ABSTRACT
Linker of nucleoskeleton and cytoskeleton (LINC) complexes connect the nucleus to the cytoskeleton in eukaryotic cells. We previously reported that the overexpression of SUN2, an inner nuclear membrane protein and LINC complex component, inhibits HIV infection between the steps of reverse transcription and nuclear import in a capsid-specific manner. We also reported that SUN2 silencing does not modulate HIV infection in several cell lines. Silencing of SUN2 was recently reported to decrease HIV infection of CD4 T cells, an effect which was suggested to result from modulation of cyclophilin A (CypA)-dependent steps of HIV infection. We confirm here that HIV infection of primary CD4 T cells is compromised in the absence of endogenous SUN2, and we extend these findings to additional viral strains. However, we find that CypA is not required for the decreased infection observed in SUN2-silenced cells and, conversely, that endogenous SUN2 is not required for the well-documented positive modulation of HIV infection by CypA. In contrast, CD4 T cells lacking SUN2 exhibit a considerable defect in proliferative capacity and display reduced levels of activation markers and decreased viability. Additionally, SUN2-silenced CD4 T cells that become infected support reduced levels of viral protein expression. Our results demonstrate that SUN2 is required for the optimal activation and proliferation of primary CD4 T cells and suggest that the disruption of these processes explains the contribution of endogenous SUN2 to HIV infection in primary lymphocytes.
IMPORTANCE Linker of nucleoskeleton and cytoskeleton (LINC) complexes connect the nucleus to the cytoskeleton. We previously reported that the overexpression of the LINC complex protein SUN2 inhibits HIV infection by targeting the viral capsid and blocking infection before the virus enters the nucleus. A recent report showed that the depletion of endogenous SUN2 in primary CD4 T cells results in decreased HIV infection and that this involves cyclophilin A (CypA), a host protein that interacts with the capsid of HIV to promote infection. We confirm that HIV infection is reduced in CD4 T cells lacking SUN2, but we find no role for CypA. Instead, SUN2 silencing results in CD4 T cells with decreased viability and much lower proliferation rates. Our results show that SUN2 is required for optimal CD4 T cell activation and proliferation and explain the reduced level of HIV infection in the absence of SUN2.
KEYWORDS: CD4 T cell, SUN2, human immunodeficiency virus
INTRODUCTION
Linker of nucleoskeleton and cytoskeleton (LINC) complexes connect the nucleus to the cytoskeleton and are evolutionarily conserved in eukaryotes from budding and fission yeasts and Caenorhabditis elegans to D. melanogaster, plants, and mammals (1). The LINC complex is defined by the interaction of KASH domain proteins and SUN domain proteins in the perinuclear space. KASH domain proteins in mammals include several Nesprins, which span the outer nuclear membrane and interact with actin filaments, microfilaments, and microtubules. In mammalian cells, homotrimers of SUN1 or SUN2 interact with KASH domain proteins and with the nuclear lamina and chromatin. This physical connection allows the transmission of forces across the nuclear envelope and plays important roles in numerous cellular processes, including the control of nuclear size and morphology; anchoring and movement of nuclei during syncytium formation, cell polarization, and muscle and neuronal development; cytoskeleton organization; chromosome tethering during mitosis and meiosis; regulation of signaling, cell division, migration, and apoptosis; and DNA repair (1–3). SUN proteins have been implicated in the pathology of certain laminopathies (4), and defects in LINC complex components are responsible for several inheritable human diseases, including Emery-Dreifuss muscular dystrophy, cerebellar ataxia, arthrogryposis, hearing loss, and dilated cardiomyopathy (5, 6).
Little is known about how the LINC complex component SUN2 might act as a cofactor or cellular defense during viral infection. An overexpression screen of putative interferon-stimulated genes (ISGs) found that SUN2 inhibited HIV-1 infection of a T cell line through unknown mechanisms (7). While we and others subsequently found that SUN2 was not induced by interferon in human cells (8, 9), we reported that its overexpression inhibits infection by HIV-1 and HIV-2 in multiple cell lines and in monocyte-derived dendritic cells (MDDCs) (8). This inhibition occurred at the time of or before viral nuclear import and could be circumvented by several naturally occurring HIV strains or by a single point mutation in the viral capsid. We also demonstrated that the host protein and HIV cofactor cyclophilin A (CypA), which modulates HIV infection in many cell types (reviewed in references 10–12), was involved in the inhibition of HIV infection by overexpressed SUN2 (8). In that same study, we found no role for endogenous SUN2 during infection of several adherent cell lines (8). In contrast, Lahaye and colleagues recently reported that endogenous SUN2 acts as a cofactor for HIV infection of primary CD4 T cells and MDDCs (9). Silencing of SUN2 led to a modest decrease in single-round HIV infection, while spreading infection was strongly impacted. Through the use of CypA inhibitors, those authors obtained results suggesting that SUN2 facilitates HIV infection by modulating the positive effects of CypA on the virus. However, the positive effects of CypA on HIV infection have long been known to alter levels of reverse transcripts, 2-long terminal repeat (LTR) circles, and integrated DNA (13–18), whereas levels of viral DNA forms were unchanged in SUN2-depleted cells despite reduced levels of productive infection (9). This raises the possibility that alternative mechanisms might explain the decreased infection of cells depleted of SUN2.
