Abstract
Background & Aims
Emerging data on the gut microbiome in autism spectrum disorder (ASD) suggest that altered host–microbe interactions may contribute to disease symptoms. Although gut microbial communities in children with ASD are reported to differ from individuals with neurotypical development, it is not known whether these bacteria induce pathogenic neuroimmune signals.
Methods
Because commensal clostridia interactions with the intestinal mucosa can regulate disease-associated cytokine and serotonergic pathways in animal models, we evaluated whether microbiome-neuroimmune profiles (from rectal biopsy specimens and blood) differed in ASD children with functional gastrointestinal disorders (ASD-FGID, n = 14) compared with neurotypical (NT) children with FGID (NT-FGID, n = 15) and without abdominal pain (NT, n = 6). Microbial 16S ribosomal DNA community signatures, cytokines, and serotonergic metabolites were quantified and correlated with gastrointestinal symptoms.
Results
A significant increase in several mucosa-associated Clostridiales was observed in ASD-FGID, whereas marked decreases in Dorea and Blautia, as well as Sutterella, were evident. Stratification by abdominal pain showed multiple organisms in ASD-FGID that correlated significantly with cytokines (interleukin [IL]6, IL1, IL17A, and interferon-γ). Group comparisons showed that IL6 and tryptophan release by mucosal biopsy specimens was highest in ASD children with abdominal pain, whereas serotonergic metabolites generally were increased in children with FGIDs. Furthermore, proinflammatory cytokines correlated significantly with several Clostridiales previously reported to associate with ASD, as did tryptophan and serotonin.
Conclusions
Our findings identify distinctive mucosal microbial signatures in ASD children with FGID that correlate with cytokine and tryptophan homeostasis. Future studies are needed to establish whether these disease-associated Clostridiales species confer early pathogenic signals in children with ASD and FGID.
Keywords: Microbiome, Microbiome–Gut–Brain Axis, Gastrointestinal Disorders, Mucosa, Serotonin
Abbreviations used in this paper: ASD, autism spectrum disorder; 5-HIAA, 5-hydroxyindoleacetic acid; 5-HT, serotonin; FGID, functional gastrointestinal disorder; GI, gastrointestinal; GM-CSF, granulocyte-macrophage colony-stimulating factor; GROα, growth-related oncogene alpha; IBS, irritable bowel syndrome; IFN, interferon; IL, interleukin; IP, interferon gamma-induced protein; MCP-1, monocyte chemoattractant protein; MIP, macrophage inflammatory protein; NT, neurotypical; OTU, operational taxonomic unit; QPGS-RIII, Questionnaire on Pediatric Gastrointestinal Symptoms-Rome III; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor
Graphical Abstract
See editorial on page 131.
Summary.
We evaluated whether microbiome-neuroimmune profiles differed in children with autism spectrum disorder and functional gastrointestinal disorders, as compared with neurotypical children. Our findings identified distinctive mucosal microbial signatures in children with autism spectrum disorder and functional gastrointestinal disorders that correlated with cytokine and tryptophan homeostasis.
Gastrointestinal (GI) dysfunction is a critical factor influencing the quality of life in individuals with autism spectrum disorder (ASD) and may contribute to behavioral challenges.1 A clinical meta-analysis of functional GI disorders (FGID) supports the notion that GI dysfunction occurs more frequently in children with ASD than in children with neurotypical development.2 GI disturbances ranging from constipation to diarrhea are widely documented in this population and are complicated further by the nonverbal and/or limited communication abilities of these children.1, 2, 3, 4, 5
Most occurrences of GI dysfunction in children with ASD are the result of functional causes, rather than being of anatomic or known physiologic origin.1, 3, 6 Such FGIDs encompass a broad range of disorders including functional constipation, nonretentive fecal incontinence, functional abdominal pain, abdominal migraines, and irritable bowel syndrome.7 The etiology of FGIDs is poorly understood in children with neurotypical development, let alone in ASD. A common cause of FGIDs is thought to emanate from disturbances to normal communications between the brain and gut—referred to as the brain-gut axis,8 and more recently as the microbiome–gut–brain axis.9, 10 Altered microbiome–gut–brain signals also have been reported in ASD, and may contribute independently toward clinical symptoms by changing microbiome composition, tryptophan–serotonin imbalance, and immunologic pathways.11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 These preliminary reports have indicated that altered gut–brain communications not only may play a role in the increased occurrence of FGIDs in ASD individuals, but could advance our understanding of potential risk factors for FGID in the ASD community.
Differences in the types and composition of various bacterial species in children with ASD have been reported.22, 23, 24, 25, 26, 27 Most studies have focused on stool specimens because it is difficult to obtain adequate numbers of GI mucosal biopsy specimens from children with ASD. This is a potentially important deficiency in the field because mucosa-associated microbes preferentially regulate host homeostasis, as is evident in the induction of T-cell immune responses, as well as maintaining serotonin biosynthesis from dietary tryptophan in the mucosa.28, 29, 30, 31, 32, 33 Because it is unethical to perform an endoscopy on children with ASD without a clinical indication, the only cohort that can be captured is generally ASD with FGID. For this reason, it is important to compare mucosal (isolated from tissue specimens) microbiome communities in children with and without ASD and FGID because stool specimens, although providing a more feasible approach to power microbiome studies, may provide misleading insights into disease regulatory circuits in ASD. A single study, with a follow-up report, remains the only characterization of the mucosal microbiome in pediatric ASD.23, 34 Although focusing mainly on phylum level changes, the key differences associated with the ASD group with FGID were seen as an increase in Clostridiales, particularly in Lachnospiraceae and Ruminococcaceae. These preliminary reports merit deeper investigation because the Clostridiales are emerging as major microbial regulators of gut-derived T-cell immune and serotonergic signals that may be associated with ASD.30, 31, 32, 33, 35
Our study compared mucosa-associated microbial communities in children with ASD with previous reports characterizing stool in this population. Furthermore, we investigated whether mucosa-associated microbes correlated with altered tryptophan–serotonin metabolism and cytokine networks in clinically distinct patient cohorts. Here, we report a unique mucosa-associated microbiome signature in children with ASD that correlates significantly with quantitative cytokine and tryptophan measurements, as well as clinical symptoms. This analysis shows functional associations that distinguish both the clinical group and GI symptoms. Because mucosal-associated microbes remain poorly defined in ASD, we advance this field by linking both previously reported and new gut-microbe interactions as possible drivers of disease-associated signaling networks in ASD and in functional abdominal pain.
Materials and Methods
Study Protocol
Study participants were recruited from the outpatient pediatric GI procedure suite at Nationwide Children’s Hospital in Columbus, Ohio. Participants were identified by chart reviews of males and females aged 3–18 years who were undergoing a lower endoscopy for one of the following symptoms: abdominal pain, altered stool patterns, or painless bright red blood per rectum. The research protocol was approved by the Nationwide Children’s Institutional Review Board, and written consent was obtained from parents of the participants.
On the day of the procedure, participants were approached for recruitment if they met the following criteria: normal height and weight growth patterns; previously normal blood work (complete blood count, complete metabolic panel, normal IgA level, and normal IgA anti–tissue transglutaminase level); and no antibiotics, steroids, or GI infection during the previous 3 months. ASD participants were required to have an Autism Diagnostic Observation Schedule–confirmed diagnosis. Non-ASD participants were screened with the Social Responsiveness Scale to ensure they did not show behaviors in the range of those that often are displayed in children with ASD. Parents of the non-ASD participants were asked to fill out the 65-item parent report questionnaire. If the Social Responsiveness Scale score was less than the standardized cut-off value of 51, the participant was considered appropriate for the study. Once consent was obtained, parents of all participants were asked to complete a Questionnaire on Pediatric Gastrointestinal Symptoms-Rome III (QPGS-RIII) version.
