ABSTRACT
Programmed cell death is a tightly controlled process critical for the removal of damaged or infected cells. Pro- and antiapoptotic proteins of the Bcl-2 family are pivotal mediators of this process. African swine fever virus (ASFV) is a large DNA virus, the only member of the Asfarviridae family, and harbors A179L, a putative Bcl-2 like protein. A179L has been shown to bind to several proapoptotic Bcl-2 proteins; however, the hierarchy of binding and the structural basis for apoptosis inhibition are currently not understood. We systematically evaluated the ability of A179L to bind proapoptotic Bcl-2 family members and show that A179L is the first antiapoptotic Bcl-2 protein to bind to all major death-inducing mammalian Bcl-2 proteins. We then defined the structural basis for apoptosis inhibition of A179L by determining the crystal structures of A179L bound to both Bid and Bax BH3 motifs. Our findings provide a mechanistic understanding for the potent antiapoptotic activity of A179L by identifying it as the first panprodeath Bcl-2 binder and serve as a platform for more-detailed investigations into the role of A179L during ASFV infection.
IMPORTANCE Numerous viruses have acquired strategies to subvert apoptosis by encoding proteins capable of sequestering proapoptotic host proteins. African swine fever virus (ASFV), a large DNA virus and the only member of the Asfarviridae family, encodes the protein A179L, which functions to prevent apoptosis. We show that A179L is unusual among antiapoptotic Bcl-2 proteins in being able to physically bind to all core death-inducing mammalian Bcl-2 proteins. Currently, little is known regarding the molecular interactions between A179L and the proapoptotic Bcl-2 members. Using the crystal structures of A179L bound to two of the identified proapoptotic Bcl-2 proteins, Bid and Bax, we now provide a three-dimensional (3D) view of how A179L sequesters host proapoptotic proteins, which is crucial for subverting premature host cell apoptosis.
KEYWORDS: ASFV, apoptosis, Bax family, Bcl-2, X-ray crystallography
INTRODUCTION
B-cell lymphoma-2 (Bcl-2) family members regulate intrinsic or mitochondrially initiated apoptosis, an intracellular programmed cell death. As major arbiters of mitochondrial integrity, Bcl-2 proteins play a key role in regulating the release of apoptosis regulators from mitochondria (1, 2). Bcl-2 family members have been subdivided into prosurvival and proapoptotic signaling proteins and are characterized by the presence of one or more Bcl-2 homology (BH) motifs that are key to their activity (3). In mammals, prosurvival members of the family include Bcl-2, Bcl-xL, Bcl-w, Mcl-1, A1, and Bcl-b (4). The proapoptotic proteins Bak and Bax are essential for executing mitochondrially initiated cell death in mammals by triggering the release of cytochrome c from the mitochondrial outer membrane (MOM) through pore formation induced by higher-order homo-oligomers of Bax or Bak (5, 6). While Bak constitutively localizes to the MOM via its C-terminal transmembrane anchor, Bax is primarily cytosolic and translocates to the MOM after an apoptotic stimulus (7). Bax and Bak activation is triggered by the activity of BH3-only proteins (7). The BH3-only proteins include Bim, Bid, Puma, Noxa, Bmf, Bik, Bad, and Hrk and function either by directly activating Bak and Bax or by sequestering and neutralizing the prosurvival Bcl-2 members. Unlike the prosurvival Bcl-2 proteins and Bax and Bak, which contain multiple BH motifs, BH3-only proteins possess only the BH3 motif, which engages the canonical ligand-binding groove on the prosurvival proteins as an α-helix (3). The BH3-only proteins are upregulated in response to cellular insults, including growth factor deprivation or exposure to cytotoxic drugs, leading to the activation of cell death mechanisms (7).
Viruses have evolved a number of molecular strategies to prevent premature host cell apoptosis, including direct caspase inhibitors and inhibitors of the intrinsic apoptosis pathway (8). Adenoviruses and Epstein-Barr virus (EBV) encode viral Bcl-2-like proteins (4, 9–11) that are required for successful viral propagation and/or persistence (12). Other viruses encode antiapoptotic proteins that display weak or no overt sequence identity with Bcl-2. A number of these distant orthologous Bcl-2 proteins are encoded by viruses that are members of the Poxviridae family, including vaccinia virus and variola virus, which both encode F1L, vaccinia virus N1L, E3L, and GAAP, and myxoma virus-encoded M11L (13–21), as well as the more recently identified fowlpox virus FPV039, orf virus ORF125, deerpox virus DPV022, and sheeppox virus SPPV14 (22–26). The use of sequence and/or structural homologs of Bcl-2 for immune modulatory purposes is not limited to the pox viruses and herpesviridae.
African swine fever virus (ASFV) is a large double-stranded DNA virus and the sole member of Asfarviridae. ASFV is responsible for a highly transmissible lethal hemorrhagic infection in domestic pigs and is endemic to East African wild pig populations (27). The genome of ASFV encodes a suite of immune modulatory proteins and virulence factors (28, 29), including the Bcl-2 like protein A179L (30). In cellular assays, A179L is a potent inhibitor of apoptosis, protects both HeLa cells and insect cells against apoptosis (31), and is localized at the mitochondria and endoplasmic reticulum (32). Functionally, A179L was shown to bind to several BH3-only proteins, including Bid and Bim, but not to Noxa, and also binds proapoptotic Bak and Bax (33). However, the affinities and the hierarchy of any of these interactions remain to be clarified. In addition to its role in apoptosis, A179L modulates autophagy via interaction with Beclin-1 and inhibits autophagosome formation under starvation conditions (32).
