ABSTRACT
Cyclic GMP-AMP synthase (cGAS) is a key DNA sensor capable of detecting microbial DNA and activating the adaptor protein stimulator of interferon genes (STING), leading to interferon (IFN) production and host antiviral responses. Cells exhibited reduced type I IFN production in response to cytosolic DNA in the absence of cGAS. Although the cGAS/STING-mediated DNA-sensing signal is crucial for host defense against many viruses, especially for DNA viruses, few viral components have been identified to specifically target this signaling pathway. Herpes simplex virus 1 (HSV-1) is a DNA virus that has evolved multiple strategies to evade host immune responses. In the present study, we found that HSV-1 tegument protein UL41 was involved in counteracting the cGAS/STING-mediated DNA-sensing pathway. Our results showed that wild-type (WT) HSV-1 infection could inhibit immunostimulatory DNA-induced activation of the IFN signaling pathway compared with the UL41-null mutant virus (R2621), and ectopic expression of UL41 decreased cGAS/STING-mediated IFN-β promoter activation and IFN-β production. Further study indicated that UL41 reduced the accumulation of cGAS to abrogate host recognition of viral DNA. In addition, stable knockdown of cGAS facilitated the replication of R2621 but not WT HSV-1. For the first time, HSV-1 UL41 was demonstrated to evade the cGAS/STING-mediated DNA-sensing pathway by degrading cGAS via its RNase activity.
IMPORTANCE HSV-1 is well known for its ability to evade host antiviral responses and establish a lifelong latent infection while triggering reactivation and lytic infection under stress. Currently, whether HSV-1 evades the cytosolic DNA sensing and signaling is still poorly understood. In the present study, we found that tegument protein UL41 targeted the cGAS/STING-mediated cellular DNA-sensing pathway by selectively degrading cGAS mRNA. Knockdown of endogenous cGAS could facilitate the replication of R2621 but not WT HSV-1. Furthermore, UL41 was shown for the first time to act directly on cGAS. Findings in this study could provide new insights into the host-virus interaction and help develop new approaches against HSV-1.
KEYWORDS: HSV-1, vhs/UL41, cGAS, DNA sensing
INTRODUCTION
The innate immune system is the first line of defense against invading pathogens and is crucial for the subsequent activation of the adaptive immune response. The first step in innate immunity is to detect the invading pathogen through various pathogen recognition receptors (PRRs) which recognize pathogen-associated molecular patterns and trigger the production of type I interferon (IFN) and other antiviral factors (1, 2). Besides Toll-like receptors in the cellular membrane or endosome, Nod-like receptors and the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) in the cytoplasm, there are also several recently discovered cytosolic DNA sensors, such as cyclic GMP-AMP (cGAMP) synthase (cGAS), gamma interferon-inducible protein 16 (IFI16), DNA-dependent activator of IFN-regulatory factors, and absent in melanoma 2 and RNA polymerase III (3–6). Among these DNA sensors, cGAS, which is a nucleotidyltransferase containing highly related structural and enzymatic features with the well-known double-stranded RNA (dsRNA)-sensing 2-5-oligoadenylate synthase (OAS), is currently considered the principal sensor of cytosolic DNA (7–9). Upon binding to DNA fragments, cGAS utilizes GTP and ATP to produce cGAMP through its enzymatic activity, and the latter activates the downstream adaptor protein stimulator of interferon gene (STING), which then recruits TANK-binding kinase 1 (TBK1) and traffics from endoplasmic reticulum to a perinuclear endosomal compartment, leading to the activation of IFN regulatory factor 3 (IRF3) and resulting in IFN-β production (8–10).
Recent studies have revealed that the cGAS/STING-mediated cytosolic DNA-sensing signal pathway is important for cellular innate responses and intrinsic resistance to DNA virus infection (11–13). However, so far little is known about the countermeasures by DNA viruses, including herpes simplex virus 1 (HSV-1), against the cGAS/STING-mediated DNA sensing signal pathway.