We sought to further understand the contribution of endogenous SUN2 to HIV infection in primary CD4 T cells. While we also observe reduced HIV infection when SUN2 is silenced, we find no evidence for an involvement of CypA. In contrast, SUN2-silenced CD4 T cells exhibit a considerable defect in proliferative capacity and display reduced levels of activation markers and decreased viability. Additionally, SUN2-silenced T cells that become infected express lower levels of viral proteins. Our results indicate that SUN2 is required for the optimal activation and proliferation of primary CD4 T cells and that disruption of these processes likely explains the contribution of endogenous SUN2 to HIV infection in primary lymphocytes.
RESULTS
SUN2 is required for optimal CD4 T cell proliferation.
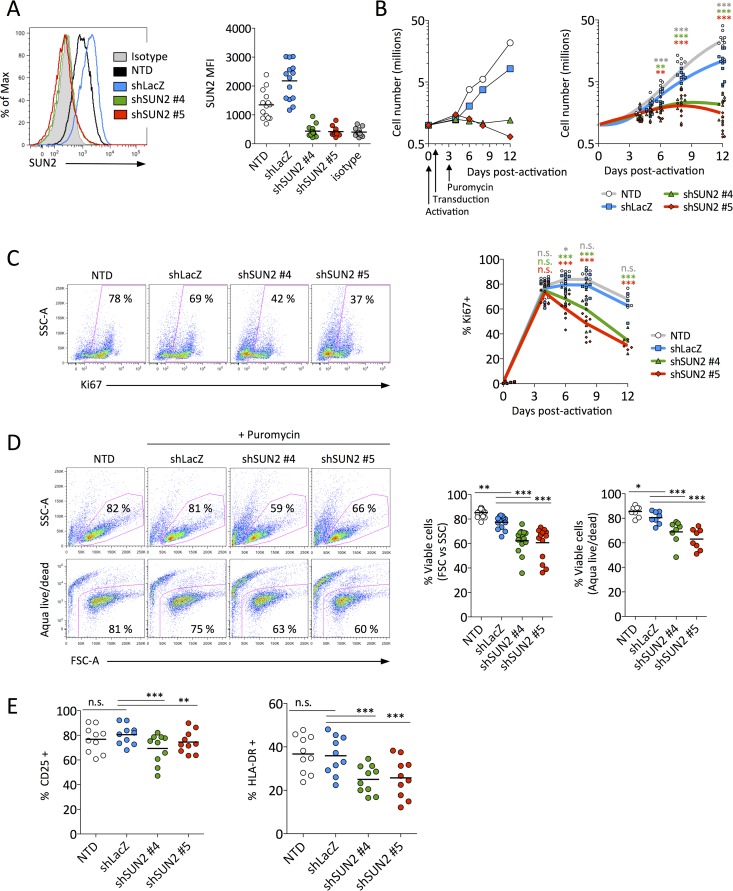
To address the role of endogenous SUN2 during HIV infection of primary CD4 T cells, we first examined the consequences of SUN2 silencing on CD4 T cell health. Freshly isolated CD4 T cells were activated with anti-CD2/CD3/CD28 beads and were then transduced with lentiviral particles encoding short hairpin RNA (shRNA) against two different regions of SUN2 (shSUN2#4 and shSUN2#5) or a control shRNA targeting LacZ (shLacZ), as reported previously (9). SUN2 was routinely undetectable by 5 to 6 days postransduction (Fig. 1A) using an antibody previously validated for the detection of endogenous SUN2 by flow cytometry (8). We observed a substantial decrease in the proliferation rate of SUN2-silenced cells compared to nontransduced (NTD) or control transduced (shLacZ) cells (Fig. 1B); whereas the number of control cells increased by ∼5-fold within a week and by 10- to 30-fold after 12 days, cells lacking SUN2 had only doubled after 1 week, at which point their numbers often reached a plateau or began to decrease. The impact of the loss of SUN2 was also observed when cells were stained for Ki67. Levels of this proliferation marker rapidly increased under all conditions following activation and transduction but were significantly decreased (Fig. 1C) starting at times when SUN2 levels were depleted (Fig. 1A).
FIG 1.
SUN2 silencing impairs CD4 T cell proliferation, activation, and viability. (A) Representative histograms of SUN2 levels in donor 4 at 5 days postransduction (left) and SUN2 levels (geometric MFI) in 14 donors at days 5 to 6 postransduction (right). (B) Proliferation of control or shSUN2-transduced CD4 T cells for representative donor 11 (left) and for 12 donors (right) relative to 1 million cells at the time of activation. (C) Percentage of cells expressing the proliferation marker Ki67 in representative donor 14 at 8 days postactivation (left) and kinetics of Ki67 expression in 8 donors (right). (D) Viability of control or shSUN2-transduced cells as measured by forward-scatter area (FSC-A) and side-scatter area (SSC-A) properties at day 5 postransduction or by using a fixable viability dye (Aqua Live/Dead) at day 6 postransduction in cells from representative donor 6 (left) or from 14 donors (FSC versus SSC) and 8 donors (Aqua Live/Dead) (right). (E) Activation status of cells from 10 donors as measured by surface levels of the intermediate activation marker CD25 (left) or the late activation marker HLA-DR (right). Note that subsequent experiments (Fig. 2 to 4) were performed using cells from the donors whose data are presented in this figure. In panels B to E, statistical analyses were performed using repeated-measures one-way analysis of variance with Dunnett's posttest to compare each condition to those for shLacZ-transduced cells. n.s., not significant.
