Abstract
Background & Aims
Cholestasis promotes endoplasmic reticulum (ER) stress in the liver, however, the effect of ER stress on hepatic bile acid metabolism is unknown. We aim to determine the effect of ER stress on hepatic bile acid synthesis and transport in mice.
Methods
ER stress was induced pharmacologically in C57BL/6J mice and human hepatoma (HepG2) cells. The hepatic expression of genes controlling bile acid synthesis and transport was determined. To measure the activity of the primary bile acid synthetic pathway, the concentration of 7α-hydroxy-4-cholesten-3-1 was measured in plasma.
Results
Induction of ER stress in mice and HepG2 cells rapidly suppressed the hepatic expression of the primary bile acid synthetic enzyme, cholesterol 7α-hydroxylase. Plasma levels of 7α-hydroxy-4-cholesten-3-1 were reduced in mice subjected to ER stress, indicating impaired bile acid synthesis. Induction of ER stress in mice and HepG2 cells increased expression of the bile salt export pump (adenosine triphosphate binding cassette [Abc]b11) and a bile salt efflux pump (Abcc3). The observed regulation of Cyp7a1, Abcb11, and Abcc3 occurred in the absence of hepatic inflammatory cytokine activation and was not dependent on activation of hepatic small heterodimer partner or intestinal fibroblast growth factor 15. Consistent with suppressed bile acid synthesis and enhanced bile acid export from hepatocytes, prolonged ER stress decreased the hepatic bile acid content in mice.
Conclusions
Induction of ER stress in mice suppresses bile acid synthesis and enhances bile acid removal from hepatocytes independently of established bile acid regulatory pathways. These data show a novel function of the ER stress response in regulating bile acid metabolism.
Keywords: Unfolded Protein Response, Cyp7a1, 7α-Hydroxy-4-Cholesten-3-1, Bile Acid Synthesis
Abbreviations used in this paper: ABC, adenosine triphosphate binding cassette; C4, 7α-hydroxy-4-cholesten-3-1; CYP7A1, cholesterol 7α-hydroxylase; DMEM, Dulbecco's modified Eagle medium; DMSO, dimethyl sulfoxide; ER, endoplasmic reticulum; ERK, extracellular signaling-regulated kinase; FGF, fibroblast growth factor; FXR, farnesoid X receptor; IL, interleukin; IRE1α, inositol requiring enzyme 1α; JNK, c-Jun-N-terminal kinase; mRNA, messenger RNA; NTCP, sodium/taurocholate cotransporter; RIDD, regulated inositol requiring enzyme 1α–dependent messenger RNA decay; SHP, small heterodimer partner; UPR, unfolded protein response
Graphical Abstract
See editorial on page 135.
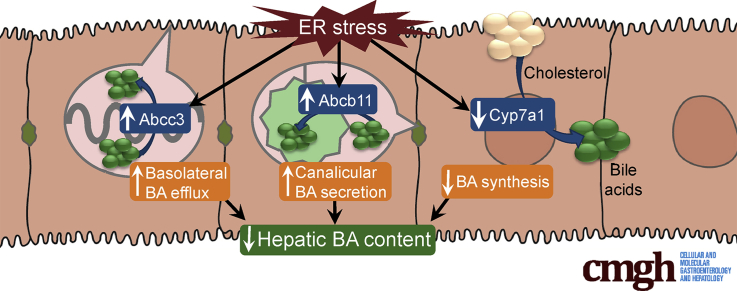
Summary.
We show that induction of endoplasmic reticulum stress in mice suppresses the primary bile acid synthetic pathway controlled by cholesterol 7α-hydroxylase and activates hepatic bile acid transporters that promote the removal of excess bile acids from the liver.
The enterohepatic circulation of bile acids is a highly efficient process by which bile acid homeostasis is maintained. Bile acids are secreted from the liver into bile via the rate-limiting adenosine triphosphate binding cassette transporter, ATP-binding cassette subfamily B member 11 (ABCB11), and subsequently are transported in bile to the lumen of the small intestine where they aid in the digestion of dietary fat, cholesterol, and fat-soluble vitamins.1, 2 Bile acids ultimately are taken up in the ileum and returned to the liver via the portal circulation where they are transported across the hepatic sinusoidal membrane by the sodium/taurocholate cotransporter (NTCP).3, 4, 5 Although the enterohepatic circulation of bile acids is a highly efficient process, de novo synthesis of bile acids must occur in the liver to replenish the small fraction of bile acids that are lost by fecal excretion. New bile acids are synthesized in the liver from cholesterol, which is catalyzed by the rate-limiting cytochrome P450 enzyme, cholesterol 7α hydroxylase (CYP7A1).6, 7, 8 Disruption of the enterohepatic circulation of bile acids can result in accumulation of bile acids within the liver, leading to cholestatic liver injury. Numerous compensatory mechanisms are activated in response to cholestasis to reduce bile acid accumulation, including suppression of bile acid uptake, increased bile acid efflux, enhanced biliary bile acid secretion, and reduced bile acid synthesis.9
Extrahepatic biliary obstruction is the classic cause of cholestasis, however, intrahepatic cholestasis can result from impairment in bile acid transporter function or disruption of the compensatory mechanisms that counteract hepatic bile acid accumulation. Systemic inflammation is a well-described cause of cholestasis in the absence of mechanical biliary obstruction.9, 10, 11, 12, 13 The pathogenesis of inflammation-induced cholestasis is incompletely defined but is thought to involve inflammatory cytokine-mediated inhibition of hepatic bile acid transporter expression.11, 12, 13 Moreover, the interplay between bile acid metabolism and cellular stress responses in the liver is complex and incompletely understood.
Activation of the unfolded protein response (UPR), a highly conserved signaling cascade induced by endoplasmic reticulum (ER) stress, has been identified as a feature of many hepatic diseases including cholestatic liver disease.14, 15, 16, 17, 18, 19, 20, 21 It is unclear whether activation of the UPR in the setting of cholestasis is a manifestation of a generalized inflammatory response or whether the UPR directly functions in the pathogenesis of cholestatic liver disease. Although the UPR initially was identified as a mechanism to maintain proper protein folding and processing, it now is clear that this signaling cascade regulates a wide range of cellular processes. In particular, the UPR increasingly is recognized as a critical regulator of hepatic lipid metabolism, which is tightly linked to bile acid metabolism.11, 13, 14, 15 Accumulating evidence has suggested that bile acids modulate ER stress, however, it is unknown whether ER stress, in turn, alters bile acid homeostasis.22, 23, 24 In this article we explore the effect of ER stress on the regulation of bile acid synthesis and transport in mice.
