Abstract
Background & Aims
Hepatic cholesterol accumulation and autophagy defects contribute to hepatocyte injury in fatty liver disease. Bile acid synthesis is a major pathway for cholesterol catabolism in the liver. This study aims to understand the molecular link between cholesterol and bile acid metabolism and hepatic autophagy activity.
Methods
The effects of cholesterol and cholesterol 7α-hydroxylase (CYP7A1) expression on autophagy and lysosome function were studied in cell models. The effects and mechanism of disrupting enterohepatic bile acid circulation on hepatic autophagy were studied in mice.
Results
The results first showed differential regulation of hepatic autophagy by free cholesterol and cholesterol ester, whereby a modest increase of cellular free cholesterol, but not cholesterol ester, impaired lysosome function and caused marked autolysosome accumulation. We found that CYP7A1 induction, either by cholestyramine feeding in mice or adenovirus-mediated CYP7A1 expression in hepatocytes, caused strong autophagy induction. Mechanistically, we showed that CYP7A1 expression markedly attenuated growth factor/AKT signaling activation of mechanistic target of rapamycin (mTOR), but not amino acid signaling to mTOR in vitro and in vivo. Metabolomics analysis further found that CYP7A1 induction not only decreased hepatic cholesterol but also altered phospholipid and sphingolipid compositions. Collectively, these results suggest that CYP7A1 induction interferes with growth factor activation of AKT/mTOR signaling possibly by altering membrane lipid composition. Finally, we showed that cholestyramine feeding restored impaired hepatic autophagy and improved metabolic homeostasis in Western diet–fed mice.
Conclusions
This study identified a novel CYP7A1–AKT–mTOR signaling axis that selectively induces hepatic autophagy, which helps improve hepatocellular integrity and metabolic homeostasis.
Keywords: Cholesterol, Cholestyramine, Fatty Liver, Nuclear Receptor
Abbreviations used in this paper: ACAT, acyl-CoA:cholesterol acyltransferase; CE, cholesterol ester; ChTM, cholestyramine; CQ, chloroquine; CYP7A1, cholesterol 7α-hydroxylase; DIO, diet-induced obesity; ER, endoplasmic reticulum; FC, free cholesterol; 4EBP-1, eukaryotic translation initiation factor 4E-binding protein 1; GSK3β, glycogen synthase kinase 3β; HMGCR, HMG-CoA reductase; LC3, microtubule-associated protein 1A/1B-light chain 3; LDLR, low-density lipoprotein receptor; LMP, lysosome membrane permeabilization; mRNA, messenger RNA; mTOR, the nutrient sensing mechanistic target of rapamycin; PI, phosphatidylinositol; PM, plasma membrane; S6, tibosomal protein S6; SREBP, sterol response element binding protein
Graphical Abstract
See editorial on page 133.
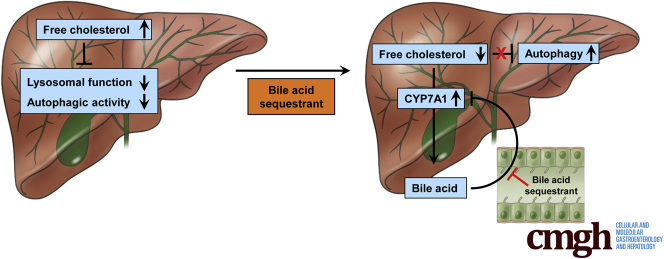
Summary.
Free cholesterol accumulation impairs lysosome function and inhibits autophagic activity in hepatocytes. Induction of bile acid synthesis attenuates AKT/mechanistic target of rapamycin (mTOR) signaling, leading to autophagy activation. Targeting the enterohepatic bile acid signaling by bile acid sequestrant induces hepatic autophagy in mice.
Emerging evidence supports that nonesterified free cholesterol (FC) accumulates in fatty livers and contributes to hepatocyte injury.1, 2, 3, 4 Impaired hepatic cholesterol homeostasis further contributes to dyslipidemia, atherosclerosis, and a higher risk of cardiovascular disease.5 Hepatic cholesterol homeostasis is coordinately controlled by input and elimination pathways. The sterol response element binding protein-2 (SREBP-2)–mediated endoplasmic reticulum (ER) cholesterol-sensing mechanism regulates cellular cholesterol input pathways.6 A decrease of ER cholesterol leads to proteolytic cleavage activation of SREBP-2, which induces the expression of 3-hydroxy-3-methylglutaryl-coenzyme A-CoA reductase (HMGCR) and low-density lipoprotein receptor (LDLR) to increase intracellular cholesterol levels. On the other hand, bile acid synthesis is the major cholesterol elimination pathway. The cholesterol 7α-hydroxylase (CYP7A1) is an ER resident enzyme that catalyzes the first and rate-limiting step in the conversion of cholesterol into bile acids.7 Bile acid synthesis has a major impact on ER cholesterol content, and CYP7A1 induction can activate the entire SREBP-2–regulated transcriptional network to regulate cellular cholesterol metabolism in the liver.8 This ER cholesterol-sensing mechanism is the molecular basis for the cholesterol-lowering drugs statins and bile acid sequestrants. In addition, a number of studies have suggested that nutrient signaling regulation of hepatic bile acid synthesis may be tightly linked to hepatic metabolic homeostasis.9, 10, 11
Autophagy is a well-conserved cellular self-degradation system that eliminates protein aggregates and damaged organelles to help maintain cellular integrity.12 Autophagy is also a catabolic process that generates nutrients and energy by degrading macromolecules in response to nutrient deprivation.13 The nutrient-sensing mechanistic target of rapamycin (mTOR) signaling plays a key role in regulating autophagy activity in response to changes of nutrient availability.13 A hepatic autophagy defect exacerbated hepatocyte injury in fatty liver diseases.14, 15, 16 Furthermore, obesity and fatty liver are associated with defective hepatic autophagy, possibly owing to hyperinsulinemia, lipid accumulation, and impaired vesicle fusion.17, 18, 19
Recent clinical and basic research have shown that targeting the enterohepatic bile acid signaling by bile acid sequestrants or the selective apical sodium-dependent bile acid transporter inhibitors represents a promising therapeutic strategy for decreasing plasma cholesterol and improving insulin sensitivity in diabetes and fatty liver disease, and attenuating bile acid toxicity in cholestasis.20, 21, 22, 23 However, the highly complex changes of hepatic and intestinal metabolic and signaling pathways caused by disrupting enterohepatic bile acid circulation still are incompletely understood. In this study, we show that free cholesterol accumulation is a major causal factor for liver autophagy impairment, and that targeting the enterohepatic bile acid signaling is an effective approach to induce hepatic autophagy via a novel CYP7A1–AKT–mTOR signaling cascade, which is potentially beneficial in improving hepatocellular integrity and metabolic homeostasis.
Materials and Methods
Reagents
Antibodies against microtubule-associated protein 1A/1B-light chain 3 (LC3B), phospho- and total tibosomal protein S6 (S6) (S240/244), eukaryotic translation initiation factor 4E-binding protein 1 (4EBP-1) (T37/46), AKT (S473), glycogen synthase kinase 3β (GSK3β) (S9), and histone 3 were purchased from Cell Signaling Technology (Danvers, MA). P62 antibody was purchased from Abnova (Walnut, CA). Actin antibody, water-soluble cholesterol (in methyl-β-cyclodextrin), cholestyramine (ChTM), acyl-CoA:cholesterol acyltransferase (ACAT) inhibitor Sandoz 58-035 chloroquine, and cholestyramine were purchased from Sigma (St. Louis, MO). Lysotracker red and anti-SREBP-2 antibody were from Thermo Scientific (Grand Island, NY). Highly purified human LDL was purchased from Lee Biosolutions (Maryland Heights, MO). The mCherry-galectin expression vector was described previously.24 Lipoprotein-depleted serum was purchased from Kalen Biomedical, LLC.
Animals
Male 8- to 10-week-old C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, ME). The Western diet (TD.88137; Harlan Teklad, East Millstone, NJ) contains 42% fat calories and 0.2% cholesterol. ChTM was mixed with diets (2%, wt/wt). For confocal microscopy, mice were injected with Ad-RFP-GFP-LC3 at 108 pfu/mouse via tail vein 3 days before death. All mice were fasted for 6 hours starting from approximately 6 AM and killed. All animals received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals. All protocols were approved by the Institutional Animal Care and Use Committee.
Cell Culture and Treatments
HepG2 cells were purchased from ATCC (Manassas, VA). Cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum or in Dulbecco's modified Eagle medium containing lipoprotein-depleted serum as indicated. Experiments were initiated when cells were 60%–80% confluent. Primary human hepatocytes were obtained from the Cell Isolation Core at Kansas University Medical Center and treatment was started within 24 hours after the cells were plated.
