ABSTRACT
The genome of the multihost bacteriophage ΦK64-1, capable of infecting Klebsiella capsular types K1, K11, K21, K25, K30, K35, K64, and K69, as well as new capsular types KN4 and KN5, was analyzed and revealed that 11 genes (S1-1, S1-2, S1-3, S2-1, S2-2, S2-3, S2-4, S2-5, S2-6, S2-7, and S2-8) encode proteins with amino acid sequence similarity to tail fibers/spikes or lyases. S2-5 previously was shown to encode a K64 capsule depolymerase (K64dep). Specific capsule-degrading activities of an additional eight putative capsule depolymerases (S2-4 against K1, S1-1 against K11, S1-3 against K21, S2-2 against K25, S2-6 against K30/K69, S2-3 against K35, S1-2 against KN4, and S2-1 against KN5) was demonstrated by expression and purification of the recombinant proteins. Consistent with the capsular type-specific depolymerization activity of these gene products, phage mutants of S1-2, S2-2, S2-3, or S2-6 lost infectivity for KN4, K25, K35, or K30/K69, respectively, indicating that capsule depolymerase is crucial for infecting specific hosts. In conclusion, we identified nine functional capsule depolymerase-encoding genes in a bacteriophage and correlated activities of the gene products to all ten hosts of this phage, providing an example of type-specific host infection mechanisms in a multihost bacteriophage.
IMPORTANCE We currently identified eight novel capsule depolymerases in a multihost Klebsiella bacteriophage and correlated the activities of the gene products to all hosts of this phage, providing an example of carriage of multiple depolymerases in a phage with a wide capsular type host spectrum. Moreover, we also established a recombineering system for modification of Klebsiella bacteriophage genomes and demonstrated the importance of capsule depolymerase for infecting specific hosts. Based on the powerful tool for modification of phage genome, further studies can be conducted to improve the understanding of mechanistic details of Klebsiella phage infection. Furthermore, the newly identified capsule depolymerases will be of great value for applications in capsular typing.
KEYWORDS: Klebsiella, bacteriophage, capsular type, capsule depolymerase, multiple host
INTRODUCTION
The genus Klebsiella, especially the species Klebsiella pneumoniae, is an important human pathogen that causes a wide range of diseases, including both community and hospital-acquired infections. It is associated with septicemia, pneumonia, and urinary tract infections (1, 2) and also is responsible for a globally emerging disease, pyogenic liver abscess complicated with metastatic meningitis and endophthalmitis (3, 4).
Klebsiella spp. typically display a layer of thick, polysaccharide-based capsule on their surfaces. The expression of diverse capsule structure caused by different sugar compositions and linkages divide them into distinct serotypes. In addition, genetic variation of capsular polysaccharide synthesis (cps) regions in various types was also indicated. At present, at least 81 capsular types have been defined, including 77 types from reference strains recognized by serological reactivity tests established during the period 1926 to 1977 and 4 new types of K. pneumoniae (KN1 to KN4) characterized by molecular genotyping and phage typing in recent years (5–9). These polysaccharide coats confer resistance to host immune defenses and hostile environments (10, 11) and are associated with increased virulence (12, 13). Moreover, capsule could also act as a primary receptor for bacteriophage, which often possess tail fibers or tail spikes containing capsule depolymerization activities (14, 15). Degradation of bacterial capsule enables the phage to gain access to the host cell surface and bind to the secondary receptor. Given the specificity of capsule depolymerases, capsule type-specific phage or depolymerases also have been used in capsular typing. Klebsiella phages have been reported since 1940 and have been used for determination of several K-types of Klebsiella (16–21). However, little was known about the host specificity determinants of these phages until the recent characterization and identification of bacteriophage-borne depolymerases (6, 22). A KN2-specific phage, 0507-KN2-1, and its capsule depolymerase were identified and used for capsular typing of the new KN2 capsular type (6). Another study documented a phage (NTUH-K2044-K1-1) and its capsule depolymerase that exhibited specificity for capsular type K1 and showed therapeutic efficacy in K1 K. pneumoniae-infected mice (22). We recently isolated a multihost Klebsiella-infecting bacteriophage, ΦK64-1 (8), which appeared to belong to the Myoviridae family of viruses based on the sequence similarity. This phage (with a genome size of 346,602 bp) has 541 probable protein-coding genes (>300 bp in length), including 11 tail fiber/spike or lyase encoding genes (designated S1-1, S1-2, S1-3, S2-1, S2-2, S2-3, S2-4, S2-5, S2-6, S2-7, and S2-8 in the present study), which may possess depolymerization activity. In a previous study, we presented the capsule depolymerase activity of K64dep encoded by S2-5 and demonstrated the efficacy for treatment of multiple-drug-resistant K. pneumoniae infections.
Given the correlation between phage-encoded capsular depolymerases and phage host specificity, we speculated that ΦK64-1 (which has been proved to infect Klebsiella K1, K11, K21, K25, K30, K35, K64, and K69 reference strains [8] and is here shown to be able to infect new types KN4 and KN5 as well) could encode other capsule depolymerases apart from K64dep. Therefore, we sought to explore the functions of the rest of tail fiber/spike or lyase encoding genes and better understand their contributions to the wide capsular type host spectrum. In the present study, we identified an additional eight ΦK64-1-encoded capsule depolymerases by assessing the specificity of proteins produced from the corresponding genes and of recombinant bacteriophage deleted for the respective loci. This result provided a correlation between the activities of the nine capsule depolymerases encoded by ΦK64-1 and the host range of this virus. These observations revealed that carriage of multiple depolymerases accounts for this phage's multiple host-specific infective activities. The newly identified capsule depolymerases are expected to facilitate the classification of these capsular types.
RESULTS
Host range of ΦK64-1.
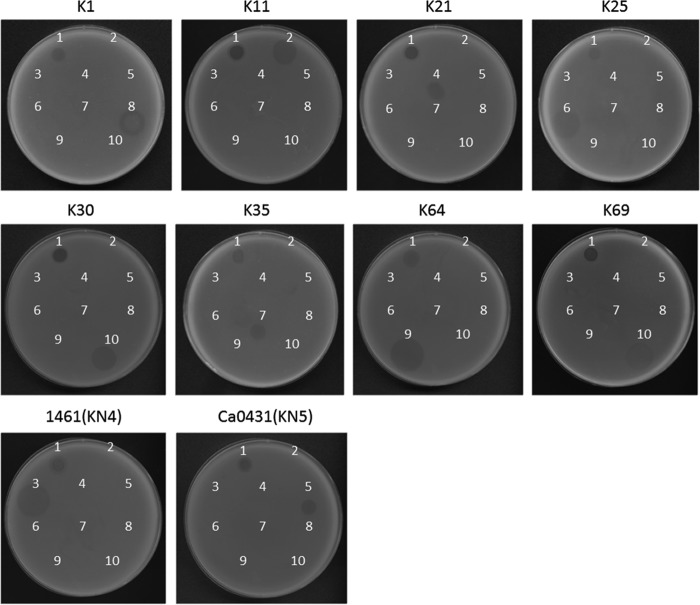
Seventy-seven reference strains, four strains with documented new capsular types (KN1, KN2, KN3, and KN4), and an additional strain (Ca0431) with a new wzc sequence (9) (accession number LC121097) that may correspond to a novel capsular type (designated KN5) of Klebsiella were used to determine the host range of ΦK64-1. The results of spot tests indicated that ΦK64-1 can infect ten strains, i.e., reference strains K1, K11, K21, K25, K30, K35, K64, and K69 and the new type strains 1461 (KN4) and Ca0431 (KN5) (Fig. 1). Additional K1 (NTUH-K2044, Canada PLA, A8126, and ATCC 35593), K21 (KCR74), K25 (KCR75), K30 (1353), KN4 (4565), and K64 (KCR2A, KCR3, KCR4, and KCR5) strains also were included in the spot tests. The results indicated that these clinical strains were infected by ΦK64-1 (Fig. 2). We therefore hypothesized that ΦK64-1 possessed capsule depolymerases able to recognize and digest at least 10 types of capsule.
FIG 1.
Spot test of wild-type and S1-2, S2-2, S2-3, and S2-6 deletion mutants of ΦK64-1. The reference strains K1, K11, K21, K25, K30, K35, K64, K69, 1461 (KN4) and Ca0431 (KN5) were grown on LB plates. Phages (106 PFU) were spotted on the plate, and the lysis zone could be observed after overnight incubation. Spots: 1, wild type; 2, S1-2 deletion mutant; 3, S2-2 deletion mutant; 4, S2-3 deletion mutant; 5, S2-6 deletion mutant.
FIG 2.