Loss of SUN2 results in decreased cell viability and lower T cell activation status.
To determine whether decreased proliferation following SUN2 silencing might be associated with a loss of viability, we first looked for changes in the cells' forward- and side-scatter properties. We observed a significant decrease in the percentage of cells exhibiting a healthy phenotype when SUN2 was depleted (Fig. 1D). Cells were also incubated with a viability dye to quantify the proportion of living cells, which additionally demonstrated a lower viability of SUN2-silenced cells than of controls (Fig. 1D). While the proportion of living cells was slightly higher for nontransduced than for control transduced cells by both measures of cell viability, this is not surprising given that puromycin selection was used on transduced cells only. We then asked whether SUN2 silencing alters the activation status of CD4 T cells. In the absence of SUN2, levels of the intermediate activation marker CD25 were modestly although statistically significantly reduced, while levels of the late activation marker HLA-DR were more strongly reduced (Fig. 1E). Control transduction did not alter the level of either activation marker compared to nontransduced cells. Together, our results demonstrate that SUN2 is required for efficient proliferation, activation, and viability of primary CD4 T cells.
Spreading HIV infection is strongly reduced in the absence of SUN2.
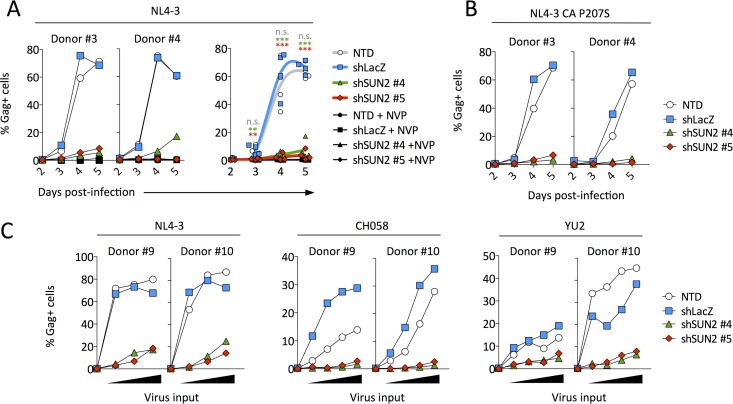
To determine the contribution of endogenous SUN2 to HIV infection, primary CD4 T cells were transduced with control or SUN2 shRNA and subsequently infected with a low virus inoculum. While transduction with control shRNA led to infection kinetics that were almost identical to those of nontransduced cells, infection was severely compromised when SUN2 was silenced (Fig. 2A). We previously identified a single-amino-acid change in the viral capsid (P207S) that leads to resistance to overexpressed SUN2 (8) and thus asked whether a virus bearing this mutation would be less susceptible to the effects of SUN2 silencing. Replication of this capsid mutant was affected to the same extent as the wild-type virus following SUN2 depletion (Fig. 2B), suggesting that endogenous SUN2 does not influence HIV in the same manner as when it is overexpressed. We also tested the ability of the transmitted/founder virus strain CH058 (19) and of YU2, an R5-tropic virus strain cloned directly from uncultured material (20), to replicate in the absence of SUN2. As shown in Fig. 2C, these strains exhibited a replication defect similar to that observed for NL4-3 infection of SUN2-depleted cells.
FIG 2.
Spreading HIV infection in SUN2-silenced CD4 T cells is greatly reduced. (A) Cells were infected overnight with 0.3 ng NL4-3 p24 per well, and the percentage of infected (Gag-positive [Gag+]) cells among the live-cell population was determined on days 2 to 5 postinfection. Results depict representative donors 3 and 4 (left) and the combined results for 4 donors (right). Statistical analysis was performed using repeated-measures one-way analysis of variance with Dunnett's posttest to compare each condition to those for shLacZ-transduced cells. n.s., not significant. (B) Infection of cells from donors 3 and 4, as described above for panel A, using the CA P207S mutant, which is resistant to overexpressed SUN2. (C) Infection of cells from donors 9 and 10 using a range of virus inputs for three different strains (1, 3, or 9 ng p24 per well for NL4-3; 1, 3, 9, or 18 ng p24 per well for CH058; and 1, 3, 9, or 27 ng p24 per well for YU2). Infection levels were measured on day 3 postinfection.
SUN2 has a modest impact on single-round HIV infection and acts independently of cyclophilin A.
