
| First | GDEAIDNVAY | rOmpB |
| Second | ***-M*AKFG | rOmpA |
An asterisk indicates identity; a dash indicates no secondary sequence call.
c240 antibody is specific to rOmpA autotransporter region.
An antipeptide antibody, c240, specific to the transmembrane domain of rOmpA, was developed in rabbits. Immunoblotting (Fig. 3) revealed that the c240 antibody was reactive with a band of 32 kDa in lysates of Sheila Smith but was absent from both Iowa and the rompA::int312 strains, which do not produce rOmpA (31, 32).
FIG 3.
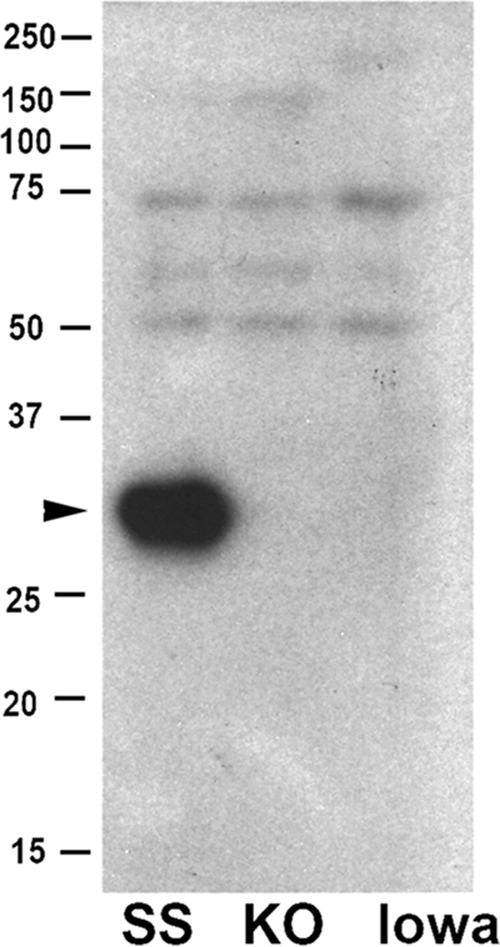
Western blot using antibody specific to rOmpA autotransporter. A Western blot of whole-cell rickettsial lysates probed with a specific antibody, c240, generated to the autotransporter region of rOmpA is shown. The peptide identified by Western blotting is specific only to strains carrying rOmpA and is the size predicted by cleavage of the autotransporter. Lanes: SS, Sheila Smith strain; KO, Sheila Smith ompA::int312 strain; Iowa, Iowa strain.
An R. rickettsii Iowa strain variant expressing rOmpA displays defects in rOmpA processing.
R. rickettsii Iowa is an avirulent spotted fever group rickettsia that was originally described as displaying marked changes in virulence, from mildly virulent to virulent to avirulent over many egg passages (33). A high-egg-passage variant (291 egg passages [EP]) was found to display a processing defect in rOmpB and the absence of rOmpA (21, 32). Recently, we discovered a seed stock from an early egg passage (4 EP) which was plaque purified on Vero cells and examined for differences. Two distinct plaque morphologies were observed (Fig. 4A): a large plaque (L) and a small plaque (S). Both were expanded for further characterization. Both the large- and small-plaque Iowa variants formed actin tails (Fig. 4B); thus, the difference in plaque size cannot be attributed to a failure to polymerize actin (9). The L clone displayed a higher growth rate than the S clone (Fig. 4C) and exhibited characteristics similar to those of the high-passage-number R. rickettsii Iowa described previously (21, 32).
FIG 4.
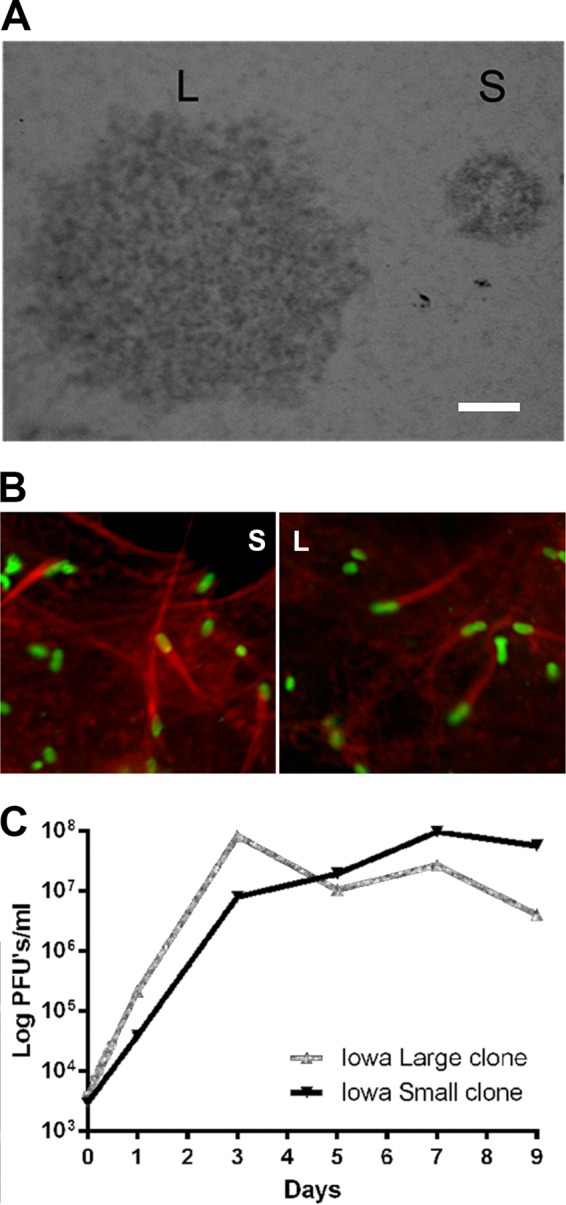
R. rickettsii Iowa low-egg-passage large- and small-plaque morphologies. (A) Vero cell monolayers infected with a low-passage-number (4 EP) Iowa strain revealed two distinct plaque types. A large-plaque variant (L) and a small-plaque variant (S) were subsequently cloned and expanded for further study. (B) Actin polymerization by the L and S variants. Monolayers of Vero cells were infected with the Iowa strain L and S variants for 24 h. Rickettsiae were detected by indirect immunofluorescence using monoclonal antibody 13-2 (1) to rOmpB; F actin was stained with 10 U/ml of rhodamine phalloidin. Bar, 1 mm. (C) Growth curve of the Iowa strain L and S variants. Rickettsiae were grown in Vero cells at 34°C, and samples were taken for cell disruption and for replating the lysates to enumerate the PFU. Data points are the means from three replicates. Error bars representing the standard errors of the mean (SEM) are not visible under the symbols.