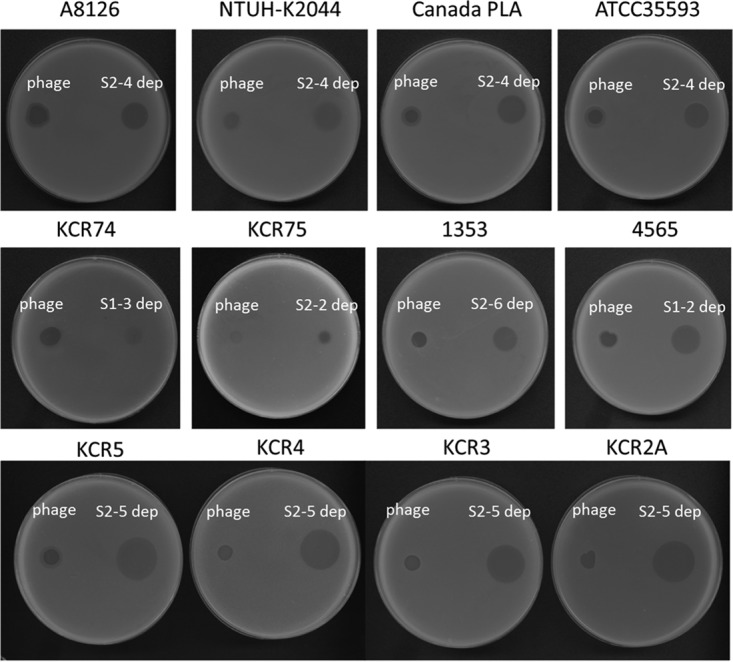
Spot test of ΦK64-1 and capsule depolymerases on additional clinical isolates of Klebsiella. Clinical isolates with K1 capsule (A8126, NTUH-K2044, Canada PLA, and ATCC 35593), with K21 capsule (KCR74), with K25 capsule (KCR75), with K30 capsule (1353), with KN4 capsule (4565), and with K64 capsule (KCR2A, KCR3, KCR4, and KCR5) were grown on LB plates. Phages or capsule depolymerase were spotted on the plate, and a plaque- or capsule depolymerase-generated semiclear spot could be be observed after overnight incubation.
Analysis of the genome sequences and morphology of ΦK64-1.
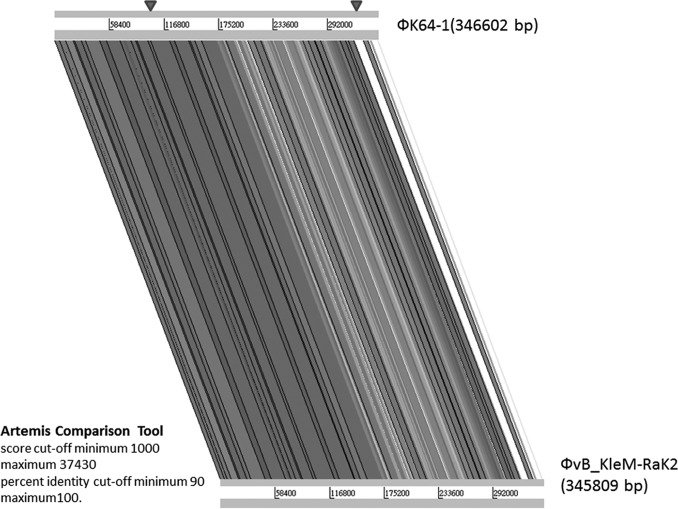
The circularly permutated genome of ΦK64-1 is 346,602 bp (accession number AB897757) with a G+C content of 31.7%. Analysis indicated the presence of 541 probable protein-coding genes (>300 bp in length) and four functional tRNA genes (tRNAArg, tRNASer, tRNASer2, and tRNAAsn) within the ΦK64-1 genome. Two genes code for RNA polymerase sigma factors were predicted; however, no homologue of RNA polymerase was detected from the genome. Nucleotide BLAST analysis revealed that ΦK64-1 exhibited high DNA similarity with a large Klebsiella-infecting myovirus with a genome size of 345,809 bp, bacteriophage vB_KleM-RaK2 (accession number JQ513383); our sequence covered 94% of the vB_KleM-RaK2 genome and showed 99% maximum DNA identity (144,392 bp/145,643 bp). Despite the high level of sequence similarity, some variable regions were indicated by comparative genomic analysis (Fig. 3). We further screened the ΦK64-1 genome for genes that encode tail fibers/spikes or lyases because these gene products were reported to have enzymatic activity against the bacterial capsule (23), which determine host range. In ΦK64-1, the products of 11 open reading frames (ORFs; S1-1 [GenBank accession number LC121100], S1-2 [LC121098], S1-3 [LC121099], S2-1 [LC121101], S2-2 [LC121102], S2-3 [LC121103], S2-4 [LC121104], S2-5 [AB897513], S2-6 [LC121105], S2-7 [LC121106], and S2-8 [LC121107]) exhibited similarity with tail fiber/spike or lyase proteins (Table 1). Among these 11 gene products, S2-2, S2-3, S2-6, and S2-8 showed a high degree of similarity to (are closely related to) proteins encoded by genes from bacteriophage vB_KleM-RaK2 (>90% amino acid identity); the predicted S2-1, S2-4, S2-5, and S2-7 proteins exhibited moderate sequence identity to gene products from bacteriophage vB_KleM-RaK2 (518/633 [82%], 231/259 [89%], 225/298 [76%], and 454/605 [75%], respectively). Notably, the S1-1, S1-2, and S1-3 proteins seemed to be unique to ΦK64-1, given that these three gene products showed only small regions of sequence similarity to proteins encoded by bacteriophage vB_KleM-RaK2 (Table 2 and Fig. 4). The morphology of purified ΦK64-1 phage particles was examined using transmission electron microscopy (TEM) (Fig. 5). The phage is characterized by an isometric head and a contractile tail and resembled Myoviridae family members. Furthermore, this phage appeared to have six long tail fibers displaying spike-like structures, which is similar to bacteriophage vB_KleM-RaK2.
FIG 3.
Genome comparative analysis of ΦK64-1 and ΦvB_KleM-RaK2. Genome comparative analysis was performed using an Artemis comparison tool (score cutoffs: minimum, 1,000, and maximum, 37,430; percent identity cutoffs: minimum, 90, and maximum, 100). Arrowheads indicate the regions in which 11 putative capsule depolymerases were located (S2-8 is in the left site; the remaining 10 genes are in the right area).
TABLE 1.
Putative tail fibers/spikes/capsule depolymerases of phage K64-1a
| ORF | Type (location [nt]) | Product size (aa) | Homologue | Accession no | Sequence identity (%)b | Activityc |
|---|---|---|---|---|---|---|
| S1-1 | Complementary (325887–327995) | 702 | Tail fiber of Klebsiella phage K11 | YP_002003830.1 | 360/590 (61) | K11* |
| S1-2 | Complementary (321593–323803) | 736 | Tail spike protein head-binding protein of Klebsiella phage 0507-KN2-1 | YP_008532048.1 | 44/113 (39) | KN4† |
| S1-3 | Complementary (323855–325810) | 651 | Tail spike protein of Salmonella phage FSL SP-063 | AGF88658.1 | 44/140 (31) | K21* |
| S2-1 | Complementary (328067–331648) | 1193 | Putative tail fiber protein of Pectobacterium phage PP1 | YP_007010682.1 | 40/133 (30) | KN5* |
| S2-2 | Complementary (331729–333483) | 584 | Phage T7 tail fiber protein of Klebsiella pneumoniae | WP_020326882.1 | 177/496 (36) | K25† |
| S2-3 | Complementary (333493–335832) | 779 | K5 lyase of Enterobacter phage K1-5 | YP_654147.1 | 72/232 (31) | K35† |
| S2-4 | Complementary (335902–338568) | 888 | Tail fiber protein of Enterobacter phage EcP1 | YP_007003187.1 | 57/221 (26) | K1* |
| S2-5 | 338917–341907 | 996 | Putative tail fiber of Enterobacter phage RTP | YP_398994.1 | 171/579 (30) | K64* |
| S2-6 | 341980–344283 | 767 | Pectate lyase superfamily protein of Klebsiella pneumoniae | WP_020801644.1 | 161/462 (35) | K30, K69† |
| S2-7 | 344312–346471 | 719 | Tail fiber domain protein of Pseudomonas savastanoi pv. savastanoi NCPPB 3335 | EFH99537.1 | 45/96 (47) | |
| S2-8 | 90154–91941 | 595 | Putative tail fiber protein of Cronobacter phage vB_CsaM_GAP32 | YP_006987359.1 | 255/510 (50) |
nt, nucleotide; aa, amino acids.
As determined by BLAST-P.
*, Evidenced by protein expression; †, evidenced by both protein expression and phage mutants.
TABLE 2.
Related genes of phage K64-1 putative tail fibers/spikes/capsule depolymerases in phage vB_KleM-RaK2a
| ORF | Product size (aa) | Most related gene in phage vB_KleM-RaK2 | Accession no. | Sequence identityb (%) |
|---|---|---|---|---|
| S1-1 | 702 | Putative tail fiber protein | YP_007007682 | 187/718 (26) |
| S1-2 | 736 | Hypothetical protein RaK2_00526 | YP_007007681 | 75/122 (61) |
| S1-3 | 651 | Putative structural protein | YP_007007685 | 30/90 (33) |
| S2-1 | 1193 | Putative tail fiber protein | YP_007007683 | 518/633 (82) |
| S2-2 | 584 | Putative structural protein | YP_007007684 | 581/584 (99) |
| S2-3 | 779 | Putative structural protein | YP_007007685 | 754/779 (97) |
| S2-4 | 888 | Putative structural protein | YP_007007686 | 231/259 (89) |
| S2-5 | 996 | Putative structural protein | YP_007007687 | 225/298 (76) |
| S2-6 | 767 | Putative structural protein | YP_007007688 | 690/767 (90) |
| S2-7 | 719 | Putative structural protein | YP_007007689 | 454/605 (75) |
| S2-8 | 595 | Hypothetical protein RaK2_00098 | YP_007007253 | 595/595 (100) |
aa, amino acids.