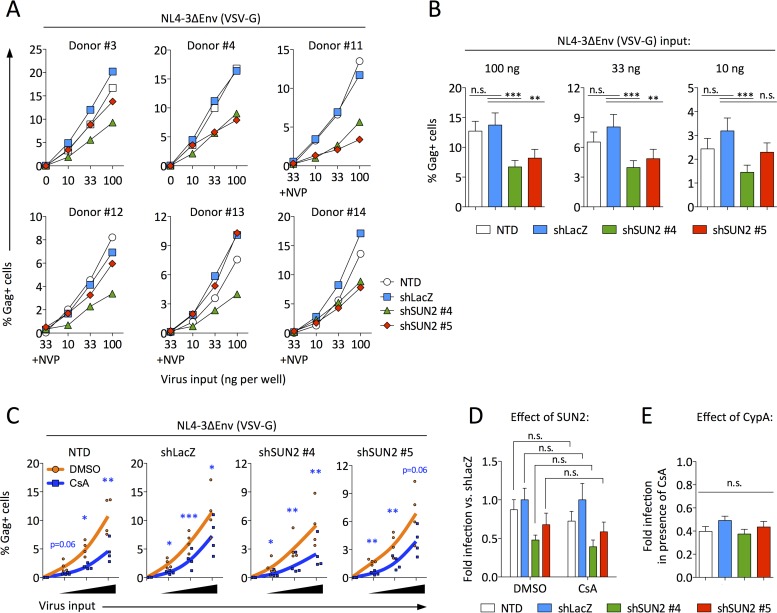
To better understand the replication defect observed upon infection of SUN2-silenced CD4 T cells, we used vesicular stomatitis virus envelope glycoprotein (VSV-G)-pseudotyped NL4-3 that does not encode Env. Cells were infected 5 days after the transduction of shRNA-encoding lentiviral particles, when the knockdown of SUN2 was near complete (Fig. 1A). Single-round infection was decreased by an average of 2-fold in the absence of SUN2 (Fig. 3A and B), consistent with data reported previously (9). We next asked whether CypA might be involved in this modest effect. Cyclosporine (CsA) acts as a competitive inhibitor of CypA binding to the viral capsid and impacts single-round infection of primary activated CD4 T cells to an identical extent as a nonimmunosuppressive CypA inhibitor (9). This does not exclude that certain differences could result depending on the CypA inhibitor used. Regardless of treatment with dimethyl sulfoxide (DMSO) or with CsA, single-round infection was reduced in SUN2 knockdown cells compared to control cells (Fig. 3C [note the different y axis scales for shSUN2 versus control cells]). To determine whether this effect was equivalent under both treatment conditions, we calculated the level of infection for each cell type relative to infection of shLacZ-transduced cells in the presence of DMSO or in the presence of CsA (Fig. 3D). These data show that the depletion of SUN2 reduced HIV infection equally regardless of CypA availability, demonstrating that CypA is not required for inhibition of infection in SUN2-silenced cells.
FIG 3.
Single-round HIV infection in SUN2-silenced CD4 T cells is modestly reduced independently of CypA. (A) Cells from donors 3, 4, and 11 to 14 were infected by spinoculation using a range of virus inputs. The percentage of infected (Gag+) cells was determined at 48 h postinfection. (B) Combined results for the 6 donors from panel A. (C) Combined results from infection of cells from donors 11 to 14, as described above for panel A, in the presence of 0.02% DMSO or 2 μM CsA. Statistical analyses of data in panels B and C were performed using repeated-measures one-way analysis of variance with Dunnett's posttest to compare each condition to those for shLacZ-transduced cells. n.s., not significant. (D) From the data shown in panel C, the effect of SUN2 was determined by calculating the infection level under each condition relative to infection of shLacZ-transduced cells, for both DMSO and CsA treatments, where a value of <1 indicates that infection was inhibited compared to infection of control cells. For each donor, infection levels were calculated and averaged across all virus input levels. Statistical analyses were performed by unpaired two-tailed t tests. n.s., not significant. (E) From the data shown in panel C, the effect of CypA was determined by calculating the ratio of the percentage of Gag+ cells following CsA treatment to the percentage of Gag+ cells following DMSO treatment, where a value of <1 indicates that CsA inhibited infection. For each donor, ratios were calculated and averaged across all virus input levels. Statistical analysis was performed using one-way analysis of variance. n.s., not significant. In panels B, D, and E, error bars represent standard errors of the means.
While the analysis shown in Fig. 3D asked whether CypA is necessary for the effects of SUN2 silencing on HIV infection, we also asked whether endogenous SUN2 is required for the well-known positive effects of CypA during HIV infection. To address the latter question, we calculated the fold infection in the presence of CsA (compared to DMSO) for each cell type (Fig. 3E). This analysis shows that CsA treatment inhibited infection ∼2-fold whether or not endogenous SUN2 was present, indicating that SUN2 is not required for CypA to positively modulate HIV infection. In other words, if SUN2 was required for the positive activities of CypA during HIV infection, we would have expected no reduction in infection following CsA treatment of SUN2-silenced cells. Together, these data suggest that the decrease in infection observed upon SUN2 silencing is not dependent on CypA and that the ability of CypA to positively modulate HIV infection does not require SUN2.
Reduced efficiency of viral gene expression in CD4 T cells lacking SUN2.