RESULTS
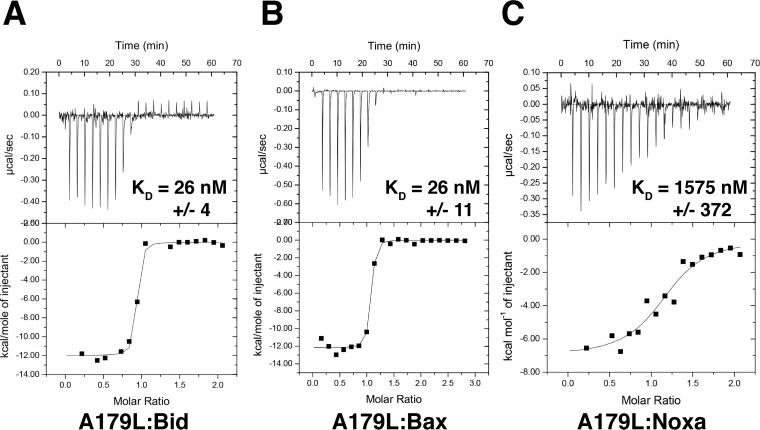
To understand the molecular and structural basis for A179L inhibition of apoptosis, we systematically examined the ability of recombinant A179L to engage peptides spanning the BH3 motif of proapoptotic Bcl-2 proteins from the pig (Sus scrofa), including those from Bid, Bim, Puma, Bmf, Bik, Bad, Noxa, and Hrk (Fig. 1A and C; Table 1). Interestingly, A179L was able to bind to all tested BH3 motifs with various affinities. Several proapoptotic BH3-only proteins were engaged with high affinity, including Bid (KD, 26 nM), Bim (KD, 6 nM), and Puma (KD, 31 nM), whereas Bmf (KD, 254 nM), Bik (KD, 190 nM), and Bad (KD, 258 nM) were bound with lower affinities. In contrast, Hrk (KD, 1487 nM) and Noxa (KD, 1575 nM) were engaged with only modest affinities. Furthermore, both Bak (KD, 29 nM) and Bax (KD, 26 nM) (Fig. 1B) were bound with high affinity, whereas Bok was bound with only weak affinity (KD, 6905 nM). These results establish A179L as the first panproapoptotic Bcl-2 binding protein (34).
FIG 1.
A179L interactions with peptides from proapoptotic Bcl-2 family members. The affinity of recombinant A179L for BH3 motif peptides (26-mers, except for a Bax 28-mer and a Bid 34-mer) was assessed using isothermal titration calorimetry (ITC). Raw heats of titration for A179L:Bid BH3 (A), A179L:Bax BH3 (B), and A179L:Noxa BH3 (C) interactions.
TABLE 1.
Isothermal titration calorimetry measurements of interactions of A179L with BH3 motif peptides from prosurvival Bcl-2 proteins
| Peptide |
KD (nM)a of peptide interaction with: |
|
|---|---|---|
| WT A179L | A179L G85A | |
| Bad | 258 ± 65 | NBb |
| Bid | 26 ± 4 | NB |
| Bim | 6 ± 5 | NB |
| Bik | 190 ± 8 | NB |
| Bmf | 254 ± 6 | NB |
| Hrk | 1,487 ± 335 | NB |
| Noxa | 1,575 ± 372 | NB |
| Puma | 31 ± 10 | 201 ± 3 |
| Bak | 29 ± 11 | NB |
| Bax | 26 ± 11 | NB |
| Bok | 6,905 ± 335 | NB |
| Beclin-1 | 1,985 ± 350 | NB |
Values are means ± SD.
NB, no binding observed.
To understand the structural basis for the promiscuity of A179L, we determined the crystal structures of A179L bound to peptides spanning the Bid and Bax BH3 motifs (Fig. 2A and D; Table 2). As expected, A179L adopts a Bcl-2 fold comprising 8 α-helices that form a globular helical bundle fold. α-Helices 2 to 5 form the canonical ligand binding groove found in prosurvival members of the family that engages the BH3 motif of proapoptotic proteins (4). However, despite the presence of BH3 motif ligands in the A179L binding grove, the region corresponding to α3 is not helical but rather adopts an extended configuration reminiscent of the region corresponding to α3 in Bcl-xL (35). Clear and continuous electron density is observed for residues 9 to 143 for A179L:Bid and 2 to 143 for A179L:Bax. A DALI analysis revealed that among all Bcl-2 proteins, complexes of Bcl-2 with Bax BH3 (Fig. 2E; PDB ID 2XA0; Fig. 3) (36) and Bcl-xL with Bid BH3 (Fig. 2B; PDB ID 4QVE; Fig. 3) (37) are most similar to A179L, with root mean square deviation (RMSD) values of 1.7 and 2.0 Å over 125 and 127 Cα atoms, respectively, and sequence identities of 23% in both cases. The structurally closest viral Bcl-2 homolog to A179L is the EBV BHRF1:Bim BH3 complex (Fig. 2F; PDB ID 2V6Q; Fig. 3), with RMSD values of 2.8 Å for both A179L complexes over 125 Cα atoms and a sequence identity of 16%.
FIG 2.
Crystal structures of A179L:Bid BH3, A179L:Bax BH3, and comparison with other BH3-complexes of Bcl-2 proteins. The view in all panels is into the hydrophobic binding groove formed by helices α2 to α5. (A) A179L (green) in complex with the Bid BH3 motif (magenta). A179L helices are labeled α1 to α8. (B) Bcl-xL (sand) in complex with the Bid BH3 motif (magenta) (37). (C) Variola virus F1L (cyan) in complex with Bid BH3 motif (magenta) (20). (D) A179L (green) in complex with the Bax BH3 motif (yellow). (E) Bcl-2 (brown) in complex with Bax BH3 motif (yellow) (36). (F) EBV BHRF1 (blue) in complex with Bim BH3 (orange) (40).
TABLE 2.