As determined by BLAST-P.
FIG 4.
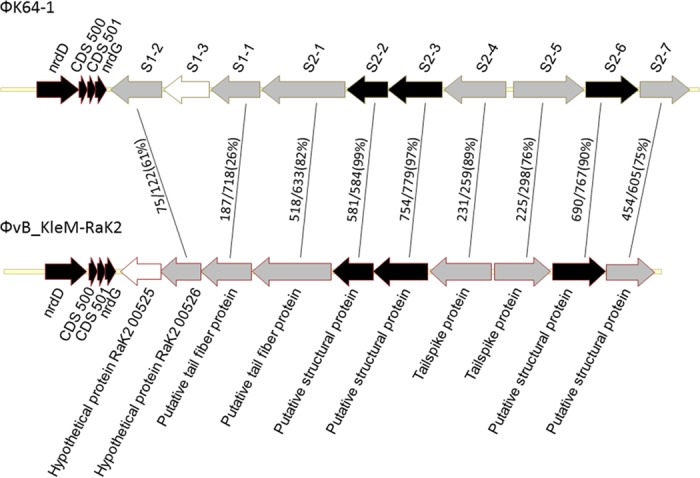
Comparison of the coding regions of putative capsule depolymerases of ΦK64-1 and ΦvB_KleM-RaK2. ORFs S1-1, S1-2, S1-3, S2-1, S2-2, S2-3, S2-4, S2-5, S2-6, and S2-7 of ΦK64-1 and corresponding genes in ΦvB_KleM-RaK2 were compared, and the amino acid sequence identities are shown (results for S2-8 and its homolog in ΦvB_KleM-RaK2 are not shown in the diagram). Gene products that shared a high degree of similarity (≥90% amino acid identity) are indicated by black arrows. Gene products that have no significant sequence similarity (and thus were unable to correlate to any genes in this region) are indicated by white arrows. Gray arrows indicate genes that exhibited <90% sequence identity to their corresponding genes.
FIG 5.
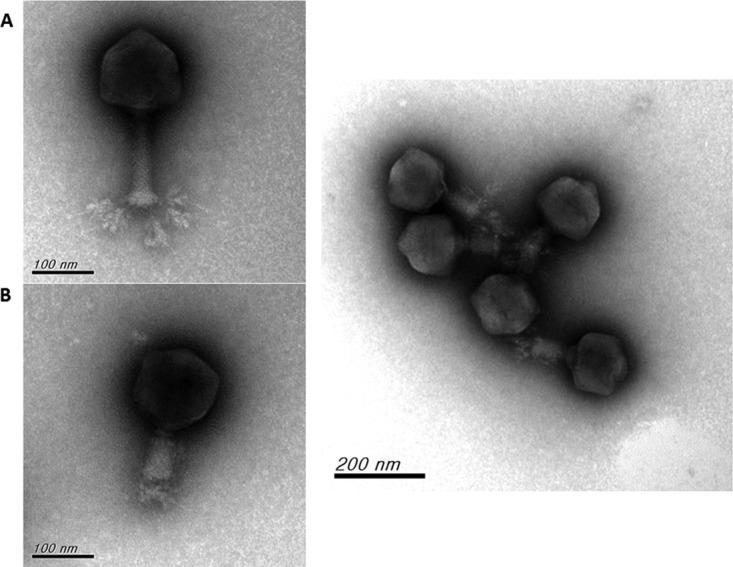
Electron micrographs of ΦK64-1. (A) ΦK64-1 phage particles with extended tail; (B) ΦK64-1 phage particles with contracted tail.
Expression of putative capsule depolymerases.
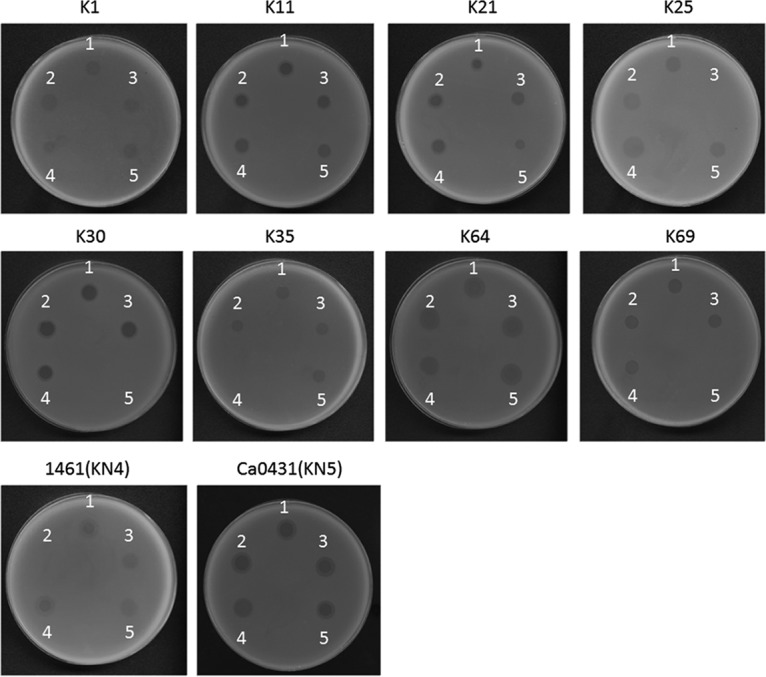
In order to determine whether these putative tail fibers/lyases had capsule-digesting activities, these ΦK64-1 genes were cloned and expressed via a pET-28c or a pcold TF DNA expression system. Capsule depolymerase activities of the resulting purified proteins then were assessed by spot tests against the ten known ΦK64-1 hosts (K1, K11, K21, K25, K30, K35, K64, K69, KN4, and KN5). Results indicated that nine of the eleven putative tail fibers/spikes/lyases generated semiclear spots on individual lawns of K1, K11, K21, K25, K30, K35, K64, K69, KN4, or KN5 bacteria. Specifically, activities were observed as follows: S2-4 against K1, S1-1 against K11, S1-3 against K21, S2-2 against K25, S2-6 against K30/K69, S2-3 against K35, S2-5 against K64, S1-2 against KN4, and S2-1 against KN5 (Fig. 6), whereas no depolymerase activity was observed in S2-7 and S2-8. The specificities of these capsule depolymerases were further clarified by testing against all other documented capsular types and additional strains, including capsular types K1 (NTUH-K2044, Canada PLA, A8126, and ATCC 35593), K21 (KCR74), K25 (KCR75), K30 (1353), KN4 (4565), and K64 (KCR2A, KCR3, KCR4, and KCR5). These results revealed that each of the nine enzymes could digest only capsule from the respective unique capsular type (as indicated above), with the sole exception of S2-6, which was able to digest capsule from strains of both type K30 and type K69 (Fig. 2 and data not shown). Therefore, these enzymes appear to be capsule type-specific depolymerases, such that the nine enzymes correspond to all ten hosts of ΦK64-1. To clarify whether proteins with activities for depolymerization of the same capsule exhibit sequence similarity, we compared the predicted amino acid sequence of S2-4 to that of a recently reported K1 capsule depolymerase, K1-ORF34 protein (22); the two gene products exhibited only limited amino acid sequence identity (205/612 [33%]) across the entire lengths of the two proteins. Moreover, we did not find any sequence conservation among the eleven putative tail fibers/spikes/lyases encoded by ΦK64-1.
FIG 6.
Spot test of capsule depolymerase. The reference strains K1, K11, K21, K25, K30, K35, K64, and K69, as well as the new type strains 1461 (KN4) and Ca0431 (KN5), were grown on LB plates. Phage (106 PFU) or capsule depolymerase (100 ng) were spotted on the plates. Spots: 1, K64-1 phage; 2, S1-1 protein; 3, S1-2 protein; 4, S1-3 protein; 5, S2-1 protein; 6, S2-2 protein; 7, S2-3 protein; 8, S2-4 protein; 9, S2-5 protein; 10, S2-6 protein.
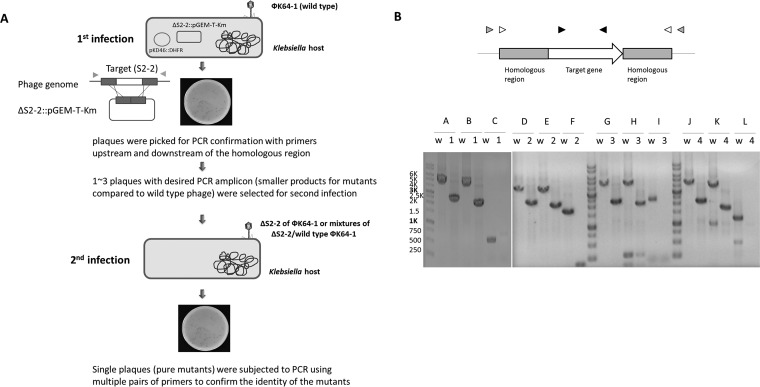
Determination of the functions of putative capsule depolymerases by deletion analysis in recombinant phage.