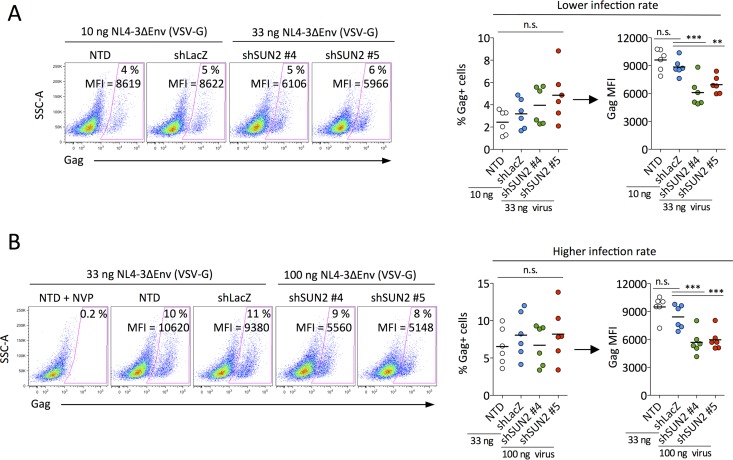
Our results demonstrate that the depletion of SUN2 in primary CD4 T cells results in significantly lower proliferation as well as decreased cell viability and a lower activation status (Fig. 1). Since it was previously shown that decreased HIV infection of SUN2-depleted cells is not associated with altered levels of integrated DNA (9), we asked whether SUN2 silencing might attenuate viral gene expression. To address this question, we compared the levels of viral gene expression per infected cell (Gag geometric mean fluorescence intensity [MFI]) following single-round infection in the presence and absence of SUN2 under conditions of equal productive infection rates. We found that cells depleted of SUN2 supported lower levels of Gag expression at both low (Fig. 4A) and higher (Fig. 4B) infection rates. Overall, our results demonstrate that the HIV-1 replication defect in SUN2-silenced CD4 T cells is not mediated by CypA and suggest that reduced infection is a likely consequence of the profound effects of SUN2 silencing on the health of CD4 T cells.
FIG 4.
SUN2-silenced CD4 T cells support reduced levels of viral gene expression. Shown is a comparison of the percentages of infected cells and levels of gene expression (Gag geometric MFI) at lower (A) and higher (B) rates of productive infection (nanograms of p24 per well used for single-round infection are indicated). Data for representative donor 4 are shown at the left, and the combined results for 6 donors are shown at the right. Gag MFI comparisons are between cell populations with equivalent percentages of infected cells. Statistical analyses were performed using repeated-measures one-way analysis of variance. Where significant differences were identified, Dunnett's posttest was used to compare each condition to those for shLacZ-transduced cells. n.s., not significant.
DISCUSSION
Recently, a role for endogenous SUN2 during CypA-dependent steps of HIV infection was proposed, whereby SUN2 silencing in primary CD4 T cells resulted in decreased levels of HIV infection (9). Based on treatment with a CypA inhibitor, the authors of that study proposed that SUN2 acts as a cofactor for HIV infection by modulating viral replication steps that are promoted by CypA (9). However, it is unclear how SUN2 might impact CypA-dependent steps of HIV replication, given that CypA modulates levels of reverse transcripts, 2-LTR circles, and integrated DNA (13–18), but no differences in levels of reverse transcripts or integrated DNA were observed in SUN2-silenced cells (9). This result would appear to exclude a role for SUN2 in the effects mediated by CypA. CypA has also been implicated in HIV integration site selection, although the disruption of CypA by CsA treatment led to integration into chromosomal regions with higher gene density and activity (21). Thus, if SUN2 impacted this activity of CypA, equal or higher levels of infection might be expected upon SUN2 silencing rather than the decrease observed by us and others (9). We report here that SUN2 silencing decreases HIV infection regardless of treatment with a CypA inhibitor; in other words, the effects of SUN2 silencing and CsA treatment on HIV infection are additive. Our results also show that loss of SUN2 does not alter the positive effects of CypA on HIV infection, since CsA treatment reduces HIV infection equally whether SUN2 is present or depleted. While the source of this discrepancy regarding a potential contribution of SUN2 to the effects of CypA are unknown, one difference is that the single-round infections performed previously used NL4-3ΔvifΔvprΔvpuΔenvΔnef encoding green fluorescent protein (GFP) (9), while we used full-length NL4-3 containing a stop codon at the start of Env.
In the same study by Lahaye and colleagues, SUN2 was reported to modulate, in a CypA-dependent manner, infection of murine but not human cells by lentiviral vectors (lentivectors) with capsids that were engineered to have artificially enhanced CypA binding affinities (9). That is, in mouse bone marrow-derived dendritic cells (BMDCs), infection by HIV-derived CA-mutated lentivectors was restricted compared to that with wild-type vectors, but their infection was rescued by either CsA treatment or SUN2 knockout. However, it is not clear whether this model reflects the situation in human cells, since the authors of that study reported that silencing of human SUN2 did not rescue infection by these same capsid mutant lentivectors (9). Furthermore, infection of mouse BMDCs with wild-type HIV-based lentivectors was not affected by treatment with CsA (9). Nonetheless, our results do not exclude that SUN2 might play a role in the ability of CypA to modulate infection of murine cells by HIV strains with engineered CypA binding affinities. Rather, our data demonstrate that infection of primary human CD4 T cells by wild-type HIV is not mediated in a CypA/SUN2-dependent fashion but that these proteins impact HIV infection independently.