Measurement of Mitochondria Bioenergetic Function
All XF assays were performed with an XF24–3 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA). Mitochondria from mouse livers were isolated by differential centrifugation methods as described previously.25 Mitochondria from 4 mice per group were pooled and the assays were performed in triplicate. Isolated liver mitochondria in mitochondrial assay solution (1× mitochondrial assay solution: 70 mmol/L sucrose, 220 mmol/L mannitol, 10 mmol/L KH2PO4, 5 mmol/L MgCl2, 2 mmol/L HEPES, 1 mmol/L ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, and 0.2% [wt/vol] fatty acid–free bovine serum albumin, pH 7.2 at 37°C) containing substrate (10 mmol/L succinate + 2 μmol/L rotenone) was plated. The sensor cartridge was loaded with port A, adenosine diphosphate (4 mmol/L final); port B, oligomycin (2.5 μg/mL final); port C, FCCP (4 μmol/L final); and port D, antimycin A (4 μmol/L final) to measure the bioenergetic profile. The respiration was measured in a coupled state in the presence of substrate (basal respiration), followed by state 3 (phosphorylating respiration, in the presence of adenosine diphosphate and substrate), state 4 (resting respiration) after conversion of adenosine diphosphate to adenosine triphosphate, state 4o (induced with the addition of oligomycin), and then maximal uncoupler-stimulated respiration (state 3u).
Metabolomics Analysis
The metabolomics analysis was performed by Metabolon, Inc (Durham, NC). Briefly, liver samples (n = 5) were prepared using the automated MicroLab STAR system from Hamilton Company (Reno, NV). Several recovery standards were added based on tissue weight before sample extraction for data normalization. The extract was analyzed on a Waters Acquity UPLC (Milford, MA) and a Thermo Scientific Q-Exactive high-resolution/accurate mass spectrometer interfaced with a heated electrospray ionization source and Orbitrap mass analyzer (Grand Island, NY). Raw data were extracted, peak-identified, and processed using Metabolon’s reference library and software. Peaks were quantified using the area under the curve.
Immunoblot
Cell or liver homogenates were incubated in 1× RIPA buffer containing 1% sodium dodecyl sulfate and protease inhibitor cocktail for 1 hour on ice, followed by sonication and centrifugation. Supernatant were used for sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Densitometry was performed with ImageJ software (National Institutes of Health, Bethesda, MD). The band intensity (normalized to actin) of 3 experiments was expressed as means ± SD values that were placed below the representative blot.
Confocal Microscopy
Cells were plated in one 10-cm culture dish. In some experiments, cells were infected with adenoviral vectors or transfected with plasmids, and then plated on chamber slides. Mice were injected with Ad-RFP-GFP-LC3 at 108 pfu/mouse via tail vein. After 3 days, mice were fasted for 6 hours since 6 AM and then killed. Cells and liver tissues were fixed with 4% paraformaldehyde. Images were acquired with a Leica DM 5500 confocal microscope (Buffalo Grove, IL).
Electron Microscopy
Fresh liver tissue was fixed in 2% glutaraldehyde followed by 1% OsO4. After dehydration, sections were stained with uranyl acetate and lead citrate. Images were taken with a JEM 1016CX electron microscope (Peabody, MA) as previously described.26
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated with Tri-reagent (ThermoFisher Scientific, Grand Island, NY). Real-time polymerase chain reaction were performed with primers and SYBR master mix (Bio-Rad, Hercules, CA). Amplification of 18S was used as an internal control. Relative messenger RNA (mRNA) expression was quantified using the comparative Ct method and expressed as 2−ΔΔCt.
Lipids and Glucose Analysis
Lipids were extracted in chloroform:methanol (2:1, vol/vol). Total cholesterol and triglyceride levels were measured with assay kits (Pointe Scientific, Canton, MI). Free cholesterol was measured with an assay kit from Thermo Scientific. The cholesterol ester value was obtained by subtracting free cholesterol from total cholesterol in the same sample. Blood glucose was measured with the Onetouch Ultra glucose monitor (LifeScan, Wayne, PA).
Recombinant Adenovirus
The construction and use of adenovirus expressing tandem RFP-GFP-LC3B for the study of autophagy flux was described previously.27 Adenovirus expressing the rat Cyp7a1 gene was a generous gift from Dr William Michael Pandak (Virginia Commonwealth University, Richmond, VA).28 Ad-null was purchased from Vector Biolabs (Philadelphia, PA). Ad-GFP was from Dr Li Wang (University of Connecticut, Storrs, CT).
Cathepsin B Activity Assay
As described previously,29 cells or liver tissues were lysed in M2 buffer (50 mmol/L Tris, pH 7.5, 130 mmol/L NaCl, 10% glycerol, 0.5% NP-40, 0.5 mmol/L EDTA, and 0.5 mmol/L ethylene glycol-bis[β-aminoethyl ether]-N,N,N′,N′-tetraacetic acid in dH2O) without protease inhibitors. Protein lysates then were incubated with cathepsin B fluorogenic substrate z-RR-AMC and fluorescence intensity was determined using a Tecan Infinite M200 Pro plate reader (Morrisville, NC). L-leucyl-L-leucine methyl ester, which accumulates in lysosomes and causes lysosome impairment, was used as a positive control.
Statistical Analysis
Results were expressed as means ± SE for animal studies. Statistical analysis was performed with the Student t test or analysis of variance. A P value less than .05 was considered statistically significant.
Access to Data
All authors had access to all the data and have reviewed and approved the final manuscript.
Results
Hepatic Autophagy Is Highly Sensitive to Inhibition by Cellular FC Loading, but not LDL Loading
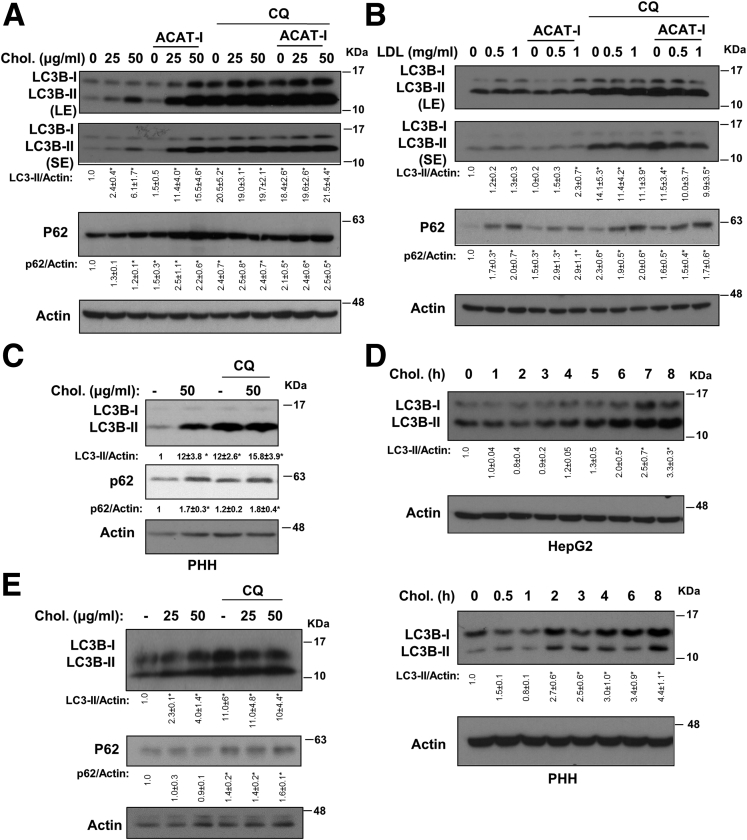
We first subjected HepG2 cells to FC loading for 16 hours and measured LC3B in an autophagic flux assay. FC loading dose-dependently increased LC3B-II and p62 protein, but did not increase LC3B-II and p62 further in the presence of chloroquine (CQ) (Figure 1A), suggesting autophagy inhibition at the lysosomal level. It is known that FC loading first enriches the plasma membrane (PM) cholesterol pool, and excessive FC is delivered to other intracellular organelles including the ER where excessive FC is esterified to cholesterol ester (CE) by the ACAT for cytosolic storage.30 LC3B-II and p62 accumulation was enhanced markedly in the presence of ACAT inhibitor (Figure 1A). In comparison, treatment of purified human LDL, which contained primarily CE, caused very modest increases of LC3B-II abundance with or without the presence of an ACAT inhibitor (Figure 1B). Similar results were obtained when cells were cultured in medium containing lipoprotein-depleted serum (not shown). The FC-induced LC3B-II accumulation was reproduced in primary human hepatocytes (Figure 1C). Treating HepG2 cells with the same concentration of the vehicle (methyl-β-cyclodextrin) did not affect LC3 and p62 protein (data not shown). FC or LDL loading did not alter the mRNA of key autophagy genes ULK1, autophagy protein 5, autophagy protein 7, and LC3B, except that LDL loading increased p62 mRNA by approximately 3-fold (data not shown), which may contribute partially to increased p62 protein levels (Figure 1B).
Figure 1.
Free cholesterol caused rapid LC3B accumulation. (A and B) Western blot: HepG2 cells were cultured in 10% fetal bovine serum and treated with the indicated concentrations of cholesterol and purified human LDL with or without ACAT inhibitor (12.5 μg/mL) for 16 hours. Some cells were treated with CQ (50 μmol/L) during the last 6 hours as indicated. LE, long exposure; SE, short exposure. (C) Primary human hepatocytes (PHH) were treated with free cholesterol for 16 hours. Some cells were treated with CQ (50 μmol/L) during the last 6 hours. (D) HepG2 cells or primary human hepatocytes were treated with 50 μg/mL cholesterol for up to 8 hours. LC3B and actin protein levels were measured by Western blot. (E) HepG2 cells were pretreated with 50 μmol/L CQ for 0.5 hours, followed by cholesterol treatment for an additional 5 hours. *Statistical significance vs control.