Crystallographic data collection and refinement statisticsa
| Characteristic (unit) | Native A179L:Bid BH3 | I3C A179L:Bax BH3 | Native A179L:Bax BH3 |
|---|---|---|---|
| Data collection | |||
| Space group | C2 | P61 | P61 |
| Cell dimensions | |||
| a, b, c (Å) | 67.76, 48.78, 56.54 | 97.13, 97.13, 39.85 | 94.13, 94.13, 40.83 |
| α, β, γ (°) | 90, 104.98, 90 | 90, 90, 120 | 90, 90, 120 |
| Wavelength (Å) | 0.9537 | 1.4586 | 0.9537 |
| Resolution (Å) | 39.11–2.6 (2.693–2.6) | 36.51–2.92 (2.99–2.92) | 36.51–2.5 (2.55–2.5) |
| Rsym or Rmerge | 0.073 (0.353) | 0.120 (0.749) | 0.050 (1.22) |
| I/σI | 8.0 (2.5) | 20.1 (3.5) | 21.1 (1.6) |
| Completeness (%) | 92.2 (94.0) | 99.9 (99.7) | 100 (100) |
| Redundancy | 1.9 (1.9) | 19.1 (19.1) | 6.8 (6.7) |
| Refinement | |||
| Resolution (Å) | 39.11–2.6 (2.693–2.6) | 36.51–2.4 (2.488–2.5) | |
| No. of reflections | 9,758 | 7,293 | |
| Rwork/Rfree | 0.193/0.254 | 0.234/0.263 | |
| No. of atoms | |||
| Protein | 1,366 | 1,316 | |
| Ligand/ion | 5 | 0 | |
| Water | 32 | 15 | |
| No. of B factors | |||
| Protein | 37.8 | 92.4 | |
| Ligand/ion | 59.7 | 0.0 | |
| Water | 36.4 | 61.1 | |
| RMSD | |||
| Bond lengths (Å) | 0.013 | 0.008 | |
| Bond angles (°) | 1.47 | 0.92 |
Values in parentheses are for the highest-resolution shell.
FIG 3.
Sequence alignment of A179L with Bcl-2 family members. The alignment was generated using MUSCLE with sequences from ASFV A179L (Uniprot accession number P42485), EBV BHRF1 (P03182), and human Bcl-xL (Q07817) and Bcl-2 (P10415).
A179L:BH3 motif interactions.
The Bid BH3 peptide binds into a surface groove formed by helices α2 to -5 of A179L (Fig. 2A), in a configuration similar to that previously observed for mammalian prosurvival Bcl-2 members such as Bcl-xL (Fig. 2B) or the unrelated viral Bcl-2 protein variola virus F1L (Fig. 2C) (20). Bax BH3 binds to A179L in an equivalent manner (Fig. 2D), and the two complexes superimpose with an RMSD of only 0.4 Å over the entire A179L backbone, which indicates their similarity.
In the A179L:Bid BH3 complex (Fig. 4, left panel), Bid utilizes the four canonical hydrophobic residues I83, L87, I90, and M94 to engage the A179L ligand binding groove. Furthermore, the conserved ionic interaction between proapoptotic BH3 motifs and prosurvival Bcl-2 proteins is formed by A179L R86 and Bid D92 is also present. An additional ionic interaction is formed by A179L E76 and Bid R81. These interactions are supplemented by hydrogen bonds between A179L N83 and Bid D92 as well as Y96 and between A179L Y46 and the main chain amide group of Bid L87.
FIG 4.
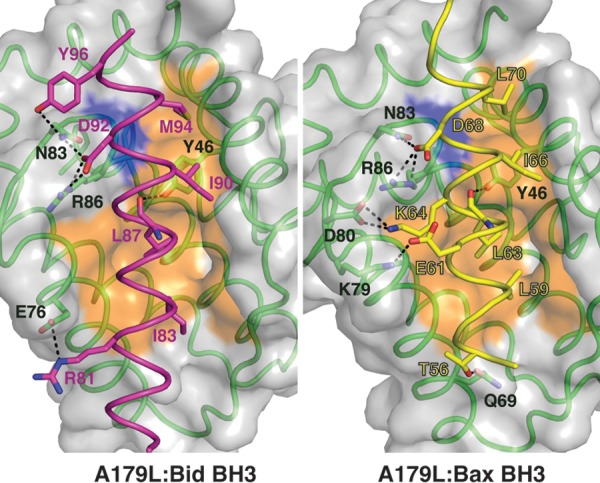
Detailed views of the A179L:BH3 peptide interfaces. (Left) The A179L surface, backbone, and floor of the binding groove are shown in gray, green, and orange, respectively, while Bid BH3 is shown in cyan. The four key hydrophobic residues of Bid (I83, L87, I90, M94) protruding into the binding groove and the conserved salt bridge formed by A179L R86 and Bid D92 are labeled, as well as residues involved in additional ionic interactions and hydrogen bonds. (Right) The A179L surface, backbone, and floor of the binding groove are shown in gray, green, and orange, respectively, while Bid BH3 is shown in yellow. The four key hydrophobic residues of Bid (L59, L63, I66, L70) protruding into the binding groove and the conserved salt bridge formed by A179L R86 and Bid D68 are labeled, as well as residues involved in additional ionic interactions and hydrogen bonds.
In the A179L:Bax BH3 complex (Fig. 4, right panel), the four hydrophobic interactions are contributed by L59, L63, I66, and L70 of the BH3 motif, whereas the salt bridge is formed by A179L R86 and Bax D68. Furthermore, the hydrogen bond between A179L Y46 and the main chain of Bax L63 as well as A179L N83 and Bax D68 previously observed in the A179L:Bid complex are also present in the form of A179L Y46:Bid L87 as well as A179L N83 and Bid D92. However, several interactions distinguish the A179L:Bax complex from the Bid BH3 complex. In A179L:Bax BH3, two additional ionic interactions are formed by A179L D80 and Bax K64 as well as A179L K79 and Bax E61. In addition, A179L Q69 and Bax T56 also form a hydrogen bond.
A key feature of A179L engagement with BH3 motif peptides is the accommodation of the conserved Gly in the BH3 motif. This is achieved via A179L G85, which forms part of the ligand binding groove formed by helix α4. This Gly is conserved across many prosurvival Bcl-2 proteins, as its lack of a side chain enables the tight packing with the Gly of the incoming BH3 motif (4). Mutation of A179L G85 to Ala severely compromises the ability of A179L to engage proapoptotic Bcl-2, with A179L G85 binding Puma BH3 with only 200 nM affinity, an ∼7-fold reduction compared to the wild type, and not displaying affinity for any other BH3 motif peptide tested (Table 1).