An R. rickettsii Iowa variant expresses rOmpA.
The small- and large-plaque variants of R. rickettsii Iowa were density gradient purified for analysis by SDS-PAGE (Fig. 5A). Multiple high-molecular-mass bands were excised for identification by mass spectrometry. Surprisingly, the small-plaque variant of the Iowa strain was found to express rOmpA. The large-plaque variant exhibited a similar profile as the high-egg-passage strain, with relatively more of the unprocessed precursor of rOmpB than the mature form. The presence of rOmpA on the Iowa small clone was confirmed by immunofluorescence (Fig. 5B) using a monoclonal antibody specific to rOmpA (27).
FIG 5.
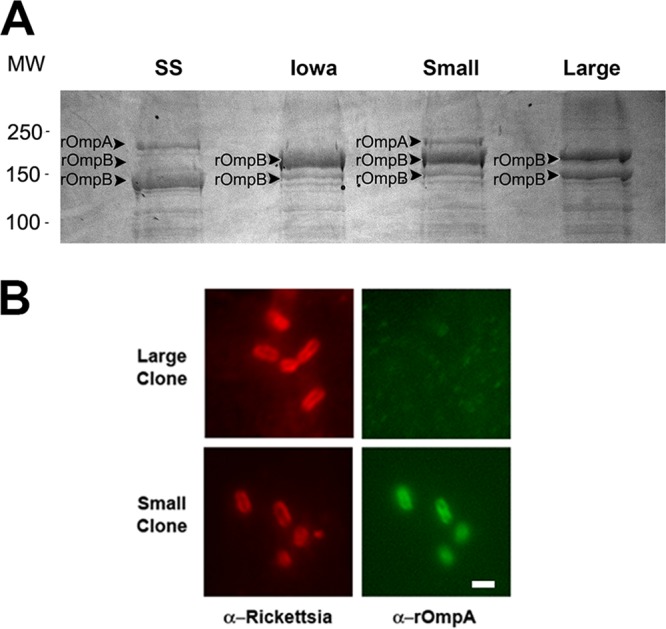
The presence of rOmpA in the small-plaque Iowa strain was confirmed by mass spectrometry and immunofluorescence. (A) Coomassie brilliant blue-stained SDS-PAGE gel of rickettsial L and S Iowa variants. The high-molecular-weight bands were excised and identified by mass spectrometry. MW, molecular weight in thousands. (B) Immunofluorescence with a monoclonal antibody to rOmpA confirms the presence of the protein in the S but not the L variant. α, anti.
Genomic sequence analysis of the Iowa large- and small-plaque variants.
The R. rickettsii Iowa large- and small-plaque variants were submitted for comparative genome sequencing. Importantly, the two variants displayed all the single-nucleotide polymorphisms that distinguished the Iowa strain from the virulent Morgan strain (34), thus confirming both as members of the R. rickettsii Iowa lineage (Table 3). In addition, the small-plaque variant contains an 11-bp insertion in RrIowa_0029, encoding the autotransporter Sca1, which disrupts the open reading frame to prematurely truncate Sca1 at amino acid 74 (Table 4). Both the large- and small-plaque variants of Iowa lack the 1-bp deletion within ompA that truncates rOmpA of the high-passage-number reference Iowa strain at amino acid 184 (and showing here as a single-base-pair insertion relative to strain Iowa). This single-base-pair insertion restores the open reading frame of rOmpA in both the large- and small-plaque variants. In the large-plaque variant, a subsequent 11-bp deletion disrupts the open reading frame and truncates rOmpA at amino acid 792 within the repeat region. The small-plaque variant, however, contains an in-frame 891-bp deletion within the repeat region that internally shortens the mature protein by 297 amino acids or approximately four repeat units but leaves the autotransporter domain intact (Fig. 6).
TABLE 3.