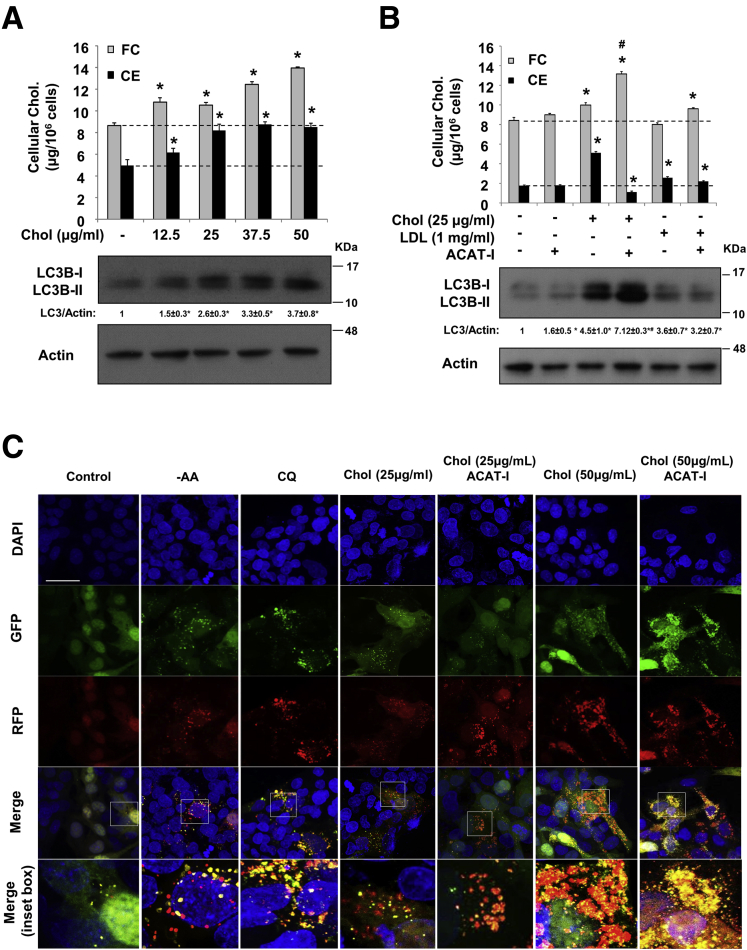
To further elaborate on these findings, we next subjected cells to FC loading in a short-term time course. FC loading time-dependently caused LC3B-II to accumulate over the course of 8 hours in HepG2 cells and human hepatocytes (Figure 1D). At the 5-hour time point, FC loading did not increase LC3B-II and p62 further in the presence of CQ in an autophagic flux assay, which confirmed rapid autophagy inhibition (Figure 1E). Because FC loading for 8 hours already caused LC3B accumulation (Figure 1D), we therefore measured cellular cholesterol levels at the 8-hour time point. FC loading at both 12.5 and 25 μg/mL increased cellular FC by approximately 20%, and increased cellular CE by approximately 25% and approximately 65%, respectively, and resulted in a dose-dependent increase of LC3B levels (Figure 2A). In contrast, LDL loading increased cellular CE by approximately 40%, but did not result in a corresponding increase in cellular FC content unless ACAT was inhibited simultaneously (Figure 2B). LDL loading caused a very mild increase of LC3B, which was not affected further by ACAT inhibitor despite an FC increase by approximately 15% (Figure 2B). These results indicate that LDL uptake, lysosomal LDL CE hydrolysis, and lysosomal FC transport are highly coordinated processes. Some of these steps, such as LDLR down-regulation, may be rate-limiting to maintain cellular cholesterol levels relatively constant. Nevertheless, addition of ACAT inhibitor completely prevented CE increase but increased FC further by an additional approximately 30%, and led to further LC3B-II accumulation in FC-treated cells (Figure 2B). These results provide important evidence to support that autophagy inhibition was owing primarily to cellular FC accumulation.
Figure 2.
Free cholesterol impaired autolysosome clearance. (A and B) HepG2 cells were treated with cholesterol, LDL or ACAT inhibitor (12.5 μg/mL) for 8 hours as indicated. Cellular cholesterol and LC3B protein levels were measured. Cholesterol assays were repeated in triplicates. Results are expressed as means ± SD. *Statistical significance vs untreated controls. Statistical significance vs 25 μg/mL cholesterol treated. (C) Confocal microscope: HepG2 cells were infected with Ad-RFP-GFP-LC3B at a multiplicity of infection of approximately 0.5 for 16 hours. Cells then were treated with cholesterol with or without ACAT inhibitor (12.5 μg/mL) for 16 hours, with CQ (50 μmol/L) for 6 hours. Scale bar: 30 μm. DAPI, 4′,6-diamidino-2-phenylindole.
FC Accumulation Impairs Lysosomal Function and Inhibits Autophagy
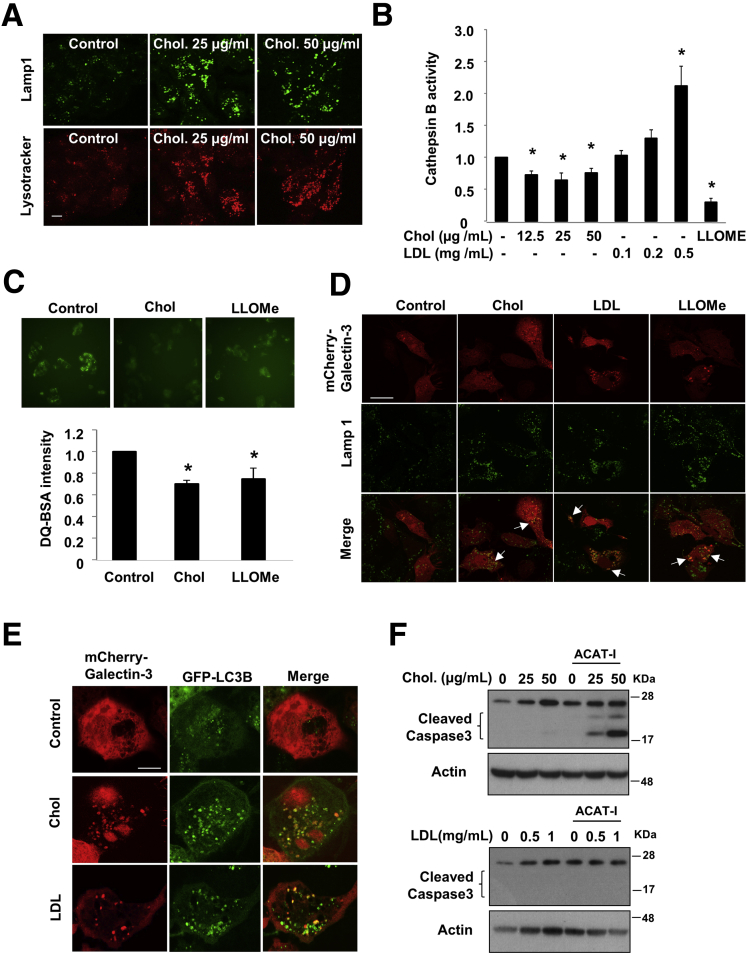
To obtain further insights on the molecular basis of FC-induced LC3B-II accumulation, we next studied autophagic flux in HepG2 cells expressing a tandem RFP-GFP-LC3B fusion protein. It was reported that GFP, but not RFP, preferentially was quenched in autolysosomes, and thus red puncta mainly represented fused autolysosomes, and merged yellow puncta mainly represented autophagosomes.31 This was first validated by amino acid starvation and by CQ treatment (Figure 2C). FC-treated cells had an increased number of yellow puncta (Figure 2C). A significant number of these yellow puncta were found to be associated closely with enlarged red puncta, which was especially prominent in cells treated with 50 μg/mL FC. Because yellow puncta are thought to represent autophagosomes, these results suggest autophagosome accumulation is owing to defective autophagosome fusion with lysosomes because lysosome function may be impaired by FC accumulation. In addition, it also is possible that decreased autolysosome clearance efficiency may contribute to the accumulation of yellow and red puncta in these cells. In support of this hypothesis, we showed that FC loading for 8 hours caused enlarged lysotracker red–positive puncta and enlarged Lamp1-positive puncta, which represent acidic late endocytic compartments including autolysosomes (Figure 3A). Furthermore, FC loading significantly decreased lysosome function as evidenced by both reduced cathepsin B activity (Figure 3B) and attenuated intracellular DQ-BSA hydrolysis (Figure 3C). In contrast, LDL loading dose-dependently increased cathepsin B activity (Figure 3B), which we speculate was because LDL endocytosis increased the total number of late endosomes/lysosomes. To gain further understanding of FC-induced lysosome impairment, we evaluated lysosome membrane permeabilization (LMP) in cells expressing mCherry-tagged galectin 3, a cytosolic and nuclear protein that localizes to the lysosomes upon LMP.24 FC loading induced galectin-3–positive puncta that co-localized with Lamp1-positive puncta (Figure 3D), suggesting that FC loading induced LMP. Furthermore, galectin-3 also co-localized with LC3B-positive puncta (Figure 3E), indicating that FC loading also might damage the GFP-LC3–positive vesicle structures. Unexpectedly, LDL loading also induced galectin-3 localization to Lamp1-positive puncta and LC3-positive puncta (Figure 3D and E). We speculate that the presence of LDL-derived FC in some late endocytic compartments may cause some degree of LMP and galectin-3 recruitment. However, only FC loading at 50 μg/mL caused weak caspase-3 cleavage, which was enhanced markedly in the presence of ACAT inhibitor (Figure 3F).