Specimen Collections
Blood and biopsy specimens were collected only from participants with a normal-appearing colonoscopy as determined by the pediatric GI specialist. A blood specimen was obtained at the time the intravenous needle was placed, prior to administration of intravenous fluids, and was held until it was determined that the participant had a normal-appearing mucosa. If the specialist confirmed there was no signs of erythema, decreased vasculature, erosions, ulcers, or other signs of infection/inflammation in the colon, then 5 biopsy specimens from the rectum were collected.
The whole blood specimen (1.5–2.5 mL) was obtained in an EDTA-containing tube, gently mixed with 5–6 inversions, and kept at room temperature until the end of the colonoscopy. The blood was divided into 250-μL aliquots and stored at -80°C until analysis. Biopsy specimens were placed immediately in 2 mL of saline on ice. Biopsy specimens were transported to a laboratory for processing within 15 minutes. One biopsy specimen was placed in a cryotube and stored at -80°C for later analysis of mucosal serotonin levels. The remaining 4 biopsy specimens were used for microbiome analysis or to create a supernatant for measurement of secreted cytokines, serotonin, and tryptophan.
Mucosal Supernatant
Mucosal supernatants were created as previously described.36, 37 Four biopsy specimens were placed in 1 mL of oxygenated Hanks' balanced salt solution (HBSS) at 37°C in a screw-top cryotube. The tube was placed in a water bath at 37°C for 25 minutes. Oxygen was administered across the surface of the HBSS via a needle placed through the screw top. After 25 minutes, the tube was spun at 7600 rpm for 10 minutes. Supernatant then was collected and stored at -80°C until analysis. Of the 4 biopsy specimens, 1 specimen was placed in a cryotube without solution and stored at -80°C for 16S ribosomal DNA analysis performed at the Texas Children’s Microbiome Center. A second biopsy specimen was placed in formalin and sent to pathology at Nationwide Children’s Hospital for review by a pathologist.
Microbiome Characterization
Frozen tissue specimens were thawed on ice, and 0.01–0.02 g were added to a MO BIO PowerBead Tube (MO BIO Laboratories, Carlsbad, CA) and vortexed for 15 minutes for gentle homogenization. Subsequent material was processed through the standard MO BIO PowerSoil extraction kit protocol (MO BIO Laboratories). Quantity and quality of the resulting nucleic acid content was confirmed by Nanodrop-1000 and Qubit (Thermo Fisher Scientific, Inc., Wilmington, DE). Amplification and sequencing of the V1V3 and V4 regions of the 16S ribosomal RNA gene was performed using the NEXTflex 16S V1V3 and V4 Amplicon-Seq Kit 2.0 (Bioo Scientific, Austin, TX) with 20 ng of input DNA, and sequences were generated on the Illumina MiSeq platform (Illumina, San Diego, CA) with a minimum of 100,000 sequences generated per sample. Sequence data were processed through the LotuS pipeline as previously described.38 Briefly, reads were de-multiplexed, and paired ends were stitched. Quality filtering was performed before operational taxonomic unit (OTU) clustering using a modified version of the UPARSE algorithm.39 Taxonomic assignment was performed with RDP as the classifier and HitDB40 and SILVA41 as the selected databases. Organisms potentially classified to the species level were based on individual OTUs of significance. OTUs failing to classify as bacteria at the kingdom level were removed before further analysis.
Cytokine Measurements
Blood and biopsy supernatants were assayed using a Human Cytokine/Chemokine Premixed 38-plex kit (EMD Millipore, Billerica, MA) in a MagPix system (Luminex Corporation, Austin, TX). The 38 analytes tested were as follows: epidermal growth factor, fibroblast growth factor 2, eotaxin, tumor growth factor (TGF)α, granulocyte colony–stimulating factor, fms-related tyrosine kinase 3 ligand (FIt-3L), granulocyte-macrophage colony–stimulating factor (GM-CSF), fractalkine, interferon (IFN)α2, IFNγ, growth related oncogene alpha (GROα), macrophage-derived chemokine, interleukin (IL)1RA, IL1α, IL1β, IL2, IL3, IL4, IL5, IL6, IL7, IL8, IL9, IL10, IL12p40, IL12p70, IL13, IL15, IL17A, soluble CD40-ligand, monocyte chemoattractant protein 1 (MCP-1), MCP-3, interferon gamma-induced protein 10, macrophage inflammatory protein 1 alpha (MIP-1α), MIP-1β, tumor necrosis factor alpha (TNFα), TNFβ, and vascular endothelial growth factor (VEGF). Raw data were acquired using xPONENT for Magpix (Luminex Corporation, Austin, TX), version 4.2, build 1324, and analyzed with MasterPlex QT version 5.0.0.73 (Hitachi MiraiBio, San Francisco, CA).
Serotonin, 5-Hydroxyindoleacetic Acid, and Tryptophan Measurements
Levels of serotonin (5-HT), tryptophan, and 5-hydroxyindoleacetic acid (5-HIAA, the major metabolite of 5-HT) were measured in biopsy supernatants after deproteinization. Levels of 5-HT and 5-HIAA were measured in biopsy specimens following sonication and deproteinization. Compounds were determined by high-performance liquid chromatography as previously described, with minor modification.42
Functional GI Questionnaire
Responses of the QPGS-RIII were used to confirm and/or determine the study group for which the participant met criteria, ASD with a functional GI disorder (ASD-FGID), neurotypical with a functional GI disorder (NT-FGID), or neurotypical without a functional GI disorder (NT). The QPGS-RIII is a 71-item parent report instrument that is used in clinical research to assess the chronicity and severity of GI symptoms and helps to characterize which particular FGID criteria of GI symptoms are met, including functional abdominal pain, irritable bowel syndrome (IBS), abdominal migraine, and functional constipation. For the ASD participants, the responses to the QPGS-RIII and the lack of physiological findings for their abdominal pain and/or altered stool patterns confirmed that the ASD participants met the criteria for an FGID. For the non-ASD patients, responses to the QPGS-RIII were used to confirm whether the participant belonged to the NT-FGID or the NT group. In addition to the criteria of abdominal pain and altered stool patterns, non-ASD participants were recruited for painless rectal bleeding. The goal of recruiting painless rectal bleeding participants was to recruit as close to normal a study group as possible, particularly a group of children with benign juvenile polyps and without any symptoms associated with FGID.
Statistical Analysis
Statistical analysis of taxonomic and functional profiles was used for the visualization and statistical analysis of OTUs.43 Principal component analysis was used to visualize overall differences between the 3 groups. Comparisons between groups (ASD-FGID, NT-FGID, and NT) were made at various taxonomic levels, including the OTU level. Taxonomic assignments for representative sequences of significant OTUs were confirmed by manual database searches and alignments. In addition, microbial differences were evaluated based on other available clinical data (eg, reported GI symptoms). Analysis of variance was performed for multiple group comparisons, and the Welch t test was performed for comparisons between 2 groups.
Pearson correlations with P values were calculated for the OTUs, cytokines, and serotonin using RStudio (RStudio, Boston, MA). The P values were corrected for multiple testing with the Benjamini–Hochberg method to control for false-discovery rate. For cytokine and serotonin measurements, group differences between ASD-FGID, NT-FGID, and NT were calculated using the Wilcoxon rank-sum test implemented in RStudio. For missing cytokine measurements, a value of 10% of the lowest measurement for that cytokine was inserted.