Nonsynonymous SNPs distinguishing Iowa and confirming the large- and small-plaque variantsa
| Coordinate | Gene | SNP |
M_aa | Product | |||
|---|---|---|---|---|---|---|---|
| Iowa | Small-plaque variant | Large-plaque variant | Morgan | ||||
| 65860 | RrIowa_0091 | A | A | A | G | S260G | Succinate dehydrogenase iron-sulfur subunit |
| 194500 | RrIowa_0228 | A | A | A | G | K13E | Hypothetical protein |
| 229483 | RrIowa_0265 | A | A | A | C | L6W | Prolyl endopeptidase family |
| 250900 | RrIowa_0290 | A | A | A | G | *68W | Hypothetical protein/ankyrin |
| 857393 | RrIowa_1080 | A | A | A | G | M257T | Arp2/3 complex activation protein/RickA |
| 882319 | RrIowa_1113 | T | T | T | G | D27A | Hypothetical protein/ankyrin repeat |
| 1036868 | RrIowa_1321 | T | T | T | C | K99E | RNase H |
| 1090471 | RrIowa_1396 | A | A | A | G | I4M | Hypothetical protein/endonuclease subunit |
| 1127496 | RrIowa_1431 | A | A | A | G | K208R | Methyltransferase |
| 1252300 | RrIowa_1582 | A | A | A | G | F25S | Type I restriction-modification system methylation subunit |
Single-nucleotide polymorphisms (SNPs) which distinguish the R. rickettsii Iowa strain from Sheila Smith, R, and Morgan strains (34). Coordinates are from the reference Iowa strain (CP000766.3). M_aa, amino acid change in Morgan relative to the Iowa strain. The asterisk indicates a stop codon.
TABLE 4.
Indels unique to Iowa variantsa
| Coordinate | Gene | Indel size (bp) |
Type | Product | |||
|---|---|---|---|---|---|---|---|
| Iowa | Small-plaque variant | Large-plaque variant | Morgan | ||||
| 20389 | RrIowa_0029 | 11 | Insertion | Sca1 | |||
| 376310 | RrIowa_0450 | 11 | Insertion | Hypothetical protein | |||
| 1128779 | RrIowa_1432 | 3 | Deletion | Hypothetical protein | |||
| 1180402 | RrIowa_1493 | 891 | Insertion | Outer membrane protein A | |||
| 1180004 | RrIowa_1493 | 11 | Deletion | Outer membrane protein A | |||
| 1181871 | RrIowa_1494 | 1 | 1 | 1 | Insertion | Outer membrane protein A | |
Insertions and deletions which distinguish the R. rickettsii Iowa strain from the Iowa strain variants and from the Morgan strain. Coordinates are from the reference Iowa strain (CP000766.3).
FIG 6.
Schematic of the truncations of the R. rickettsii rOmpA autotransporter from the high- and low-passage-number variants of the Iowa strain in comparison to that from the virulent Sheila Smith strain.
Defective processing of rOmpA by the Iowa strain.
Because the small-plaque variant of the Iowa strain expresses rOmpA, we were able to examine whether it displayed a deficiency in the processing of rOmpA similar to that observed for rOmpB (21). Immunoblotting with monoclonal antibody 13-3 (27) identified a single, mature rOmpA band of R. rickettsii Sheila Smith and Hino but identified two high-molecular-mass species in the Iowa small-plaque variant. This profile is reminiscent of that of rOmpB in the Iowa strain, which exhibits both the unprocessed precursor and the proteolytically processed mature form. As previously shown (32) and confirmed by DNA sequencing, the high-egg-passage Iowa strain and the large clone do not express rOmpA (Fig. 7A). Immunoblotting with the c240 antibody against the autotransporter domain of rOmpA confirmed the predominant presence of the unprocessed precursor form of rOmpA in the small-plaque variant with a lesser amount of the cleaved β fragment (Fig. 7B). No unprocessed precursor is detected in either of the two virulent strains examined, and only the cleaved β fragment is observed. All of the Iowa variants are defective in the processing of rOmpB (Fig. 7C).
FIG 7.
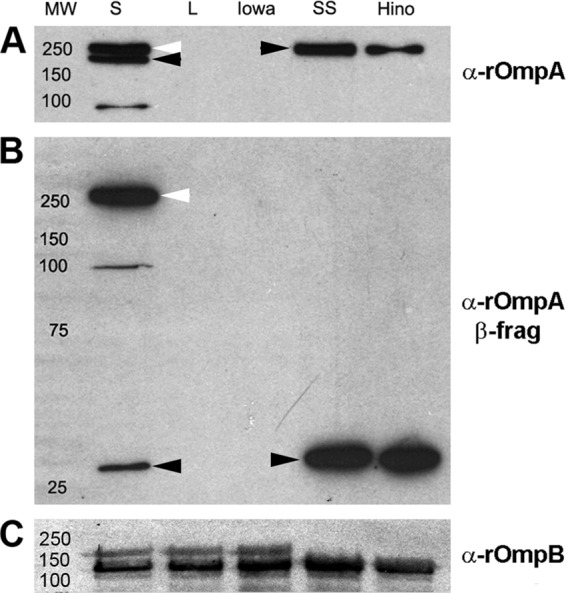
Defect of processing of rOmpA in the R. rickettsii low-egg-passage Iowa small-plaque variant. (A) Immunoblot of the low-egg-passage Iowa small (S)- and large (L)-plaque variants, high-egg-passage Iowa, and virulent R. rickettsii Sheila Smith (SS) or Hino strains with a monoclonal antibody (13-3) against the rOmpA passenger domain (α, anti). The unprocessed precursor forms are indicated by white arrowheads, and the proteolytically processed passenger domains are indicated by black arrowheads. (B) Immunoblot of strains described for panel A using the rabbit polyclonal antibody against the rOmpA β fragment. The unprocessed precursor forms are indicated by white arrowheads, and the proteolytically processed autotransporter domains are indicated by black arrowheads. (C) Immunoblot against rOmpB using a polyclonal rabbit antibody against the passenger domain of rOmpB demonstrating the deficiency in rOmpB cleavage by all of the Iowa variants. It is important to note that this particular antibody preferentially recognizes the processed, passenger domain of rOmpB despite its being a relatively minor species in the Iowa strain.