Figure 3.
Free cholesterol loading impaired lysosome function. (A) HepG2 cells were treated with cholesterol for 8 hours, and stained with either Lamp1 or lysotracker red. Scale bar: 10 μm. (B) Cathepsin B activity was determined in HepG2 cells treated with the indicated concentration of cholesterol or LDL for 16 hours. L-leucyl-L-leucine methyl ester (LLOMe) was used as a positive control. Assays were repeated in triplicates. Results are expressed as means ± SD. *Statistical significance vs untreated controls. (C) DQ-BSA assay in HepG2 cells treated with 25 μg/mL cholesterol for 16 hours. LLOMe was used as a positive control. (D) HepG2 cells were transfected with plasmids expressing mCherry–galactin-3. Confocal imaging of mCherry–galectin-3 and Lamp1 co-localization (indicated by white arrows) in HepG2 cells treated with 25 μg/mL cholesterol or 0.5 mg/mL LDL for 16 hours. LLOMe was used as a positive control. Scale bar: 30 μm. (E) HepG2 cells transfected with plasmids expressing mCherry–galactin-3 and GFP-LC3 were treated with 0.5 mg/mL LDL or 25 μg/mL cholesterol for 16 hours, and images were taken with a confocal microscopy. Scale bar: 10 μm. (F) HepG2 cells were treated with cholesterol or LDL with or without ACAT inhibitor for 16 hours. Cleaved caspase 3 was detected in Western blot. Chol, cholesterol.
CYP7A1 Induction Increased Hepatic Autophagy
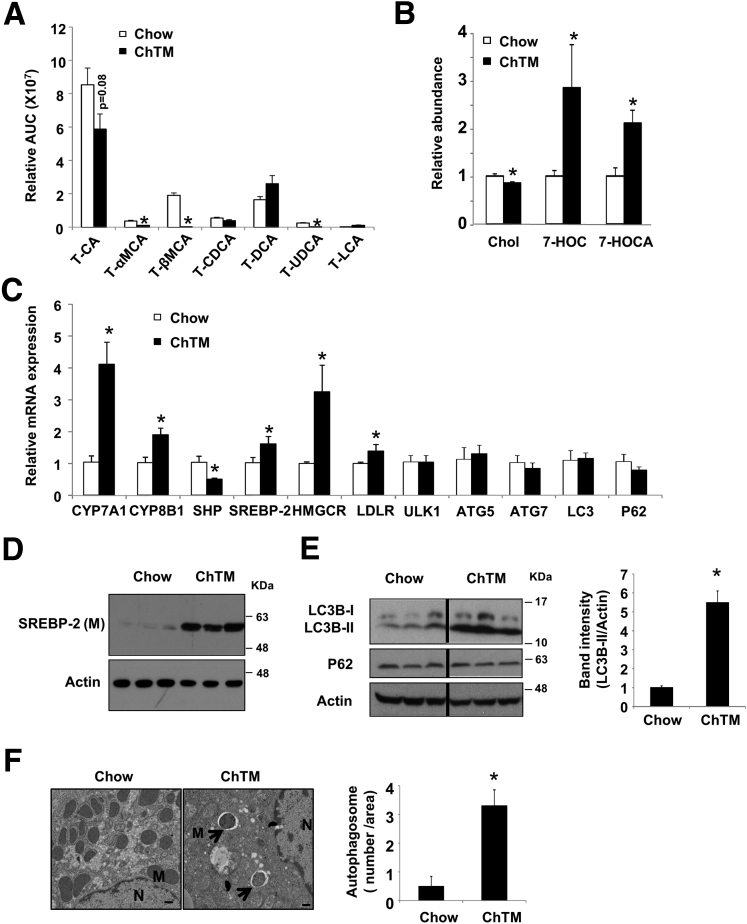
Bile acid synthesis is the major ER cholesterol catabolic mechanism specific to hepatocytes. Given that autophagy was highly sensitive to cellular FC increase, we next asked if active removal of intracellular FC via stimulating bile acid synthesis could modulate autophagy in the liver. To address this question, we fed mice a ChTM-containing chow diet for a short period of 6 days, which reduced most species of tauro-conjugated bile acids in the liver (Figure 4A). ChTM feeding increased liver CYP7A1 and CYP8B1 mRNA and decreased farnesoid X receptor target gene small heterodimer partner (SHP) mRNA (Figure 4C). ChTM feeding significantly decreased FC by approximately 15% and increased bile acid synthesis intermediates 7-hydroxycholesterol and 7α-hydroxy-3-oxo-4-cholestenoate (Figure 4B). CYP7A1 induction caused ER cholesterol depletion as evidenced by SREBP-2 cleavage and higher HMGCR and LDLR mRNA levels (Figure 4C and D). SREBP-2 precursor was not detected by the antibody used. Under these conditions, ChTM resulted in approximately 5-fold higher LC3B-II protein without p62 protein accumulation in mouse livers (Figure 4E). Furthermore, electron microscopy analysis showed significantly increased numbers of double-membrane autophagosomes in the liver of ChTM-fed mice (Figure 4F). ChTM feeding did not induce the mRNA levels of key autophagy genes (Figure 4C), suggesting that ChTM induction of autophagy was independent of transcriptional control of autophagy genes.
Figure 4.
ChTM feeding induced hepatic autophagy in lean mice. Male C57BL/6J mice were fed a chow diet or a chow diet containing 2% ChTM for 6 days. All mice were briefly fasted for 6 hours starting at 6 AM and then killed. (A and B) Hepatic tauro-conjugated bile acids, cholesterol, 7-hydroxycholesterol (7-HOC), and 7α-hydroxy-3-oxo-4-cholestenoate (7-HOCA). All results are expressed as means ± SE (n = 5). *Statistical significance vs chow-fed controls. CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; LCA, lithocholic acid; MCA, muricholic acid; T, tauro; UDCA, ursodeoxycholic acid. (C) Hepatic mRNA expression. (D) Hepatic SREBP-2 mature protein. (E) Hepatic LC3B, P62, and actin protein levels. (F) Electron microscopy analysis of liver sections. N, nucleus; M, mitochondria. Arrows denote double-membrane autophagosomes. Scale bar: 500 nm. Right: average number of autophagosomes per image area at 3000× magnification from 5–10 images per group. A representative image from each group is shown. AUC, area under the curve.
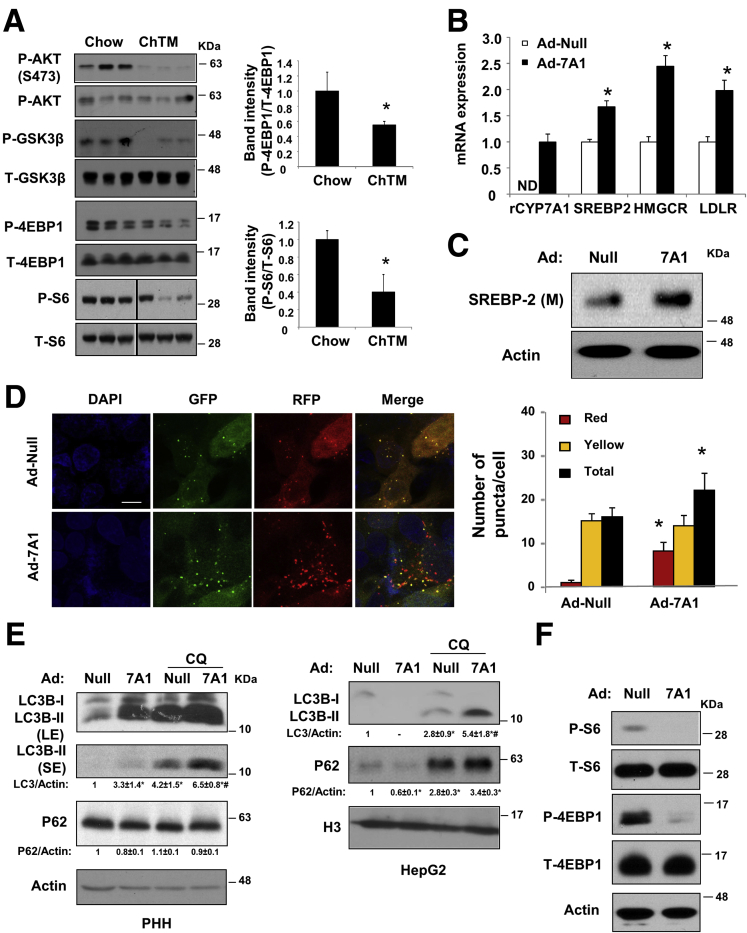
We next measured liver mTOR signaling, which is a master regulator of autophagy initiation. The liver phosphorylation levels of mTOR targets S6 and 4EBP-1 decreased by approximately 50% in ChTM-fed mice (Figure 5A). We also found that the basal phosphorylation levels of AKT and GSK3β, an AKT target, were significantly lower in ChTM-fed mice (Figure 5A). These results provide a signaling basis that linked increased autophagy to decreased mTOR activity in ChTM-fed mice. To distinguish if autophagy induction by ChTM was a result of increased CYP7A1 expression or decreased hepatic bile acid concentration, we next turned to CYP7A1 overexpressing HepG2 cells in vitro. As expected, overexpression of a rat CYP7A1 in HepG2 cells increased SREBP-2 mature form as well as the mRNA of HMGCR and LDLR (Figure 5B and C).28 CYP7A1 overexpression markedly induced autophagic flux in both HepG2 cells and primary human hepatocytes (Figure 5D and E). Consistent with our in vivo observation in ChTM-fed mice, CYP7A1 overexpression markedly decreased phosphorylation of S6 and 4EBP-1 (Figure 5F). Taken together, these results showed an interesting link between cellular CYP7A1 induction and mTOR signaling that controls autophagy activity.