HSV-1 belongs to the Alphaherpesvirinae subfamily, and it is a typical double-stranded DNA virus that encodes over 80 proteins. As a master of immune evasion, HSV-1 has evolved multiple strategies to counteract host antiviral responses (14). Although much progress has been made in understanding the mechanisms of HSV-1-mediated immune evasion, most of these findings focused on Toll-like receptor- and RLR-mediated signaling pathways, and our understanding of how HSV-1 evades cellular DNA sensing pathways is still quite limited. However, a recent study showed that HSV-1 d109 infection did not cause robust cGAMP production, indicating that there might be at least one viral component capable of inhibiting the cGAS-mediated cytosolic DNA-sensing pathway (15).
HSV-1 tegument protein UL41, also known as the virion host shutoff protein, is an endoribonuclease with the activity of mRNA-specific RNase and plays a role in HSV-1-mediated evasion of host antiviral responses (16–21). UL41 triggers selective degradation of host mRNAs to facilitate viral infection. In the present study, we found that ectopic expression of UL41 inhibited cGAS/STING-mediated activation of the IFN pathway. Moreover, compared with the UL41-null mutant virus (R2621), wild-type (WT) HSV-1 infection could inhibit immunostimulatory DNA (ISD)-induced activation of IFN signaling pathway. Further study showed that UL41 reduced the accumulation of cGAS mRNA and downregulated its protein. In addition, stable knockdown of cGAS facilitated the replication of R2621 but not WT HSV-1. For the first time, HSV-1 UL41 was demonstrated to abrogate the cGAS/STING-mediated signal pathway by degrading cGAS via its RNase activity.
RESULTS
HSV-1 UL41 inhibits IFN-β activation by the cGAS/STING pathway.
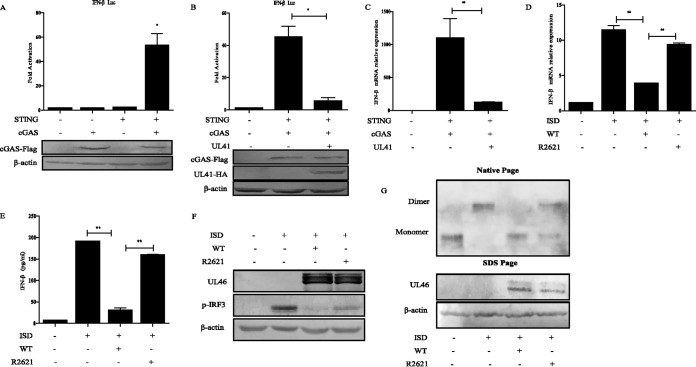
Previous studies have reported that cGAS/STING-mediated type I IFN activation plays a crucial role against HSV-1 infection (8, 9). To evaluate if individual viral protein could regulate the cGAS/STING pathway, we screened for viral proteins that could inhibit IFN-β promoter activation triggered by cotranfection of cGAS and STING. In HEK293T cells, ectopic expression of cGAS or a minimal amount of STING alone could not activate the IFN-β promoter, whereas cotransfection of cGAS and STING plasmids significantly activated the IFN-β promoter (Fig. 1A). Based on this model, HEK293T cells were cotransfected with empty vector or expression plasmids encoding HSV-1 proteins along with cGAS-Flag, STING-hemagglutinin (HA), and IFN-β promoter construct and subjected to dual-luciferase reporter (DLR) assays to detect IFN-β promoter activity. We found that cotransfection of UL41-HA could significantly inhibit the activation of the IFN-β promoter (Fig. 1B). Next, we validated the DLR result by measuring IFN-β mRNA levels through quantitative reverse transcription-PCR (qRT-PCR), and similar results were obtained (Fig. 1C). These data indicated that UL41 downregulated the cGAS/STING-mediated activation of the IFN pathway.
FIG 1.