Results
Clinical Groups
A total of 38 children ages 3–18 years (median age, 9 y) initially consented. Three of the 38 were found to have an abnormal colonoscopy, 2 with increased inflammation, and 1 with a pinworm infection. Therefore, 35 children were included in the final analysis and are described in Table 1. The cohort included 3 female and 32 male children and, after review of all data, subjects were assigned to the following groups: ASD-FGID (n = 14), NT-FGID (n = 15), and NT (n = 6). The ASD-FGID group included all male children with a median age of 8.5 years (age range, 4–13 y), the NT-FGID group included 3 female and 12 male children with a median age of 10.5 years (age range, 3–18 y), and the NT group included all male children with a median age of 5.5 years (age range, 3–14 y). Age differences were not statistically different between the clinical groups, and stratification by age group did not show any similarities with the microbial profiles of the 3 groups under investigation. When stratifying specifically by reported abdominal pain, the following groups were used: ASD-FGID with no pain reported (n = 5), ASD-FGID with abdominal pain (n = 9), NT with no pain reported (n = 10), and NT with abdominal pain (n = 11). Subjects were placed in the groups with no pain if they did not report abdominal pain via QPGS-RIII, and the 15 subjects in the groups with no pain included those with functional constipation (n = 7), nonretentive fecal incontinence (n = 1), functional dyspepsia (n = 1), and those with no GI-related findings per the QPGS-RIII report (n = 6).
Table 1.
Age and GI Symptoms Present in Each Group: ASD-FGID, NT-FGID, and NT
| Variable | ASD-FGID (n = 14), n (%) | NT-FGID (n = 15), n (%) | NT (n = 6), n (%) | Total (N = 35), N (%) |
|---|---|---|---|---|
| Median age, y (range) | 8.5 (4–13) | 10.5 (3–18) | 5.5 (3–14) | 9 (3–18) |
| Abdominal pain | 9 (64) | 11 (73) | 0 | 20 (57) |
| Irritable bowel syndrome | 7 (50) | 7 (47) | 0 | 14 (40) |
| Abdominal migraine | 4 (29) | 3 (20) | 0 | 7 (20) |
| Aerophagia | 5 (36) | 1 (7) | 0 | 6 (17) |
| Functional constipation | 4 (29) | 5 (33) | 0 | 9 (26) |
Mucosa-Associated Microbiome Communities in ASD
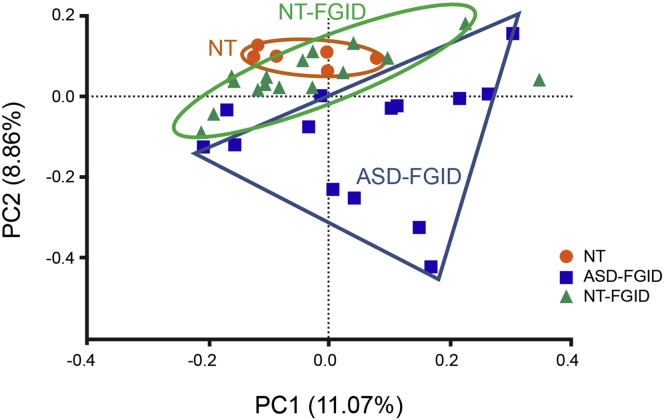
Principal component analysis showed clear separation between the ASD-FGID group and the NT-FGID and NT groups (Figure 1). Although the ASD-FGID subjects clustered separately from the NT groups, there was overlap between the NT-FGID and NT groups, with the NT group forming a distinct cluster within the larger NT-FGID group. Of note, medications were not found to affect the microbiome or cytokine profile significantly, and no age-related microbiome was identified that could explain this separation in ASD individuals.
Figure 1.
Separate clusters for the ASD-FGID (blue squares), NT-FGID (green triangles), and NT (orange circles) groups are evident via principal component analysis.
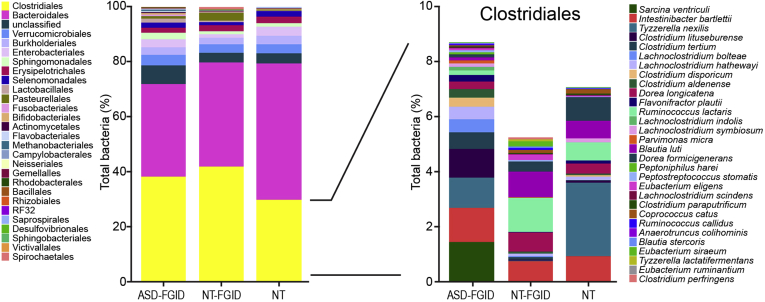
Overall microbiome profiles of each group illustrated subtle but noticeable differences (Figure 2). The most abundant orders across all groups included members of Clostridiales, Bacteroidales, Verrucomicrobiales, Burkholderiales, and Enterobacteriales. Resolution to genus level was moderately efficient in determining differences between groups, but analysis at the OTU level provided a greater degree of resolution and elucidated key differences in the ASD-FGID group. This deeper analysis was facilitated by the UPARSE algorithm as one of the state-of-the art systems biology tools for accurate identification and analysis of microbial OTUs.
Figure 2.
Microbiome profiles per group (ASD-FGID, NT-FGID, and NT) show subtle differences in overall composition at the order level (left). Greater differentiation between the groups was observed when comparisons were made at the OTU level. Right: Clostridiales OTUs that were further identified to the species level. The separation of the ASD-FGID group from the NT-FGID and NT groups was highly driven by both increases and decreases in specific Clostridiales.
The cohort was compared based on age group (<6 y, 7–12 y, and 13–18 y), and the separation of the ASD-FGID, NT-FGID, and NT groups could not be attributed to age. Subtle differences were noted among the age groups, however, with increases in OTUs most closely resembling Alistipes putredinis (P = .003) and Clostridium perfringens (P = .029) in the 13–18 years group, a decrease in Parabacteriodes distasonis (P = .036) in the 13–18 years group, and an increase in Ruminococcus species (P = .024) in the group with children younger than 6 years. Only 3 females were recruited into the study, and all were in the NT-FGID group with 1 in each age group. No difference was identified based on sex when comparing all neurotypical participants.
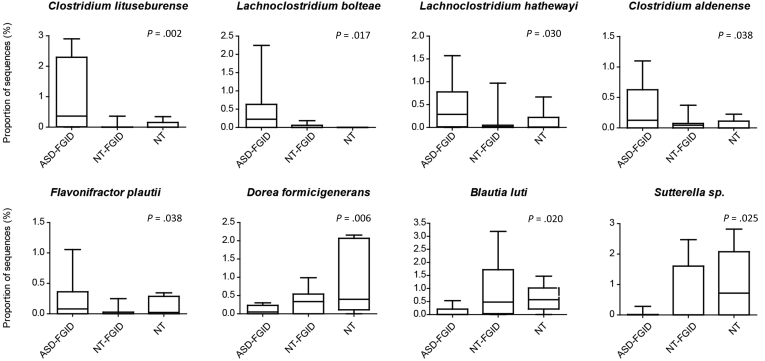
With focused analysis at the OTU level, an increase in Clostridiales, specifically sequences identified as Clostridium lituseburense (P = .002), Lachnoclostridium bolteae (P = .017), Lachnoclostridium hathewayi (P = .030), Clostridium aldenense (P = .038), and Flavonifractor plautii (P = .038), was observed in the ASD-FGID group compared with the NT-FGID and NT groups (Figure 3). Decreases in 2 other Clostridiales OTUs, Dorea formicigenerans (P = .006) and Blautia luti (P = .020), as well as reduced Sutterella species (P = .025), also was seen in the ASD-FGID group. When removing a single subject in the NT-FGID group with an overwhelming amount of Terrisporobacter as an outlier, the presence of Terrisporobacter species (P = .045) also was associated significantly with the ASD-FGID group. In addition, statistically significant differences were identified between the NT-FGID and NT groups, with increases in several OTUs (Faecalibacterium prausnitzii, Roseburia intestinalis, Oscillospira valericigenes, and Bilophila wadsworthia) observed in the NT-FGID group relative to the NT group.
Figure 3.
Differences in OTUs related to specific bacteria associated with ASD were identified in the mucosal microbial community. Overall, an increase in Clostridiales (C lituseburense, P = .002; L bolteae, P = .017; L hathewayi, P = .030; C aldenense, P = .038; and F plautii, P = .038) was seen in the ASD-FGID group compared with the NT-FGID and NT groups. A decrease in relative abundance in the ASD-FGID group was seen for D formicigenerans (P =.006), B luti (P = .020), and Sutterella species (P = .025). Plots depict the maximum and minimum (whiskers), upper and lower quartile limits (box), and median (horizontal line).