We randomly selected four genes (the ORFs encoding S1-2, S2-2, S2-3, or S2-6) to investigate their roles in infection of ΦK64-1 by generating deletion mutants (deleted individually) (Fig. 7B). The positive rate of the desired mutants was shown in Table S1 in the supplemental material. As the results indicate, the frequency varied in different mutants (from 9 to 80% in the first infection). Since wild-type/mutant mixtures existed when the first infection was carried out (i.e., wild-type and mutant amplicons coexist in a single plaque [data not shown]), we selected one to three mutant-positive plaques and performed an additional, second infection to obtain single plaques again and finally confirmed the pure mutants with multiple primers. Having these mutants, we used spot tests and an efficiency-of-plating (EOP) assay to determine the effect of these deletions on phage infectivity toward different hosts. Spot tests revealed that phage harboring ΔS1-2, ΔS2-2, ΔS2-3, and ΔS2-6 lost infectivity for KN4, K25, K35, and K30/K69, respectively, but retained infectivity for other hosts (Fig. 1). The EOP assay also showed that ΔS1-2, ΔS2-2, ΔS2-3, and ΔS2-6 lost the ability to infect specific hosts, as spot tests revealed, and the ability to infect other types was not significantly altered compared to that of the wild type (Table 3, P ≥ 0.05 [Student t test]).These data are consistent with the protein expression and activity experiment (above) that demonstrated that S1-2, S2-2, S2-3, and S2-6 are capsule depolymerases with specificity for KN4, K25, K35, and K30/K69, respectively. These results further demonstrated that these capsule depolymerases are crucial for the infection of specific hosts by ΦK64-1.
FIG 7.
Construction of the S1-2, S2-2, S2-3, and S2-6 deletion mutants. (A) Schematic depiction of the process for the generation of phage mutants. Two plasmids were transformed into the Klebsiella host, one is pKD46-DHFR, which can increase the recombination rate, the other is a plasmid that carried a selective deletion of the ORF while leaving the flanking regions intact (ΔS2-2::pGEM-T-Km is an example for the deletion of S2-2). After phage infection (refer to as the first infection), single plaques were picked from the plate for PCR confirmation to detect the presence of mutant DNA. Furthermore, a second infection was performed to obtain pure mutants because wild-type/mutant mixtures may exist in a single plaque when the first infection was performed. Finally, the single plaques from the second infection were subjected to PCR using multiple pairs of primers to confirm the identity of the mutant phage. (B) PCR confirmation of S1-2, S2-2, S2-3, and S2-6 deletion mutants. The upper panel shows a diagram of the genome region of target gene and primers. Three pairs of primers for each mutant were used in PCR confirmation. The primer pair indicated by black arrowheads was used to check that the target gene was absent in the mutant bacteriophage. White and gray arrowheads denote the two primer pairs used to confirm the gene alignment of this region after deletion. The lower panel shows the PCR results for the wild type and mutants using different pairs of primers. Lanes: w, wild type; 1, S1-2 mutant; 2, S2-2 mutant; 3, S2-3 mutant; 4, S2-6 mutant. A, D, G, and J: gray arrowhead primers were used (S1-2 gF and S1-2+48 inverse F, S2-2_g+1061F and S2-2_g+1208R, S2-3_g+1159F and S2-3_g+1087R, and S2-6+1068gF and S2-6+1050gR for the corresponding target genes S1-2, S2-2, S2-3, and S2-6, respectively); B, E, H and K: white arrowhead primers were used (S1-2 201F1 and S1-2 4434 R2, S2-2+1034F and S2-2+1007R, S2-3+1079F and S2-3+1005R, and S2-6+793F and S2-6+965R for the target genes S1-2, S2-2, S2-3, and S2-6, respectively); C, F, I, and L: black arrowhead primers were used (S1-2 A2 and S1-2 PR, 11_S2_ORF2F PF and 11_S2_ORF2R PR, 11_S2_ORF3F PF and 11_S2_ORF3R PR, and S2-6 5B and S2-6 5A for the target genes S1-2, S2-2, S2-3, and S2-6, respectively). The expected size of the PCR amplicons were (in bp) 4,554, 2,343, 4,253, 2,042, 553, none, 4,021, 2,429, 3,791, 2,199, 1,753, none, 4,583, 2,244, 4,421, 2,082, 2,340, none, 4,230, 2,116, 3,869, 1,755, 1,255, and none for A-w, A-1, B-w, B-1, C-w, C-1, D-w, D-2, E-w, E-2, F-w, F-2, G-w, G-3, H-w, H-3, I-w, I-3, J-w, J-4, K-w, K-4, L-w, and L-4, respectively.
TABLE 3.
EOPs of wild-type ФK64-1, ΔS1-2, ΔS2-2, ΔS2-3, and ΔS2-6 on different hostsa
| Host | EOP (%) |
||||
|---|---|---|---|---|---|
| WT | ΔS1-2 | ΔS2-2 | ΔS2-3 | ΔS2-6 | |
| NTUH-K2044 (K1) | 100 | 100 | 100 | 100 | 100 |
| K11 | 113 | 113 | 106 | 114 | 125 |
| K21 | 113 | 107 | 100 | 129 | 125 |
| K25 | 82 | 93 | 0 | 121 | 125 |
| K30 | 94 | 113 | 88 | 93 | 0 |
| K35 | 113 | 88 | 100 | 0 | 133 |
| K64 | 88 | 106 | 82 | 114 | 133 |
| K69 | 114 | 88 | 100 | 107 | 0 |
| KN4 | 93 | 0 | 76 | 107 | 125 |
| KN5 | 131 | 94 | 100 | 93 | 117 |
WT, wild type. Phage preparations were made, and the titers were determined using NTUH-K2044 (K1); the EOP was 100%. The infectivity of wild-type or mutant phages toward different hosts are shown by the EOP value, a ratio indicating how well the phage infects different hosts compared to NTUH-K2044 (K1). Three independent experiments were performed, and the averages are shown. The strains used in this experiment were NTUH-K2044 (K1), reference strain K11, 6668E (K21), VGHN4 (K25), reference strain K30, reference strain K35, reference strain K64, reference strain K69, 4565 (KN4), and Ca0431 (KN5).
Each ΦK64-1 virion possesses nine tail fiber/capsule depolymerase proteins.
In order to clarify whether each of the ΦK64-1 particles contains nine tail fiber proteins or whether different populations of phage particles containing different depolymerases are produced after infection, we performed an adsorption assay by preincubating ΦK64-1 with the ten hosts individually, and then we determined the decreased titer for the ten hosts (23). For example, since phage particle containing K1 tail fiber would attach to K1 bacteria, the reduced titer would be similar on different hosts when each viral particle contains nine tail fiber proteins. Our results indicated that preincubation of ΦK64-1 with its hosts for 5 min resulted in an ∼100-fold decrease of virions when titered on different strains (see Table S2 in the supplemental material), whereas no phage particle loss was observed when ΦK64-1 preincubated with a non-ΦK64-1 host (A4528-K2 strain) (data not shown). The results of adsorption also revealed that the reduced titers on different strains has no significant difference after preincubation with K1 (P ≥ 0.05 [Student t test]), indicating that each of the phage particles contained all tail fiber/capsule depolymerase proteins. Similar results were observed when the phage was preincubated with K11, K21, K25, K30, K35, K64, K69, KN4, and KN5 strains, and the virus titers were determined for these strains.
DISCUSSION
The large (346,602-bp) genome of bacteriophage K64-1 exhibited homology to a giant Klebsiella-infecting myovirus, bacteriophage vB_KleM-RaK2 (24). DNA sequences and virion morphology revealed that ΦK64-1 is a member of the Myoviridae family, which belongs to the Caudovirales, an order of viruses also known as tailed bacteriophages with double-stranded DNA genomes. ΦK64-1 encodes 11 proteins that exhibit sequence similarity to tail fiber/spike or lyase. Interestingly, with the exception of S2-8, these genes are clustered in a region of ∼25 kb. The phenomenon is consistent with a previous observation that genes with similar functions are usually arranged in a modular or cassette configuration in phage genomes (25–27). The corresponding region (∼23 kb) of bacteriophage vB_KleM-RaK2 also contains multiple tail fiber/spike encoding genes. It is worth noting that some genes within the region of ΦK64-1 exhibit a high level of sequence similarity to genes from the region of bacteriophage vB_KleM-RaK2, while others share very limited sequence identity. Therefore, the variety of this region between the two phages may result from the occurrence of recombination events. Since it appeared that this region determines host specificity, we speculated that bacteriophage vB_KleM-RaK2 is also a multispecificity phage with a host spectrum different from that of ΦK64-1, although the capsular types of Klebsiella hosts of bacteriophage vB_KleM-RaK2 are still unknown.