Figure 5.
CYP7A1 overexpression attenuated AKT/mTOR signaling and induced autophagy flux in vitro. (A) Male C57BL/6J mice were fed a chow diet or a chow diet containing 2% ChTM for 6 days. Hepatic phosphorylated and total S6, 4EBP-1, AKT, and GSK3β. ∗Statistical significance vs Chow. (B and C) HepG2 cells were infected with Ad-null or Ad-CYP7A1 at a multiplicity of infection of 10 for 16 hours, and mRNA expression and SREBP-2 mature protein levels were measured. ∗Statistical significance vs Ad-Null. (D) Confocal microscopy: HepG2 cells were infected with Ad-RFP-GFP-LC3B at a multiplicity of infection of approximately 0.5 and Ad-CYP7A1 or Ad-null at a multiplicity of infection of 10 for 16 hours. Puncta number was quantified manually in approximately 50 cells. Results are expressed as means ± SE. *Statistical significance vs controls. Scale bar: 10 μm. (E) Western blot: primary human hepatocytes (PHH) or HepG2 cells were infected with Ad-null or Ad-CYP7A1 at a multiplicity of infection of approximately 10 for 16 hours. Some cells then were treated with CQ (50 μmol/L) for an additional 6 hours. LE, long exposure; SE, short exposure; H3, histone 3, as loading control. *Statistical significance vs Ad-Null. #Statistical significance vs Ad-Null+CQ. (F) HepG2 cells were infected with Ad-null or Ad-CYP7A1 at a multiplicity of infection of 10 for 16 hours in serum-containing medium. Phosphorylated and total S6 and 4EBP-1 were measured. DAPI, 4′,6-diamidino-2-phenylindole.
CYP7A1 Induction Attenuated Growth Factor Signaling to mTOR, but not Amino Acid Signaling to mTOR
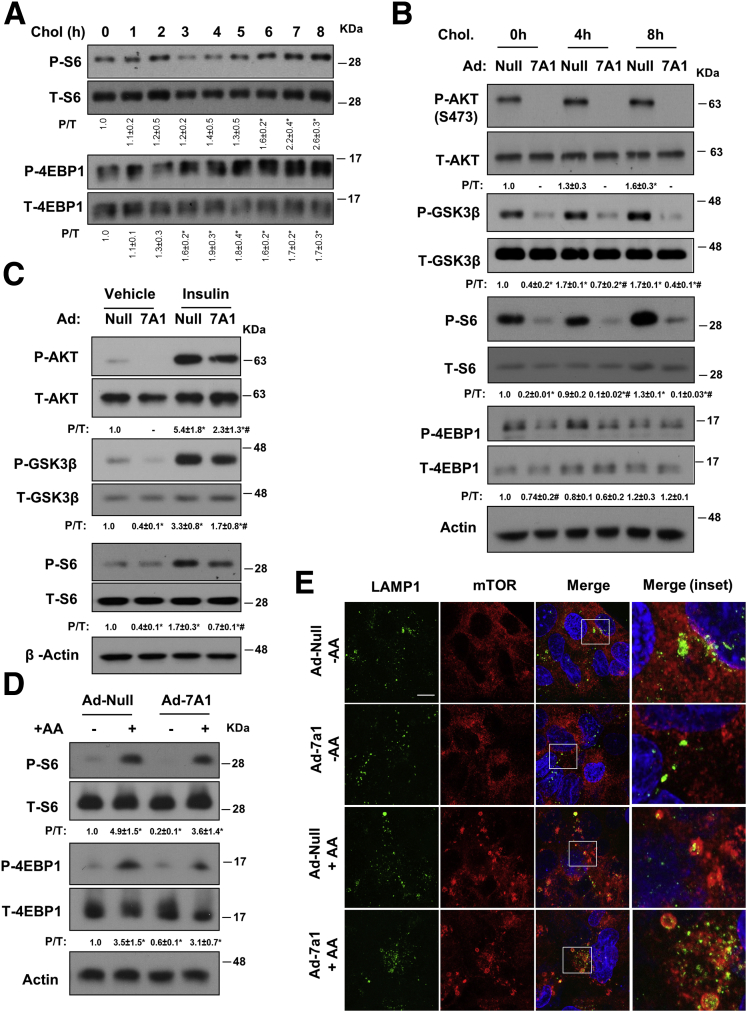
Growth factor signaling from the cell surface and amino acid signaling on the lysosomes are 2 major nutrient-sensing mechanisms activating mTOR,13 and thus were investigated further. We first determined if changes in mTOR signaling were linked to changes in cellular cholesterol levels, and found FC loading time-dependently enhanced the phosphorylation of S6 and 4EBP1, as well as AKT and its target GSK3β (Figure 6A and B). These cholesterol stimulatory effects on AKT and mTOR signaling largely were prevented by CYP7A1 overexpression (Figure 6B). These results were in line with several recent studies showing that cellular cholesterol abundance and cholesterol PM trafficking are essential in insulin/AKT signaling activation via proposed lipid raft microdomains.32, 33 Indeed, we found that CYP7A1 overexpression also could attenuate insulin-induced phosphorylation of AKT, GSK3β, and S6 in HepG2 cells (Figure 6C). In contrast, CYP7A1 overexpression did not prevent amino acid reactivation of mTOR (Figure 6D) or amino acid–induced mTOR recruitment to the Lamp1-positive puncta (Figure 6E). These results support that increasing CYP7A1 expression induces autophagy via attenuating AKT signaling to mTOR from the cell surface but independent of interference with amino acid signaling to mTOR on the lysosomes.
Figure 6.
CYP7A1 overexpression attenuated insulin signaling to mTOR. (A) HepG2 cells were cultured in serum-containing medium and treated with 50 μg/mL cholesterol for up to 8 hours. P/T, the ratio of phosphorylated and total protein was calculated based on densitometry. *Statistical significance vs untreated controls. (B) Cells were infected with Ad-Null or Ad-CYP7A1 at a multiplicity of infection of 10 for 16 hours in serum-containing medium. Some cells were treated with 50 μg/mL cholesterol for the indicated time. *Statistical significance vs Ad-Null at 0 hours. (C) HepG2 cells were infected with Ad-Null or Ad-CYP7A1 at a multiplicity of infection of 10 for 16 hours in serum-containing medium. All cells were cultured in serum-free medium for 8 hours first, and some cells then were treated with 10 nmol/L insulin for 30 minutes. *Statistical significance vs Ad-Null+vehicle. #Statistical significance vs Ad-Null+insulin. (D and E) HepG2 cells were infected with Ad-null or Ad-CYP7A1 at a multiplicity of infection of 10 for 16 hours in serum-containing medium. Cells then were amino acid–starved by culturing in Earle's Balanced Salt Solution (EBSS) for 2 hours. Amino acids (mixture of essential and nonessential amino acids) then were added back for 30 minutes. (D) Phosphorylated and total S6 and 4EBP1 were measured by Western blot. *Statistical significance vs Ad-Null. (E) Co-localization of Lamp1 and mTOR was studied by confocal microscopy. (Inset) Boxed sections in merged images. Chol, cholesterol.