Early-passage Iowa strains show no evidence of virulence in an animal model.
Because the Iowa strain had been reported to exhibit fluctuating virulence through early laboratory passages (33), we tested the early large- and small-plaque variants of the Iowa strain for comparison to high-passage-number Iowa and the virulent Sheila Smith strain in a guinea pig model of infection (Fig. 8). None of the Iowa variants elicited fever in this model, regardless of the presence of rOmpA. All variants infected the animals, as demonstrated by seroconversion. Inoculation with the same mass of formalin-killed rickettsiae as used for infection did not lead to seroconversion, indicating that replication had occurred.
FIG 8.
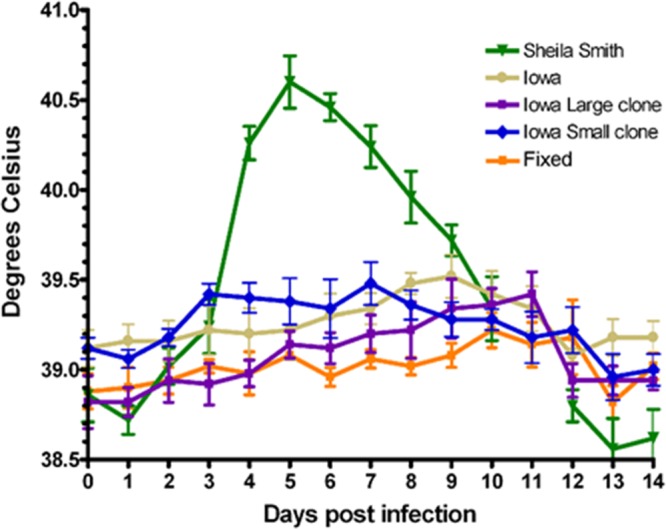
Determination of virulence of R. rickettsii Iowa low-egg-passage large- and small-plaque variants in guinea pigs. Female Hartley guinea pigs were inoculated intraperitoneally with 1,000 PFU. Animal temperatures were monitored for 14 days. Each point represents the mean temperature for five animals ± SEM.
DISCUSSION
Autotransporters represent a class of proteins which are transported to the outer membrane of Gram-negative bacteria without external sources of energy (25). Autotransporters are a large and diverse family of proteins but share a structure with an N-terminal secretory signal peptide, a passenger domain containing diverse sequences conveying an array of functions, and a conserved carboxy-terminal transmembrane (β) domain which functions to translocate the passenger domain across the bacterial outer membrane (18–20). The passenger domains are frequently associated with virulence functions (24). R. rickettsii encodes at least four autotransporters: rOmpA, rOmpB, Sca1, and Sca2. rOmpA and rOmpB are protective immunogens, and both of these proteins have been implicated in adhesion and/or entry based on either inhibition by specific antibodies (13, 15, 35) or altered interactions of Escherichia coli expressing these with host cells (12, 14). Sca2 is a nucleator of actin and is involved in the actin-based motility common to spotted fever group rickettsiae (9, 10). Here we show that like the β fragment of rOmpB (21, 30), the β fragment of rOmpA is proteolytically cleaved from the precursor and that all putative autotransporters of R. rickettsii possess an identical GDE sequence at the predicted cleavage site.
The similarities of the rOmpA and rOmpB β domains had previously prevented identification of the rOmpA β fragment. rOmpB is the most predominant protein on the rickettsial outer membrane and has a molar concentration roughly 9 times higher than that of rOmpA (3, 36–38). The rOmpA and rOmpB β fragments have nearly identical masses (32.1 kDa and 31.7 kDa, respectively) and pIs (9.37 and 9.48) and thus migrate to the same position on both one- and two-dimensional gels, obscuring the rOmpA β fragment. It has been suspected that rOmpA may be processed, since the observed size of rOmpA is significantly smaller than its predicted form. The evidence for an rOmpA postprocessing event accounts for this discrepancy in size.
Rickettsia species have reduced genomes (7, 39) reflected in the fragmented distribution and pseudogenization of Sca proteins within the genus (8), yet rOmpA is conserved within the spotted fever group and rOmpB is conserved throughout the Rickettsia genus. Furthermore, there is evidence that both rOmpA and rOmpB evolve under positive selection (8). However, rOmpA is not essential for replication in vitro (32) and does not contribute to virulence in a guinea pig model of infection (31). The fact that rOmpA is evolutionarily conserved among spotted fever group rickettsiae in nature suggests that it may have an essential role in other aspects of rickettsial natural history, such as survival in the tick host.