The QPGS-RIII responses also were reviewed to extract data on specific GI symptoms including IBS (n = 14), functional constipation (n = 9), aerophagia (n = 6), and abdominal migraine (n = 7). An increase in C aldenense (P = .049) was associated with IBS, and decreases in several organisms were associated with functional constipation including F plautii (P = .039), Bacteroides eggerthii (P = .024), Bacteroides uniformis (P = .040), F prausnitzii (P = .013), and Clostridium clariflavum (P = .031). Aerophagia also was associated with an increase in C aldenense (P = .030), as well as with a decrease in a variety of other organisms including B luti (P = .003), Bifidobacterium adolescentis (P = .019), Eubacterium ventriosum (P = .045), Anoxystipes fissicatena (P = .024), Coprococcus comes (P = .042), Eubacterium ramulus (P = .006), and Phascolarctobacterium faecium (P = .049). Decreases in Akkermansia muciniphila (P = .038), Coprococcus catus (P = .007), Odoribacter splanchnicus (P = .050), Clostridium lactatifermentans (P = .033), and Ruminococcus lactaris (P = .037) were associated with the presence of abdominal migraine. Notably, there was no significant overlap between organisms associated with specific GI symptoms and organisms contributing to the separation of the ASD and NT groups.
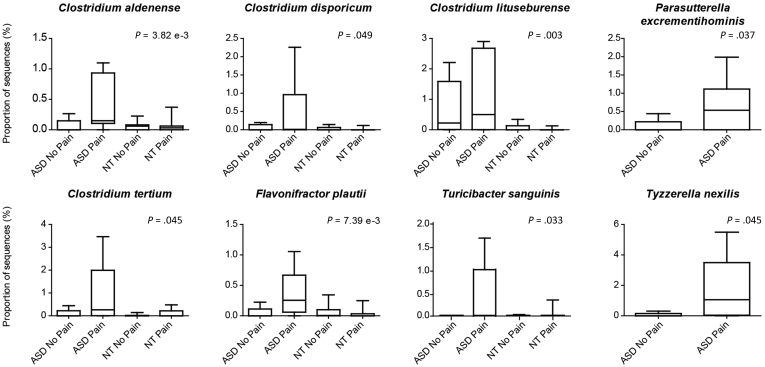
Stratification by abdominal pain showed 6 additional organisms (OTUs) closely related to pain predominantly in the ASD-FGID group when compared across all 4 pain groups: Turicibacter sanguinis (P = .033), C aldenense (P = .004), C lituseburense (P = .003), F plautii (P = .007), Clostridium disporicum (P = .049), and Clostridium tertium (P = .045) (Figure 4). When delving more deeply into a comparison between the ASD-FGID group with pain and the ASD-FGID group with no pain, C aldenense (P = .033) and F plautii (P = .042) again were associated significantly with the ASD-FGID group with pain. In addition, Tyzzerella species (P = .045) and Parasutterella excrementihominis (P = .037) also emerged as significantly associated with the ASD-FGID group with pain compared with the ASD-FGID group with no pain.
Figure 4.
Bacteria associated with pain in ASD-FGID were identified. When comparing across all groups, a significant increase in OTUs identified as C aldenense (P = .004), C disporicum (P = .049), C lituseburense (P = .003), C tertium (P = .045), F plautii (P = .007), and T sanguinis (P = .033) was noted in the cohort of ASD children in whom abdominal pain had been reported. Comparisons solely between the 2 ASD groups, with and without abdominal pain, also showed 2 additional organisms of interest that were increased in the ASD pain group: P excrementihominis (P = .037) and T nexilis (P = .046). Plots depict the maximum and minimum (whiskers), upper and lower quartile limits (box), and median (horizontal line).
Mucosal Tryptophan and Serotonin System
Cytokine and serotonergic correlations with microbial OTUs were performed using the Wilcoxon rank-sum test to compare differences between groups and showed multiple significant findings (Table 2). Decreased tryptophan levels detected in the supernatant of the GI biopsy specimens from the ASD-FGID group were found to be significant when compared with either the NT-FGID (P = .006) or the NT group (P = .009). Of further relevance to the serotonin pathway, 5-HIAA (the primary metabolite of serotonin) was significant with comparisons of ASD-FGID with NT (P = .009, increased 5-HIAA in the ASD-FGID biopsy specimens) and NT-FGID with NT (P = .043, decreased 5-HIAA in the supernatant of the NT-FGID group). An increase in 5-HIAA detected in biopsy specimens also was associated with the presence of abdominal pain (P = .039).
Table 2.
Group-Wise Comparisons of Median Values of Serotonin and Cytokines (From Blood) With the Following Group Designations: ASD-FGID, NT-FGID, and NT
| Serotonin/cytokine | ASD-FGID vs NT-FGID, medians | ASD-FGID vs NT, medians | NT-FGID vs NT, medians | GI pain vs no GI pain, medians | Significant results |
|---|---|---|---|---|---|
| 5-HT (biopsy) | 6065.82 vs 4613.16 | 6065.82 vs 8302.54 | 4613.16 vs 8302.54 | 4908.99 vs 8189.65 | |
| 5-HT (supernatant) | 20.04 vs 17.04 | 20.04 vs 21.14 | 17.04 vs 21.14 | 17.16 vs 19.40 | |
| 5-HT (blood) | 204.38 vs 167.08 | 204.38 vs 241.48 | 167.08 vs 241.48 | 177.49 vs 221.81 | |
| 5-HIAA (biopsy) | 1258.02 vs 1175.67 | 1258.02 vs 679.34a | 1175.67 vs 679.34 | 1316.48 vs 959.75a | ↑ ASD-FGID ↑ GI pain |
| 5-HIAA (supernatant) | 18.26 vs 24.79 | 18.26 vs 33.68 | 24.79 vs 33.68a | 19.44 vs 28.47 | ↑ NT |
| 5-HIAA/5-HT (biopsy) | 0.22 vs 0.22 | 0.22 vs 0.10 | 0.22 vs 0.10a | 0.27 vs 0.14a | ↑ NT-FGID ↑ GI pain |
| 5-HIAA/5-HT (supernatant) | 1.17 vs 1.36 | 1.17 vs 1.51 | 1.36 vs 1.51 | 1.29 vs 1.47 | |
| TRP (blood) | 6866.75 vs 6345.15 | 6866.75 vs 6901.17 | 6345.15 vs 6901.17 | 6560.40 vs 6720.00 | |
| TRP (supernatant) | 118.21 vs 172.19a | 118.21 vs 236.76a | 172.19 vs 236.76 | 149.67 vs 173.51 | ↓ ASD-FGID |
| GROα | 1140.29 vs 1652.57a | 1140.29 vs 2146.31 | 1652.57 vs 2146.31 | 1402.25 vs 1559.75 | ↓ ASD-FGID |
| MCP-1 | 1553.71 vs 1162.24 | 1553.71 vs 979.38a | 1162.24 vs 979.38 | 1468.16 vs 963.45a | ↑ ASD-FGID ↑ GI pain |
| Eotaxin | 261.86 vs 224.92 | 261.86 vs 185.83a | 224.92 vs 185.83 | 256.96 vs 197.06a | ↑ ASD-FGID ↑ GI pain |
| IL9 | 2.50 vs 2.42 | 2.50 vs 6.71 | 2.42 vs 6.71 | 2.50 vs 2.84 | |
| Fractalkine | 129.83 vs 145.29 | 129.83 vs 148.21 | 145.29 vs 148.21 | 132.87 vs 153.63 | |
| IL5 | 3.25 vs 3.03 | 3.25 vs 3.01 | 3.03 vs 3.01 | 3.51 vs 2.83 | |
| IL6 | 1.12 vs 0.96 | 1.12 vs 3.55 | 0.96 vs 3.55 | 1.11 vs 1.12 | |
| MDC | 635.14 vs 587.58 | 635.14 vs 788.22 | 587.58 vs 788.22 | 599.07 vs 664.13 | |
| FGF2 | 156.00 vs 198.56 | 156.00 vs 208.48 | 198.56 vs 208.48 | 180.13 vs 194.50 | |
| Flt-3L | 0.729 vs 0.729 | 0.729 vs 0.729 | 0.729 vs 0.729 | 0.729 vs 0.729 | |
| IFN-α2 | 33.61 vs 41.61 | 33.61 vs 79.53a | 41.61 vs 79.53a | 37.36 vs 55.50 | ↓ ASD-FGID ↑ NT |
NOTE. Boldface indicates significant relationships.