Materials and Methods
Animals and Treatments
Male C57BL/6J mice (8–10 weeks of age) were purchased from Jackson Laboratories (Bar Harbor, ME). The mice were in the light 14 hours per day and in darkness 10 hours per day everyday of their life (per routine animal housing protocol) and were given free access to standard laboratory chow and water. To induce acute ER stress, mice were treated with a single intraperitoneal (IP) injection of tunicamycin (0.5 mg/kg) and were killed 6 hours later. To induce prolonged ER stress, mice were treated with daily injections of tunicamycin (0.1 mg/kg IP) for 5 days (cumulative dose, 0.5 mg/kg IP). Control mice were treated with vehicle (10% dimethyl sulfoxide [DMSO] IP). Blood was collected by cardiac puncture and immediately centrifuged to collect the plasma. The livers were excised rapidly, flushed with ice-cold saline, sectioned, and snap-frozen in liquid nitrogen. The small intestine was removed, flushed with ice-cold saline, and the terminal 5-cm segment was snap-frozen in liquid nitrogen. The livers and small intestine were stored at -80°C until analysis. Liver histology was prepared by the Northwestern University Mouse Histology and Phenotyping Laboratory. All animal protocols were approved by the Northwestern University Institutional Animal Care and Use Committee.
Plasma and Hepatic Biochemical Analysis
Hepatic bile acid concentration was measured using a BioQuant total bile acid colorimetric assay (BQ Kits, San Diego, CA). Plasma alanine aminotransferase level (Teco Diagnostics, Anaheim, CA) was measured using a spectrophotometric assay per the manufacturer’s protocol. Total cholesterol content in liver homogenate was measured using an Infinity spectrophotometric assay (Fisher Scientific, Middletown, VA). Plasma 7α-hydroxy-4-cholesten-3-1 (C4) measurement was performed at the Mayo Clinic Immunochemical Core Lab (Rochester, MN).
High-Performance Liquid Chromatography Assay
Hepatic bile acid composition was measured by high-performance liquid chromatography as previously described.25 Samples were spiked with glycocholic acid as an internal standard to control for extraction efficiency. The content of tauromuricholic acid, taurocholic acid, and taurochenodeoxycholic acid was calculated from a standard curve and reported as the percentage of total hepatic bile acids.
Cell Culture
Human hepatoma (HepG2) cells (ATCC, Mannasas, VA) were cultured in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum and maintained at 37°C in 5% CO2. Cells were grown to 80% confluence in 6-well plates and treated with 12 μmol/L tunicamycin, 100 nmol/L thapsigargin (Sigma-Aldrich, St. Louis, MO), 5 mmol/L DL-homocysteine (Sigma-Aldrich), or vehicle (DMSO/saline) in serum-free DMEM for 6 hours. To determine whether the effects of tunicamycin are dependent on c-Jun-N-terminal kinase (JNK) or extracellular signaling-regulated kinase (ERK), HepG2 cells were treated with the JNK inhibitor SP600125 (Sigma-Aldrich) at a concentration of 25 μmol/L or the MAPK/ERK kinase inhibitor PD184352 (Santa Cruz Biotechnology, Dallas, TX) at 1 μmol/L or vehicle (DMSO/saline) as previously described.26 One hour after exposure to SP600125 or PD184352, cells were treated with tunicamycin (12 μmol/L; Sigma-Aldrich) in serum-free DMEM and incubated for an additional 6 hours. Successful inhibition of JNK and ERK activation was confirmed by Western blot analysis as previously described.26
Analysis of Gene and Protein Expression
Total RNA from frozen liver, ileum, or cultured HepG2 cells was isolated using TRIzol reagent (Ambion Life Technologies, Carlsbad, CA), and real-time quantitative polymerase chain reaction was performed as previously described.27, 28 Total protein was isolated from frozen liver samples and Western blotting was performed as previously described.27, 28 Protein detection was performed using polyclonal rabbit antibodies to CYP7A1 (Proteintech, Rosemont, IL) and glyceraldehyde-3-phosphate dehydrogenase (Cell Signaling Technology, Danvers, MA). Bound antibody was detected using goat anti-rabbit polyclonal horseradish-peroxidase antibody (Cell Signaling Technology) and developed using enhanced chemiluminescence Western Blotting Substrate (Cell Signaling Technology). Representative Western blots of pooled samples are shown. Densitometry was performed on individual samples using ImageJ software (available: imagej.nih.gov/ij/; National Institutes of Health, Bethesda, MD).
Statistical Analysis
Data are presented as means ± SD. Comparisons between groups were performed using the Student t test analysis.
All authors had access to the study data and reviewed and approved the final manuscript.
Results
ER Stress Suppresses the Primary Bile Acid Synthetic Pathway
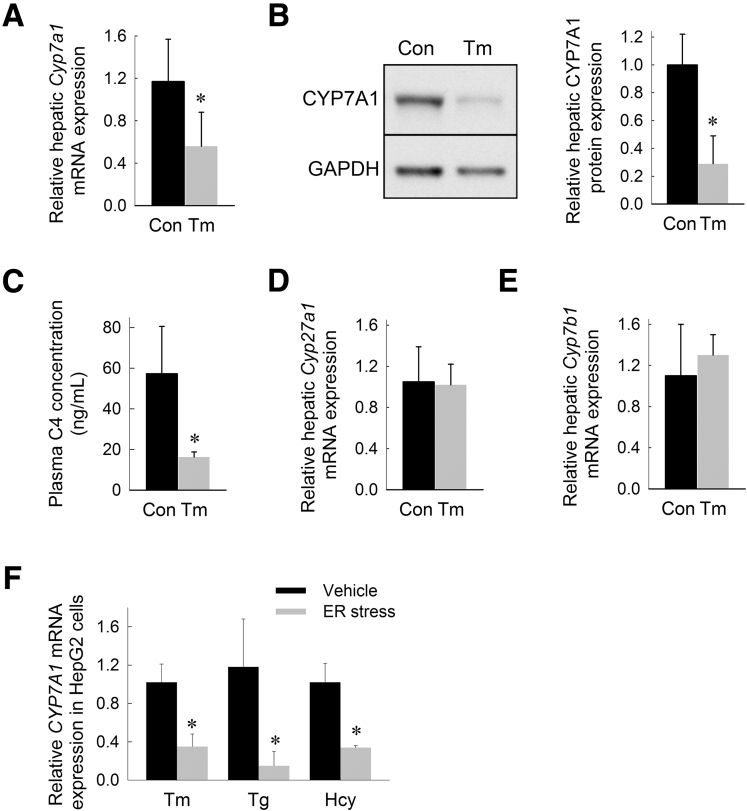
CYP7A1 is the primary bile acid synthetic enzyme controlling the rate-limiting step in the conversion of hepatic cholesterol to bile acids.29, 30 To determine the effect of ER stress on hepatic Cyp7a1 expression, mice were treated with tunicamycin (0.5 mg/kg IP), a well-established ER stress–inducing agent in mice.31, 32, 33 Mice treated with tunicamycin showed robust hepatic UPR activation at 6 hours as evidenced by induction of glucose-regulated protein 78 kilodaltons, an ER chaperone and master regulator of the UPR, spliced X-box binding protein 1, a major mediator of the inositol requiring enzyme 1α (IRE1α) branch of the UPR, and CCAAT/enhancer binding protein homologous protein, a regulator of ER stress-induced apoptosis34, 35, 36, 37 (Table 1). Induction of hepatic ER stress resulted in significant suppression of hepatic Cyp7a1 messenger RNA (mRNA) and CYP7A1 protein expression (Figure 1A and B). We next determined whether ER stress suppresses CYP7A1-dependent bile acid synthesis. The plasma concentration of C4, a stable intermediate generated in the synthesis of bile acids from cholesterol, is an indicator of the activity of the CYP7A1-dependent bile acid synthetic pathway. Consistent with the observed suppression of CYP7A1, induction of ER stress in mice reduced the plasma concentration of C4 (Figure 1C). Although CYP7A1 regulates the major pathway of bile acid synthesis, bile acid synthesis also occurs via the alternative (acidic) pathway of bile acid synthesis involving sterol 27-hydroxylase and oxysterol 7-α-hydroxylase.38, 39, 40 We found that neither sterol 27-hydroxylase nor oxysterol 7α hydroxylase mRNA expression was altered by induction of ER stress (Figure 1D).