In extension of our previous work (8) showing that K64dep (S2-5 in our genome sequence) is a K64 capsule depolymerase, we demonstrated here the capsule-degrading activities of another eight proteins, including K1 capsule depolymerase (S2-4), K11 capsule depolymerase (S1-1), K21 capsule depolymerase (S1-3), K25 capsule depolymerase (S2-2), K30/K69 capsule depolymerase (S2-6), K35 capsule depolymerase (S2-3), KN4 capsule depolymerase (S1-2), and KN5 capsule depolymerase (S2-1) among the 11 putative tail fibers/spikes or lyases. The products of two genes, S2-7 and S2-8, did not exhibit capsule depolymerization activity in the expression and assay systems used here. Even expression using the pcold TF DNA vector (known to facilitate efficient production of soluble proteins) did not permit definition of a capsule depolymerase activity. However, we cannot rule out the possibility that these two proteins may exhibit enzyme activities if we optimize expression conditions. According to genomic analysis, ΦK64-1 exhibits high similarity with bacteriophage vB_KleM-RaK2. S2-7 protein shows 75% amino acid identity with ORF534, and S2-8 is identical to ORF098 from bacteriophage vB_KleM-RaK2. Both ORF534 and ORF098 were proved to be structural proteins in bacteriophage vB_KleM-RaK2 by tandem mass spectrometry analysis, suggesting that S2-7 and S2-8 should be structural proteins. Therefore, whether these genes encode structural proteins without a relevant enzymatic function, or whether these proteins possess depolymerization activities toward other unknown capsular types await further analyses. Moreover, the 11 putative tail fibers/spikes/lyases from ΦK64-1 exhibited no sequence conservation, suggesting that these capsule depolymerases seem to be structurally distinct. Even for proteins that are able to digest capsule of the same type (S2-4 and a previously reported K1 capsule depolymerase, the K1-ORF34 protein), only limited sequence identity was observed. It is possible that these enzymes depolymerize the polysaccharides via different mechanisms or target different cleavage sites. Future work will be needed to determine the characteristics of these enzymes.
According to the adsorption results, we proposed that the nine tail fiber/spike proteins coexist in each phage particle. In addition, although depolymerase activities have not yet been determined in S2-7 and S2-8, the two tail fiber proteins are presumably present in ΦK64-1 virion as well. Compared to bacteriophage vB_KleM-RaK2, which has 10 tail spike/tail fiber proteins in its virus particle, ΦK64-1 may contain 11 tail spike/tail fiber proteins, and we have proved that 9 of them possess depolymerase activities. Moreover, among the 11 proteins, 8 (i.e., S1-1, S1-2, S1-3, S2-1, S2-4, S2-5, S2-6, and S2-8) that exhibit similarity to tail spike proteins may be associated with the presence of spike-like structure studded on tail fibers. However, how these proteins are assembled and how they are arranged on the tail structure of ΦK64-1 remains to be clarified.
Recombineering is a powerful tool for modification of bacteriophage genomes. Notably, deletion of specific genes can be used to clarify the role of individual loci in phage biology. Some methods have been described for recombineering lytically growing phages. One technique, which uses phage lambda as a model system, involves phage infection, competent cell preparation, and electroporation of recombineering DNA substrates (28, 29). The second technique is bacteriophage recombineering of electroporated DNA (BRED), which was first described for mycobacteriophage engineering (30) and subsequently applied to construction of coliphage mutants (31). In BRED, bacterial cells inducibly expressing recombination functions are electroporated with a combination of phage DNA template and a targeting substrate. Moreover, CRISPR-Cas system is also exploited to edit the genome of phages by increasing recombination efficiencies (32) or used to counterselect nonedited phage genomes (33). In the present work, we modified the reported methods to generate Klebsiella bacteriophage mutants for further examination. Briefly, genetic engineering of ΦK64-1 was conducted using a Klebsiella host carrying two plasmids: one plasmid contained the mutated genes, and the second plasmid (pKD46) expressed λ-Red recombinases under inducing conditions; production of the recombinases increased the recombination efficiency. After infection of the host by the target phage, homologous recombination can occur, resulting in the generation of the desired mutant phage. In the present study, we successfully constructed deletion mutants of ΦK64-1 using this modified approach. To the best of our knowledge, this work represents the first example of the generation of Klebsiella phage mutants by recombineering.
To clarify the role of these genes in phage infection, purified mutant phages were subjected to host range testing. ΔS1-2, ΔS2-2, ΔS2-3, and ΔS2-6 mutations resulted in a loss of infectivity for KN4, K25, K35, and K30/K69, respectively. These results were consistent with the specificity of the capsule depolymerization activities of S1-2, S2-2, S2-3, and S2-6 for the respective hosts. These results further demonstrated the importance of specific capsule depolymerase activities for infecting bacteria with the corresponding capsule types. Notably, K30 and K69 are known to share very similar capsule structures, differing only in the linkage between β-d-Gal-p and the pyruvyl group (34, 35). Thus, we presume that S2-6's capsule depolymerase activity recognizes a structure shared between the K30 and K69 capsules; thus, the loss of S2-6 would result in the inability to infect both K30 and K69 type strains.
Since degradation of the bacterial capsule by depolymerases enables phage to penetrate the capsule and to gain access to receptors on the cell surface, variation in the capsular structure may be a mechanism for host evasion of bacteriophage infection. However, bacteriophage also have developed strategies to overcome these defenses. Previous studies documented a dual-specificity coliphage (K1-5) that can infect and grow on either K1 or K5 strains of Escherichia coli; ΦK1-5 encodes two different tail fiber proteins that confer this extended host spectrum (23). The present work provides an extreme example of a multihost bacteriophage: ΦK64-1 encodes multiple capsule depolymerases that contribute to the observed capsule type-specific host spectrum. Acquisition of various capsule depolymerases may confer an evolutionary advantage for bacteriophage that grows in an environment with a mixture of bacteria that possess different capsular types.
The difficulties in determining capsular types in Klebsiella by serological diagnosis have been noted in several studies (36–38). Consequently, several techniques for molecular capsular typing were developed to circumvent these problems (7, 9, 39, 40). In addition, phage-borne capsular polysaccharide depolymerases recently were used in capsular typing of Klebsiella (6, 22). The identification of capsule depolymerases in ΦK64-1 with specificity for K1, K11, K21, K25, K30/K69, K35, K64, KN4, and KN5 could serve as the basis for further rapid and simple approaches for the characterization of these capsular types.
In conclusion, we identified eight capsule depolymerases in the multihost bacteriophage K64-1. Together with the K64dep (S2-5), which was characterized in our previous study, these enzymes represent a total of nine capsule depolymerases, with activities consistent with the ten known hosts of ΦK64-1. Our study not only provides an example of carriage of multiple depolymerases in a phage with a wide capsular type host spectrum but also establishes a recombineering system for modification of Klebsiella bacteriophage genomes. Based on this powerful tool for modification of phage genome, more studies can be conducted to improve our understanding of the mechanistic details of phage infection in this genus. In addition, we expect to find practical applications for the newly identified capsule depolymerases in Klebsiella capsular typing.
MATERIALS AND METHODS
Bacterial strains.
Strains representing 82 capsular types were used for host range determination, which include 77 Klebsiella reference strains purchased from the Statens Serum Institute (Copenhagen, Denmark), four new type strains reported previously (A1517, Ca0507, N386, and 1461 represent KN1, KN2, KN3, and KN4, respectively) (6–9), and another strain, Ca0431, exhibited a novel type (currently identified and designated KN5). Additional K1 (NTUH-K2044, Canada PLA, A8126, and ATCC 35593), K21 (KCR74, 6668E), K25 (KCR75, VGHN4), K30 (1353), KN4 (4565), and K64 (KCR2A, KCR3, KCR4, and KCR5) strains were also used (Table 4) (8, 9, 22).
TABLE 4.