FGF2, fibroblast growth factor 2; TRP, tryptophan.
Significant at P < .05.
Microbial-Tryptophan/Serotonin Correlations
Several organisms that were found in significantly increased abundance in the ASD-FGID group compared with the NT-FGID and NT groups also were found to correlate with the serotonin pathway (Table 3). Specifically, Erysipelotrichaceae, C lituseburense, and Terrisporobacter species were found to correlate with tryptophan quantitated in the supernatant of the tissue biopsy specimens from the ASD-FGID cohort. In addition, L bolteae, L hathewayi, and F plautii correlated directly with levels of serotonin in tissue biopsy specimens.
Table 3.
Correlation of Organisms With Increased Abundance in the ASD Group With Tryptophan or Serotonin
| Specimen | Organism | Serotonin pathway | r | P valuea |
|---|---|---|---|---|
| Supernatant | Erysipelotrichaceae | Tryptophan | 0.625 | .004 |
| Supernatant | C lituseburense | Tryptophan | 0.671 | .025 |
| Supernatant | Terrisporobacter species | Tryptophan | 0.913 | .000 |
| Biopsy | L bolteae | 5-HT | 0.802 | .002 |
| Biopsy | L hathewayi | 5-HT | 0.686 | .003 |
| Biopsy | F plautii | 5-HT | 0.856 | .000 |
After multiple testing correction (Benjamini–Hochberg).
Mucosal Cytokine Profiles
In comparing the ASD-FGID with the NT-FGID group, decreased GROα was significant (P = .043). The ASD-FGID with NT comparison produced several additional significant cytokine relationships including increases in MCP-1 (P = .005), eotaxin (P = .032), and IFN-α2 (P = .038). Upon separation of the cohort (all 3 groups) based on abdominal pain, increases in MCP-1 (P = .033) and eotaxin (P = .028) were associated significantly with the presence of abdominal pain.
Microbial-Cytokine Correlations
Multiple cytokines correlated strongly with bacteria associated with the ASD-FGID cohort (Table 4). Organisms increased in the ASD-FGID cohort that correlated positively with cytokines included C aldenense (IL7), C lituseburense (IL1RA, IL12p70, IL1α, IL5, IL6, IP10, MIP-1α, MIP-1β, and VEGF), L bolteae (IFN-α2), L hathewayi (IL15, IL9, IL1β, and IL7), F plautii (granulocyte-macrophage colony–stimulating factor, IL9), and Terrisporobacter species (VEGF, IL1α, and TNF-β). Organisms decreased in abundance in the ASD-FGID cohort that correlated positively with T helper 1 regulatory cytokines included D formicigenerans (IL12 p70) and Sutterella species (IL12 p70).
Table 4.
Cytokine Correlations With Bacteria Significantly Associated With the ASD-FGID Group
| Specimen | Organism | Cytokine | R | P valuea | ASD-FGID |
|---|---|---|---|---|---|
| Serum | C aldenense | IL7 | 0.652 | .011 | ↑ |
| Biopsy | C lituseburense | IL1RA | 0.739 | .009 | ↑ |
| Serum | C lituseburense | IL12p70 | 0.959 | .000 | ↑ |
| Serum | C lituseburense | IL17A | 0.703 | .027 | ↑ |
| Serum | C lituseburense | IL1α | 0.996 | .000 | ↑ |
| Serum | C lituseburense | IL5 | 0.963 | .000 | ↑ |
| Serum | C lituseburense | IL6 | 0.972 | .000 | ↑ |
| Serum | C lituseburense | IP-10 | 0.685 | .019 | ↑ |
| Serum | C lituseburense | MIP-1α | 0.813 | .001 | ↑ |
| Serum | C lituseburense | MIP-1β | 0.895 | .000 | ↑ |
| Serum | C lituseburense | VEGF | 0.743 | .012 | ↑ |
| Serum | L bolteae | IFN-α2 | 0.752 | .024 | ↑ |
| Serum | L hathewayi | IL15 | 0.582 | .035 | ↑ |
| Serum | L hathewayi | IL9 | 0.729 | .016 | ↑ |
| Serum | L hathewayi | IL1β | 0.572 | .033 | ↑ |
| Serum | L hathewayi | IL7 | 0.643 | .013 | ↑ |
| Serum | F plautii | GM-CSF | 0.783 | .003 | ↑ |
| Serum | F plautii | IL9 | 0.708 | .050 | ↑ |
| Biopsy | Terrisporobacter species | VEGF | 0.694 | .031 | ↑ |
| Serum | Terrisporobacter species | IL1α | 0.969 | .000 | ↑ |
| Serum | Terrisporobacter species | TNF-β | 0.897 | .020 | ↑ |
| Serum | D formicigenerans | IL12p70 | 0.766 | .000 | ↓ |
| Serum | Sutterella species | IL12p70 | 0.892 | .023 | ↓ |
After multiple testing correction (Benjamini–Hochberg).
Beyond the organisms associated with ASD, additional OTUs more closely associated with pain in the ASD-FGID group also correlated strongly with cytokines including C disporicum (IFN-γ, IL12p70, IL17A, IL5, IL6, IP-10, MIP-1α, MIP-1β, and VEGF), C tertium (IL1RA, IFN-γ, IL12p70, IL17A, IL1α, IL5, IL6, MIP-1α, and MIP-1β), P excrementihominis (GROα), and Tyzzerella nexilis (sCD40L) (Table 5).
Table 5.
Cytokines Significantly Associated With Abdominal Pain Within the ASD-FGID Group Compared With Groups With No Abdominal Pain
| Specimen | Organism | Cytokine | R | P valuea |
|---|---|---|---|---|
| Serum | C disporicum | IFNγ | 0.817 | .010 |
| Serum | C disporicum | IL12p70 | 0.978 | .000 |
| Serum | C disporicum | IL17A | 0.961 | .000 |
| Serum | C disporicum | IL5 | 0.990 | .000 |
| Serum | C disporicum | IL6 | 0.966 | .001 |
| Serum | C disporicum | IP-10 | 0.818 | .009 |
| Serum | C disporicum | MIP-1α | 0.911 | .000 |
| Serum | C disporicum | MIP-1β | 0.951 | .000 |
| Serum | C disporicum | VEGF | 0.873 | .008 |
| Biopsy | C tertium | IL1RA | 0.752 | .024 |
| Serum | C tertium | IFNγ | 0.911 | .000 |
| Serum | C tertium | IL12p70 | 0.992 | .000 |
| Serum | C tertium | IL17A | 0.975 | .000 |
| Serum | C tertium | IL1α | 0.988 | .000 |
| Serum | C tertium | IL5 | 0.976 | .000 |
| Serum | C tertium | IL6 | 0.971 | .000 |
| Serum | C tertium | MIP-1α | 0.920 | .000 |
| Serum | C tertium | MIP-1β | 0.895 | .000 |
| Serum | C tertium | VEGF | 0.832 | .012 |
| Biopsy | P excrementihominis | GROα | 0.784 | .002 |
| Serum | T nexilis | sCD40L | 0.778 | .014 |
After multiple testing correction (Benjamini–Hochberg).