If SUN2 does not modulate HIV infection via CypA, what might be an alternative mechanism? Productive infection of CD4 T cells, which represent the major target for HIV-1, requires cellular activation (22–24). T cell activation provides a permissive state for infection not only because of the inactivation of restriction factors such as SAMHD1 (25, 26) but also due to the upregulation or activation of transcriptional initiation and elongation factors critical for HIV replication, including NF-κB, AP-1, NFAT, and cyclin T1/CDK9 (27–31). Thus, our finding of decreased activation, viability, and proliferation of SUN2-silenced CD4 T cells provides insight into the reduction of HIV infection in these cells. Consistent with this notion is our observation that SUN2-depleted cells that become infected support lower levels of viral gene expression, which could be related to the availability of critical transcription initiation and elongations factors. This might contribute to the 2-fold decrease in single-round infection observed by us and others (9) in SUN2-silenced CD4 T cells, by shifting otherwise Gaglow cells below the population-level detection limit. It is possible that additional steps of the viral replication cycle are also rendered less efficient due to the overall decreased health of activated CD4 T cells that lack SUN2. Future work will determine whether SUN2 might act directly on the virus in addition to the effects described here.
Our previous characterization of HIV-1 and HIV-2 infection in the presence of overexpressed SUN2 demonstrated that the effect is specific for the viral capsid, that it occurs between the steps of reverse transcription and nuclear entry, that it requires the SUN2 nucleoplasmic domain, and that it involves CypA (8). However, we report here that SUN2 silencing in primary CD4 T cells modulates HIV infection independently of CypA. These data can be reconciled if the overexpression and depletion of SUN2 impact HIV infection through different mechanisms. Indeed, this appears to be the case, since SUN2 silencing in several cell lines had no effect on HIV infection, even when the overexpression of SUN2 inhibited infection in the same cell types (8). In contrast, we now show that the depletion of SUN2 affects the health of primary CD4 T cells, suggesting alternate CypA-independent mechanisms that could account for the decreased HIV infection in these cells.
Overall, our results demonstrate that SUN2 is important for the proliferation, activation, and viability of primary CD4 T cells and suggest that SUN2 does not promote CypA-dependent steps of HIV replication.
MATERIALS AND METHODS
Cells, viruses, and reagents.
Blood from anonymous healthy donors was obtained from the Etablissement Français du Sang (EFS) (the national blood bank of France), from which peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll centrifugation. CD4+ lymphocytes were isolated from CD14-negative PBMCs by selection against CD14 followed by selection for CD4 (catalog numbers 130-050-201 and 130-045-101, respectively; Miltenyi Biotec). Immediately after isolation, CD4 T cells were activated with CD2/CD3/CD28 microbeads (catalog number 130-091-441; Miltenyi Biotec) in multiple wells of 96-well U-bottom plates at 200,000 cells per well in 100 μl by using 1 microbead per 4 cells. Activated CD4 T cells were maintained in RPMI containing 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin (referred to as supplemented RPMI) plus 100 U/ml interleukin-2 (IL-2) (R&D Systems). 293T cells were obtained from the American Type Culture Collection (ATCC) and were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 1% penicillin-streptomycin. HIV-1 NL4-3, NL4-3ΔEnv, YU2, and CH058 were produced by transfection of 293T cells (in the presence of a VSV-G expression vector in the case of NL4-3ΔEnv). The CA P207S mutation was introduced into pNL4-3 by site-directed mutagenesis. Nevirapine (NVP) was obtained from the NIH AIDS Research and Reference Reagent Program and was used at 5 μM. CsA was obtained from Sigma (catalog number 30024) and was used at 2 μM as reported previously (9). Protamine sulfate was obtained from Sigma (catalog number P4020) and was used at 8 μg/ml.
Transduction of CD4 T cells.
Silencing of SUN2 in primary CD4 T cells was based on a method described previously (9), with several modifications. Lentiviral particles encoding shRNA targeting LacZ or SUN2 and pseudotyped simultaneously with both HIV (X4-tropic) and VSV-G envelope proteins were produced by calcium phosphate transfection of 293T cells in T-160 flasks. Plasmids for transfection were used as follows: 32 μg for pLKO.1-puro-shLacZ, pLKO.1-puro-shSUN2#4 (TRCN0000143335), or pLKO.1-puro-shSUN2#5 (TRCN0000143336); 20 μg for psPAX2; 4 μg for pHXB2-env; and 4 μg for pCMV-VSV-G. All plasmids were a kind gift of X. Lahaye and N. Manel. Lentiviral particles were filtered through a 0.45-μm-pore-size membrane, concentrated by ultracentrifugation, and assayed for p24 content by an enzyme-linked immunosorbent assay (ELISA).