HSV-1 UL41 inhibits IFN-β activation by the cGAS/STING pathway. (A) HEK293T cells were cotransfected with IFN-β promoter plasmid (IFN-β-Luc; 200 ng), Renilla luciferase (pRL-TK; 50 ng) reporter plasmid, and various plasmids (15 ng of cGAS-Flag or 2.5 ng of STING-HA or STING-HA and cGAS-Flag combined). Luciferase activity was measured 24 h posttransfection in the cell lysates. (B) HEK293T cells were cotransfected with IFN-β-Luc reporter, pRL-TK, cGAS-Flag, and STING-HA along with empty vector (200 ng), or UL41-HA plasmid (200 ng). Luciferase activity was measured 24 h posttransfection in the cell lysates. The expression of cGAS and UL41 were analyzed by WB using anti-Flag, anti-HA, and anti-β-actin (as a control) MAbs. (C) HEK293T cells were cotransfected with cGAS-Flag, STING-HA along with empty vector, or UL41-HA plasmid. At 24 h posttransfection, cells were harvested and subjected to qRT-PCR analysis. (D and E) HFFs were infected with WT HSV-1or R2621 at an MOI of 5 for 2 h, then cell medium was replaced with DMEM containing 10% FBS, and ISD (4 μg/ml) was transfected using Lipofectamine LTX according to the manufacturer's recommendations. Cells were harvested and subjected to qRT-PCR analysis at 7 h posttransfection (D) or ELISA analysis at 18 h posttransfection (E). (F and G) HFFs were infected and transfected as described for panel D, and then cells were harvested at 7 h posttransfection and subjected to WB analysis to detect IRF3 phosphorylation (F) or native PAGE to detect IRF3 dimerization (G). The data represent results from one of the triplicate experiments. Error bars represent SDs of three independent experiments. Statistical analysis was performed using Student's t test with GraphPad Prism 5.0 software. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01).
Human foreskin fibroblasts (HFFs) express many DNA sensors, including cGAS, and have been used to study their functions (15, 22). Immunostimulatory DNA (ISD) is a double-stranded DNA 60-mer oligonucleotide derived from the HSV-1 genome and has a high capacity to induce IFN-β production (23). To determine whether UL41 could affect the production of IFN-β induced by ISD, HFFs were infected with wild-type (WT) or UL41-null mutant (R2621) HSV-1 for 2 h before ISD transfection. Then cells were harvested at 7 h posttransfection and subjected to qRT-PCR to analyze IFN-β mRNA. As shown in Fig. 1D, infection of HFFs with WT HSV-1, but not R2621, significantly inhibited ISD induced IFN-β mRNA. We further performed enzyme-linked immunosorbent assay (ELISA) analysis to evaluate concentrations of IFN-β in cell culture supernatants, and similar results were obtained (Fig. 1E). These data suggested that WT HSV-1 infection decreased ISD-induced production of IFN-β via UL41.
IRF3 is a key transcription factor in the type I IFN signaling pathway, and phosphorylation and dimerization of IRF3 is a hallmark of IFN-mediated early antiviral responses. We next examined the effect of UL41 on ISD-mediated activation of IRF3. HFFs were infected with WT HSV-1 or R2621 for 2 h before ISD transfection, and robust phosphorylation and dimerization of IRF-3 were observed following ISD stimulation. We found that infection of WT HSV-1 abrogated ISD-induced phosphorylation of IRF-3, while infection of R2621 did not (Fig. 1F). Similarly, ISD triggered IRF-3 dimerization was markedly reduced under WT HSV-1 but not R2621 infection (Fig. 1G). Taken together, these results demonstrated that UL41 inhibited IFN-β activation by the cGAS/STING pathway.
UL41 inhibits the IFN signaling pathway at the level upstream of STING.