Table 1.
Activation of the Hepatic and Intestinal Unfolded Protein Response in Mice Treated With Tunicamycin for 6 Hours or 5 Days
| Relative hepatic expression |
Relative ileal expression |
|||
|---|---|---|---|---|
| 6 hours | 5 days | 6 hours | 5 days | |
| Grp78/BiP | 47.4a ± 20.5 | 19.3a ± 15.9 | 1.2 ± 0.5 | 8.4a ± 6.0 |
| Xbp1s | 5.7a ± 1.9 | 5.1a ± 4.9 | 1.1 ± 0.4 | 0.5a ± 0.2 |
| Chop | 197.7a ± 79.6 | 20.5a ± 21.1 | 1.2 ± 0.6 | 10.9a ± 4.8 |
NOTE. Hepatic and ileal mRNA expression levels relative to vehicle-treated controls. Means (n = 6) ± SD.
Chop, CCAAT/enhancer binding protein homologous protein; Grp78/BiP, glucose-regulated protein 78 kilodaltons; Xbp1s, X-box binding protein 1.
P < .05 vs vehicle-treated control at same time point.
Figure 1.
ER stress suppresses the primary bile acid synthetic pathway in mice. (A) Relative hepatic Cyp7a1 mRNA expression, (B) Western blot of hepatic CYP7A1 protein levels with corresponding densitometry analysis, (C) plasma C4 concentration (ng/mL), (D) relative hepatic sterol 27-hydroxylase (Cyp27a1) mRNA expression, and (E) relative hepatic oxysterol 7α hydroxylase (Cyp7b1) mRNA expression in mice treated with tunicamycin (Tm) for 6 hours. (F) Relative CYP7A1 mRNA levels in HepG2 cells treated with Tm, thapsigargin (Tg), or homocysteine (Hcy) for 6 hours. Representative Western blot of hepatic protein from 6 individually treated mice pooled per lane. Gene expression and plasma analysis are shown as the means (n = 6) ± SD. For cell culture experiments, gene expression is reported as the means of 6 identically treated replicates. *P < .05 vs vehicle-injected controls. Con, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
There are significant differences in the expression levels and regulation of the CYP7A1 gene in mice and human beings. Most notably, human beings lack an Liver X Receptor-response element in the CYP7A1 promoter, rendering human CYP7A1 unresponsive to dietary cholesterol.41, 42, 43 To exclude the possibility that the effects of ER stress on Cyp7a1 expression are specific to mice, we determined the effect of pharmacologic ER stress on CYP7A1 expression in a human hepatoma cell line (HepG2). Paralleling our findings in vivo, induction of ER stress in HepG2 cells suppressed CYP7A1 expression (Figure 1E). To ensure that these effects are not specific to tunicamycin, we also treated HepG2 cells with 2 alternative pharmacologic ER stress–inducing agents, thapsigargin and homocysteine, for 6 hours. CYP7A1 expression was suppressed by 87% and 67% in HepG2 cells treated with thapsigargin and homocysteine, respectively (Figure 1E).
Suppression of Cyp7a1 by ER Stress Is Independent of Farnesoid X Receptor–Dependent Bile Acid Feedback Inhibition Pathways
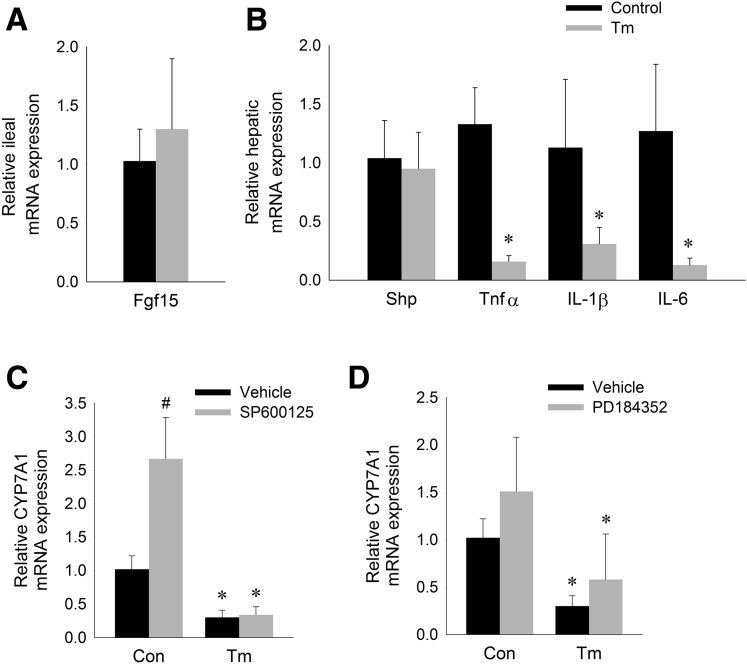
A major mechanism of Cyp7a1 regulation is via feedback inhibition from bile acids.44, 45, 46 Specifically, bile acids bind to the farnesoid X receptor (FXR) in the ileum, stimulating release of fibroblast growth factor (FGF) 15/19 from the ileocyte, which subsequently acts in the liver to suppress Cyp7a1 transcription.47, 48 Intestinal ER stress, achieved through oral administration of tunicamycin to mice, has been shown to induce ileal Fgf15 expression.49 We found that administration of intraperitoneal tunicamycin did not induce intestinal ER stress at 6 hours (Table 1). Consistent with an absence of intestinal UPR activation, we found no induction of ileal Fgf15 expression in mice treated with intraperitoneal tunicamycin for 6 hours (Figure 2A). These data indicate that ER stress suppresses hepatic Cyp7a1 independent of ileal Fgf15 activation.