DISCUSSION
Many large DNA viruses utilize immune modulatory factors to subvert Bcl-2-mediated apoptosis, often by encoding structural and functional mimics of Bcl-2 (8). Although these viral proteins vary in sequence significantly, they are strikingly similar on a structural level (4). However, despite the high level of structural similarity, the detailed mechanisms of action vary considerably across the different viruses examined to date (4), with myxomavirus M11L-mediated inhibition being primarily mediated by Bax/Bak sequestration (19), whereas vaccinia virus F1L requires only Bim sequestration (38) and EBV BHRF1 targets Bim (39) and Bak (40).
In this study, we have shown that A179L is able to engage all major mammalian proapoptotic Sus scrofa BH3-only proteins as well as Bak and Bax. This level of promiscuity across proapoptotic Bcl-2 is unusual and not seen elsewhere in any other prosurvival Bcl-2 protein identified to date (4). Previous studies have identified a subset of proapoptotic interaction partners by using yeast-two-hybrid assays; however, the biological significance of each interaction remains to be clarified (33). Considering the striking promiscuity of A179L, it is interesting to speculate as to whether or not the host selection by both arthropod vectors, such as the soft tick, and host mammals, including the pig (41), may have played a role in shaping A179L into an ultrapromiscuous Bcl-2 protein that is able to subvert two very different intrinsic apoptosis machineries (42).
Among the mammalian prosurvival Bcl-2 proteins, Bcl-2, Bcl-xL, and Bcl-w are able to bind a substantial number of BH3-only proteins but do not engage Noxa (43, 44). Furthermore, only Bcl-xL from this group of three molecules engages Bak with high affinity. In contrast, mammalian A1 (45) and Mcl-1 do not bind Bad BH3 but do engage Noxa with high affinity (43). Bcl-b harbors an unusually restricted binding BH3 motif profile among the mammalian prosurvival Bcl-2 proteins and is able to bind only Bim and Bax (34). Among the viral Bcl-2 proteins, the poxviral SPPV14 is promiscuous (25), binding all BH3-only proteins except Bad, Bik, and Noxa, whereas herpesviral M11 is also highly promiscuous but does not bind Bad and Bik (46).
To understand the structural basis for this observed promiscuity of A179L, we determined the crystal structures of A179L bound to peptides that span the BH3 motifs of Sus scrofa (boar) Bid and Bax BH3. Our analysis revealed that A179L utilizes the canonical ligand binding groove found in all other prosurvival Bcl-2 proteins and that similar to other Bcl-2-BH3 interactions, A179L engages four key hydrophobic residues on the same face of an α-helix of the BH3 motif interacting in hydrophobic pockets. In addition, a highly conserved ionic interaction between the carboxylate of an Asp from the BH3 motif and the guanidinium ion of Arg86 from A179L is present. However, despite the presence of key conserved structural hallmarks of Bcl-2-BH3 interaction, we identified a number of features that distinguish A179L ligand engagement from other members of the family. These include additional ionic interactions formed by A179L E76 and Bid R81 and by A179L D80 and Bax K64 as well as A179L K79 and Bax E61, which are likely to provide considerable specificity. Interestingly, the A179L E76:ssBid R81 ionic interaction is also found in the variola virus F1L:Bid complex in the form of F1L E143:Bid R83 (20), unlike the Bcl-xL, Mcl-1, A1, and Bcl-w complexes with Bid, where this interaction is absent (37, 45, 47, 48). In contrast, the two additional ionic interactions present in the A179L:Bax BH3 complex formed by D80:K64 and K79:E61 are also conserved in the Bcl-2:Bax BH3 complex as D140:R64 and R139:E61 (36), whereas in the poxviral DPV022:Bax BH3 complex (26) and Mcl-1:Bax complex they are absent (49). In Bcl-xL:Bax BH3, only one of the two ionic interactions is present in the form of Bax K64 with Bcl-xL E129 and D133 (49).
In addition to specific interactions, the overall configuration of the A179L ligand binding groove also suggests a possible underlying reason for its broad specificity. The region corresponding to the α3 helix in Bcl-2 proteins that forms one side of the A179L ligand binding groove is not helical and adapts an extended configuration. The lack of rigid secondary structure suggests that this side of the binding groove harbors some flexibility, a likely requirement to be able to engage such a broad range of prodeath Bcl-2 ligands. This is similar to what is observed in several mammalian prosurvival Bcl-2 proteins, in particular Bcl-xL and Bcl-2 (Fig. 2B and E), that engage a large number of proapoptotic Bcl-2 proteins. In contrast, the structures of viral Bcl-2 proteins bound to ligands typically display well-ordered and highly structured α3 helices, as seen in BHRF1 (40) (Fig. 2F), F1L (20, 38) (Fig. 2C), M11L (19), and DPV022 (26), which all have much more restricted proapoptotic Bcl-2 protein binding profiles and may thus require less intrinsic flexibility within the binding groove to allow high-affinity interactions with a smaller subset of prodeath ligands.
To validate the binding mode of A179L and BH3 motif ligands and to probe structural constraints to ligand binding, we also examined the BH3 motif binding of an A179L mutant, G85A. A179L G85A completely lost affinity for all BH3 motif peptides tested except Puma BH3, which was bound with 200 nM affinity, representing an ∼7-fold reduction in affinity compared to wild-type A179L. Interestingly, a previous study reported that an A179L G85A mutant lost all prosurvival activity in transfected human myeloid leukemia K562 cells, suggesting that inhibition of Puma is not a relevant mechanism of action for A179L-mediated inhibition of apoptosis (31).
We have identified A179L as a highly promiscuous prodeath Bcl-2 binding protein that is able to engage BH3 motif peptides of the proapoptotic Bcl-2 family members in vitro and defined the structural basis of prodeath Bcl-2 binding by determining the crystal structures of A179L bound to both Bid and Bax BH3 motifs. These findings provide a mechanistic understanding for the potent antiapoptotic activity of A179L and serve as a platform for more-detailed investigations into the role of A179L during apoptosis inhibition and the importance of individual proapoptotic Bcl-2 members during suppression of apoptosis during ASFV infection and viral persistence.