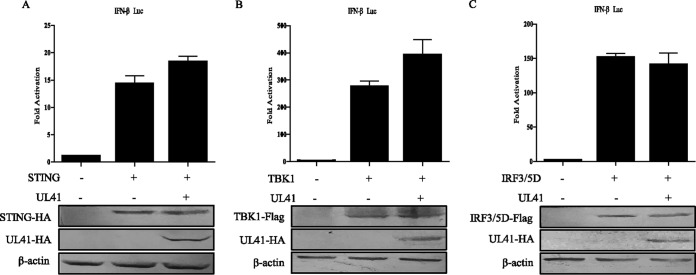
To determine at what level in the pathway UL41 blocks IFN-β activation, HEK293T cells were cotransfected with empty vector or UL41 plasmids along with IFN-β-Luc reporter and plasmids expressing important adaptor proteins in the pathway, including STING, TBK1 kinase, and the active form of IRF3 (IRF3/5D). Although ectopic expression of a minimal amount of STING in HEK293T cells did not activate the IFN-β promoter, transfection of a large amount of STING could successfully lead to IFN-β promoter activation (24, 25) (Fig. 2A). All expression constructs resulted in a 15- to 280-fold induction of IFN-β-Luc reporter activity (Fig. 2). We found that ectopic expression of UL41 did not affect the activation of IFN-β promoter driven by STING, TBK1, or IRF3/5D (Fig. 2). Collectively, these results suggested that UL41 inhibited the IFN signaling pathway at the level upstream of STING.
FIG 2.
UL41 inhibits the IFN signaling pathway at the level upstream of STING. (A to C) HEK293T cells were cotransfected with IFN-β-Luc reporter, pRL-TK, and STING (A), TBK1 (B), or IRF3/5D (C) along with empty vector or UL41-HA plasmid. Cells were harvested 24 h after transfection and subjected to DLR assay. The expression of STING, TBK1, IRF3/5D, and UL41 was analyzed by WB analysis. The data represent results from one of the triplicate experiments. Error bars represent standard deviations of three independent experiments. Statistical analysis was performed using Student's t test with GraphPad Prism 5.0 software.
HSV-1 tegument protein UL41 downregulates the expression of cGAS.
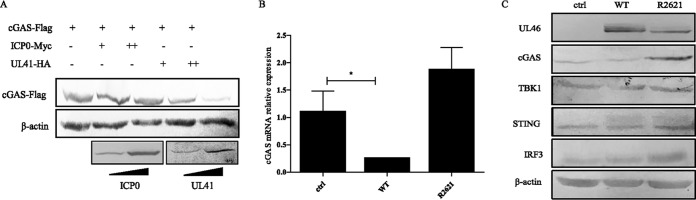
The aforementioned data lead us to hypothesize that UL41 might act directly on cGAS. We and others have shown that UL41 selectively degrades both viral and cellular mRNAs containing an AU-rich element (ARE) in its 3′ untranslated region (UTR) (17, 18, 20, 26). Meanwhile, ICP0, an HSV-1 immediate early protein, directs the proteasomal degradation of several cellular antiviral proteins (27, 28). To determine whether ectopic expression of UL41 or ICP0 could decrease the expression of cGAS, HEK293T cells were cotransfected with cGAS-Flag and UL41-HA or ICP0-Myc plasmids, and then cells were harvested and subjected to Western blot (WB) analysis. As shown in Fig. 3A, HSV-1 tegument protein UL41, not ICP0, downregulated the expression of cGAS in a dose-dependent manner.
FIG 3.
HSV-1 tegument protein UL41 downregulates the expression of cGAS. (A) HEK293T cells were cotransfected with cGAS-Flag and UL41-HA or ICP0-Myc plasmids. At 24 h posttransfection, cells were harvested and subjected to WB analysis. (B and C) HFFs were infected with WT HSV-1 or R2621 at an MOI of 5, and then cells were harvested 20 h postinfection and subjected to RT-PCR (B) or WB analysis with antibodies against UL46, cGAS, TBK1, STING, IRF3, or β-actin (C). Uninfected HFFs were used as a control (ctrl). The data represent results from one of the triplicate experiments. Statistical analysis was performed using Student's t test with GraphPad Prism 5.0 software. *, 0.01 < P < 0.05).
UL41 is an endoribonuclease with a substrate specificity similar to that of RNase A and mediates selective degradation of both viral and cellular mRNAs (16, 19). We next examined whether cGAS mRNA was downregulated under HSV-1 infection. HFFs were infected with WT HSV-1 or R2621 for 20 h, and then cells were harvested and subjected to qRT-PCR to analyze cGAS mRNA. As shown in Fig. 3B, WT HSV-1, but not R2621, significantly reduced the accumulation of cGAS mRNA (P = 0.02). Similarly, HFFs were infected with WT HSV-1 or R2621 and subjected to WB analysis. The data showed that compared with R2621, WT HSV-1 abrogated cGAS expression (Fig. 3C).