Twenty-four hours after the activation of CD4 T cells (see above), medium was replaced with a final volume per well of 100 μl supplemented RPMI containing 100 U/ml IL-2, 8 μg/ml protamine, and 10 ng p24 of shRNA lentiviral particles (wells contained 200,000 cells at the time of activation). Cells were spinoculated for 2 h at 1,200 × g at 37°C and then left to rest in an incubator at 37°C for 1 h, and the medium was then replaced with 200 μl supplemented RPMI containing 100 U/ml IL-2. Nontransduced cells were subjected to an identical protocol in the absence of lentiviral particles. Two days following spinoculation, 50 μl medium per well was replaced with 50 μl supplemented RPMI containing 100 U/ml IL-2 and 8 μg/ml puromycin (final puromycin concentration, 2 μg/ml), except for nontransduced cells, to which puromycin was not added in this or subsequent steps. The following day (24 h post-puromycin treatment and 3 days postspinoculation), cells were routinely pooled, counted, and returned to culture at 100,000 cells per well.
Infections.
For spreading infections in Fig. 2A and B, 100,000 cells per well (96-well U-bottom plate) were infected on day 3 postransduction by the addition of 0.3 ng p24 per well of NL4-3 (or NL4-3 CA P207S) in a final volume of 100 μl supplemented RPMI containing 100 U/ml IL-2, 8 μg/ml protamine sulfate, and 2 μg/ml puromycin. After overnight incubation, virus-containing medium was removed, and cells were washed with phosphate-buffered saline (PBS), split into three replicate wells, and resuspended in supplemented RPMI containing 100 U/ml IL-2 and 2 μg/ml puromycin. Where used, NVP was present at 5 μM during overnight infection and was maintained throughout the infection. Spreading infections shown in Fig. 2C were performed identically, except that infection was performed on day 4 postransduction by the addition of a range of virus inputs, as indicated in the figure legend, and cells were fixed after 3 days. Infections were analyzed by flow cytometry.
For single-round infections, 100,000 cells per well (96-well U-bottom plate) were infected on day 5 postransduction by the addition of 10 ng, 33 ng, or 100 ng p24 VSV-G-pseudotyped NL4-3ΔEnv in a final volume of 100 μl supplemented RPMI containing 100 U/ml IL-2, 8 μg/ml protamine sulfate, and 2 μg/ml puromycin. Infection was done by spinoculation for 2 h at 1,200 × g at 37°C; cells were left to rest in an incubator at 37°C for 1 h and were then washed with PBS and resuspended in 200 μl supplemented RPMI containing 100 U/ml IL-2 and 2 μg/ml puromycin. Where used, DMSO (0.02%), CsA (2 μM), or NVP (5 μM) was present during spinoculation and was replenished after spinoculation. Cells were fixed after 48 h and analyzed by flow cytometry.
Flow cytometry.
We used antibodies against HIV-1 Gag (KC57-fluorescein isothiocyanate [FITC] or KC57-Rd1; Beckman Coulter) (1/500 dilution), SUN2 (EPR6557; Abcam) (1/500 dilution), Ki67 (FITC conjugated, catalog number 556026; BD Biosciences) (1/50 dilution), CD25 (phycoerythrin [PE] conjugated, catalog number 34011; BD Biosciences) (1/25 dilution), and HLA-DR (FITC conjugated, catalog number IM0463U; IOtest) (1/25 dilution) and matched isotype controls. Secondary Cy5-conjugated goat anti-rabbit antibody (catalog number A10523; Thermo Fisher) (1/500 dilution) was used for the detection of SUN2. Living cells were stained for surface CD25 and HLA-DR and were subsequently fixed with 4% paraformaldehyde (PFA) for 5 min. For intracellular staining, cells were fixed with 4% PFA for 5 min and then permeabilized with saponin (Gag) or with Triton X-100 (0.9% for 7 min for SUN2 and Ki67). Viability was quantified by incubation with Live/Dead fixable Aqua dye (catalog number L34957; Thermo Fisher) (1/1,000 dilution). Samples were acquired on a FACSCanto II instrument (BD Biosciences) and analyzed using Flow Jo software.
Statistical analyses.
All statistical analyses (identified in the figure legends) were performed with GraphPad Prism 5.0.
Ethics statement.
This study was carried out in accordance with guidelines established by the Institut Pasteur. All primary human cells were derived from blood of anonymous healthy donors, which was obtained from the EFS (the national blood bank of France). Donors provided written informed consent to the EFS at the time of blood donation.
ACKNOWLEDGMENTS
We thank N. Manel and X. Lahaye for kindly providing plasmids for production of shSUN2 and shLacZ virus-like particles and for discussions. We thank Alex Compton, Timothée Bruel, and Nicoletta Casartelli for critical readings of the manuscript and all members of the Virus and Immunity Unit for helpful discussions. We also thank the EFS and the NIH AIDS Research and Reference Reagent Program for providing reagents.