Discussion
Over the past 15 years, multiple bacterial organisms have been suspected of playing a role in the development of GI symptoms in patients with autism. A number of biomarkers that could connect to microbial profiles also have been identified in ASD, including alterations in serotonin levels, IL1β, and IL6.35, 44, 45, 46, 47 Early stool-based culture and real-time polymerase chain reaction–based studies reported an increased abundance of Clostridium species in the GI tract of children with autism,48, 49, 50 but more recent studies failed to reach a consensus as to whether an ASD-specific microbial signature exists.24, 51 Stool-based microbiome characterization in several cohorts identified specific genera that differed in ASD vs control subjects, but most previous studies conducted their analysis at higher taxonomic levels, with little published data at the genus or species (ie, OTU) level.22, 25, 26, 27 In addition, prior attempts to characterize the gut microbiome in ASD by next-generation sequencing have had multiple limitations related to selection of the control group, heterogeneous ASD cohorts, and limited data regarding GI phenotype of subjects. A single tissue-based study in pediatric ASD has been published with a total sample size of 22 subjects (ASD, n = 15; NT, n = 7) and a limited age range of 3–5 years. Although the analysis focused mainly on higher taxonomic levels, an increase in Clostridiales, especially Lachnospiraceaea and Ruminococcaceae, was observed.23 This is highly consistent with the microbiome profiles we detected, in which the strongest association in the ASD-FGID group resulted from an increase in Clostridiales.
The ability to pinpoint key differences in the mucosal microbiome of these children was in part owing to improvements in technology. Improved sequencing technology and deeper sequencing, particularly in a low-biomass sample such as tissue, enabled the data to be better classified. In addition, updated software algorithms and databases provided more accurate classification, and critical evaluation at the OTU level, with individual representative sequence searches for confirmation of identification, contributed to the higher resolution of this data set compared with prior studies. This may explain why previous studies with analyses founded at the family level or higher taxonomically were unable to identify microbial signatures within ASD consistently. Beyond technical advances, appropriate stratification of groups also may have been critical in this study, in which the ASD group had a verified GI phenotype with symptoms profound enough to warrant evaluation via colonoscopy.
Overall, our identification of clostridial species aligns with those previous autism studies that have identified microbiome alterations. Finegold et al48 originally described increased Clostridium in ASD in 2002, and ultimately identified a novel species, Clostridium (Lachnoclostridium) bolteae, associated with ASD, specifically in those with GI symptoms. Real-time polymerase chain reaction analysis targeting C bolteae confirmed the increased abundance in ASD,49 but subsequent studies at other institutions have not explicitly reported the detection of C bolteae in ASD cohorts; although few studies have focused strongly on GI phenotypes. The identification of C bolteae and strong association with ASD described in these initial analyses are important to the evolving views of the microbiome–gut–brain axis in ASD. A recent publication also discussed the significant increase in C bolteae in otherwise healthy individuals after antibiotic administration,52 creating further interest in the potential role of the organism and antibiotic treatment in children with ASD. This may be particularly relevant in light of our study finding a potential association requiring confirmation between this organism and the serotonin system, which has long been implicated in ASD.45, 53
The association between the microbiome and abdominal pain also has been studied in the context of several GI disorders. Prior research in pediatric IBS identified increases in γ-proteobacteria, specifically Haemophilus parainfluenzae, and a Ruminococcus-like organism.54 The ASD–FGID cohort also showed a significantly increased abundance of a Ruminococcaceae organism that could not be classified further, as well as, upon removal of a single outlier, a trend toward increased abundance of H parainfluenzae. In addition, Turicibacter species was associated with abdominal pain in the ASD-FGID cohort and was reported previously in pediatric patients with IBS who failed to respond to a low fermentable oligosaccharides, disaccharides, monosaccharides, and polyols diet.55
An increase in inflammatory cytokines has been reported previously in ASD, and increased cytokines have been reported in association with a regressive autism phenotype with significant communication challenges and aberrant behaviors.56, 57 With mounting evidence showing an impact on brain development and behavior in rodents,35 IL6 and IL17A were of particular interest in this cohort. IL6 and IL17A are both implicated in the maternal immune activation models that result in autism-like and schizophrenia-like phenotypes in rodents.58, 59 Furthermore, increased IL6 has been seen in clinical cohorts of ASD.13, 14, 17, 60 Although no significant group differences or trends were seen for IL17A, this cytokine correlated with Clostridia species that were enriched in ASD-FGID patients. IL6 was highest in the ASD-FGID group, a trend that was exaggerated further in the subset of the ASD-FGID reporting abdominal pain. IL6 also was correlated significantly with 2 Clostridium species (C disporicum and C tertium) that were associated with ASD and abdominal pain. Similarly, IFN-γ has been noted as increased in individuals with ASD,13, 16, 17, 61 and IFN-γ was increased in ASD and correlated with C disporicum and C tertium. These trends potentially link clostridial species with increased inflammation and abdominal pain in ASD, and although correlative at this stage, provide the direction needed for future studies.
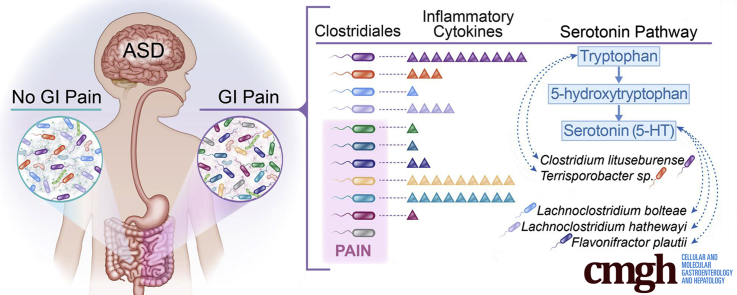
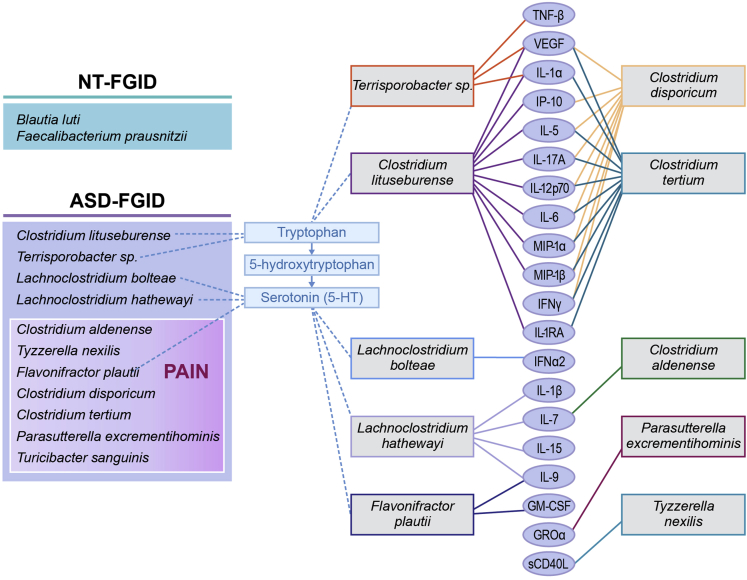
Also intriguing is the correlation of microbiome composition with tryptophan and serotonin levels. Tryptophan detected in the supernatant of the GI biopsy specimen was significantly different in the ASD–FGID group compared with either the NT-FGID or the NT group. Figure 5 shows the serotonin pathway and relevant organisms associated with ASD in our cohort, as well as cytokines that correlated with those specific OTUs of interest. Three organisms correlated strongly with tryptophan levels (Erysipelotrichaceae, C lituseburense, and Terrisporobacter species). L bolteae, as well as L hathewayi and F plautii, showed an association with serotonin levels. Long-standing findings have indicated increased whole-blood 5-HT levels in a subset of approximately 30% of children with autism spectrum disorder,44, 45, 46, 47 as well as decreased levels of the serotonin metabolite, melatonin, in a substantial subpopulation.62 Interestingly, whole-blood 5-HT levels correlate with lower GI symptoms in ASD.63 Furthermore, a recent study showed that an ASD-associated serotonin transporter variant leads to altered gut development and function.64 Our microbial correlations provide further evidence for the relevance of the tryptophan–serotonin pathway in ASD,62 although our sample size did not allow us to focus on an ASD subgroup. Of particular interest are recent reports that metabolic products of the Clostridiales regulate the production of serotonin in enteroendocrine cells in the GI tract via transcriptional regulation of Tph1.30, 31 Our finding that these organisms and the serotonergic pathway are linked to functional pain in ASD merits further investigation. By linking the mucosal microbiome, serotonin, and inflammatory cytokines with GI symptoms in ASD, this study builds on previous work in multiple areas of autism research. The identification of increased abundance of key organisms known to be associated with a specific ASD GI phenotype as well as the description of other organisms of potential importance in not only ASD, but abdominal pain in ASD, may be important for the future development of microbial biomarkers and treatments focused on the microbiome.