Figure 2.
Suppression of CYP7A1 by ER stress is independent of known bile acid regulatory pathways. (A) Relative ileal Fgf15 mRNA expression and (B) relative hepatic Shp, tumor necrosis factor (Tnf)α, Il1β, and Il6 mRNA expression in mice treated with tunicamycin (Tm) for 6 hours. Means (n = 6) ± SD. *P < .05 vs control. (C and D) Relative CYP7A1 mRNA levels in HepG2 cells treated with (C) Tm ± JNK-inhibitor (SP600125) or (D) tunicamycin ± MAPK/ERK kinase inhibitor (PD184352) for 6 hours. Means (n = 6) ± SD. *P < .05 vs control cells not treated with Tm, #P < .05 vs control cells not treated with inhibitor.
Although activation of intestinal FXR-FGF15/19 signaling now is considered the major mechanism of bile acid feedback inhibition of Cyp7a1 transcription, bile acids also inhibit Cyp7a1 via activation of FXR within the liver, leading to induction of the small heterodimer partner (Shp). We found that induction of ER stress in mice did not increase hepatic Shp expression, indicating that tunicamycin does not suppress Cyp7a1 via a FXR-SHP–dependent mechanism (Figure 2B).
Suppression of Cyp7a1 by ER Stress Is Independent of Hepatic Inflammatory Cytokine Activation
Suppression of Cyp7a1 expression is a feature of the hepatic inflammatory response.11, 12 Activation of cytokines such as tumor necrosis factor α, interleukin (IL)1β, and IL6 is associated with liver injury and has been shown to suppress hepatic Cyp7a1 expression in vitro and in vivo.11, 50, 51, 52, 53 Therefore, we considered the possibility that suppression of Cyp7a1 expression by ER stress is a manifestation of a broader stress response associated with inflammatory cytokine activation. We found no significant inflammatory cytokine activation at 6 hours after induction of ER stress (Figure 2B). On the contrary, acute exposure to ER stress suppressed hepatic Tnfα, Il1β, and Il6 expression. These data indicate that suppression of bile acid synthesis by ER stress is independent of inflammatory cytokine activation.
Suppression of CYP7A1 Expression by ER Stress Is Not Dependent on JNK and ERK Mitogen-Activated Protein Kinase Activation
Activation of JNK has been implicated in the inhibition of CYP7A1 by inflammatory cytokines.52, 54, 55 Furthermore, it is well established that JNK activation occurs in response to ER stress.26, 56, 57 To confirm whether ER stress–induced CYP7A1 suppression is dependent on JNK activation, we determined the effects of tunicamycin on HepG2 cells treated with the JNK inhibitor SP600125. Inhibition of JNK signaling in HepG2 cells did not prevent suppression of CYP7A1 by tunicamycin (Figure 2C).
FGF15/19-mediated suppression of CYP7A1 expression is dependent on ERK activation.58, 59 We assessed whether ER stress–induced CYP7A1 suppression is dependent on ERK activation. Inhibition of ERK in HepG2 cells using PD184352 did not prevent ER stress–induced suppression of CYP7A1, thus providing additional evidence that the effect of ER on hepatic bile acid synthesis is not dependent on FGF15/19 activation (Figure 2D).
ER Stress Regulates Hepatic Bile Acid Transporters in Mice
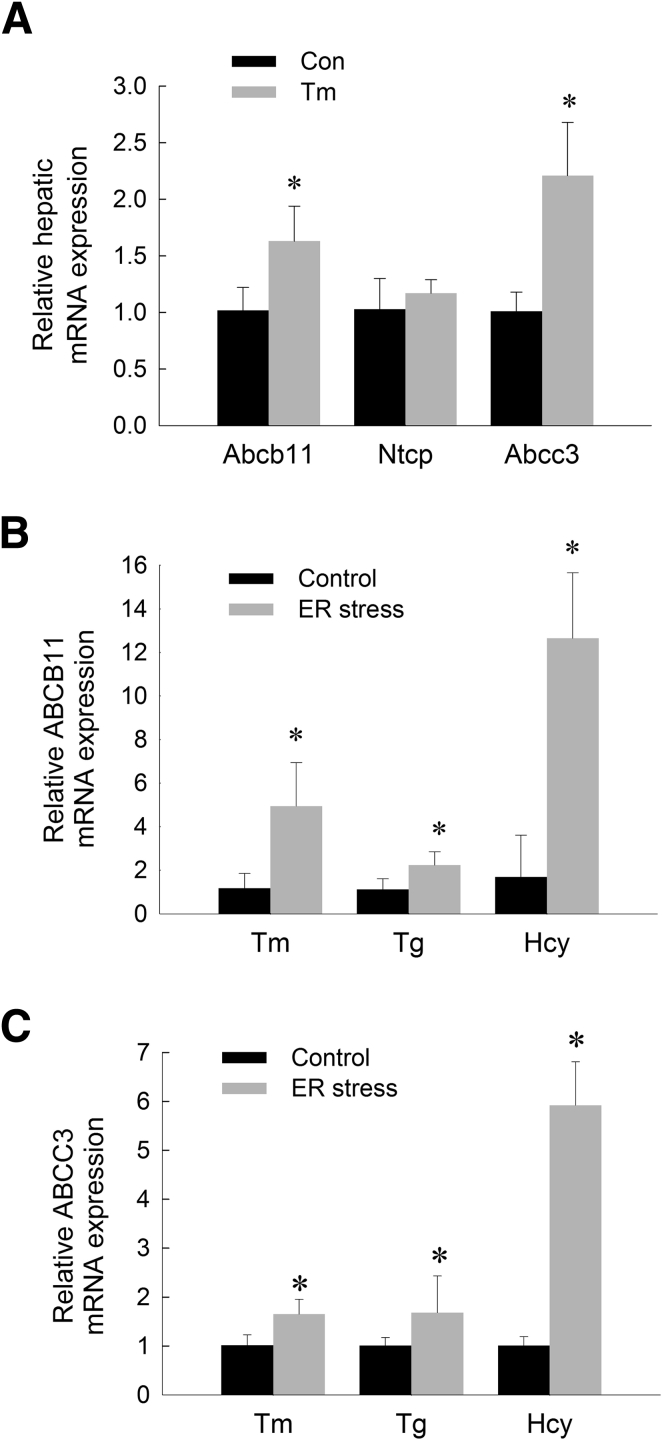
Suppression of Cyp7a1 expression is a major compensatory mechanism to prevent bile acid accumulation in the setting of cholestatic liver injury. Several other compensatory mechanisms are activated in the liver in response to cholestasis, including suppression of bile acid uptake, induction of bile acid efflux, and enhanced biliary bile acid secretion.9 We next examined the effects ER stress on these anticholestatic mechanisms. Acute ER stress increased hepatic expression of adenosine triphosphate binding cassette (Abc)b11, the canalicular bile salt export pump, yet had no significant effect on the hepatic expression of Ntcp, the basolateral transporter controlling sinusoidal uptake of bile acids into hepatocytes (Figure 3A). The multidrug resistance-associated protein 3 (ABCC3) is a basolateral bile acid efflux pump that is thought to have a protective role in the setting of cholestasis by removing bile acids from the cholestatic liver.60, 61 The expression of Abcc3 was increased in response to acute ER stress (Figure 3A).