D.A.D. was supported by Sidaction. Work in the laboratory of O.S. was supported by grants from the ANRS, Sidaction, the AREVA Foundation, the Vaccine Research Institute, the Labex IBEID program, the FP7 program HIT Hidden HIV (Health-F3-2012-305762), and the Institut Pasteur. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Rothballer A, Kutay U. 2013. The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma 122:415–429. doi: 10.1007/s00412-013-0417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Starr DA, Fridolfsson HN. 2010. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang W, Worman HJ, Gundersen GG. 2015. Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol 208:11–22. doi: 10.1083/jcb.201409047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C-Y, Chi Y-H, Mutalif RA, Starost MF, Myers TG, Anderson SA, Stewart CL, Jeang K-T. 2012. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell 149:565–577. doi: 10.1016/j.cell.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isermann P, Lammerding J. 2013. Nuclear mechanics and mechanotransduction in health and disease. Curr Biol 23:R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Méjat A, Misteli T. 2010. LINC complexes in health and disease. Nucleus 1:40–52. doi: 10.4161/nucl.1.1.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donahue DA, Amraoui S, Di Nunzio F, Kieffer C, Porrot F, Opp S, Diaz-Griffero F, Casartelli N, Schwartz O. 2016. SUN2 overexpression deforms nuclear shape and inhibits HIV. J Virol 90:4199–4214. doi: 10.1128/JVI.03202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lahaye X, Satoh T, Gentili M, Cerboni S, Silvin A, Conrad C, Ahmed-Belkacem A, Rodriguez EC, Guichou J-F, Bosquet N, Piel M, Le Grand R, King MC, Pawlotsky J-M, Manel N. 13 April 2016. Nuclear envelope protein SUN2 promotes cyclophilin-A-dependent steps of HIV replication. Cell Rep doi: 10.1016/j.celrep.2016.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sokolskaja E, Luban J. 2006. Cyclophilin, TRIM5, and innate immunity to HIV-1. Curr Opin Microbiol 9:404–408. doi: 10.1016/j.mib.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Hilditch L, Towers GJ. 2014. A model for cofactor use during HIV-1 reverse transcription and nuclear entry. Curr Opin Virol 4:32–36. doi: 10.1016/j.coviro.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell EM, Hope TJ. 2015. HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol 13:471–483. doi: 10.1038/nrmicro3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braaten D, Luban J. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J 20:1300–1309. doi: 10.1093/emboj/20.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braaten D, Franke EK, Luban J. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol 70:3551–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song C, Aiken C. 2007. Analysis of human cell heterokaryons demonstrates that target cell restriction of cyclosporine-resistant human immunodeficiency virus type 1 mutants is genetically dominant. J Virol 81:11946–11956. doi: 10.1128/JVI.00620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ylinen LMJ, Schaller T, Price A, Fletcher AJ, Noursadeghi M, James LC, Towers GJ. 2009. Cyclophilin A levels dictate infection efficiency of human immunodeficiency virus type 1 capsid escape mutants A92E and G94D. J Virol 83:2044–2047. doi: 10.1128/JVI.01876-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinkasserer A, Harrison R, Billich A, Hammerschmid F, Werner G, Wolff B, Peichl P, Palfi G, Schnitzel W, Mlynar E. 1995. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus type 1 (HIV-1): interference with early and late events in HIV-1 replication. J Virol 69:814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Iaco A, Luban J. 2014. Cyclophilin A promotes HIV-1 reverse transcription but its effect on transduction correlates best with its effect on nuclear entry of viral cDNA. Retrovirology 11:11. doi: 10.1186/1742-4690-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R, Haynes BF, Shaw GM, Hahn BH, Kappes JC. 2012. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol 86:2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol 65:3973–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hué S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. 2011. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog 7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margolick JB, Volkman DJ, Folks TM, Fauci AS. 1987. Amplification of HTLV-III/LAV infection by antigen-induced activation of T cells and direct suppression by virus of lymphocyte blastogenic responses. J Immunol 138:1719–1723. [PubMed] [Google Scholar]
- 23.Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J 9:1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDougal JS, Mawle A, Cort SP, Nicholson JK, Cross GD, Scheppler-Campbell JA, Hicks D, Sligh J. 1985. Cellular tropism of the human retrovirus HTLV-III/LAV. I. Role of T cell activation and expression of the T4 antigen. J Immunol 135:3151–3162. [PubMed] [Google Scholar]
- 25.White TE, Brandariz-Nuñez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. 2013. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruffin N, Brezar V, Ayinde D, Lefebvre C, Schulze Zur Wiesch J, van Lunzen J, Bockhorn M, Schwartz O, Hocini H, Lelievre J-D, Banchereau J, Levy Y, Seddiki N. 2015. Low SAMHD1 expression following T-cell activation and proliferation renders CD4+ T cells susceptible to HIV-1. AIDS 29:519–530. doi: 10.1097/QAD.0000000000000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garriga J, Peng J, Parreño M, Price DH, Henderson EE, Graña X. 1998. Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene 17:3093–3102. doi: 10.1038/sj.onc.1202548. [DOI] [PubMed] [Google Scholar]
- 28.Herrmann CH, Carroll RG, Wei P, Jones KA, Rice AP. 1998. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J Virol 72:9881–9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams SA, Greene WC. 2007. Regulation of HIV-1 latency by T-cell activation. Cytokine 39:63–74. doi: 10.1016/j.cyto.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pessler F, Cron RQ. 2004. Reciprocal regulation of the nuclear factor of activated T cells and HIV-1. Genes Immun 5:158–167. doi: 10.1038/sj.gene.6364047. [DOI] [PubMed] [Google Scholar]
- 31.Macian F. 2005. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol 5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]