MATERIALS AND METHODS
Protein expression and purification.
Synthetic cDNA encoding codon-optimized A179L (Uniprot accession number P42485) lacking 31 C-terminal residues (referred to as A179LΔC31) was cloned into the bacterial expression vector pGEX-6p-1 (Bioneer). Recombinant A179LΔC31, a construct with the 31 C-terminal residues truncated, was expressed in BL21 star cells using the autoinduction method (50) for 24 h at 25°C with shaking.
The cells were harvested by centrifugation at 6,000 rpm (JLA 9.1000 rotor, Beckman Coulter Avanti J-E) for 20 min and resuspended in 50 ml lysis buffer A (50 mM Tris [pH 8.5], 150 mM NaCl, and 1 mM EDTA) supplemented with lysozyme. The cells were lysed using one cycle at a pressure of 35 kPa in a Constant Systems Benchtop cell disrupter (Constant Systems Ltd.). The lysate was transferred into SS34 tubes for further centrifugation at 16,000 rpm (JA-25.50 rotor, Beckman Coulter Avanti J-E) for 20 min. The supernatant was loaded onto a 2-ml glutathione Sepharose 4B resin column (GE Healthcare) equilibrated with buffer A. After sample application, the column was washed with five column volumes of buffer A followed by HRV 3C protease cleavage on the column overnight at 4°C.
The column was washed with 5 column volumes of buffer A to remove the liberated target protein, and the protein was concentrated using a centrifugal concentrator with a 10-kDa molecular mass cutoff (Amicon Ultra 15) to a final volume of 1 ml. Concentrated A179L was subjected to size exclusion chromatography using a Superdex S200 10/300 column mounted on an ÄKTApure system (GE Healthcare) equilibrated in 25 mM HEPES (pH 7.5)–150 mM NaCl, where it eluted as a single peak. The final sample purity was estimated to be >95% based on SDS-PAGE analysis. Appropriate fractions were pooled and concentrated using a centrifugal concentrator with a 10-kDa molecular mass cutoff (Amicon Ultra 15) to a final concentration of 5.3 mg/ml.
Differential scanning fluorimetry.
Thermal melt analyses of wild-type A179L and A179L G85A were performed as previously described (51) using a Bio-Rad CFX96 reverse transcription (RT)-PCR instrument and analyzed using the program Meltdown (52).
Measurement of dissociation constants.
Binding affinities were measured using a MicroCal iTC200 system (GE Healthcare) at 25°C using A179L in 25 mM HEPES (pH 7.5)–150 mM NaCl at a final concentration of 20 μM. BH3 motif peptide ligands were used at a concentration of 200 μM and titrated using 19 injections of 2.0 μl of ligand. All affinity measurements were performed in triplicate. Protein concentrations were measured using a Nanodrop UV spectrophotometer (Thermo Scientific) at a wavelength of 280 nm. Peptide concentrations were calculated based on the dry peptide weight after synthesis. The BH3 motif peptides used were commercially synthesized and were purified to a final purity of 95% (GenScript). The following sequences were used for peptide synthesis: ss_Bim, DMRPEIWIAQELRRIGDEFNAYYPRR (Uniprot accession code C1KGB6; residues 139 to 162); ss_Puma, EEQWAREIGAQLRRMADDLNALYERR (accession code XM_013998532.1; residues 179 to 204); ss_Bik, SSEAPHHLAMQLASIADDLELRLLLP (accession code XM_005663754.2; residues 59 to 74); ss_Hrk, RSSAAQLTAARLKAIGDELHQRTMWR (accession code XM_003359104.3; residues 26 to 51); ss_Bax, VPQDASTKKLSECLKRIGDELDSNMELQ (Uniprot accession code F1RIQ4; residues 50 to 77); ss_Bak, PSSTMGQVGRQLAIIGDDINRRYDSE (Uniprot accession code F1RZR2; residues 69 to 94); ss_Bid, SESQEAVIRDIARHLARIGDRMEYGIRPGLVDSL (Uniprot accession code Q4JHS0; residues 73 to 106); ss_Bad, ILWAAQRYGRELRRMSDEFQGSFKKG (Uniprot accession code F1RQ09; residues 112 to 137); ss_Bmf, QHRAEVQIAGKLQCIADQFHRLHMQQ (Uniprot accession code M3VH85; residues 125 to 150); ss_Noxa, PPDPEVECAIQFRRIGDKLNFRQKLL (Uniprot accession code F1SMU1; residues 76 to 102).
Crystallization and data collection.
Complexes of A179L with Bid BH3 and Bax BH3 were prepared as previously described (53). Briefly, A179L complexes were reconstituted by adding ss_Bid BH3 motif or ss_Bax BH3 motif peptides at a 1:1.25 molar ratio to A179L. The reconstituted complexes were concentrated to 5 mg ml−1 using a 3-kDa molecular mass cutoff centrifugal concentrator (Millipore), flash-cooled, and stored under liquid nitrogen. High-throughput sparse matrix screening was carried out using 96-well sitting-drop trays (Swissci) and the vapor-diffusion method at 20°C at the CSIRO Collaborative Crystallization Centre (C3), Melbourne, Australia. The initial crystallization conditions used were from commercially available screening kits (Shotgun Screen, PACT, and JCSG from C3 CSIRO).
Crystals of A179L:Bax BH3 were obtained at 5 mg ml−1 using the sitting-drop method at 20°C in 1.6 M ammonium sulfate–0.1 M sodium acetate (pH 4.2), using streak seeding. The crystals were flash-cooled at −173°C in mother liquor supplemented with 20% (vol/vol) ethylene glycol. The A179L:Bax BH3 complex formed single hexagonal crystals belonging to space group P61 in the hexagonal crystal system.