Figure 3.
ER stress regulates the expression of hepatic bile acid transporters in mice and HepG2 cells. (A) Relative hepatic mRNA levels of Abcb11, Ntcp, and Abcc3 in mice treated with tunicamycin (Tm) for 6 hours. Relative mRNA expression of (B) ABCB11 and (C) ABCC3 in HepG2 cells treated with Tm, thapsigargin (Tg), or homocysteine (Hcy) for 6 hours. Means (n = 6) ± SD. *P < .05 vs controls
Having shown that hepatic Abcb11 and Abcc3 are activated in mice subjected to ER stress, we next measured ABCB11 and ABCC3 expression in human hepatoma cells (HepG2) treated with tunicamycin. Paralleling our findings in vivo, induction of ER stress with tunicamycin, thapsigargin, or homocysteine in HepG2 cells increased expression of ABCB11 and ABCC3 (Figure 3B and C).
Prolonged ER Stress Reduces the Hepatic Bile Acid Content in Mice
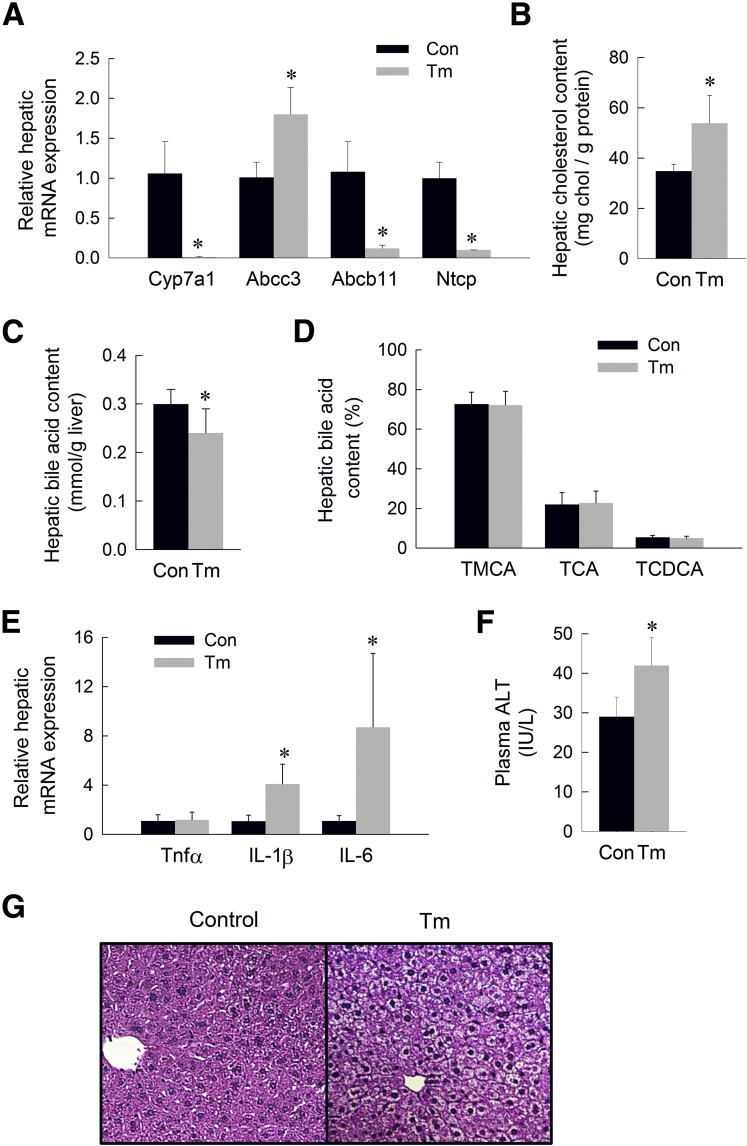
We have shown that induction of ER stress suppresses the primary bile acid synthetic pathway and enhances expression of transporters responsible for the removal of bile acids from the liver. These pathways seemingly would converge to reduce the hepatic bile acid content over time. To test this hypothesis, we examined the effect of prolonged ER stress on hepatic bile acid content. Mice were treated with daily injections of low-dose tunicamycin (0.1 mg/kg IP) for 5 days. Similar to the pattern observed in mice acutely after induction of ER stress, mice subjected to prolonged ER stress showed induction of the hepatic UPR associated with marked suppression of Cyp7a1 and induction of Abcc3 (Table 1 and Figure 4A). Consistent with suppressed CYP7A1-dependent bile acid synthesis, hepatic cholesterol content was increased and hepatic bile acid content was decreased in response to prolonged ER stress (Figure 4B and C). The relative composition of the hepatic bile acids was unaffected by ER stress (Figure 4D). Unlike acute ER stress, prolonged ER stress suppressed hepatic expression of Abcb11 and Ntcp, bile acid transporters known to be suppressed by hepatic inflammation (Figure 4A). We found that prolonged ER stress resulted in induction of the inflammatory cytokines Il1β and Il6, suggesting activation of a more generalized hepatic inflammatory response in the setting of sustained ER stress (Figure 4E). Consistent with the development of hepatic inflammation, mice subjected to prolonged ER stress showed a mild increase in plasma alanine aminotransferase level (Figure 4F) and early ballooning degeneration on H&E-stained liver sections (Figure 4G). There was no overt hepatic steatosis or cholestasis evident histologically. Prolonged ER stress also resulted in induction of the intestinal UPR, suggesting activation of a broader systemic stress response over time (Table 1).
Figure 4.