Figure 5.
The identification of distinct microbial profiles of the NT-FGID, ASD-FGID, and ASD-FGID with abdominal pain groups was influenced greatly by the differences in relative abundances of the organisms listed (left). Several of these bacteria also were found to correlate with the serotonin pathway (center), as well as with multiple cytokines (right).
In summary, GI phenotypes in ASD are highly variable, and heterogeneity of the autism population can mask significant signals related to GI symptomology. Here, we present distinct microbiome and cytokine measures in the rectal mucosa of children with ASD and confirmed FGIDs compared with neurotypical children both with and without FGIDs. From rectal biopsy specimens obtained during endoscopy, deep molecular analysis of the mucosal microbiome in ASD-FGID showed distinct microbial communities at the predicted species level. Correlations of Clostridiales, including multiple Clostridium species previously associated with ASD, with tryptophan, serotonin, and inflammatory cytokine levels yielded a unique multi-omic profile specific to ASD-FGID and ASD-FGID with abdominal pain. Cytokines indicative of inflammation correlated strongly with several bacteria associated with ASD-FGID, as was the case with tryptophan levels, and these potential associations will be confirmed in future work. These data suggest the presence of a specific microbiome profile in ASD with the identification of organisms previously strongly associated within similar cohorts, albeit in stool-based studies. We advance this concept by showing that these ASD-related organisms interact with the intestinal mucosa and are associated with altered neuroimmune signaling. Although these initial findings are correlative, these data form the framework for future studies targeting tryptophan–serotonin metabolism and inflammatory pathways in FGID in ASD.
Acknowledgment
The authors wish to acknowledge Karen Prince for her significant graphic arts contribution to this article as well as Sue Venable for cytokine measurements.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This project was supported by the Health Resources and Services Administration of the US Department of Health and Human Services under cooperative agreement UA3 MC11054–Autism Intervention Research Network on Physical Health. This information or content and conclusions are those of the author and should not be construed as the official position or policy of, nor should any endorsements be inferred by, the Health Resources and Services Administration, Department of Health and Human Services, or the US Government. This work was conducted through the Autism Speaks Autism Treatment Network serving as the Autism Intervention Research Network on Physical Health. This work also was supported by Autism Speaks (GI and Neurobehavioral Processes grant 9455), the National Institute of Diabetes, Digestive, and Kidney Diseases (UH2 DK093990, UH3 DK083990, and DK56338), and the National Institute of Allergy and Infectious Diseases (RO1AI100914 and U01 AI124290-01).
References
- 1.Buie T., Campbell D.B., Fuchs G.J., 3rd Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics. 2010;125(Suppl 1):S1–S18. doi: 10.1542/peds.2009-1878C. [DOI] [PubMed] [Google Scholar]
- 2.McElhanon B.O., McCracken C., Karpen S. Gastrointestinal symptoms in autism spectrum disorder: a meta-analysis. Pediatrics. 2014;133:872–883. doi: 10.1542/peds.2013-3995. [DOI] [PubMed] [Google Scholar]
- 3.Gorrindo P., Williams K.C., Lee E.B. Gastrointestinal dysfunction in autism: parental report, clinical evaluation, and associated factors. Autism Res. 2012;5:101–108. doi: 10.1002/aur.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horvath K., Papadimitriou J.C., Rabsztyn A. Gastrointestinal abnormalities in children with autistic disorder. J Pediatr. 1999;135:559–563. doi: 10.1016/s0022-3476(99)70052-1. [DOI] [PubMed] [Google Scholar]
- 5.Fulceri F., Morelli M., Santocchi E. Gastrointestinal symptoms and behavioral problems in preschoolers with autism spectrum disorder. Dig Liver Dis. 2016;48:248–254. doi: 10.1016/j.dld.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Furuta G.T., Williams K., Kooros K. Management of constipation in children and adolescents with autism spectrum disorders. Pediatrics. 2012;130(Suppl 2):S98–S105. doi: 10.1542/peds.2012-0900H. [DOI] [PubMed] [Google Scholar]
- 7.Hyman P.E., Milla P.J., Benninga M.A. Childhood functional gastrointestinal disorders: neonate/toddler. Gastroenterology. 2006;130:1519–1526. doi: 10.1053/j.gastro.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 8.Mayer E.A., Tillisch K. The brain-gut axis in abdominal pain syndromes. Annu Rev Med. 2011;62:381–396. doi: 10.1146/annurev-med-012309-103958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer E.A., Savidge T., Shulman R.J. Brain-gut microbiome interactions and functional bowel disorders. Gastroenterology. 2014;146:1500–1512. doi: 10.1053/j.gastro.2014.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy R.L., Olden K.W., Naliboff B.D. Psychosocial aspects of the functional gastrointestinal disorders. Gastroenterology. 2006;130:1447–1458. doi: 10.1053/j.gastro.2005.11.057. [DOI] [PubMed] [Google Scholar]
- 11.Daly E., Ecker C., Hallahan B. Response inhibition and serotonin in autism: a functional MRI study using acute tryptophan depletion. Brain. 2014;137:2600–2610. doi: 10.1093/brain/awu178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croonenberghs J., Wauters A., Devreese K. Increased serum albumin, gamma globulin, immunoglobulin IgG, and IgG2 and IgG4 in autism. Psychol Med. 2002;32:1457–1463. doi: 10.1017/s0033291702006037. [DOI] [PubMed] [Google Scholar]
- 13.Croonenberghs J., Bosmans E., Deboutte D. Activation of the inflammatory response system in autism. Neuropsychobiology. 2002;45:1–6. doi: 10.1159/000048665. [DOI] [PubMed] [Google Scholar]
- 14.Jyonouchi H., Sun S., Le H. Proinflammatory and regulatory cytokine production associated with innate and adaptive immune responses in children with autism spectrum disorders and developmental regression. J Neuroimmunol. 2001;120:170–179. doi: 10.1016/s0165-5728(01)00421-0. [DOI] [PubMed] [Google Scholar]
- 15.Jyonouchi H., Geng L., Streck D.L. Immunological characterization and transcription profiling of peripheral blood (PB) monocytes in children with autism spectrum disorders (ASD) and specific polysaccharide antibody deficiency (SPAD): case study. J Neuroinflammation. 2012;9:4. doi: 10.1186/1742-2094-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Chauhan A., Sheikh A.M. Elevated immune response in the brain of autistic patients. J Neuroimmunol. 2009;207:111–116. doi: 10.1016/j.jneuroim.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chez M.G., Dowling T., Patel P.B. Elevation of tumor necrosis factor-alpha in cerebrospinal fluid of autistic children. Pediatr Neurol. 2007;36:361–365. doi: 10.1016/j.pediatrneurol.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Croen L.A., Grether J.K., Yoshida C.K. Antidepressant use during pregnancy and childhood autism spectrum disorders. Arch Gen Psychiatry. 2011;68:1104–1112. doi: 10.1001/archgenpsychiatry.2011.73. [DOI] [PubMed] [Google Scholar]
- 19.Kane M.J., Angoa-Perez M., Briggs D.I. Mice genetically depleted of brain serotonin display social impairments, communication deficits and repetitive behaviors: possible relevance to autism. PLoS One. 2012;7:e48975. doi: 10.1371/journal.pone.0048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson G., Maes M. Redox regulation and the autistic spectrum: role of tryptophan catabolites, immuno-inflammation, autoimmunity and the amygdala. Curr Neuropharmacol. 2014;12:148–167. doi: 10.2174/1570159X11666131120223757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Mahony S.M., Clarke G., Borre Y.E. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res. 2015;277:32–48. doi: 10.1016/j.bbr.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 22.Finegold S.M., Dowd S.E., Gontcharova V. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Williams B.L., Hornig M., Buie T. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One. 2011;6:e24585. doi: 10.1371/journal.pone.0024585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gondalia S.V., Palombo E.A., Knowles S.R. Molecular characterisation of gastrointestinal microbiota of children with autism (with and without gastrointestinal dysfunction) and their neurotypical siblings. Autism Res. 2012;5:419–427. doi: 10.1002/aur.1253. [DOI] [PubMed] [Google Scholar]