The processing of autotransporters in R. rickettsii and the display of mature proteins on the outer membrane are likely important events in rickettsial pathogenesis. rOmpB is expressed as a precursor of approximately 168 kDa that is then cleaved into a mature 120-kDa passenger domain and 32-kDa β fragment (21, 30). A fully attenuated strain of R. rickettsii, Iowa, contains a defect in the processing of rOmpB and is the only known strain to date that carries this processing defect (34). Observations of the pre- and postprocessed forms of rOmpB in Iowa revealed that the immature, unprocessed protein is the dominant form in vivo (21).
The discovery of an early-egg-passage aliquot was fortuitous. When the Iowa strain was first described (33), plaque purification procedures for rickettsiae had not yet been developed. Plaque assay of this early-egg-passage seed stock identified two distinct plaque types that were confirmed as the Iowa strain by the presence of unique single-nucleotide polymorphisms. One of these variants expressed a nearly intact rOmpA, although it exhibited an in-frame 891-bp deletion in the repeat region of the gene that caused it to be somewhat truncated. The other variant, like the high-egg-passage Iowa strain originally described (33), was truncated within the repeat region at a site downstream of the high-egg-passage reference Iowa strain. These observations suggest that the loss of rOmpA occurred relatively soon after isolation and further imply that the presence of an unprocessed rOmpA may be deleterious to the rickettsiae.
rOmpA is not essential for replication in cell culture or mammals (31, 32), and the unprocessed form appears to be selected against in laboratory passage. The finding that Sca1 was truncated in one of the Iowa variants suggests that the presence of any of the autotransporters in an unprocessed form may be deleterious to the rickettsiae. Notably, rOmpB has not been observed to be truncated or disrupted in the Iowa strain, although it is processed inefficiently. rOmpB is conserved among all rickettsiae of both the spotted fever and typhus groups and thus may be essential (8). Interestingly, whatever defect is present in the putative outer membrane protease that is apparently responsible for the processing of both rOmpB and rOmpA, it appears to not completely block proteolytic processing but only reduces the efficiency. We speculate that the small amount of correctly processed rOmpB may be sufficient to allow growth in cell culture and that complete disruption of the putative outer membrane protease may be toxic.
Autotransporters may be postprocessed either through autocatalysis or cleavage by an outer membrane protease. The findings here suggest that both rOmpA and rOmpB are cleaved through an external process and not through autocatalysis. The amino acid sequence of rOmpB from Iowa does not contain amino acid sequence differences from other processed forms of rOmpB from the closely related virulent Morgan, Hauke, and Hino strains (34), none of which contain any demonstrable error in rOmpB processing. Therefore, the possibility that a genetic mutation has impaired the autocatalytic activity of rOmpB in Iowa is remote. Currently, the only known strain defective for the postprocessing of rOmpB is Iowa, which is also defective in the production of rOmpA. Autotransporters may not only have functions in pathogenesis such as host activity modulation, motility, and cytotoxicity but may also themselves function as proteases that process other transporters (40). We had previously questioned whether the absence of rOmpA from the Iowa strain might account for the lack of processing of rOmpB (32). An isogenic rompA mutant (31) in the highly virulent Sheila Smith strain is able to process rOmpB in a manner identical to the wild type. Therefore, the ability of R. rickettsii to process rOmpB does not seem to depend on the presence of rOmpA. Furthermore, sequence analysis of the four intact Sca proteins in R. rickettsii revealed a motif with a consensus GDE amino acid sequence immediately downstream of the predicted cleavage sites, suggesting that a specific outer membrane protease may be responsible for the processing of all the R. rickettsii autotransporters.
MATERIALS AND METHODS
Rickettsial strains and purification.
R. rickettsii strains Sheila Smith and Iowa (33) were grown and propagated in Vero cells in M199 medium and purified by Renografin density gradient centrifugation (41). Density gradient-purified bacteria were then serially diluted and plated onto Vero cells and overlaid with M199 agar to determine the number of PFU by plaque assay (32). Direct rickettsial counts were performed in parallel by acridine orange staining and filtration onto black Nuclepore filters essentially as previously described (43).
Comparative genome sequencing.
R. rickettsii genomic DNA from approximately 1 × 1010 purified organisms was extracted using an UltraClean microbial DNA isolation kit (Mo Bio). Approximately 2 μg of genomic DNA was provided to the Rocky Mountain Laboratories Genomics Core Facility for comparative genome sequencing. DNAs from the R. rickettsii Iowa large-plaque and small-plaque variants were compared to R. rickettsii reference strain Iowa (CP000766). Libraries were run as 2 × 300 bp paired-end reads on an Illumina MiSeq sequencer, which produced ∼10 million reads per sample. Reads were trimmed for TruSeq Adapter sequence and trimmed and filtered for low-quality sequence using the FASTX-Toolkit. The resulting reads were mapped to the R. rickettsia strain Iowa genome (CP000766.3) using Bowtie2 (44). The assembly was corrected using the Pilon (45) genome assembly improvement tool.
Bioinformatic analysis of rOmpA.
rOmpA (A1G_06990), rOmpB (A1G_06030), Sca1 (A1G_00130), and Sca2 (A1G_00670) sequences were from the R. rickettsii Sheila Smith genome (RefSeq NC_009882.1). Global pairwise alignment of rOmpA and rOmpB was performed using EMBOSS Stretcher with the Needleman-Wunsch algorithm for alignment optimization. Hydropathy plots of the amino acid sequences of rOmpA and rOmpB from the annotated autotransporter domain were performed using the Kyte-Doolittle moving-scale average (46). Structural prediction and modeling of the rOmpA, rOmpB, Sca1, and Sca2 autotransporter domains were accomplished using the Phyre2 suite (47). Multilocus sequence alignment of all Sheila Smith Sca proteins was accomplished using Clustal Omega (48).