Effect of prolonged ER stress on bile acid metabolism. (A) Relative hepatic mRNA expression of Cyp7a1, Abcc3, Abcb11, and Ntcp, (B) hepatic cholesterol content (mg chol/g liver), (C) hepatic bile acid content (mmol/g liver), (D) hepatic bile acid composition (% of total hepatic bile acids), (E) relative hepatic mRNA expression of tumor necrosis factor (Tnf)α, IL1β, and IL6, (F) plasma alanine aminotransferase (ALT) level (IU/L), and (G) H&E-stained liver sections of mice treated with tunicamycin for 5 days. Means (n = 6) ± SD. *P < .05 vs vehicle-injected controls. Con, control; TCA, taurocholic acid; TCDCA, taurochenodeoxycholic acid; Tm, tunicamycin; TMCA, tauromuricholic acid.
Discussion
Bile acids perform critical physiologic functions, however, when present in excess these molecules promote hepatotoxicity. In response to hepatocellular stress, protective mechanisms are activated to reduce hepatic accumulation of toxic bile acids. We show that ER stress activates anticholestatic mechanisms and decreases hepatic bile acid content. Potentially the most critical ER stress–induced anticholestatic mechanism we have identified is suppression of the CYP7A1-dependent bile acid synthetic pathway. We found that suppression of CYP7A1 occurs early after induction of ER stress, in the absence of inflammatory cytokine activation. In addition, we showed that CYP7A1 suppression by ER stress occurred in the absence of hepatic Shp or ileal Fgf15 activation. These data show that CYP7A1 suppression by ER stress was independent of established FXR-dependent or cytokine-mediated pathways. Moreover, these data strongly suggest that the observed effects of acute ER stress on hepatic bile acid metabolism were a direct result of ER stress and not simply a manifestation of a broader hepatic inflammatory response.
We found that ER stress also regulated the expression of hepatic basolateral and canalicular transporters. ER stress acutely increased hepatic expression of Abcc3, a basolateral bile salt efflux pump, and Abcb11, the canalicular bile salt export pump. These findings suggest that, as part of a coordinated protective response, the hepatocyte removed toxic bile acids from the liver in response to ER stress. Consistent with this assertion was the finding that prolonged ER stress lead to a reduction in hepatic bile acid content.
Contrary to the acute response to ER stress, we found that sustained ER stress led to suppression of Abcb11 and Ntcp. Furthermore, we found that sustained ER stress promoted a hepatic inflammatory response associated with cytokine activation. Suppression of Abcb11 and Ntcp has been a well-established feature of the hepatic inflammatory response,12, 13, 62, 63, 64, 65 and we speculate that suppression of these transporters with prolonged ER stress is secondary to inflammatory cytokine activation.
We have speculated that suppression of CYP7A1 is a direct result of UPR activation yet it remains unclear which element(s) of this highly intricate signaling cascade may be responsible for the observed phenotype. The UPR is initiated through 3 ER transmembrane receptors, PKR-like ER kinase, activating transcription factor 6 α, and IRE1α and 1 master chaperone, glucose-regulated protein 78 kilodaltons. The IRE1α pathway is the most evolutionarily conserved branch of the UPR and is considered the major mediator of the adaptive response to cellular stress.66, 67, 68 It therefore is reasonable to hypothesize that the IRE1α arm of the UPR may mediate an adaptive response aimed at maintaining bile acid homeostasis. Furthermore, although all 3 branches of the UPR have been shown to modulate metabolic pathways, the IRE1α pathway has emerged as the branch most strongly implicated in regulating hepatic lipid metabolism, which is linked intimately to bile acid metabolism.69, 70, 71 Activated IRE1α, functioning as an endonuclease, induces nonconventional splicing of the transcription factor X-box binding protein 1, which has been implicated in the transcription of a myriad of genes involved in proteostasis and metabolic processes. It has become increasingly apparent, however, that the major regulatory effect of IRE1α activation is attributable to regulated IRE1α-dependent mRNA decay (RIDD).72 A rapidly increasing number of mRNAs and microRNAs involved in a diverse array of cellular processes have been identified as RIDD targets.72, 73, 74, 75 At least 2 such mRNAs encode ER-localized cytochrome P450 enzymes, Cyp1a2 and Cyp2e1.76 These data raise the possibility that in the setting of ER stress, Cyp7a1 mRNA may be targeted for destruction via RIDD.
The present work implicates ER stress as a novel regulator of the major bile acid synthetic pathway. Although the classic pathophysiologic consequence of disrupted bile acid homeostasis is cholestatic liver injury, altered bile acid metabolism now is known to impact numerous physiologic processes and may have broader implications for human metabolic disease. Impaired bile acid metabolism has been implicated in the pathogenesis of diabetes and obesity, both of which are associated with ER stress.77, 78, 79 The possibility that altered bile acid homeostasis is a mechanistic link between ER stress and the development of metabolic disease warrants further investigation.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This publication was supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases grants R01DK093807 and K08DK095992, an AGA Research Scholar Award, the George Lockerbie Liver Cancer Foundation, the Max Goldenberg Foundation, and a National Institutes of Health/National Center for Advancing Translational Sciences Clinical and Translational Science Awards grant UL1TR000135. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
References
- 1.Gerloff T., Stieger B., Hagenbuch B. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem. 1998;273:10046–10050. doi: 10.1074/jbc.273.16.10046. [DOI] [PubMed] [Google Scholar]
- 2.Chiang J.Y. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson P.A., Lan T., Rao A. Bile acid transporters. J Lipid Res. 2009;50:2340–2357. doi: 10.1194/jlr.R900012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazaridis K.N., Pham L., Tietz P. Rat cholangiocytes absorb bile acids at their apical domain via the ileal sodium-dependent bile acid transporter. J Clin Invest. 1997;100:2714–2721. doi: 10.1172/JCI119816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson P.A., Hubbert M., Haywood J. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J Biol Chem. 2005;280:6960–6968. doi: 10.1074/jbc.M412752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pikuleva I.A. Cytochrome P450s and cholesterol homeostasis. Pharmacol Ther. 2006;112:761–773. doi: 10.1016/j.pharmthera.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Kramer W., Buscher H.P., Gerok W. Bile salt binding to serum components. Taurocholate incorporation into high-density lipoprotein revealed by photoaffinity labelling. Eur J Biochem. 1979;102:1–9. doi: 10.1111/j.1432-1033.1979.tb06257.x. [DOI] [PubMed] [Google Scholar]