Klebsiella strains used in this study
| Capsular type | Strain | Species | Source or referencea |
|---|---|---|---|
| K1 | A5054 | K. pneumoniae | Reference strain |
| NTUH-K2044 | K. pneumoniae | 22 | |
| Canada PLA | K. pneumoniae | 22 | |
| A8126 | K. pneumoniae | 22 | |
| ATCC 35593 | K. pneumoniae | 22 | |
| K2 | B5055 | K. pneumoniae | Reference strain |
| K3 | C5046 | K. pneumoniae | Reference strain |
| K4 | D5050 | K. pneumoniae subsp. ozaenae | Reference strain |
| K5 | E5051 | K. pneumoniae subsp. ozaenae | Reference strain |
| K6 | F052 | K. pneumoniae subsp. ozaenae | Reference strain |
| K7 | Aerogenes 4140 | K. pneumoniae | Reference strain |
| K8 | Klebsiella 1015 | K. pneumoniae | Reference strain |
| K9 | Klebsiella 1056 | K. pneumoniae | Reference strain |
| K10 | Klebsiella 919 | K. pneumoniae | Reference strain |
| K11 | Klebsiella 390 | K. pneumoniae | Reference strain |
| K12 | Klebsiella 313 | K. pneumoniae | Reference strain |
| K13 | Klebsiella 1470 | K. pneumoniae | Reference strain |
| K14 | 138 | K. (Raoultella) planticola | Reference strain |
| K15 | Mich. 61 | K. pneumoniae | Reference strain |
| K16 | 2069/49 | K. pneumoniae | Reference strain |
| K17 | 2005/49 | K. pneumoniae | Reference strain |
| K18 | 1754/49 | K. pneumoniae | Reference strain |
| K19 | 293/50 | K. pneumoniae | Reference strain |
| K20 | 889/50 | K. pneumoniae | Reference strain |
| K21 | 1702/49 | K. pneumoniae | Reference strain |
| KCR74 | K. pneumoniae | 8 | |
| 6668E | K. pneumoniae | NTUH | |
| K22 | 1996/49 | K. pneumoniae | Reference strain |
| K23 | 2812/50 | K. pneumoniae | Reference strain |
| K24 | 1680/49 | K. pneumoniae | Reference strain |
| K25 | 2002/49 | K. pneumoniae | Reference strain |
| KCR75 | K. pneumoniae | 8 | |
| VGHN4 | K. pneumoniae | VGH | |
| K26 | 5884 | K. oxytoca | Reference strain |
| K27 | 6613 | K. pneumoniae | Reference strain |
| K28 | 5758 | K. pneumoniae | Reference strain |
| K29 | 5725y | K. oxytoca | Reference strain |
| K30 | 7824 | K. pneumoniae | Reference strain |
| 1353 | K. pneumoniae | NTUH | |
| K31 | 6258 | K. pneumoniae | Reference strain |
| K32 | 6837 | K. (Raoultella) ornithinolytica | Reference strain |
| K33 | 6168 | K. pneumoniae | Reference strain |
| K34 | 7522 | K. pneumoniae | Reference strain |
| K35 | 7444 | K. (Raoultella) planticola | Reference strain |
| K36 | 8306 | K. pneumoniae | Reference strain |
| K37 | 8238 | K. pneumoniae | Reference strain |
| K38 | 8414 | K. pneumoniae | Reference strain |
| K39 | 7749 | K. pneumoniae | Reference strain |
| K40 | 8588 | K. pneumoniae | Reference strain |
| K41 | 6177 | K. michiganensis | Reference strain |
| K42 | 1702 | K. pneumoniae | Reference strain |
| K43 | 2482 | K. pneumoniae | Reference strain |
| K44 | 7730 | K. (Raoultella) ornithinolytica | Reference strain |
| K45 | 8464 | K. pneumoniae | Reference strain |
| K46 | 5281 | K. pneumoniae | Reference strain |
| K47 | 9682 | K. pneumoniae | Reference strain |
| K48 | 1196 | K. variicola | Reference strain |
| K49 | 6115 | K. variicola | Reference strain |
| K50 | 1303/50 | K. pneumoniae II-B | Reference strain |
| K51 | 4715/50 | K. pneumoniae | Reference strain |
| K52 | 5759/50 | K. pneumoniae | Reference strain |
| K53 | 1756/51 | K. variicola | Reference strain |
| K54 | Stanley | K. variicola | Reference strain |
| K55 | 3985/51 | K. pneumoniae | Reference strain |
| K56 | 3534/51 | K. variicola | Reference strain |
| K57 | 4425/51 | K. variicola | Reference strain |
| K58 | 636/52 | K. variicola | Reference strain |
| K59 | 2212/52 | K. michiganensis | Reference strain |
| K60 | 4463/52 | K. pneumoniae II-B | Reference strain |
| K61 | 5710/52 | K. pneumoniae | Reference strain |
| K62 | 5711/52 | K. pneumoniae | Reference strain |
| K63 | 5845/52 | K. pneumoniae | Reference strain |
| K64 | NCTC 8172 | K. pneumoniae | Reference strain |
| KCR2A | K. pneumoniae | 8 | |
| KCR3 | K. pneumoniae | 8 | |
| KCR4 | K. pneumoniae | 8 | |
| KCR5 | K. pneumoniae | 8 | |
| K65 | SW4 | K. (Raoultella) terrigena | Reference strain |
| K66 | 438(3a) | K. michiganensis | Reference strain |
| K67 | 264(1) | K. (Raoultella) terrigena | Reference strain |
| K68 | 265(1) | K. (Raoultella) terrigena | Reference strain |
| K69 | 889 | K. (Raoultella) terrigena | Reference strain |
| K70 | 167 | K. michiganensis | Reference strain |
| K71 | 4349 | K. variicola | Reference strain |
| K72 | 1205 | K. (Raoultella) ornithinolytica | Reference strain |
| K74 | 371 | K. oxytoca | Reference strain |
| K79 | 325 | K. (Raoultella) planticola | Reference strain |
| K80 | 708 | K. pneumoniae II-B | Reference strain |
| K81 | 370 | K. pneumoniae | Reference strain |
| K82 | 3454-70 | K. pneumoniae | Reference strain |
| KN1 | A1517 | K. pneumoniae | 7 |
| KN2 | Ca0507 | K. pneumoniae | 6 |
| KN3 | N386 | K. pneumoniae | 8 |
| 1461 | K. pneumoniae | 9 | |
| KN4 | 4565 | K. pneumoniae | NTUH |
| KN5 | Ca0431 | K. pneumoniae | 6 |
NTUH, National Taiwan University Hospital; VGH, Taipei Veterans General Hospital.
EOP assay.
An efficiency-of-plating (EOP) assay was used to quantitate the ability of phage to infect different hosts as previous described (18). The EOP value is a ratio indicating how well a bacteriophage plates on different strains compared to the host which the phage preparation made from.
Determination of host range of phage and capsule depolymerase activity.
Spot tests (41) were performed to observe whether bacteria are permissive for phage infection; the assay also was applied to verify the activity of capsule depolymerases. Briefly, a 9-cm-diameter Luria-Bertani (LB) agar plate was overlaid with top agar that had been inoculated with 200 μl of a fresh bacterial culture. Aliquots of 106 PFU phage or 100 ng of a suspension of purified recombinant capsule depolymerase were spotted onto the plate after the top agar layer had solidified. After overnight incubation at 37°C, lytic or semiclear spots were observed.
Genome sequence analysis.
Coding sequences were predicted by MolGen bioinformatics webtools (http://www.molgenrug.nl/index.php/bioinformatics) and annotated by NCBI-protein BLAST. Comparative genomic analysis was performed using the Artemis comparison tool (Sanger Institute, Hinxton, United Kingdom) using the score cutoffs of a minimum of 1,000 and a maximum of 37,430, and percent identity cutoffs of a minimum of 90 and a maximum of 100. tRNAscan-SE 1.21 (http://lowelab.ucsc.edu/tRNAscan-SE/) was used to search for tRNAs (42).
Expression and purification of putative capsule depolymerases.
To yield N-terminally (His)6-tagged proteins, ORFs (including stop codons) were inserted into a pET-28c expression vector (Novagen, Madison, WI) or a pcold TF DNA expression system (TaKaRa, Tokyo, Japan; this system can increase protein solubility) via flanking NheI and XhoI (S1-1, S1-2, S1-3, S2-1, S2-3, S2-4, S2-6, and S2-8) or NdeI and XhoI (S2-2 and S2-7) restriction sites. The primers used for construction of the expression vectors are listed in Table 5. PCR amplifications were performed with the Long and Accurate PCR system (TaKaRa). The cycling program was 96°C for 3 min, followed by 30 cycles of 96°C for 30 s, 50°C for 15 s, and 72°C for 3 min. The products were ligated into expression vectors after restriction enzyme digestion. The resulting plasmids were transformed into E. coli BL21(DE3), and expression was induced by incubation with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 15°C overnight. The resulting His-tagged proteins were purified using nickel beads (GE Healthcare, Uppsala, Sweden) according to the manufacturer's instructions.
TABLE 5.