Membrane fractionation.
Membrane fractionation of R. rickettsii was as described previously with some modifications (42, 49, 50). Briefly, density gradient-purified R. rickettsii bacteria were pelleted, and approximately 5 × 109 bacteria were washed in TE (50 mM Tris [pH 7.5], 5 mM EDTA) with 1 mM dithiothreitol (DTT) and resuspended in buffer L (50 mM Tris [pH 7.5], 5 mM EDTA, 1 mM DTT, 250 mM sucrose, 50 μg/ml DNase) supplemented with X ProBlock Gold bacterial 2D protease inhibitor cocktail (Goldbio, St. Louis, MO). Cells were then ruptured in a cell disruption vessel by nitrogen cavitation (Parr Instrument Company, Moline, IL). After bacterial lysis, disrupted cells were centrifuged at 4,000 rpm for 10 min at 4°C in order to remove intact cells. The crude lysate was then pelleted by centrifugation in an SW41 swinging-bucket rotor at 175,000 × g for 1 h at 4°C. The membrane pellet was resuspended in buffer L and then layered over a sucrose gradient from 35 to 50%, and the gradient was centrifuged in an SW41 rotor for 16 h at 4°C. Fractions were collected in 500-μl aliquots. The outer membrane fraction was pooled and run on a 10% polyacrylamide gel.
Mass spectrometry of rickettsial proteins.
The outer membrane fraction of R. rickettsii Sheila Smith was separated by SDS-PAGE and stained with a SilverQuest silver stain kit. Whole-cell lysates of R. rickettsii Sheila Smith, Iowa, and the large- and small-plaque variants of early-egg-passage Iowa were run on SDS-PAGE gels and stained with Coomassie brilliant blue. The bands of interest were excised, and the gel slices were processed essentially as described previously (51). Matrix-assisted laser desorption ionization–time of flight spectra were collected using an Applied Biosystems 4800 MS/MS MALDI TOF/TOF analyzer.
Antibody production.
A polyclonal rabbit antibody to the β fragment of rOmpA was contractually developed (Thermo Scientific, Atlanta, GA). The distal carboxy-terminal end of the rOmpA protein was selected as a potential target, and a custom peptide with the sequence KQSKTSFDVGVGVTAKHK was generated. The peptide is initiated ∼240 amino acids downstream of the annotated autotransporter domain and was thus labeled “c240.” Two rabbits were immunized, and antibody from the terminal bleed for each animal was then antigen affinity purified.
Immunoblotting, immunoprecipitation, and immunofluorescence.
Immunoblot analyses with monoclonal antibodies (MAbs) 13-2 (anti-rOmpB) and 13-3 (anti-rOmpA) or affinity-purified c240 antibody (27) were performed as previously described (31).
Immunofluorescence was performed with MAbs 13-2 and 13-3 and rabbit polyclonal antibody c240. Vero cells were infected with rickettsiae at a multiplicity of infection (MOI) of 5 overnight at 37°C in M199 medium. Monolayers were fixed in 3.7% paraformaldehyde and permeabilized with phosphate-buffered saline (PBS) with 0.01% Triton X-100 and 0.05% sodium dodecyl sulfate (SDS). Fixed coverslips were stained with a rabbit anti-R. rickettsii antibody and either MAb 13-2 or 13-3, washed, and detected with anti-mouse antibody–Alexa Fluor 488 or anti-rabbit antibody–Alexa Fluor 594. Images were acquired on a Nikon Eclipse 80i microscope with a 60× 1.4-numerical-aperture oil immersion objective and a Nikon DS-Qi1Mc camera.
N-terminal sequencing.
Outer membrane fractions or whole-cell lysates of R. rickettsii Sheila Smith were transferred to a polyvinylidene difluoride (PVDF) membrane as described above. The membrane was stained with amido black (0.1% amido black 10B, 25% isopropanol, 10% acetic acid) for 10 min, followed by destaining with several changes of distilled water. The appropriate 32-kDa bands were cut and submitted for N-terminal amino acid sequence determination by Edman degradation at the University of Nebraska Protein Structure Core Facility (Omaha, NE).
Guinea pig inoculations.
Female Hartley strain guinea pigs (average weight, 400 g) were purchased from Charles River Laboratories, MA, and housed in an animal biosafety level 3 laboratory under a protocol approved by the Rocky Mountain Laboratories Animal Care and Use Committee (protocol 2009-42). Guinea pigs were inoculated intraperitoneally with R. rickettsii strains Sheila Smith, Iowa, and Iowa large- and small-plaque variants at 1,000 PFU. Guinea pigs were inoculated intraperitoneally with the same amount of formaldehyde-fixed strain Sheila Smith or an equal volume of diluent control. Animal temperatures were monitored for 14 days after infection. The animals were sacrificed on day 30 after collection of sera via heart puncture under deep anesthesia.
Accession number(s).
Genome sequence and annotation files have been deposited in GenBank under accession numbers CP018914 for the R. rickettsii Iowa small-plaque variant and CP018913 for the R. rickettsii Iowa large-plaque variant.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the NIAID/NIH.
We thank Craig Martens and the RML Genomics Core/Research Technologies Section for the genomic sequencing and analysis.