- 8.Chiang J.Y. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J Hepatol. 2004;40:539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Geier A., Wagner M., Dietrich C.G. Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochim Biophys Acta. 2007;1773:283–308. doi: 10.1016/j.bbamcr.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 10.Omenetti S., Brogi M., Goodman W.A. Dysregulated intrahepatic CD4+ T-cell activation drives liver inflammation in ileitis-prone SAMP1/YitFc mice. Cell Mol Gastroenterol Hepatol. 2015;1:406–419. doi: 10.1016/j.jcmgh.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trauner M., Fickert P., Stauber R.E. Inflammation-induced cholestasis. J Gastroenterol Hepatol. 1999;14:946–959. doi: 10.1046/j.1440-1746.1999.01982.x. [DOI] [PubMed] [Google Scholar]
- 12.Green R.M., Beier D., Gollan J.L. Regulation of hepatocyte bile salt transporters by endotoxin and inflammatory cytokines in rodents. Gastroenterology. 1996;111:193–198. doi: 10.1053/gast.1996.v111.pm8698199. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann G., Cheung A.K., Piquette-Miller M. Inflammatory cytokines, but not bile acids, regulate expression of murine hepatic anion transporters in endotoxemia. J Pharmacol Exp Ther. 2002;303:273–281. doi: 10.1124/jpet.102.039404. [DOI] [PubMed] [Google Scholar]
- 14.Yang L., Jhaveri R., Huang J., Qi Y. Endoplasmic reticulum stress, hepatocyte CD1d and NKT cell abnormalities in murine fatty livers. Lab Invest. 2007;87:927–937. doi: 10.1038/labinvest.3700603. [DOI] [PubMed] [Google Scholar]
- 15.Wang D., Wei Y., Pagliassotti M.J. Saturated fatty acids promote endoplasmic reticulum stress and liver injury in rats with hepatic steatosis. Endocrinology. 2006;147:943–951. doi: 10.1210/en.2005-0570. [DOI] [PubMed] [Google Scholar]
- 16.Tardif K.D., Mori K., Siddiqui A. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J Virol. 2002;76:7453–7459. doi: 10.1128/JVI.76.15.7453-7459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji C., Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. 2003;124:1488–1499. doi: 10.1016/s0016-5085(03)00276-2. [DOI] [PubMed] [Google Scholar]
- 18.Kim D.S., Jeong S.K., Kim H.R. Effects of triglyceride on ER stress and insulin resistance. Biochem Biophys Res Commun. 2007;363:140–145. doi: 10.1016/j.bbrc.2007.08.151. [DOI] [PubMed] [Google Scholar]
- 19.Bochkis I.M., Rubins N.E., White P. Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nat Med. 2008;14:828–836. doi: 10.1038/nm.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamaki N., Hatano E., Taura K. CHOP deficiency attenuates cholestasis-induced liver fibrosis by reduction of hepatocyte injury. Am J Physiol Gastrointest Liver Physiol. 2008;294:G498–G505. doi: 10.1152/ajpgi.00482.2007. [DOI] [PubMed] [Google Scholar]
- 21.Malhi H., Kaufman R.J. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozcan U., Yilmaz E., Ozcan L. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein H., Payne C.M., Bernstein C. Activation of the promoters of genes associated with DNA damage, oxidative stress, ER stress and protein malfolding by the bile salt, deoxycholate. Toxicol Lett. 1999;108:37–46. doi: 10.1016/s0378-4274(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 24.Szalowska E., Stoopen G., Groot M.J. Treatment of mouse liver slices with cholestatic hepatotoxicants results in down-regulation of Fxr and its target genes. BMC Med Genom. 2013;6:39. doi: 10.1186/1755-8794-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figge A., Lammert F., Paigen B. Hepatic overexpression of murine Abcb11 increases hepatobiliary lipid secretion and reduces hepatic steatosis. J Biol Chem. 2004;279:2790–2799. doi: 10.1074/jbc.M307363200. [DOI] [PubMed] [Google Scholar]
- 26.Olivares S., Green R.M., Henkel A.S. Endoplasmic reticulum stress activates the hepatic activator protein 1 complex via mitogen activated protein kinase-dependent signaling pathways. PLoS One. 2014;9:e103828. doi: 10.1371/journal.pone.0103828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henkel A.S., Elias M.S., Green R.M. Homocysteine supplementation attenuates the unfolded protein response in a murine nutritional model of steatohepatitis. J Biol Chem. 2009;284:31807–31816. doi: 10.1074/jbc.M109.017970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henkel A.S., Anderson K.A., Dewey A.M. A chronic high-cholesterol diet paradoxically suppresses hepatic CYP7A1 expression in FVB/NJ mice. J Lipid Res. 2011;52:289–298. doi: 10.1194/jlr.M012781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen J.C. Contribution of cholesterol 7alpha-hydroxylase to the regulation of lipoprotein metabolism. Curr Opin Lipidol. 1999;10:303–307. doi: 10.1097/00041433-199908000-00003. [DOI] [PubMed] [Google Scholar]
- 30.Davis R.A., Miyake J.H., Hui T.Y. Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP. J Lipid Res. 2002;43:533–543. [PubMed] [Google Scholar]
- 31.Ozcan U., Cao Q., Yilmaz E. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 32.Vecchi C., Montosi G., Zhang K. ER stress controls iron metabolism through induction of hepcidin. Science. 2009;325:877–880. doi: 10.1126/science.1176639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winnay J.N., Boucher J., Mori M.A. A regulatory subunit of phosphoinositide 3-kinase increases the nuclear accumulation of X-box-binding protein-1 to modulate the unfolded protein response. Nat Med. 2010;16:438–445. doi: 10.1038/nm.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uemura A., Oku M., Mori K. Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J Cell Sci. 2009;122:2877–2886. doi: 10.1242/jcs.040584. [DOI] [PubMed] [Google Scholar]
- 35.McCullough K.D., Martindale J.L., Klotz L.O. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zinszner H., Kuroda M., Wang X. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirsova P., Gores G.J. Death receptor-mediated cell death and proinflammatory signaling in nonalcoholic steatohepatitis. Cell Mol Gastroenterol Hepatol. 2015;1:17–27. doi: 10.1016/j.jcmgh.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell D.W. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 39.Ren S., Marques D., Redford K. Regulation of oxysterol 7alpha-hydroxylase (CYP7B1) in the rat. Metabolism. 2003;52:636–642. doi: 10.1053/meta.2003.50106. [DOI] [PubMed] [Google Scholar]
- 40.Vlahcevic Z.R., Stravitz R.T., Heuman D.M. Quantitative estimations of the contribution of different bile acid pathways to total bile acid synthesis in the rat. Gastroenterology. 1997;113:1949–1957. doi: 10.1016/s0016-5085(97)70015-5. [DOI] [PubMed] [Google Scholar]
- 41.Chen J., Cooper A.D., Levy-Wilson B. Hepatocyte nuclear factor 1 binds to and transactivates the human but not the rat CYP7A1 promoter. Biochem Biophys Res Commun. 1999;260:829–834. doi: 10.1006/bbrc.1999.0980. [DOI] [PubMed] [Google Scholar]