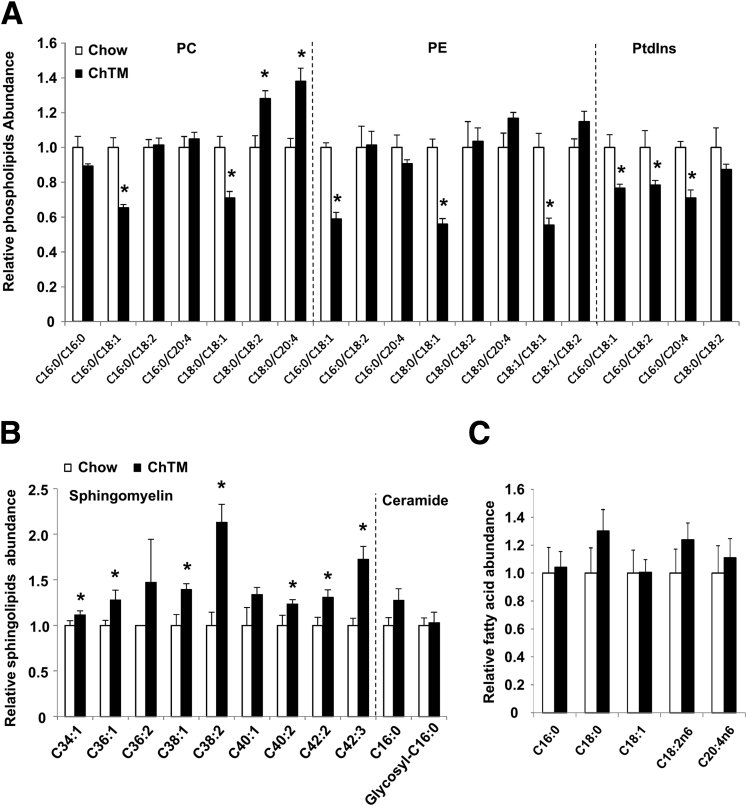
Finally, we asked if ChTM-induced cholesterol reduction indirectly could alter liver phospholipids and sphingolipids that also modulate cellular signal transduction. We noticed that the major phosphoatidylcholine and phosphoatidylethanolamine species (C16:0/C18:1 and C18:0/C18:1) were decreased in the liver of ChTM-fed mice (Figure 7A). Furthermore, phosphoatidylinositol, which serves as the substrates for phosphatidylinositol 3 kinase, showed a consistent trend of decreasing (Figure 7A). In contrast, most of the sphingomyelin species, which are known to interact with a special pool of cholesterol in the PM, were increased significantly in the liver of ChTM-fed mice (Figure 7B). Liver C16:0 ceramides were not altered (Figure 7B), and no corresponding changes in major fatty acid species were noticed (Figure 7C).
Figure 7.
ChTM altered hepatic phospholipid and sphingolipid profiles in mice. (A–C) Male C57BL/6J mice were fed a chow diet or a chow diet containing 2% ChTM for 6 days. All mice were briefly fasted for 6 hours starting at 6 AM and then killed. Liver phospholipids, sphingolipids, and fatty acids were determined by metabolomics analysis. PC, phosphoatidylcholine; PE, phosphoatidylethanolamine; PtdIns, phosphoatidylinositol. Results are expressed as means ± SE (n = 5). *Statistical significance vs chow-fed controls.
Cholestyramine Feeding Improved Metabolic Homeostasis and Restored Hepatic Autophagy in Diet-Induced Obesity Mice
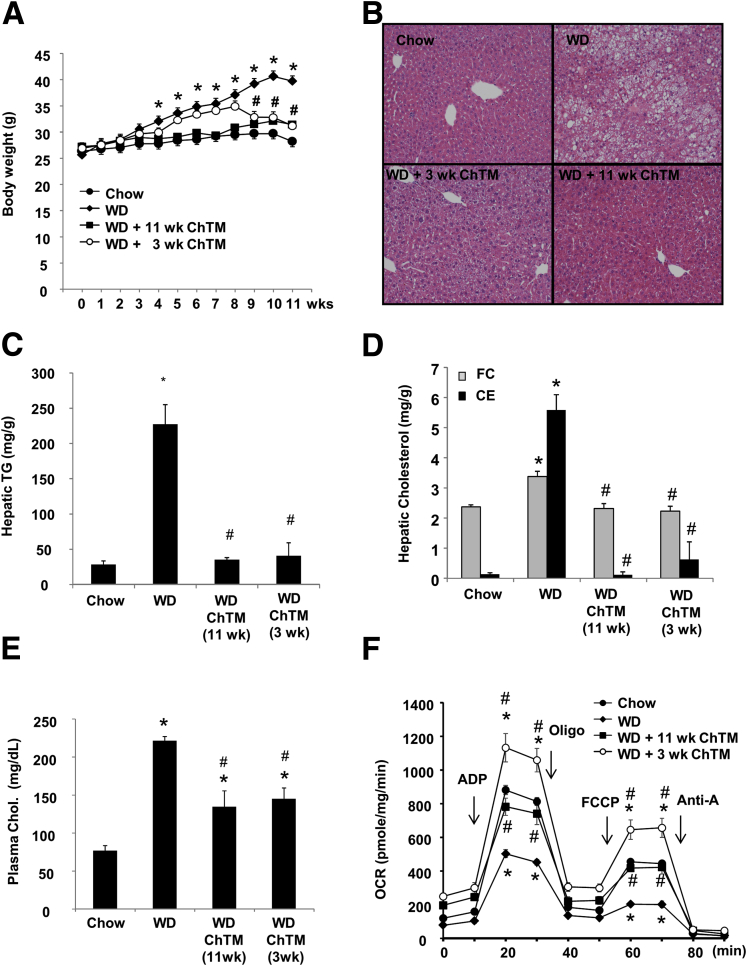
In the last set of experiments, we tested if CYP7A1 induction by ChTM could restore hepatic autophagy in diet-induced obesity (DIO) mice. Adding ChTM to a Western diet largely prevented DIO (Figure 8A). Accordingly, ChTM significantly attenuated hepatic steatosis and hypercholesterolemia (Figure 8B–E). It is worth noting that Western diet feeding markedly enriched the hepatic CE pool, but only increased hepatic FC by approximately 40% (Figure 8D), further supporting the critical role of ACAT in preventing hepatic FC accumulation. It has been reported that FC accumulation can cause mitochondrial dysfunction.2 DIO mice showed decreased mitochondrial bioenergetic function, and adding ChTM significantly improved mitochondrial bioenergetic function (Figure 8F).
Figure 8.
ChTM improved metabolic homeostasis in Western diet–fed mice. Male C57BL/6J mice were fed chow diet, Western diet (WD), or Western diet mixed with 2% ChTM for 11 weeks. The fourth group of mice were fed WD for 8 weeks, and then fed WD containing 2% ChTM for an additional 3 weeks. All mice were fasted for 6 hours starting at 6 AM and then killed. (A) Body weight. (B) H&E staining of liver sections. (C) Hepatic triglyceride (TG) level. (D) Hepatic FC and CE. (E) Plasma total cholesterol. (F) Analysis of bioenergetics function of isolated liver mitochondria using the Seahorse XF analyzer. All results are expressed as means ± SE (n = 4). *Statistical significance vs chow-fed controls. #Statistical significance vs WD-fed mice. ADP, adenosine diphosphate; Anti-A, Antimycin A; Chol, cholesterol; OCR, oxygen consumption rate; Oligo, Oligomycin.
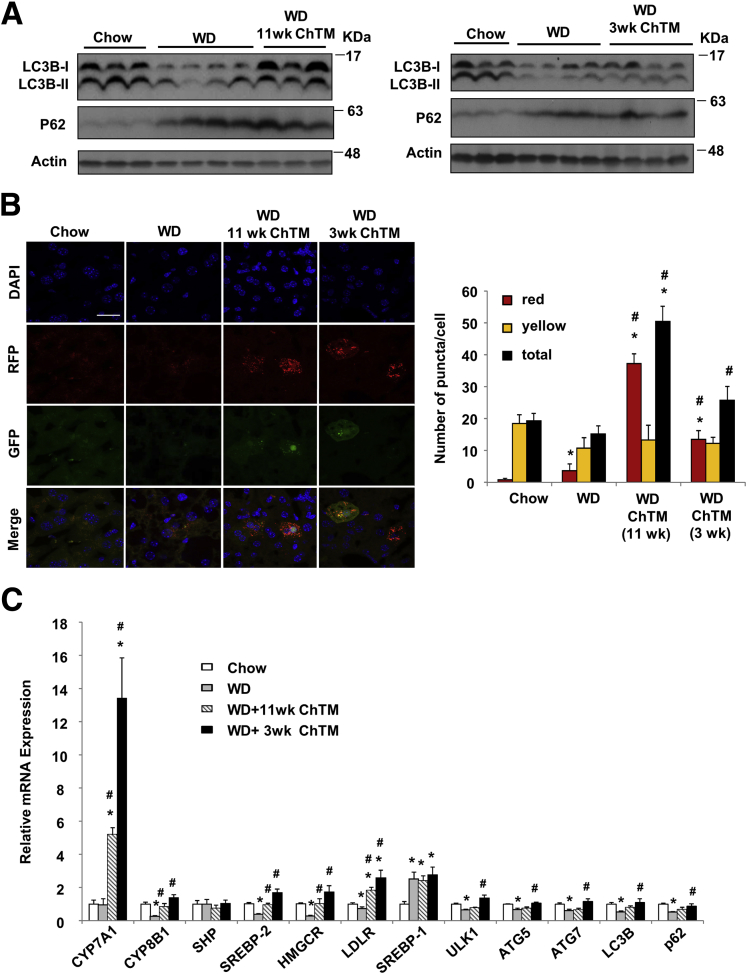
Current studies suggest that hepatic autophagy may be impaired at both initiation and lysosome fusion steps in obesity and fatty livers.17, 18, 19 We found that DIO mice had quantitatively less hepatic LC3B protein and higher P62 protein levels (Figure 9A). ChTM feeding increased LC3B-II protein but did not alter p62 protein compared with that of DIO mice (Figure 9A). We attempted to evaluate in vivo hepatic autophagic flux in mice by intraperitoneal injection of CQ, but this approach failed to increase liver LC3B or p62 levels (not shown). An alternative approach to analyze hepatic autophagic flux in RFP-GFP-LC3B–expressing mice found that the liver sections from ChTM-fed mice showed a significantly increased number of red puncta but an unaltered number of yellow puncta (Figure 9B), which suggests increased autophagy flux. Gene expression analysis showed that hepatic SREBP-2, HMGCR, and LDLR mRNA levels were repressed in DIO mice as a result of cholesterol accumulation, and were induced in ChTM-fed mice as a result of CYP7A1 induction (Figure 9C). The mRNA levels of key autophagy genes were decreased significantly by approximately 30%–50% in DIO mice. The mRNA expressions of many autophagy genes were higher in ChTM-fed mice, especially in 3-week ChTM-fed mice (Figure 9C). Recent studies have reported farnesoid X receptor regulation of autophagy gene transcription.34, 35 The involvement of this mechanism may be ruled out because ChTM feeding did not affect autophagy gene mRNA levels in chow-fed mice (Figure 4C).