Primers used in this study
| Primer | Sequence (5′–3′) | Purpose |
|---|---|---|
| S1-1-Nhe1-Sac1-F | TGGGCTAGCGAGCTCGCAAATAAATTAACACAGCCAAAAGG | S1-1 expression |
| S1-1-Xho1-R(stop) | GGCTTTATTCTCGAGTTATCCAGCTAATATAAAAGAAACC | S1-1 expression |
| S1-2-NheI-PF | ATATGAGGTTAAGGCTAGCACAAATAGTTTAATACAACC | S1-2 expression |
| S1-2-XhoI-PR | ATATGGAGGCTCTCGAGTTAGCTATTGAATGATATTAC | S1-2 expression |
| S1-3-NheI-PF | TATATTTGGAGAAGCTAGCTAATGTCTACTGAATTAACAC | S1-3 expression |
| S1-3-XhoI-PR | TCATCAAAACTCGAGTTATATTAAAAATAGTCTAATATAAC | S1-3 expression |
| S2-1-Nhe1-Sac1-F | AGGGCTAGCGAGCTCGCATTTAAATTTAAAGGCTCACTATC | S2-1 expression |
| S2-1-Xho1-R(stop) | GGGGCTTTTTCTCGAGTTAACCAGACACTTGAATATTAAATG | S2-1 expression |
| S2-2-Nde1-F | TAGGATTAACATATGGGAAATTTTATACAACCTAAAG | S2-2 expression |
| S2-2-Xho1-R(stop) | GCTTTATTTTTCTCGAGTTATGCACCTCTAATATAAG | S2-2 expression |
| S2-3-Nhe1-Sac1-F | GAGGCTAGCGAGCTCATAAACGGATTAATTCAACCAAAAGGC | S2-3 expression |
| S2-3-Xho1-R(stop) | TCCCATATTCTCGAGCTATTTTTGTAATTGTTTTTC | S2-3 expression |
| S2-4-NheI-PF | ACAGCAAATTAAGCTAGCAAATGGAAACAGAGGGTTTAAC | S2-4 expression |
| S2-4-XhoI-PR | AGGAGGCTTCTCGAGTTATAATGATATTTGCCAATATATAG | S2-4 expression |
| S2-5-NheI-PF | TGCAAACTAAGAGGCTAGCACATGTCTTTAAGTAATTTAAG | S2-5 expression |
| S2-5-XhoI-PR | ACCCGAAGGTGCTTCTCGAGTTACTGTAAATAAATTCCTG | S2-5 expression |
| S2-6-Nhe1-Sac1-F | GAGGCTAGCGAGCTCTCATTAATTCAACTTTCACCAAGTAATG | S2-6 expression |
| S2-6-Xho1-R(stop) | CCTATTTATTTCTCGAGTTACCAAGTATTTATAGATAC | S2-6 expression |
| S2-7-Nde1-F | GATTTATCACATATGTCATTAACTAATTTAAACTC | S2-7 expression |
| S2-7-Xho1-R(stop) | GGCTTTTTTCTCGAGTTATATAGTTAAGAAACTTAC | S2-7 expression |
| S2-8-Nhe1-Sac1-F | AGGGCTAGCGAGCTCAGTATTAGTTTTAATCACCCCCAGAATAC | S2-8 expression |
| S2-8-XhoI-PR | TTATTTTCGTTCAACTCGAGTTATCGACCGATTGCTTGCC | S2-8 expression |
| S1-2 201F1 | GGCATATTACGTATGCGTTC | S1-2 mutant construct and check |
| S1-2 4434R2 | GTTTCTGGATATGCAGTTCG | S1-2 mutant construct and check |
| S1-2 gF | GGTTTTACACATTTCACAACTG | S1-2 mutant check |
| S1-2+48 inverse F | AAAACAAATTTTATCCGGCG | S1-2 mutant check |
| S1-2 A2 | TGGTGGTCAAATACATAGAC | S1-2 mutant check |
| S1-2 PR | TTAGCTATTGAATGATATTAC | S1-2 mutant check |
| S2-2+1034F | TGCTTATTCAAGTACTGACG | S2-2 mutant construct and check |
| S2-2+1007R | CTATCCAAGAAAACTAATACTTC | S2-2 mutant construct and check |
| S2-2-125IF | GTACAAATACACCTACTGGG | S2-2 mutant construct |
| S2-2-35IR | GAAGTTGAACCTTTAGGTTG | S2-2 mutant construct |
| S2-2_g+1061F | GCCGCCAAGTAACAACGAAT | S2-2 mutant check |
| S2-2_g+1208R | ATCACCTTCTTGTAGTGGTG | S2-2 mutant check |
| 11_S2_ORF2F PF | ATATGGGAAATTTTATACAAC | S2-2 mutant check |
| 11_S2_ORF2R PR | GCACCTCTAATATAAGCTTG | S2-2 mutant check |
| S2-3+1079F | TCAGGGGTGGTATGGTGTAG | S2-3 mutant construct and check |
| S2-3+1005R | CCGTTACGGTCAATCAATTCCC | S2-3 mutant construct and check |
| S2-3_inverseF | TAGGATTAAAATATGGGAAATTTTATAC | S2-3 mutant construct |
| S2-3_inverseR | TACATTAACCTCAATTTACAATATAC | S2-3 mutant construct |
| S2-3_g+1159F | CACAGGTGATGTTAGCGGAA | S2-3 mutant check |
| S2-3_g+1087R | GAGGATATTCTTCCGTTTCG | S2-3 mutant check |
| 11_S2_ORF3F PF | TAAATGATAAACGGATTAATTC | S2-3 mutant check |
| 11_S2_ORF3R PR | TTTTGTAATTGTTTTTCAATTTC | S2-3 mutant check |
| S2-6+793F | TTGTTCATTTGTGGGGTTAG | S2-6 mutant construct and check |
| S2-6+965R | TACATTATCACCTGCTGGCC | S2-6 mutant construct and check |
| S2-6 inverseF | CACACTGAGAACCAAATATG | S2-6 mutant construct |
| S2-6 inverseR | TTAATAAACCTCGTTATAAAG | S2-6 mutant construct |
| S2-6+1068gF | TCGAACCGTGGGGATTTAAAG | S2-6 mutant check |
| S2-6+1050gR | ACCAAGTGTCAAACTACCAC | S2-6 mutant check |
| S2-6 5B | CAACTATATGTCCTCGACCA | S2-6 mutant check |
| S2-6 5A | GTATGGAGTGGTGTTGGTGT | S2-6 mutant check |
Phage deletion mutant construction.
The genome of ΦK64-1 was modified based on previously described methods (28, 30). First, in order to increase the recombination rate of homologous regions in Klebsiella, a pKD46-DHFR plasmid was constructed. In brief, a dihydrofolate reductase-encoding gene, dhfr, which confers trimethoprim resistance, was amplified from EZ-Tn5 <DHFR-1>Tnp transposome (Epicentre, Madison, WI) with the Long and Accurate PCR system. Cycling conditions were as follows: 96°C for 3 min, followed by 30 cycles of 96°C for 30 s, 54°C for 15 s, and 72°C for 2 min. The amplified products were ligated to ApaLI-digested pKD46, a plasmid that encodes components of the λ-Red recombinase system (31). The resulting plasmid was electroporated into the hosts of ΦK64-1 (e.g., K1 Klebsiella strains) and selected on LB agar supplemented with trimethoprim at 75 μg/ml at 30°C. Second, each target depolymerase-encoding ORF (along with flanking regions) was amplified by PCR and cloned into a modified pGEM-T Easy vector (modified by insertion of a kanamycin resistance encoding cassette into the NdeI site [7]), followed by inverse PCR and self-ligation. Each resulting plasmid carried a selective deletion of the ORF while leaving the flanking regions intact (e.g., ΔS2-2::pGEM-T-Km). The resulting plasmids were purified and transformed into the host Klebsiella strains carrying pKD46-DHFR. Transformants harboring both the deletion plasmid (e.g., ΔS2-2::pGEM-T-Km) and pKD46-DHFR were selected on LB agar supplemented with kanamycin at 50 μg/ml and trimethoprim at 75 μg/ml at 30°C. The modified host was cultured in LB medium containing 1 mM arabinose, which induced expression of the recombinase system. Log-phase cultures of the induced bacteria (optical density at 600 nm = 0.5) were coincubated with various titers of ΦK64-1 for 30 min at 30°C. Mixtures of phage and bacteria were inoculated into top agar and then overlaid on LB agar plates. After overnight incubation at 30°C, single plaques were picked from the plate with moderate number of plaques (∼30) for PCR confirmation to detect the presence of phage-borne mutant DNA resulting from recombination (first infection). Because wild-type/mutant mixtures may exist in a single plaque when the first infection was carried out, ca. 1 to 3 mutant-positive plaques were selected for the second infection to obtain single plaques again. The recombinant phage were further coincubated with Klebsiella for 30 min at 37°C, followed by use of the agar overlay method for isolation of a pure phage (second infection). The single plaques were subjected to PCR to confirm the identity of the mutant phage-forming plaques. Subsequently, ΦK64-1 deletion mutants were isolated and validated by PCR using multiple pairs of primers (Table 5 and Fig. 7).
TEM.
Phages were purified by CsCl density gradient centrifugation. The phage suspension was layered on the top of CsCl step gradient (densities, 1.1 and 1.7 g/ml) and centrifuged using a SW 41 Ti swinging bucket rotor at 66,000 × g for 16 h at 4°C. After ultracentrifugation, the phages were collected from visible hazy blue/white bands using syringe with a 23G needle, and the majority of the CsCl was removed by buffer exchange in double-distilled H2O using Amicon Ultra centrifugal filter (100,000 MWCO; Millipore). Purified phage samples were applied on the carbon-coated nitrocellulose grids, followed by negative staining with 2% uranyl acetate, and then examined in a Hitachi H-7100 transmission electron microscope.
Preincubation (adsorption) assay to determine whether all ΦK64-1 phage particles contain nine tail fibers/depolymerases.
A phage preparation of ∼108 PFU was made using NTUH-K2044 (K1) strain, and the titer was determined on the 10 hosts (K1, K11, K21, K25, K30, K35, K64, K69, KN4, and KN5). Adsorption assay (23) was performed by preincubating ΦK64-1 with 5 × 107 CFU of the 10 hosts and A4528-K2 strain (as a control) individually. After preincubation for 5 min at room temperature, the mixture was filtered using a 0.45-μm-pore-size hydrophilic polyethersulfone membrane. The filtrate titers were then determined for all 10 hosts. The phage titer was quantified by a spot titer culture assay (43).
Accession number(s).
Newly determined sequences were deposited in GenBank under accession numbers LC121097 to LC121107.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from Ministry of Science and Technology, National Taiwan University, National Taiwan University Hospital, China Medical University, and the Liver Disease Prevention and Treatment Research Foundation in Taiwan.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02457-16.