- 25.Kang D.W., Park J.G., Ilhan Z.E. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8:e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Angelis M., Piccolo M., Vannini L. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8:e76993. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L., Christophersen C.T., Sorich M.J. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism. 2013;4:42. doi: 10.1186/2040-2392-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W., Chen L., Zhou R. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol. 2014;52:398–406. doi: 10.1128/JCM.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li G., Yang M., Zhou K. Diversity of duodenal and rectal microbiota in biopsy tissues and luminal contents in healthy volunteers. J Microbiol Biotechnol. 2015;25:1136–1145. doi: 10.4014/jmb.1412.12047. [DOI] [PubMed] [Google Scholar]
- 30.Reigstad C.S., Salmonson C.E., Rainey J.F., 3rd Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015;29:1395–13403. doi: 10.1096/fj.14-259598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yano J.M., Yu K., Donaldson G.P. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atarashi K., Tanoue T., Ando M. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohnmacht C., Park J.H., Cording S. Mucosal immunology. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science. 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 34.Williams B.L., Hornig M., Parekh T. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. MBio. 2012;3 doi: 10.1128/mBio.00261-11. e00261–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi G.B., Yim Y.S., Wong H. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–939. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbara G., Stanghellini V., De Giorgio R. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology. 2004;126:693–702. doi: 10.1053/j.gastro.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 37.Barbara G., Wang B., Stanghellini V. Mast cell-dependent excitation of visceral-nociceptive sensory neurons in irritable bowel syndrome. Gastroenterology. 2007;132:26–37. doi: 10.1053/j.gastro.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 38.Hildebrand F., Tadeo R., Voigt A.Y. LotuS: an efficient and user-friendly OTU processing pipeline. Microbiome. 2014;2:30. doi: 10.1186/2049-2618-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 40.Ritari J., Salojarvi J., Lahti L. Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database. BMC Genomics. 2015;16:1056. doi: 10.1186/s12864-015-2265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quast C., Pruesse E., Yilmaz P. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson G.M., Feibel F.C., Cohen D.J. Determination of serotonin in whole blood, platelet-rich plasma, platelet-poor plasma and plasma ultrafiltrate. Life Sci. 1987;40:1063–1070. doi: 10.1016/0024-3205(87)90568-6. [DOI] [PubMed] [Google Scholar]
- 43.Parks D.H., Tyson G.W., Hugenholtz P. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boccuto L., Chen C.F., Pittman A.R. Decreased tryptophan metabolism in patients with autism spectrum disorders. Mol Autism. 2013;4:16. doi: 10.1186/2040-2392-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller C.L., Anacker A.M., Veenstra-VanderWeele J. The serotonin system in autism spectrum disorder: from biomarker to animal models. Neuroscience. 2016;321:24–41. doi: 10.1016/j.neuroscience.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masi A., Quintana D.S., Glozier N. Cytokine aberrations in autism spectrum disorder: a systematic review and meta-analysis. Mol Psychiatry. 2015;20:440–446. doi: 10.1038/mp.2014.59. [DOI] [PubMed] [Google Scholar]
- 47.Gabriele S., Sacco R., Persico A.M. Blood serotonin levels in autism spectrum disorder: a systematic review and meta-analysis. Eur Neuropsychopharmacol. 2014;24:919–929. doi: 10.1016/j.euroneuro.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Finegold S.M., Molitoris D., Song Y. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis. 2002;35:S6–S16. doi: 10.1086/341914. [DOI] [PubMed] [Google Scholar]
- 49.Song Y., Liu C., Finegold S.M. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol. 2004;70:6459–6465. doi: 10.1128/AEM.70.11.6459-6465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parracho H.M., Bingham M.O., Gibson G.R. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J Med Microbiol. 2005;54:987–991. doi: 10.1099/jmm.0.46101-0. [DOI] [PubMed] [Google Scholar]
- 51.Son J.S., Zheng L.J., Rowehl L.M. Comparison of fecal microbiota in children with autism spectrum disorders and neurotypical siblings in the Simons simplex collection. PLoS One. 2015;10:e0137725. doi: 10.1371/journal.pone.0137725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raymond F., Ouameur A.A., Deraspe M. The initial state of the human gut microbiome determines its reshaping by antibiotics. ISME J. 2016;10:707–720. doi: 10.1038/ismej.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cook E.H., Leventhal B.L. The serotonin system in autism. Curr Opin Pediatr. 1996;8:348–354. doi: 10.1097/00008480-199608000-00008. [DOI] [PubMed] [Google Scholar]
- 54.Saulnier D.M., Riehle K., Mistretta T.A. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chumpitazi B.P., Cope J.L., Hollister E.B. Randomised clinical trial: gut microbiome biomarkers are associated with clinical response to a low FODMAP diet in children with the irritable bowel syndrome. Aliment Pharmacol Ther. 2015;42:418–427. doi: 10.1111/apt.13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashwood P., Krakowiak P., Hertz-Picciotto I. Associations of impaired behaviors with elevated plasma chemokines in autism spectrum disorders. J Neuroimmunol. 2011;232:196–199. doi: 10.1016/j.jneuroim.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashwood P., Krakowiak P., Hertz-Picciotto I. Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav Immun. 2011;25:40–45. doi: 10.1016/j.bbi.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsiao E.Y., McBride S.W., Chow J. Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc Natl Acad Sci U S A. 2012;109:12776–12781. doi: 10.1073/pnas.1202556109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu W.L., Adams C.E., Stevens K.E. The interaction between maternal immune activation and alpha 7 nicotinic acetylcholine receptor in regulating behaviors in the offspring. Brain Behav Immun. 2015;46:192–202. doi: 10.1016/j.bbi.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsiao E.Y., Patterson P.H. Activation of the maternal immune system induces endocrine changes in the placenta via IL-6. Brain Behav Immun. 2011;25:604–615. doi: 10.1016/j.bbi.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh V.K. Plasma increase of interleukin-12 and interferon-gamma. Pathological significance in autism. J Neuroimmunol. 1996;66:143–145. doi: 10.1016/0165-5728(96)00014-8. [DOI] [PubMed] [Google Scholar]
- 62.Pagan C., Delorme R., Callebert J. The serotonin-N-acetylserotonin-melatonin pathway as a biomarker for autism spectrum disorders. Transl Psychiatry. 2014;4:e479. doi: 10.1038/tp.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marler S., Ferguson B.J., Lee E.B. Brief report: whole blood serotonin levels and gastrointestinal symptoms in autism spectrum disorder. J Autism Dev Disord. 2016;46:1124–1130. doi: 10.1007/s10803-015-2646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Margolis K.G., Li Z., Stevanovic K. Serotonin transporter variant drives preventable gastrointestinal abnormalities in development and function. J Clin Invest. 2016;126:2221–2235. doi: 10.1172/JCI84877. [DOI] [PMC free article] [PubMed] [Google Scholar]