Figure 9.
ChTM restored hepatic autophagy flux in Western diet–fed mice. The experimental procedure was the same as described in Figure 8. (A) Western blot: hepatic LC3B and P62 protein levels. (B) Some mice were injected with Ad-RFP-GFP-LC3B at 108 pfu/mouse 3 days before death. Mice were briefly fasted for 4 hours staring at 6 AM before death. Confocal microscopy was used for the analysis of LC3 fluorescent puncta in liver sections. The average puncta number was quantified manually in approximately 40–80 cells/group. Scale bar: 30 μm. (C) Hepatic mRNA expression. All results are expressed as means ± SE (n = 4). *Statistical significance vs chow-fed controls. #Statistical significance vs WD-fed mice. DAPI, 4′,6-diamidino-2-phenylindole.
Discussion
This study shows the differential impacts of FC and CE accumulation on hepatic autophagy and lysosome function, and provides mechanistic understanding of how FC accumulation causes autophagy defect in hepatocytes. PM is a major reservoir for cellular FC and intracellular membrane structures contain much less FC.36 Altered membrane lipid content was suggested to interfere with autophagic vesicular fusion,19 but how cellular cholesterol impacts autophagy in hepatocytes still is not fully clear. Our data suggest that FC accumulation did not stimulate autophagic flux, did not significantly block autophagosome/lysosome fusion, but primarily impaired autolysosome function and clearance, as evidenced by morphologic changes of autolysosomes, decreased autolysosome hydrolytic activity, LC3B-II accumulation, and increased LMP. It was intriguing that a very small increase of total cellular FC significantly can inhibit autophagy, especially when ACAT-mediated cholesterol esterification was blocked. Given that most intracellular organelles are relatively “cholesterol-poor,”36 a modest increase or depletion of cellular FC, such as by 10%–20% as observed by this study, may have a much more dramatic impact on intracellular organelle FC content. In addition, our finding in hepatocytes is in contrast to previous findings in macrophages where cholesterol accumulation was shown to induce autophagy to attenuate cholesterol toxicity by increasing cholesterol efflux and sphingomyelin synthesis,37, 38 suggesting a cell type–specific response to cholesterol. Under pathologic conditions in which hepatic cholesterol homeostasis severely was impaired, FC accumulation may damage organelles, impair autophagy, and sensitize the hepatocyte to injury.
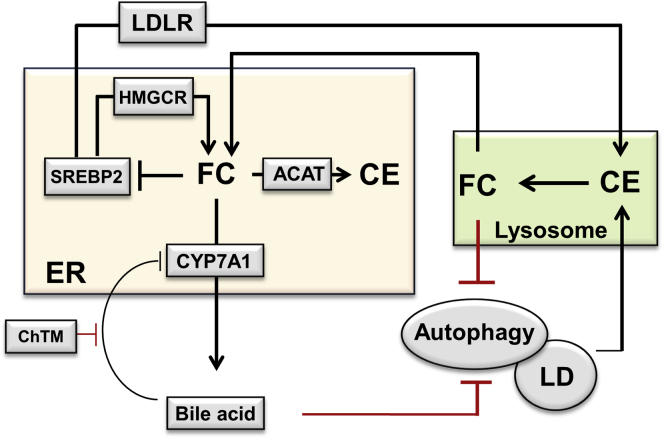
Another major finding from this study was the identification of a novel link between CYP7A1 induction, mTOR signaling, and autophagy activity, which seemed to depend on cellular cholesterol catabolism. Our results are in line with the recently reported link between cellular cholesterol trafficking and mTOR activation, although experimental methods that reduced cellular cholesterol also impaired mTOR activation in nonliver cells.39, 40 Another recent study showed that inhibiting Niemann–Pick-type C1- like 1 by ezetimibe caused lysosome FC accumulation, reduced mTOR recruitment to the lysosomes, and activated autophagy in hepatocytes and the small intestinal epithelia.41 However, although we initially speculated that CYP7A1 induction possibly may alter lysosome membrane cholesterol and subsequently decrease lysosomal mTOR localization and activation, it did not turn out to be the case. In contrast, we found that cellular cholesterol enrichment can enhance basal AKT and mTOR signaling, and that CYP7A1 overexpression could strongly attenuate both basal and insulin activation of AKT and mTOR. These results support a more plausible explanation that increased ER cholesterol catabolism somehow interfered with AKT activation at the PM. Furthermore, because cholesterol in the membrane rafts is known to be essential in PI3K/AKT activation32, 33 and a significant amount of PM cholesterol traffics to the ER even at basal conditions,42 it is even tempting to speculate that increased CYP7A1-mediated ER cholesterol catabolism decreases the PM cholesterol pool, leading to attenuated AKT signaling. It is well recognized that cholesterol dynamically interacts with sphingolipids and phospholipids in various membrane structures. Our analysis of the overall hepatic lipid profile further showed that CYP7A1 induction by ChTM not only decreased FC, but also altered hepatic phospholipids and sphingolipids. It is interesting to note that ChTM feeding decreased many liver phospholipid species (including phosphoatidylinositol) that serve as PI3K substrates. In addition, liver sphingomyelins, which sequester a pool of cholesterol in the PM, were higher in ChTM-fed mice. Although the molecular basis for increased sphingomyelins currently is unclear, it has been shown that cholesterol depletion increased the abundance of sphingomyelins in purified PM fractions of cultured cells in vitro.43 We report ChTM-induced changes in phospholipid and sphingolipid composition in mouse livers, and how such changes may broadly affect various cellular signal transduction pathways is worth further investigation. From a general perspective, however, cells somehow may sense cellular cholesterol levels to modulate mTOR activity, possibly because cholesterol is an essential nutrient for cellular growth and function. Because cholesterol deficiency may be detrimental to cell function and growth, autophagy activation upon CYP7A1 induction may be an adaptive response to mobilize stored cholesterol in the lipid droplets (Figure 10).
Figure 10.
Proposed roles of hepatic bile acid metabolism and autophagy in the regulation of cellular cholesterol homeostasis. Hepatic cholesterol homeostasis is regulated by the SREBP-2–dependent cholesterol-sensing mechanism. Hepatic cholesterol accumulation inhibits SREBP-2 activity and the expression of SREBP-2 target genes LDLR and HMGCR, leading to reduced cholesterol uptake and de novo cholesterol synthesis. In the ER, excessive FC is converted to CE for storage in lipid droplets. This study suggests that excessive hepatic FC accumulation impairs lysosome function and inhibits autophagy. On the contrary, induction of CYP7A1 by ChTM causes increased ER cholesterol catabolism, relative ER cholesterol depletion, and SREBP-2 activation, leading to induction of LDLR and HMGCR expression to replenish cellular cholesterol. This study showed that increased hepatic CYP7A1 and decreased hepatic bile acids caused by ChTM also induce autophagy, which may stimulate the hydrolysis of CE in the lipid droplet (LD) via lipophagy, leading to increased lysosomal FC transport to the ER as an adaptive response to CYP7A1-induced ER cholesterol depletion.