To demonstrate whether UL41 specifically targets cGAS, we performed WB analysis to examine the expression of key molecules in the cGAS-mediated cytosolic DNA-sensing pathway (STING, TBK1, and IRF3) during viral infection. As shown in Fig. 3C, WT HSV-1 infection failed to reduce endogenous STING, TBK1, or IRF3. Taken together, our observations substantiated that HSV-1 infection selectively reduced the accumulation of cGAS mRNA and the protein expression of cGAS via UL41.
cGAS mediates the defense against the replication of UL41-null HSV-1.
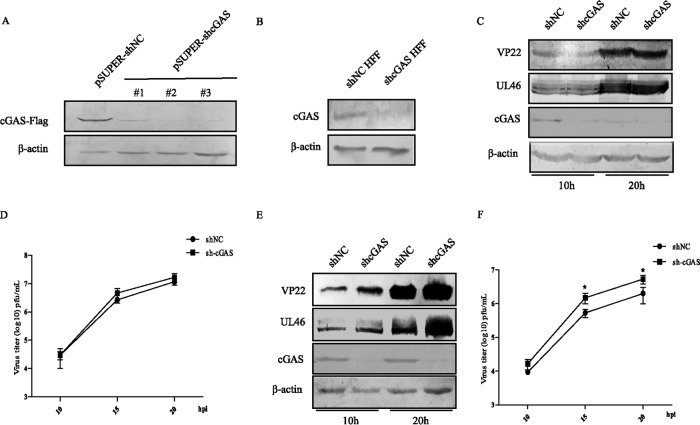
Previous studies have revealed that the activation of cGAS elicits a potent antiviral response, while the aforementioned data suggested that HSV-1 infection inhibited cGAS/STING-mediated activation of the IFN-β pathway by downregulating cGAS via UL41 (12, 29). Therefore, to better delineate the antiviral role of cGAS in HSV-1 replication, we constructed the stably transfected HFF-shNC and HFF-shcGAS cells. First, WB analysis was performed to evaluate the knockdown efficiency of cGAS RNA interference (RNAi) plasmids. HEK293T cells were cotransfected with cGAS-Flag and pSUPER-shNC or pSUPER-shcGAS plasmids, and then cells were harvested and subjected to WB analysis (Fig. 4A). HFFs were transfected with pSUPER-shcGAS#2 or pSUPER-shNC plasmid to screen for stably transfected cell lines. As shown in Fig. 4B, the expression of endogenous cGAS in HFF-shcGAS cells was markedly decreased compared with that in HFF-shNC cells.
FIG 4.
cGAS mediates the defense against the replication of UL41-null HSV-1. (A) HEK293T cells were cotransfected with cGAS-Flag and pSUPER-shNC or the indicated pSUPER-shcGAS plasmids, and cells were harvested 48 h posttransfection and subjected to WB analysis. (B) Western blot analysis of endogenous cGAS using cell lysates of HFF-shNC and HFF-shcGAS cells. (C to F) The stably transfected HFF-shNC and HFF-shcGAS cells were infected with WT HSV-1 or R2621 at an MOI of 2, then harvested at the indicated time points postinfection, and subjected to WB analysis (C and E) or viral plaque assay on Vero cells (D and F). The data represent results from one of the triplicate experiments. Statistical analysis was performed using Student's t test with GraphPad Prism 5.0 software. *, 0.01 < P < 0.05).
Then, the stably transfected HFF-shNC and HFF-shcGAS cells were infected with WT HSV-1 or R2621 and harvested at the desired time points for WB analysis and viral plaque assay. The levels of expression of UL46 and VP22 in HFF-shNC and HFF-shcGAS cells were similar upon WT HSV-1 infection, while protein levels of UL46 and VP22 were increased in HFF-shcGAS cells compared with HFF-shNC cells upon R2621 infection (Fig. 4C and E). Similarly, viral plaque assay also demonstrated that knockdown of cGAS did not affect the replication of WT HSV-1 but facilitated the replication of R2621 (Fig. 4D and F). Collectively, these data indicated that cGAS mediated the defense against the replication of R2621.