REFERENCES
- 1.Hackstadt T. 1996. The biology of rickettsiae. Infect Agents Dis 5:127–143. [PubMed] [Google Scholar]
- 2.Walker DH, Fishbein DB. 1991. Epidemiology of rickettsial diseases. Eur J Epidemiol 7:237–245. doi: 10.1007/BF00145672. [DOI] [PubMed] [Google Scholar]
- 3.Policastro PF, Hackstadt T. 1994. Differential activity of Rickettsia rickettsii ompA and ompB promoter regions in a heterologous reporter gene system. Microbiology 140:2941–2949. doi: 10.1099/13500872-140-11-2941. [DOI] [PubMed] [Google Scholar]
- 4.Ching W-M, Dasch GA, Carl M, Dobson ME. 1990. Structural analyses of the 120-kDa serotype protein antigens of typhus group rickettsiae, p 334–351. In Hechemy KE, Paretsky D, Walker DH, Mallavia LP (ed), Rickettsiology: current issues and perspectives. The New York Academy of Sciences, New York, NY. [DOI] [PubMed] [Google Scholar]
- 5.Sleytr UB, Messner P. 1983. Crystalline surface layers on bacteria. Annu Rev Microbiol 37:311–339. doi: 10.1146/annurev.mi.37.100183.001523. [DOI] [PubMed] [Google Scholar]
- 6.McLeod MP, Qin X, Karpathy SE, Gioia J, Highlander SK, Fox GE, McNeill TZ, Jiang H, Muzny D, Jacob LS, Hawes AC, Sodergren E, Gill R, Hume J, Morgan M, Fan G, Amin AG, Gibbs RA, Hong C, Yu XJ, Walker DH, Weinstock GM. 2004. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol 186:5842–5855. doi: 10.1128/JB.186.17.5842-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogata H, Audic S, Renesto-Audiffren P, Fournier PE, Barbe V, Samson D, Roux V, Cossart P, Weissenbach J, Claverie JM, Raoult D. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- 8.Blanc G, Ngwamidiba M, Ogata H, Fournier PE, Claverie JM, Raoult D. 2005. Molecular evolution of Rickettsia surface antigens: evidence of positive selection. Mol Biol Evol 22:2073–2083. doi: 10.1093/molbev/msi199. [DOI] [PubMed] [Google Scholar]
- 9.Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. 2010. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun 78:2240–2247. doi: 10.1128/IAI.00100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haglund CM, C JE, Skau CT, Kovar DR, Welch MD. 2010. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol 12:1057–1063. doi: 10.1038/ncb2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park H, Lee JH, Gouin E, Cossart P, Izard T. 2011. The rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin. J Biol Chem 286:35096–35103. doi: 10.1074/jbc.M111.263855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan GY, Cardwell MM, Hermanas TM, Uchiyama R, Martinez JJ. 2009. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell Microbiol 11:629–644. doi: 10.1111/j.1462-5822.2008.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Walker DH. 1998. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb Pathog 24:289–298. doi: 10.1006/mpat.1997.0197. [DOI] [PubMed] [Google Scholar]
- 14.Hillman RD Jr, Baktash YM, Martinez JJ. 2013. OmpA-mediated rickettsial adherence to and invasion of human endothelial cells is dependent upon interaction with alpha2beta1 integrin. Cell Microbiol 15:727–741. doi: 10.1111/cmi.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao Y, Wen B, Chen M, Niu D, Zhang J, Qiu L. 2005. Analysis of immunoprotectivity of the recombinant OmpA of Rickettsia heilongjiangensis. Ann N Y Acad Sci 1063:261–265. doi: 10.1196/annals.1355.042. [DOI] [PubMed] [Google Scholar]
- 16.Riley SP, Goh KC, Hermanas TM, Cardwell MM, Chan YG, Martinez JJ. 2010. The Rickettsia conorii autotransporter protein Sca1 promotes adherence to nonphagocytic mammalian cells. Infect Immun 78:1895–1904. doi: 10.1128/IAI.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardwell MM, Martinez JJ. 2009. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect Immun 77:5272–5280. doi: 10.1128/IAI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dautin N, Bernstein HD. 2007. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol 61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- 19.Grijpstra J, Arenas J, Rutten L, Tommassen J. 2013. Autotransporter secretion: varying on a theme. Res Microbiol 164:562–582. doi: 10.1016/j.resmic.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Benz I, Schmidt MA. 2011. Structures and functions of autotransporter proteins in microbial pathogens. Int J Med Microbiol 301:461–468. doi: 10.1016/j.ijmm.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Hackstadt T, Messer R, Cieplak W, Peacock MG. 1992. Evidence for the proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect Immun 60:159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suhr M, Benz I, Schmidt MA. 1996. Processing of the AIDA-I precursor: removal of AIDAc and evidence for the outer membrane anchoring as a beta-barrel structure. Mol Microbiol 22:31–42. doi: 10.1111/j.1365-2958.1996.tb02653.x. [DOI] [PubMed] [Google Scholar]