Native diffraction data for A179L:BaxBH3 were collected on the MX2 beamline at the Australian Synchrotron using an ADSC Quantum 315r CCD detector (Area Detector Systems Corporation, Poway, CA, USA) with an oscillation range of 1.0° per frame and a wavelength of 0.9537 Å. Diffraction data were integrated using XDS (54) and scaled using Aimless (55). A heavy atom derivative was obtained by soaking crystals in mother liquor supplemented with 0.15 M I3C (56) and 20% ethylene glycol for 120 s prior to flash-cooling. Derivative data were collected at a wavelength of 1.4586 Å, integrated using XDS (54), and scaled using Aimless (55). The structure was phased by single-wavelength anomalous diffraction (SAD) using iodine ions. Five sites were located using Shelx (57) and phased using AutoSol in Phenix (58, 59) with a Figure of Merit of 0.303. The initial map was of insufficient quality for automatic chain tracing; consequently, the A179L:Bax complex was built manually using Coot (60) and refined using Phenix (58) and Refmac (61). A179L:Bax BH3 crystals contained one chain of A179L and one chain of ss_Bax BH3 in the asymmetry unit, with a calculated solvent content of 52.8%, and the final model was refined to an Rwork/Rfree of 21.0/25.3 with 96.8% of residues in the favored region of the Ramachandran plot and no outliers.
Crystals of A179L:Bid BH3 were obtained at a protein concentration of 5 mg ml−1 using the sitting-drop method at 20°C in 27.5% (wt/vol) polyethylene glycol (PEG) 4000, 0.11 M ammonium sulfate, and 0.1 M sodium cacodylate (pH 5.6). The crystals were flash-cooled at −173°C in mother liquor supplemented with 20% (vol/vol) ethylene glycol. The A179L:Bax BH3 complex formed clusters of hexagonal plate-like crystals belonging to space group C2 in the monoclinic crystal system.
Native diffraction data were acquired for the complex A179LBid on the MX2 beamline at the Australian Synchrotron and using an ADSC Quantum 315r CCD detector (Area Detector Systems Corporation, Poway, CA, USA) with an oscillation range of 1.0° per frame and a wavelength of 0.9537 Å. Diffraction data were integrated using XDS (54) and scaled using Aimless (55). The structure was phased by molecular replacement using Phaser (62) with the previously determined structure of A179L:Bax BH3 as a search model. The final A179L:Bid complex was built manually and refined as for A179L:BaxBH3. A179L:Bid BH3 crystals contained one chain of A179L and one chain of ss_Bid BH3 in the asymmetry unit, with a calculated solvent content of 42.7%, and the final model was refined to an Rwork/Rfree of 19.3/25.4 with 98.2% of residues in the favored region of the Ramachandran plot and no outliers. Details of the data collection and refinement statistics are summarized in Table 2. All images were generated using the PyMOL Molecular Graphics system, version 1.8, from Schrödinger, LLC. All software was accessed using the SBGrid suite (63). All raw diffraction images were deposited on the SBGrid Data Bank (64) using their PDB accession numbers.
Accession number(s).
Coordinate files have been deposited in the Protein Data Bank under accession codes 5UA4 and 5UA5.
ACKNOWLEDGMENTS
We thank staff members at the MX beamlines at the Australian Synchrotron for help with X-ray data collection, the CSIRO C3 Collaborative Crystallization Centre for assistance with crystallization, and the Comprehensive Proteomics Platform at La Trobe University for core instrument support.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/JVI.02338-16.
REFERENCES
- 1.Vaux DL, Haecker G, Strasser A. 1994. An evolutionary perspective on apoptosis. Cell 76:777–779. doi: 10.1016/0092-8674(94)90350-6. [DOI] [PubMed] [Google Scholar]
- 2.Czabotar PE, Lessene G, Strasser A, Adams JM. 2014. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 3.Kvansakul M, Hinds MG. 2014. The structural biology of BH3-only proteins. Methods Enzymol 544:49–74. doi: 10.1016/B978-0-12-417158-9.00003-0. [DOI] [PubMed] [Google Scholar]
- 4.Kvansakul M, Hinds MG. 2013. Structural biology of the Bcl-2 family and its mimicry by viral proteins. Cell Death Dis 4:e909. doi: 10.1038/cddis.2013.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czabotar PE, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD, Lee EF, Yao S, Robin AY, Smith BJ, Huang DC, Kluck RM, Adams JM, Colman PM. 2013. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Shamas-Din A, Brahmbhatt H, Leber B, Andrews DW. 2011. BH3-only proteins: orchestrators of apoptosis. Biochim Biophys Acta 1813:508–520. doi: 10.1016/j.bbamcr.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Youle RJ, Strasser A. 2008. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 8.Galluzzi L, Brenner C, Morselli E, Touat Z, Kroemer G. 2008. Viral control of mitochondrial apoptosis. PLoS Pathog 4:e1000018. doi: 10.1371/journal.ppat.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White E, Sabbatini P, Debbas M, Wold WS, Kusher DI, Gooding LR. 1992. The 19-kilodalton adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumor necrosis factor alpha. Mol Cell Biol 12:2570–2580. doi: 10.1128/MCB.12.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson S, Huen D, Rowe M, Dawson C, Johnson G, Rickinson A. 1993. Epstein-Barr virus-coded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc Natl Acad Sci U S A 90:8479–8483. doi: 10.1073/pnas.90.18.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiou SK, Tseng CC, Rao L, White E. 1994. Functional complementation of the adenovirus E1B 19-kilodalton protein with Bcl-2 in the inhibition of apoptosis in infected cells. J Virol 68:6553–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altmann M, Hammerschmidt W. 2005. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol 3:e404. doi: 10.1371/journal.pbio.0030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartlett N, Symons JA, Tscharke DC, Smith GL. 2002. The vaccinia virus N1L protein is an intracellular homodimer that promotes virulence. J Gen Virol 83:1965–1976. doi: 10.1099/0022-1317-83-8-1965. [DOI] [PubMed] [Google Scholar]