REFERENCES
- 1.Abbot SL. 2003. Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and other Enterobacteriaceae, p 684–700. In Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH (ed), Manual of clinical microbiology, 8th ed ASM Press, Washington, DC. [Google Scholar]
- 2.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohmori S, Shiraki K, Ito K, Inoue H, Ito T, Sakai T, Takase K, Nakano T. 2002. Septic endophthalmitis and meningitis associated with Klebsiella pneumoniae liver abscess. Hepatol Res 22:307–312. doi: 10.1016/S1386-6346(01)00153-X. [DOI] [PubMed] [Google Scholar]
- 4.Tsai FC, Huang YT, Chang LY, Wang JT. 2008. Pyogenic liver abscess as endemic disease, Taiwan. Emerg Infect Dis 14:1592–1600. doi: 10.3201/eid1410.071254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ørskov I, Fife-Asbury MA. 1977. New Klebsiella capsular antigen K82 and the deletion of five of those previously assigned. Int J Syst Bacteriology 27:386–387. doi: 10.1099/00207713-27-4-386. [DOI] [Google Scholar]
- 6.Hsu CR, Lin TL, Pan YJ, Hsieh PF, Wang JT. 2013. Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PLoS One 8:e70092. doi: 10.1371/journal.pone.0070092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan YJ, Fang HC, Yang HC, Lin TL, Hsieh PF, Tsai FC, Keynan Y, Wang JT. 2008. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J Clin Microbiol 46:2231–2240. doi: 10.1128/JCM.01716-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan YJ, Lin TL, Lin YT, Su PA, Chen CT, Hsieh PF, Hsu CR, Chen CC, Hsieh YC, Wang JT. 2015. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob Agents Chemother 59:1038–1047. doi: 10.1128/AAC.03560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan YJ, Lin TL, Chen YH, Hsu CR, Hsieh PF, Wu MC, Wang JT. 2013. Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PLoS One 8:e80670. doi: 10.1371/journal.pone.0080670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts IS. 1995. Bacterial polysaccharides in sickness and in health: the 1995 Fleming Lecture. Microbiology 141(Pt 9):2023–2031. [DOI] [PubMed] [Google Scholar]
- 11.Cross AS. 1990. The biologic significance of bacterial encapsulation. Curr Top Microbiol Immunol 150:87–95. [DOI] [PubMed] [Google Scholar]
- 12.Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. 2006. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 193:645–654. doi: 10.1086/499968. [DOI] [PubMed] [Google Scholar]
- 13.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stummeyer K, Schwarzer D, Claus H, Vogel U, Gerardy-Schahn R, Muhlenhoff M. 2006. Evolution of bacteriophages infecting encapsulated bacteria: lessons from Escherichia coli K1-specific phages. Mol Microbiol 60:1123–1135. doi: 10.1111/j.1365-2958.2006.05173.x. [DOI] [PubMed] [Google Scholar]
- 15.Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. 2010. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol 59:145–155. [PubMed] [Google Scholar]
- 16.Adams MH, Park BH. 1956. An enzyme produced by a phage-host cell system. II. The properties of the polysaccharide depolymerase. Virology 2:719–736. [DOI] [PubMed] [Google Scholar]
- 17.Gaston MA, Ayling-Smith BA, Pitt TL. 1987. New bacteriophage typing scheme for subdivision of the frequent capsular serotypes of Klebsiella spp. J Clin Microbiol 25:1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pieroni P, Rennie RP, Ziola B, Deneer HG. 1994. The use of bacteriophages to differentiate serologically cross-reactive isolates of Klebsiella pneumoniae. J Med Microbiol 41:423–429. doi: 10.1099/00222615-41-6-423. [DOI] [PubMed] [Google Scholar]
- 19.Rieger-Hug D, Stirm S. 1981. Comparative study of host capsule depolymerases associated with Klebsiella bacteriophages. Virology 113:363–378. doi: 10.1016/0042-6822(81)90162-8. [DOI] [PubMed] [Google Scholar]
- 20.Thurow H, Niemann H, Rudolph C, Stirm S. 1974. Host capsule depolymerase activity of bacteriophage particles active on Klebsiella K20 and K24 strains. Virology 58:306–309. doi: 10.1016/0042-6822(74)90166-4. [DOI] [PubMed] [Google Scholar]
- 21.Rakieten ML, Eggerth AH, Rakieten TL. 1940. Studies with bacteriophages active against mucoid strains of bacteria. J Bacteriol 40:529–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin TL, Hsieh PF, Huang YT, Lee WC, Tsai YT, Su PA, Pan YJ, Hsu CR, Wu MC, Wang JT. 2014. Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J Infect Dis 210:1734–1744. doi: 10.1093/infdis/jiu332. [DOI] [PubMed] [Google Scholar]
- 23.Scholl D, Rogers S, Adhya S, Merril CR. 2001. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J Virol 75:2509–2515. doi: 10.1128/JVI.75.6.2509-2515.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simoliunas E, Kaliniene L, Truncaite L, Zajanckauskaite A, Staniulis J, Kaupinis A, Ger M, Valius M, Meskys R. 2013. Klebsiella phage vB_KleM-RaK2: a giant singleton virus of the family Myoviridae. PLoS One 8:e60717. doi: 10.1371/journal.pone.0060717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brussow H, Hendrix RW. 2002. Phage genomics: small is beautiful. Cell 108:13–16. doi: 10.1016/S0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- 26.Kaliniene L, Klausa V, Zajanckauskaite A, Nivinskas R, Truncaite L. 2011. Genome of low-temperature T4-related bacteriophage vB_EcoM-VR7. Arch Virol 156:1913–1916. doi: 10.1007/s00705-011-1084-y. [DOI] [PubMed] [Google Scholar]
- 27.Nolan JM, Petrov V, Bertrand C, Krisch HM, Karam JD. 2006. Genetic diversity among five T4-like bacteriophages. Virol J 3:30. doi: 10.1186/1743-422X-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marinelli LJ, Hatfull GF, Piuri M. 2012. Recombineering: a powerful tool for modification of bacteriophage genomes. Bacteriophage 2:5–14. doi: 10.4161/bact.18778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oppenheim AB, Rattray AJ, Bubunenko M, Thomason LC, Court DL. 2004. In vivo recombineering of bacteriophage lambda by PCR fragments and single-strand oligonucleotides. Virology 319:185–189. doi: 10.1016/j.virol.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Marinelli LJ, Piuri M, Swigonova Z, Balachandran A, Oldfield LM, van Kessel JC, Hatfull GF. 2008. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS One 3:e3957. doi: 10.1371/journal.pone.0003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feher T, Karcagi I, Blattner FR, Posfai G. 2012. Bacteriophage recombineering in the lytic state using the lambda red recombinases. Microb Biotechnol 5:466–476. doi: 10.1111/j.1751-7915.2011.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martel B, Moineau S. 2014. CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res 42:9504–9513. doi: 10.1093/nar/gku628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiro R, Shitrit D, Qimron U. 2014. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol 11:42–44. doi: 10.4161/rna.27766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hackland PL, Parolis H, Parolis LA. 1988. A structural investigation of the capsular polysaccharide of Klebsiella K69. Carbohydr Res 172:209–216. doi: 10.1016/S0008-6215(00)90855-3. [DOI] [PubMed] [Google Scholar]
- 35.Lindberg B, Lindh F, Lonngren J, Sutherland IW. 1979. Structural studies of the capsular polysaccharide of Klebsiella type 30. Carbohydr Res 76:281–284. doi: 10.1016/0008-6215(79)80031-2. [DOI] [PubMed] [Google Scholar]
- 36.Fung CP, Hu BS, Chang FY, Lee SC, Kuo BI, Ho M, Siu LK, Liu CY. 2000. A 5-year study of the seroepidemiology of Klebsiella pneumoniae: high prevalence of capsular serotype K1 in Taiwan and implication for vaccine efficacy. J Infect Dis 181:2075–2079. doi: 10.1086/315488. [DOI] [PubMed] [Google Scholar]
- 37.Jenney AW, Clements A, Farn JL, Wijburg OL, McGlinchey A, Spelman DW, Pitt TL, Kaufmann ME, Liolios L, Moloney MB, Wesselingh SL, Strugnell RA. 2006. Seroepidemiology of Klebsiella pneumoniae in an Australian tertiary hospital and its implications for vaccine development. J Clin Microbiol 44:102–107. doi: 10.1128/JCM.44.1.102-107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsay RW, Siu LK, Fung CP, Chang FY. 2002. Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch Intern Med 162:1021–1027. doi: 10.1001/archinte.162.9.1021. [DOI] [PubMed] [Google Scholar]
- 39.Brisse S, Issenhuth-Jeanjean S, Grimont PA. 2004. Molecular serotyping of Klebsiella species isolates by restriction of the amplified capsular antigen gene cluster. J Clin Microbiol 42:3388–3398. doi: 10.1128/JCM.42.8.3388-3398.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, Decre D. 2013. wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol 51:4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verma V, Harjai K, Chhibber S. 2009. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: a potential therapeutic agent. Curr Microbiol 59:274–281. doi: 10.1007/s00284-009-9430-y. [DOI] [PubMed] [Google Scholar]
- 42.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck NK, Callahan K, Nappier SP, Kim H, Sobsey MD, Meschke JS. 2009. Development of a spot-titer culture assay for quantifying bacteria and viral indicators. J Rapid Methods Automation Microbiol 17:455–464. doi: 10.1111/j.1745-4581.2009.00182.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.