- 42.Chen J.Y., Levy-Wilson B., Goodart S. Mice expressing the human CYP7A1 gene in the mouse CYP7A1 knock-out background lack induction of CYP7A1 expression by cholesterol feeding and have increased hypercholesterolemia when fed a high fat diet. J Biol Chem. 2002;277:42588–42595. doi: 10.1074/jbc.M205117200. [DOI] [PubMed] [Google Scholar]
- 43.Agellon L.B., Drover V.A., Cheema S.K. Dietary cholesterol fails to stimulate the human cholesterol 7alpha-hydroxylase gene (CYP7A1) in transgenic mice. J Biol Chem. 2002;277:20131–20134. doi: 10.1074/jbc.C200105200. [DOI] [PubMed] [Google Scholar]
- 44.Chiang J.Y., Kimmel R., Weinberger C. Farnesoid X receptor responds to bile acids and represses cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J Biol Chem. 2000;275:10918–10924. doi: 10.1074/jbc.275.15.10918. [DOI] [PubMed] [Google Scholar]
- 45.Sinal C.J., Tohkin M., Miyata M. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 46.Lu T.T., Makishima M., Repa J.J. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 47.Kim I., Ahn S.H., Inagaki T. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48:2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 48.Kerr T.A., Saeki S., Schneider M. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu M., Li J., Maruyama R. FGF19 (fibroblast growth factor 19) as a novel target gene for activating transcription factor 4 in response to endoplasmic reticulum stress. Biochem J. 2013;450:221–229. doi: 10.1042/BJ20121393. [DOI] [PubMed] [Google Scholar]
- 50.De Fabiani E., Mitro N., Anzulovich A.C. The negative effects of bile acids and tumor necrosis factor-alpha on the transcription of cholesterol 7alpha-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: a novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J Biol Chem. 2001;276:30708–30716. doi: 10.1074/jbc.M103270200. [DOI] [PubMed] [Google Scholar]
- 51.Feingold K.R., Spady D.K., Pollock A.S. Endotoxin, TNF, and IL-1 decrease cholesterol 7 alpha-hydroxylase mRNA levels and activity. J Lipid Res. 1996;37:223–228. [PubMed] [Google Scholar]
- 52.Li T., Jahan A., Chiang J.Y. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 2006;43:1202–1210. doi: 10.1002/hep.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luther J., Garber J.J., Khalili H. Hepatic injury in nonalcoholic steatohepatitis contributes to altered intestinal permeability. Cell Mol Gastroenterol Hepatol. 2015;1:222–232. doi: 10.1016/j.jcmgh.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta S., Stravitz R.T., Dent P. Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001;276:15816–15822. doi: 10.1074/jbc.M010878200. [DOI] [PubMed] [Google Scholar]
- 55.Jahan A., Chiang J.Y. Cytokine regulation of human sterol 12alpha-hydroxylase (CYP8B1) gene. Am J Physiol Gastrointest Liver Physiol. 2005;288:G685–G695. doi: 10.1152/ajpgi.00207.2004. [DOI] [PubMed] [Google Scholar]
- 56.Urano F., Wang X., Bertolotti A. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 57.Hu P., Han Z., Couvillon A.D. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol Cell Biol. 2006;26:3071–3084. doi: 10.1128/MCB.26.8.3071-3084.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kong B., Wang L., Chiang J.Y. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56:1034–1043. doi: 10.1002/hep.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song K.H., Li T., Owsley E. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49:297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zelcer N., Saeki T., Bot I. Transport of bile acids in multidrug-resistance-protein 3-overexpressing cells co-transfected with the ileal Na+-dependent bile-acid transporter. Biochem J. 2003;369:23–30. doi: 10.1042/BJ20021081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soroka C.J., Lee J.M., Azzaroli F. Cellular localization and up-regulation of multidrug resistance-associated protein 3 in hepatocytes and cholangiocytes during obstructive cholestasis in rat liver. Hepatology. 2001;33:783–791. doi: 10.1053/jhep.2001.23501. [DOI] [PubMed] [Google Scholar]
- 62.Trauner M., Arrese M., Lee H. Endotoxin downregulates rat hepatic ntcp gene expression via decreased activity of critical transcription factors. J Clin Invest. 1998;101:2092–2100. doi: 10.1172/JCI1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donner M.G., Schumacher S., Warskulat U. Obstructive cholestasis induces TNF-alpha- and IL-1-mediated periportal downregulation of Bsep and zonal regulation of Ntcp, Oatp1a4, and Oatp1b2. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1134–G1146. doi: 10.1152/ajpgi.00079.2007. [DOI] [PubMed] [Google Scholar]
- 64.Siewert E., Dietrich C.G., Lammert F. Interleukin-6 regulates hepatic transporters during acute-phase response. Biochem Biophys Res Commun. 2004;322:232–238. doi: 10.1016/j.bbrc.2004.07.102. [DOI] [PubMed] [Google Scholar]
- 65.Geier A., Dietrich C.G., Voigt S. Cytokine-dependent regulation of hepatic organic anion transporter gene transactivators in mouse liver. Am J Physiol Gastrointest Liver Physiol. 2005;289:G831–G841. doi: 10.1152/ajpgi.00307.2004. [DOI] [PubMed] [Google Scholar]
- 66.Calfon M., Zeng H., Urano F. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 67.Lin J.H., Li H., Yasumura D. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olivares S., Henkel A.S. Hepatic Xbp1 gene deletion promotes endoplasmic reticulum stress-induced liver injury and apoptosis. J Biol Chem. 2015;290:30142–30151. doi: 10.1074/jbc.M115.676239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee A.H., Scapa E.F., Cohen D.E. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang S., Chen Z., Lam V. IRE1alpha-XBP1s induces PDI expression to increase MTP activity for hepatic VLDL assembly and lipid homeostasis. Cell Metab. 2012;16:473–486. doi: 10.1016/j.cmet.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.So J.S., Hur K.Y., Tarrio M. Silencing of lipid metabolism genes through IRE1alpha-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 2012;16:487–499. doi: 10.1016/j.cmet.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hollien J., Weissman J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 73.Upton J.P., Wang L., Han D. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hollien J., Lin J.H., Li H. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maurel M., Chevet E., Tavernier J. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 2014;39:245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 76.Hur K.Y., So J.S., Ruda V. IRE1alpha activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med. 2012;209:307–318. doi: 10.1084/jem.20111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lefebvre P., Cariou B., Lien F. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 78.Choi S.M., Kim Y., Shim J.S. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology. 2013;57:2458–2468. doi: 10.1002/hep.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watanabe M., Horai Y., Houten S.M. Lowering bile acid pool size with a synthetic farnesoid X receptor (FXR) agonist induces obesity and diabetes through reduced energy expenditure. J Biol Chem. 2011;286:26913–26920. doi: 10.1074/jbc.M111.248203. [DOI] [PMC free article] [PubMed] [Google Scholar]