- 14.Fischer SF, Ludwig H, Holzapfel J, Kvansakul M, Chen L, Huang DC, Sutter G, Knese M, Hacker G. 2006. Modified vaccinia virus Ankara protein F1L is a novel BH3-domain-binding protein and acts together with the early viral protein E3L to block virus-associated apoptosis. Cell Death Differ 13:109–118. doi: 10.1038/sj.cdd.4401718. [DOI] [PubMed] [Google Scholar]
- 15.Graham KA, Opgenorth A, Upton C, McFadden G. 1992. Myxoma virus M11L ORF encodes a protein for which cell surface localization is critical in manifestation of viral virulence. Virology 191:112–124. doi: 10.1016/0042-6822(92)90172-L. [DOI] [PubMed] [Google Scholar]
- 16.Wasilenko ST, Stewart TL, Meyers AF, Barry M. 2003. Vaccinia virus encodes a previously uncharacterized mitochondrial-associated inhibitor of apoptosis. Proc Natl Acad Sci U S A 100:14345–14350. doi: 10.1073/pnas.2235583100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooray S, Bahar MW, Abrescia NG, McVey CE, Bartlett NW, Chen RA, Stuart DI, Grimes JM, Smith GL. 2007. Functional and structural studies of the vaccinia virus virulence factor N1 reveal a Bcl-2-like antiapoptotic protein. J Gen Virol 88:1656–1666. doi: 10.1099/vir.0.82772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douglas AE, Corbett KD, Berger JM, McFadden G, Handel TM. 2007. Structure of M11L: a myxoma virus structural homolog of the apoptosis inhibitor, Bcl-2. Protein Sci 16:695–703. doi: 10.1110/ps.062720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kvansakul M, van Delft MF, Lee EF, Gulbis JM, Fairlie WD, Huang DC, Colman PM. 2007. A structural viral mimic of prosurvival Bcl-2: a pivotal role for sequestering proapoptotic Bax and Bak. Mol Cell 25:933–942. doi: 10.1016/j.molcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Marshall B, Puthalakath H, Caria S, Chugh S, Doerflinger M, Colman PM, Kvansakul M. 2015. Variola virus F1L is a Bcl-2-like protein that unlike its vaccinia virus counterpart inhibits apoptosis independent of Bim. Cell Death Dis 6:e1680. doi: 10.1038/cddis.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gubser C, Bergamaschi D, Hollinshead M, Lu X, van Kuppeveld FJ, Smith GL. 2007. A new inhibitor of apoptosis from vaccinia virus and eukaryotes. PLoS Pathog 3:e17. doi: 10.1371/journal.ppat.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banadyga L, Gerig J, Stewart T, Barry M. 2007. Fowlpox virus encodes a Bcl-2 homologue that protects cells from apoptotic death through interaction with the proapoptotic protein Bak. J Virol 81:11032–11045. doi: 10.1128/JVI.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westphal D, Ledgerwood EC, Hibma MH, Fleming SB, Whelan EM, Mercer AA. 2007. A novel Bcl-2-like inhibitor of apoptosis is encoded by the parapoxvirus ORF virus. J Virol 81:7178–7188. doi: 10.1128/JVI.00404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banadyga L, Lam SC, Okamoto T, Kvansakul M, Huang DC, Barry M. 2011. Deerpox virus encodes an inhibitor of apoptosis that regulates Bak and Bax. J Virol 85:1922–1934. doi: 10.1128/JVI.01959-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto T, Campbell S, Mehta N, Thibault J, Colman PM, Barry M, Huang DC, Kvansakul M. 2012. Sheeppox virus SPPV14 encodes a Bcl-2-like cell death inhibitor that counters a distinct set of mammalian proapoptotic proteins. J Virol 86:11501–11511. doi: 10.1128/JVI.01115-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burton DR, Caria S, Marshall B, Barry M, Kvansakul M. 2015. Structural basis of Deerpox virus-mediated inhibition of apoptosis. Acta Crystallogr D Biol Crystallogr 71:1593–1603. doi: 10.1107/S1399004715009402. [DOI] [PubMed] [Google Scholar]
- 27.Costard S, Mur L, Lubroth J, Sanchez-Vizcaino JM, Pfeiffer DU. 2013. Epidemiology of African swine fever virus. Virus Res 173:191–197. doi: 10.1016/j.virusres.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez JM, Moreno LT, Alejo A, Lacasta A, Rodriguez F, Salas ML. 2015. Genome sequence of African swine fever virus BA71, the virulent parental strain of the nonpathogenic and tissue-culture adapted BA71V. PLoS One 10:e0142889. doi: 10.1371/journal.pone.0142889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanez RJ, Rodriguez JM, Nogal ML, Yuste L, Enriquez C, Rodriguez JF, Vinuela E. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- 30.Brun A, Rivas C, Esteban M, Escribano JM, Alonso C. 1996. African swine fever virus gene A179L, a viral homologue of bcl-2, protects cells from programmed cell death. Virology 225:227–230. doi: 10.1006/viro.1996.0592. [DOI] [PubMed] [Google Scholar]
- 31.Revilla Y, Cebrian A, Baixeras E, Martinez C, Vinuela E, Salas ML. 1997. Inhibition of apoptosis by the African swine fever virus Bcl-2 homologue: role of the BH1 domain. Virology 228:400–404. doi: 10.1006/viro.1996.8395. [DOI] [PubMed] [Google Scholar]
- 32.Hernaez B, Cabezas M, Munoz-Moreno R, Galindo I, Cuesta-Geijo MA, Alonso C. 2013. A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Curr Mol Med 13:305–316. doi: 10.2174/156652413804810736. [DOI] [PubMed] [Google Scholar]