DISCUSSION
The innate immune system utilizes various PRRs to detect viral structures, such as nucleic acids, and initiates host immune responses leading to the production of type I IFNs and proinflammatory cytokines (6). During the past decade, increasing numbers of cellular DNA sensors have been identified and characterized, which greatly broadened our knowledge on host-virus interaction; however, several DNA sensors exert distinct functions in different cell types or in response to different viruses (5, 30). Among these cellular DNA sensors, cGAS and IFI16 can both sense HSV-1 DNA and trigger host immune responses upon infection (8, 23). However, a recent study by Diner et al. reported that cGAS, not IFI16, is required for activation of the downstream STING/TBK1/IRF3 pathway (31). In addition, small hairpin RNA (shRNA)-mediated knockdown or knockout of cGAS using CRISPR/Cas9 technology could severely compromise IFN-β induction in response to transfected DNA or HSV-1 infection (8, 31).
Given the importance of cGAS/STING-mediated activation of IFN pathway in host antiviral responses, it is reasonable to presume that DNA viruses should evolve certain mechanisms to block this signal pathway. In the present study, we found that compared with R2621, WT HSV-1 infection significantly inhibited cGAS/STING-mediated IFN-β induction. Knockdown of cGAS facilitated R2621 replication but did not affect WT HSV-1 replication. It is worth noting that R2621 infection could also counteract cGAS/STING-mediated activation of IFN pathway, although to a lesser extent than that of WT HSV-1. This observation is consistent with our screening result; for example, US3, VP16, and UL42, which have previously been proved to inhibit IFN production and NF-κB activation by targeting IRF3 or p65 (32–35), could also inhibit cGAS/STING-mediated IFN-β activation. Given the fact that IRF3 and p65 are both crucial downstream factors in the cGAS/STING-mediated DNA-sensing signal pathway, it is plausible that US3, VP16, and UL42 inhibit cytosolic DNA-sensing signal in a similar manner. Collectively, these data indicate that there are other viral proteins which might also be involved in this immune evasion.
There is growing evidence that herpesviruses have evolved multiple strategies to inhibit cGAS/STING-mediated activation of the IFN pathway. Ma et al. reported that one Kaposi's sarcoma-associated herpesvirus (KSHV) protein, viral interferon regulatory factor 1, could inhibit the cGAS/STING-mediated signal pathway by blocking the STING-TBK1 interaction (24). The KSHV tegument protein ORF52 directly interacted with and inhibited cGAS enzymatic activity (36). Our previous study has demonstrated that VP24 inhibited the cGAS-STING-mediated IFN-β signaling pathway by blocking the interaction between TBK1 and IRF3 (37). In addition, a recent study by Christensen et al. demonstrated that HSV-1 immediate early protein ICP27 interacted with TBK1 and STING and abrogated activation of IRF3 (38).
Nevertheless, our current knowledge on the countermeasure of DNA viruses against the cGAS/STING-mediated DNA-sensing signal pathway is still limited, and no viral protein has been identified to act directly on cGAS. In this study, we found that UL41 targeted and reduced cGAS expression by degrading its mRNA. In addition, we and other researchers have demonstrated that UL41 is also involved in HSV-1-mediated immune evasion by targeting several IFN-stimulated genes (ISGs) (17, 20, 21). Therefore, UL41 is a broad innate immune inhibitor and can block cGAS/STING-mediated IFN signaling at multiple steps.