Finally, this study provides proof-of-concept that targeting the enterohepatic bile acid circulation induces hepatic autophagy. Importantly, bile acid synthesis exclusively occurs in the hepatocytes, which makes the identification of the CYP7A1–autophagy axis uniquely significant. Clinically, bile acid sequestrants have been used effectively to treat hypercholesterolemia and improve insulin sensitivity.20 This should decrease the risk of cardiovascular disease, the leading cause of mortality in patients with diabetes and fatty livers. Current mechanistic understanding suggests that bile acid sequestrants decrease LDL cholesterol by targeting hepatic bile acid synthesis and LDL uptake, and improve peripheral insulin sensitivity by inducing intestine glucagon-like peptide-1.44, 45 This study suggests that induction of hepatic autophagy by bile acid sequestrants also may be beneficial in improving cholesterol homeostasis and insulin sensitivity. Similar to bile acid sequestrants, new studies have shown that the potent and selective apical sodium-dependent bile acid transporter inhibitors also prevented hypercholesterolemia,21 improved insulin sensitivity,46 and attenuated bile acid toxicity in cholestasis in animal models.22, 23 It may be interesting to test whether apical sodium-dependent bile acid transporter inhibitors could induce hepatic autophagy by the same principle. Future studies will determine if targeting enterohepatic bile acid signaling may have therapeutic implications in other forms of genetic and acquired liver diseases that would benefit from selective induction of hepatic autophagy.
Acknowledgments
The authors gratefully thank Dr Tamotsu Yoshimori (Department of Genetics, Osaka University, Suita, Osaka, Japan) for providing the mCherry-galectin-3 expression plasmid.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported in part by an American Diabetes Association Junior Faculty Award (T.L.); the National Institutes of Health grant 1R01DK102487-01 (T.L); the National Center for Research Resources (5P20RR021940-07), and the National Institute of General Medical Sciences (8 P20 GM103549-07) of the National Institutes of Health (P.K.); The National Institute on Alcohol Abuse and Alcoholism grants R01 AA020518 and R01 DK102142 (W.-X.D); and the National Institutes of Health grants DK44442 and DK58379 (J.Y.L.C).
References
- 1.Puri P., Baillie R.A., Wiest M.M. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46:1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 2.Mari M., Caballero F., Colell A. Mitochondrial free cholesterol loading sensitizes to TNF- and Fas-mediated steatohepatitis. Cell Metab. 2006;4:185–198. doi: 10.1016/j.cmet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Wouters K., van Gorp P.J., Bieghs V. Dietary cholesterol, rather than liver steatosis, leads to hepatic inflammation in hyperlipidemic mouse models of nonalcoholic steatohepatitis. Hepatology. 2008;48:474–486. doi: 10.1002/hep.22363. [DOI] [PubMed] [Google Scholar]
- 4.Van Rooyen D.M., Larter C.Z., Haigh W.G. Hepatic free cholesterol accumulates in obese, diabetic mice and causes nonalcoholic steatohepatitis. Gastroenterology. 2011;141:1393–1403. doi: 10.1053/j.gastro.2011.06.040. 1403 e1391–e1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anstee Q.M., Targher G., Day C.P. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol. 2013;10:330–344. doi: 10.1038/nrgastro.2013.41. [DOI] [PubMed] [Google Scholar]
- 6.Brown M.S., Goldstein J.L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 7.Li T., Chiang J.Y. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 2014;66:948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li T., Francl J.M., Boehme S. Regulation of cholesterol and bile acid homeostasis by the cholesterol 7alpha-hydroxylase/steroid response element-binding protein 2/microRNA-33a axis in mice. Hepatology. 2013;58:1111–1121. doi: 10.1002/hep.26427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li T., Francl J.M., Boehme S. Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity. J Biol Chem. 2012;287:1861–1873. doi: 10.1074/jbc.M111.305789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T., Owsley E., Matozel M. Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology. 2010;52:678–690. doi: 10.1002/hep.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrell J.M., Chiang J.Y. Short-term circadian disruption impairs bile acid and lipid homeostasis in mice. Cell Mol Gastroenterol Hepatol. 2015;1:664–677. doi: 10.1016/j.jcmgh.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marino G., Niso-Santano M., Baehrecke E.H. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell R.C., Yuan H.X., Guan K.L. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu K., Czaja M.J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20:3–11. doi: 10.1038/cdd.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh R., Kaushik S., Wang Y. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding W.X., Li M., Chen X. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang L., Li P., Fu S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H.Y., Han J., Cao S.Y. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koga H., Kaushik S., Cuervo A.M. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staels B., Kuipers F. Bile acid sequestrants and the treatment of type 2 diabetes mellitus. Drugs. 2007;67:1383–1392. doi: 10.2165/00003495-200767100-00001. [DOI] [PubMed] [Google Scholar]
- 21.Huff M.W., Telford D.E., Edwards J.Y. Inhibition of the apical sodium-dependent bile acid transporter reduces LDL cholesterol and apoB by enhanced plasma clearance of LDL apoB. Arterioscler Thromb Vasc Biol. 2002;22:1884–1891. doi: 10.1161/01.atv.0000035390.87288.26. [DOI] [PubMed] [Google Scholar]
- 22.Miethke A.G., Zhang W., Simmons J. Pharmacological inhibition of apical sodium-dependent bile acid transporter changes bile composition and blocks progression of sclerosing cholangitis in multidrug resistance 2 knockout mice. Hepatology. 2016;63:512–523. doi: 10.1002/hep.27973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baghdasaryan A., Fuchs C.D., Osterreicher C.H. Inhibition of intestinal bile acid absorption improves cholestatic liver and bile duct injury in a mouse model of sclerosing cholangitis. J Hepatol. 2016;64:674–681. doi: 10.1016/j.jhep.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 24.Maejima I., Takahashi A., Omori H. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013;32:2336–2347. doi: 10.1038/emboj.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers G.W., Brand M.D., Petrosyan S. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One. 2011;6:e21746. doi: 10.1371/journal.pone.0021746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding W.X., Li M., Biazik J.M. Electron microscopic analysis of a spherical mitochondrial structure. J Biol Chem. 2012;287:42373–42378. doi: 10.1074/jbc.M112.413674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H., Peng Y.F., Ni H.M. Basal autophagy and feedback activation of Akt are associated with resistance to metformin-induced inhibition of hepatic tumor cell growth. PLoS One. 2015;10:e0130953. doi: 10.1371/journal.pone.0130953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandak W.M., Schwarz C., Hylemon P.B. Effects of CYP7A1 overexpression on cholesterol and bile acid homeostasis. Am J Physiol Gastrointest Liver Physiol. 2001;281:G878–G889. doi: 10.1152/ajpgi.2001.281.4.G878. [DOI] [PubMed] [Google Scholar]
- 29.Manley S., Ni H.M., Kong B. Suppression of autophagic flux by bile acids in hepatocytes. Toxicol Sci. 2014;137:478–490. doi: 10.1093/toxsci/kft246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radhakrishnan A., Goldstein J.L., McDonald J.G. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni H.M., Bockus A., Wozniak A.L. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011;7:188–204. doi: 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lasserre R., Guo X.J., Conchonaud F. Raft nanodomains contribute to Akt/PKB plasma membrane recruitment and activation. Nat Chem Biol. 2008;4:538–547. doi: 10.1038/nchembio.103. [DOI] [PubMed] [Google Scholar]
- 33.Mollinedo F., Gajate C. Lipid rafts as major platforms for signaling regulation in cancer. Adv Biol Regul. 2015;57:130–146. doi: 10.1016/j.jbior.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Lee J.M., Wagner M., Xiao R. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seok S., Fu T., Choi S.E. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516:108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., Robinet P., Smith J.D. ORMDL orosomucoid-like proteins are degraded by free-cholesterol-loading-induced autophagy. Proc Natl Acad Sci U S A. 2015;112:3728–3733. doi: 10.1073/pnas.1422455112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao X., Sluimer J.C., Wang Y. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab. 2012;15:545–553. doi: 10.1016/j.cmet.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng J., Ohsaki Y., Tauchi-Sato K. Cholesterol depletion induces autophagy. Biochem Biophys Res Commun. 2006;351:246–252. doi: 10.1016/j.bbrc.2006.10.042. [DOI] [PubMed] [Google Scholar]
- 40.Xu J., Dang Y., Ren Y.R. Cholesterol trafficking is required for mTOR activation in endothelial cells. Proc Natl Acad Sci U S A. 2010;107:4764–4769. doi: 10.1073/pnas.0910872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamura T., Ohsaki Y., Suzuki M. Inhibition of Niemann-Pick-type C1-like1 by ezetimibe activates autophagy in human hepatocytes and reduces mutant alpha1-antitrypsin Z deposition. Hepatology. 2014;59:1591–1599. doi: 10.1002/hep.26930. [DOI] [PubMed] [Google Scholar]
- 42.Brown M.S., Goldstein J.L. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 43.Das A., Brown M.S., Anderson D.D. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife. 2014;3:e02882. doi: 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harach T., Pols T.W., Nomura M. TGR5 potentiates GLP-1 secretion in response to anionic exchange resins. Sci Rep. 2012;2:430. doi: 10.1038/srep00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofmann A.F. Bile acid sequestrants improve glycemic control in type 2 diabetes: a proposed mechanism implicating glucagon-like peptide 1 release. Hepatology. 2011;53:1784. doi: 10.1002/hep.24100. [DOI] [PubMed] [Google Scholar]
- 46.Chen L., Yao X., Young A. Inhibition of apical sodium-dependent bile acid transporter as a novel treatment for diabetes. Am J Physiol Endocrinol Metab. 2012;302:E68–E76. doi: 10.1152/ajpendo.00323.2011. [DOI] [PubMed] [Google Scholar]