- 33.Galindo I, Hernaez B, Diaz-Gil G, Escribano JM, Alonso C. 2008. A179L, a viral Bcl-2 homologue, targets the core Bcl-2 apoptotic machinery and its upstream BH3 activators with selective binding restrictions for Bid and Noxa. Virology 375:561–572. doi: 10.1016/j.virol.2008.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rautureau GJ, Yabal M, Yang H, Huang DC, Kvansakul M, Hinds MG. 2012. The restricted binding repertoire of Bcl-B leaves Bim as the universal BH3-only prosurvival Bcl-2 protein antagonist. Cell Death Dis 3:e443. doi: 10.1038/cddis.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Dai S, Zhu Y, Marrack P, Kappler JW. 2003. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity 19:341–352. doi: 10.1016/S1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 36.Ku B, Liang C, Jung JU, Oh BH. 2011. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res 21:627–641. doi: 10.1038/cr.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajan S, Choi M, Baek K, Yoon HS. 2015. Bh3 induced conformational changes in Bcl-Xl revealed by crystal structure and comparative analysis. Proteins 83:1262–1272. doi: 10.1002/prot.24816. [DOI] [PubMed] [Google Scholar]
- 38.Campbell S, Thibault J, Mehta N, Colman PM, Barry M, Kvansakul M. 2014. Structural insight into BH3 domain binding of vaccinia virus antiapoptotic F1L. J Virol 88:8667–8677. doi: 10.1128/JVI.01092-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desbien AL, Kappler JW, Marrack P. 2009. The Epstein-Barr virus Bcl-2 homolog, BHRF1, blocks apoptosis by binding to a limited amount of Bim. Proc Natl Acad Sci U S A 106:5663–5668. doi: 10.1073/pnas.0901036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kvansakul M, Wei AH, Fletcher JI, Willis SN, Chen L, Roberts AW, Huang DC, Colman PM. 2010. Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1. PLoS Pathog 6:e1001236. doi: 10.1371/journal.ppat.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleiboeker SB, Scoles GA. 2001. Pathogenesis of African swine fever virus in Ornithodoros ticks. Anim Health Res Rev 2:121–128. [PubMed] [Google Scholar]
- 42.Tanaka M, Liao M, Zhou J, Nishikawa Y, Xuan X, Fujisaki K. 2007. Molecular cloning of two caspase-like genes from the hard tick Haemaphysalis longicornis. J Vet Med Sci 69:85–90. doi: 10.1292/jvms.69.85. [DOI] [PubMed] [Google Scholar]
- 43.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. 2005. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 44.Kvansakul M, Hinds MG. 2015. The Bcl-2 family: structures, interactions and targets for drug discovery. Apoptosis 20:136–150. doi: 10.1007/s10495-014-1051-7. [DOI] [PubMed] [Google Scholar]
- 45.Smits C, Czabotar PE, Hinds MG, Day CL. 2008. Structural plasticity underpins promiscuous binding of the prosurvival protein A1. Structure 16:818–829. doi: 10.1016/j.str.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 46.Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X, Kim KS, Jung JU, Oh BH. 2008. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog 4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denisov AY, Chen G, Sprules T, Moldoveanu T, Beauparlant P, Gehring K. 2006. Structural model of the BCL-w-BID peptide complex and its interactions with phospholipid micelles. Biochemistry 45:2250–2256. doi: 10.1021/bi052332s. [DOI] [PubMed] [Google Scholar]
- 48.Liu Q, Moldoveanu T, Sprules T, Matta-Camacho E, Mansur-Azzam N, Gehring K. 2010. Apoptotic regulation by MCL-1 through heterodimerization. J Biol Chem 285:19615–19624. doi: 10.1074/jbc.M110.105452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czabotar PE, Lee EF, Thompson GV, Wardak AZ, Fairlie WD, Colman PM. 2011. Mutation to Bax beyond the BH3 domain disrupts interactions with prosurvival proteins and promotes apoptosis. J Biol Chem 286:7123–7131. doi: 10.1074/jbc.M110.161281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 51.Seabrook SA, Newman J. 2013. High-throughput thermal scanning for protein stability: making a good technique more robust. ACS Comb Sci 15:387–392. doi: 10.1021/co400013v. [DOI] [PubMed] [Google Scholar]
- 52.Rosa N, Ristic M, Seabrook SA, Lovell D, Lucent D, Newman J. 2015. Meltdown: a tool to help in the interpretation of thermal melt curves acquired by differential scanning fluorimetry. J Biomol Screen 20:898–905. doi: 10.1177/1087057115584059. [DOI] [PubMed] [Google Scholar]
- 53.Kvansakul M, Czabotar PE. 2016. Preparing samples for crystallization of Bcl-2 family complexes. Methods Mol Biol 1419:213–229. doi: 10.1007/978-1-4939-3581-9_16. [DOI] [PubMed] [Google Scholar]
- 54.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 56.Beck T, Krasauskas A, Gruene T, Sheldrick GM. 2008. A magic triangle for experimental phasing of macromolecules. Acta Crystallogr D Biol Crystallogr 64:1179–1182. doi: 10.1107/S0907444908030266. [DOI] [PubMed] [Google Scholar]
- 57.Sheldrick GM. 2010. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr 66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. 2008. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 62.McCoy AJ. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P. 2013. Collaboration gets the most out of software. Elife 2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyer PA, Socias S, Key J, Ransey E, Tjon EC, Buschiazzo A, Lei M, Botka C, Withrow J, Neau D, Rajashankar K, Anderson KS, Baxter RH, Blacklow SC, Boggon TJ, Bonvin AM, Borek D, Brett TJ, Caflisch A, Chang CI, Chazin WJ, Corbett KD, Cosgrove MS, Crosson S, Dhe-Paganon S, Di Cera E, Drennan CL, Eck MJ, Eichman BF, Fan QR, Ferre-D'Amare AR, Fromme JC, Garcia KC, Gaudet R, Gong P, Harrison SC, Heldwein EE, Jia Z, Keenan RJ, Kruse AC, Kvansakul M, McLellan JS, Modis Y, Nam Y, Otwinowski Z, Pai EF, Pereira PJ, Petosa C, Raman CS, Rapoport TA, Roll-Mecak A, Rosen M, Rudenko G, Schlessinger J, Schwartz T, Shamoo Y, Sondermann H, Tao Y, Tolia N, Tsodikov OV, Westover KD, Wu H, Foster I, Maia F, Gonen T, Kirchhausen T, Crosas M, Slitz P. 2016. Data publication with the structural biology data grid supports live analysis. Nat Commun 7:10882. doi: 10.1038/ncomms10882. [DOI] [PMC free article] [PubMed] [Google Scholar]