We also found that ectopic expression of UL41 did not significantly affect STING-, TBK1-, or IRF3/5D-mediated activation of the IFN-β promoter, and WT HSV-1 infection did not affect the expression of STING, TBK1, or IRF3. Esclatine et al. predicted that the virion host shutoff protein, UL41, selectively degraded both viral and cellular mRNAs containing an ARE in its 3′ untranslated region (UTR) (26). In addition, AREs represent a common determinant of RNA stability and play a critical role in the regulation of gene expression in mammalian cells (39). According to an online ARE database (http://rna.tbi.univie.ac.at/AREsite), we found three ARE core motifs (ATTTA) in the 3′ UTR of cGAS but no motif in STING or IRF3 and only one motif in TBK1 (40), which might explain why HSV-1 infection decreases cGAS expression without affecting other adaptors in the signaling pathway. However, why UL41 specifically targets ARE-containing RNAs and the RNA degradation machinery deserves further investigation.
In summary, we have shown in this study for the first time that HSV-1 tegument protein UL41 counteracts the cGAS/STING-mediated DNA-sensing pathway by directly targeting cGAS mRNA for degradation. WT HSV-1, but not R2621, infection could inhibit ISD-induced activation of IFN signaling pathway. Furthermore, stable knockdown of cGAS significantly facilitated the replication of R2621 but not WT HSV-1. Findings in this study will expand our knowledge on the molecular mechanisms by which HSV-1 counteracts the antiviral innate immunity and provide new insights into the host-virus interaction.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
HEK293T, HFF, and Vero cell lines were obtained from the American Type Culture Collection (Manassas, VA) and cultured in Dulbecco's modified Eagle medium (DMEM; Gibco-BRL) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml of penicillin and streptomycin. The wild-type (WT) HSV-1 F strain was propagated in Vero cells and titrated as described previously (41). The protease inhibitor mixture cocktail was purchased from Thermo Fisher Scientific (MA). Radioimmunoprecipitation assay (RIPA) lysis buffer was purchased from Beyotime (Shanghai, China). Mouse anti-Myc, anti-HA, and anti-Flag monoclonal antibodies (MAbs) were purchased from Abmart (Shanghai, China). Goat anti-cGAS polyclonal antibody (pAb) and mouse anti-β-actin MAb were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-IRF3, anti-STING, anti-TBK1, anti-UL46, and anti-VP22 pAbs were made by GL Biochem Ltd. (Shanghai, China). Phospho-IRF3 (Ser396) rabbit MAb was purchased from Cell Signaling Technology (Danvers, MA).
Plasmid construction.
All enzymes used for cloning procedures were purchased from Vazyme (Nanjing, China). Small hairpin RNA specific for cGAS (shcGAS) and scrambled small hairpin RNA as a negative control (shNC) were cloned into pSUPER.retro.puro vector (Oligoengine, LA) to yield pSUPER-shcGAS and pSUPER-shNC plasmids, respectively, as described in our previous study (21). Commercial reporter plasmid pRL-TK (RL stands for Renilla luciferase, and TK stands for thymidine kinase) was purchased from Promega Corporation (Madison, WI). Other gift plasmids used include the following: cGAS-Flag (8), pcDNA3.1-Flag-TBK1 (42), IRF3/5D (43), and IFN-β promoter reporter plasmid (44).
Establishment of cGAS stable knockdown HFFs.
HFFs were transfected with pSUPER-shcGAS or pSUPER-shNC for 48 h, and then puromycin was added to cells at a concentration of 500 ng/ml. The stably transfected HFF-shNC and HFF-shcGAS cells were then cultured with puromycin (250 ng/ml).
WB analysis.
Western blot (WB) analysis was performed as previously described (28).
RNA isolation and qRT-PCR.
Total RNA was extracted using TRIzol (Invitrogen, CA) according to the manufacturer's manual. Samples were digested with DNase I and subjected to reverse transcription as previously described. The cDNA was used as the template for qRT-PCR to test the levels of IFN-β or cGAS mRNA, and 18S rRNA was used as an internal reference as previously described (17).
Transfection and DLR assays.
HFFs were transfected with Lipofectamine LTX (Invitrogen, CA) according to the manufacturer's recommendations. HEK293T cells were cotransfected with reporter plasmids, such as IFN-β-Luc and internal control plasmid pRL-TK, with or without expression plasmids as indicated, by standard calcium phosphate precipitation (45). At 24 h posttransfection, luciferase assays were performed with a dual-luciferase reporter (DLR) assay kit (Promega, Madison, WI) as described in our previous studies (41, 46).