- 23.Charbonneau ME, Janvore J, Mourez M. 2009. Autoprocessing of the Escherichia coli AIDA-I autotransporter: a new mechanism involving acidic residues in the junction region. J Biol Chem 284:17340–17351. doi: 10.1074/jbc.M109.010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson IR, Nataro JP. 2001. Virulence functions of autotransporter proteins. Infect Immun 69:1231–1243. doi: 10.1128/IAI.69.3.1231-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pohlner J, Halter R, Beyreuther K, Meyer TF. 1987. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 26.Anderson BE, McDonald GA, Jones DC, Regnery RL. 1990. A protective protein antigen of Rickettsia rickettsii has tandemly repeated, near-identical sequences. Infect Immun 58:2760–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anacker RL, Mann RE, Gonzales C. 1987. Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. J Clin Microbiol 25:167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilmore RD Jr, Hackstadt T. 1991. DNA polymorphism in the conserved 190 kDa antigen gene repeat region among spotted fever group rickettsiae. Biochim Biophys Acta 1097:77–80. doi: 10.1016/0925-4439(91)90027-7. [DOI] [PubMed] [Google Scholar]
- 29.Crocquet-Valdes PA, Weiss K, Walker DH. 1994. Sequence analysis of the 190-kDa antigen-encoding gene of Rickettsia conorii (Malish 7 strain). Gene 140:115–119. doi: 10.1016/0378-1119(94)90740-4. [DOI] [PubMed] [Google Scholar]
- 30.Gilmore RD Jr, Cieplak W, Policastro PF, Hackstadt T. 1991. The 120 kilodalton outer membrane protein (rOmp B) of Rickettsia rickettsii is encoded by an unusually long open reading frame: evidence for protein processing from a large precursor. Mol Microbiol 5:2361–2370. doi: 10.1111/j.1365-2958.1991.tb02082.x. [DOI] [PubMed] [Google Scholar]
- 31.Noriea NF, Clark TR, Hackstadt T. 2015. Targeted knockout of the Rickettsia rickettsii OmpA surface antigen does not diminish virulence in a mammalian model system. mBio 6:e00323-15. doi: 10.1128/mBio.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Porcella SF, Hackstadt T. 2008. Genomic comparison of virulent Rickettsia rickettsii Shiela Smith and avirulent Rickesttia rickettsii Iowa. Infect Immun 76:542–550. doi: 10.1128/IAI.00952-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox HR. 1941. Cultivation of rickettsiae of the rocky mountain spotted fever, typhus and Q fever groups in the embryonic tissues of developing chicks. Science 94:399–403. doi: 10.1126/science.94.2444.399. [DOI] [PubMed] [Google Scholar]
- 34.Clark TR, Noriea NF, Bublitz DC, Ellison DW, Martens C, Lutter EI, Hackstadt T. 2015. Comparative genome sequencing of Rickettsia rickettsii strains that differ in virulence. Infect Immun 83:1568–1576. doi: 10.1128/IAI.03140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anacker RL, List RH, Mann RE, Hayes SF, Thomas LA. 1985. Characterization of monoclonal antibodies protecting mice against Rickettsia rickettsii. J Infect Dis 151:1052–1060. doi: 10.1093/infdis/151.6.1052. [DOI] [PubMed] [Google Scholar]
- 36.Ching WM, Wang H, Jan B, Dasch GA. 1996. Identification and characterization of epitopes on the 120-kilodalton surface protein antigen of Rickettsia prowazekii with synthetic peptides. Infect Immun 64:1413–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dasch GA. 1981. Isolation of species-specific protein antigens of Rickettsia typhi and Rickettsia prowazekii for immunodiagnosis and immunoprophylaxis. J Clin Microbiol 14:333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer EL, Martin ML, Mallavia L. 1974. Ultrastucture of the surface of Rickettsia prowazeki and Rickettsia akari. Appl Microbiol 28:713–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersson JO, Anderson SG. 1999. Genome degradation is an ongoing process in Rickettsia. Mol Biol. Evol 16:1178–1191. doi: 10.1093/oxfordjournals.molbev.a026208. [DOI] [PubMed] [Google Scholar]
- 40.van Ulsen P, van Alphen L, ten Hove J, Fransen F, van der Ley P, Tommassen J. 2003. A neisserial autotransporter NalP modulating the processing of other autotransporters. Mol Microbiol 50:1017–1030. doi: 10.1046/j.1365-2958.2003.03773.x. [DOI] [PubMed] [Google Scholar]
- 41.Weiss E, Coolbaugh JC, Williams JC. 1975. Separation of viable Rickettsia typhi from yolk sac and L cell host components by renografin density gradient centrifugation. Appl Microbiol 30:456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao Y, Johnson HM, Bazemore-Walker CR. 2012. Improved enrichment and proteomic identification of outer membrane proteins from a Gram-negative bacterium: focus on Caulobacter crescentus. Proteomics 12:251–262. doi: 10.1002/pmic.201100288. [DOI] [PubMed] [Google Scholar]
- 43.Moos A, Hackstadt T. 1987. Comparative virulence of intra- and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect Immun 55:1144–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 47.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikaido H. 1994. Isolation of outer membranes. Methods Enzymol 235:225–234. doi: 10.1016/0076-6879(94)35143-0. [DOI] [PubMed] [Google Scholar]
- 50.Beis K, Whitfield C, Booth I, Naismith JH. 2006. Two-step purification of outer membrane proteins. Int J Biol Macromol 39:10–14. doi: 10.1016/j.ijbiomac.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clifton DR, Fields KA, Grieshaber S, Dooley CA, Fischer E, Mead D, Carabeo RA, Hackstadt T. 2004. A chlamydial type III translocated protein is trosine phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A 101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]