ELISA.
Concentrations of IFN-β in cell culture supernatants were determined by using the VeriKine human IFN-β ELISA kit from PBL Assay Science (Piscataway, NJ) according to the manufacturer's instructions.
Native PAGE.
Native polyacrylamide gel electrophoresis (PAGE) was performed using ReadyGels (7.5%; Bio-Rad) as described in our previous study (32). In brief, gels were prerun with 25 mM Tris and 192 mM glycine, pH 8.4, with 1% deoxycholate (DOC) in the cathode chamber for 30 min at 40 mA. Samples in native sample buffer (10 μg of protein, 62.5 mM Tris-Cl [pH 6.8], 15% glycerol, and 1% DOC) were size fractionated by electrophoresis at 25 mA and transferred to nitrocellulose membranes for WB analysis.
Statistical analysis.
Data are represented as means ± standard deviations (SDs) when indicated, and Student's t test was used for all statistical analyses with GraphPad Prism 5.0 software. Differences between groups were considered significant when the P value was <0.05.
ACKNOWLEDGMENTS
We thank Zhijian J. Chen for cGAS-Flag plasmid, Rongtuan Lin for STING-HA plasmid, Takashi Fujita for IFN-β-Luc, and Bernard Roizman for R2621 virus.
Work in the Zheng laboratory relevant to this article was supported by grants from the National Natural Science Foundation of China (81371795 and 81571974) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (YX13400214).
REFERENCES
- 1.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Cunha A. 2012. Innate immunity: pathogen and xenobiotic sensing—back to basics. Nat Rev Immunol 12:400. doi: 10.1038/nri3237. [DOI] [PubMed] [Google Scholar]
- 3.Hornung V. 2014. SnapShot: nucleic acid immune sensors, part 1. Immunity 41:868–868.e861. doi: 10.1016/j.immuni.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 4.O'Neill LA, Bowie AG. 2010. Sensing and signaling in antiviral innate immunity. Curr Biol 20:R328–R333. doi: 10.1016/j.cub.2010.01.044. [DOI] [PubMed] [Google Scholar]
- 5.Unterholzner L. 2013. The interferon response to intracellular DNA: why so many receptors? Immunobiology 218:1312–1321. doi: 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, Chen ZJ. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 7.Cai X, Chiu YH, Chen ZJ. 2014. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell 54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 8.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa H, Barber GN. 2011. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell Mol Life Sci 68:1157–1165. doi: 10.1007/s00018-010-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paijo J, Doring M, Spanier J, Grabski E, Nooruzzaman M, Schmidt T, Witte G, Messerle M, Hornung V, Kaever V, Kalinke U. 2016. cGAS senses human cytomegalovirus and induces type I interferon responses in human monocyte-derived cells. PLoS Pathog 12:e1005546. doi: 10.1371/journal.ppat.1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barber GN. 2014. STING-dependent cytosolic DNA sensing pathways. Trends Immunol 35:88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Su C, Zhan G, Zheng C. 2016. Evasion of host antiviral innate immunity by HSV-1, an update. Virol J 13:38. doi: 10.1186/s12985-016-0495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orzalli MH, Broekema NM, Diner BA, Hancks DC, Elde NC, Cristea IM, Knipe DM. 2015. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc Natl Acad Sci U S A 112:E1773–E1781. doi: 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everly DN Jr, Feng P, Mian IS, Read GS. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J Virol 76:8560–8571. doi: 10.1128/JVI.76.17.8560-8571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen G, Wang K, Wang S, Cai M, Li ML, Zheng C. 2014. Herpes simplex virus 1 counteracts viperin via its virion host shutoff protein UL41. J Virol 88:12163–12166. doi: 10.1128/JVI.01380-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su C, Zhang J, Zheng C. 2015. Herpes simplex virus 1 UL41 protein abrogates the antiviral activity of hZAP by degrading its mRNA. Virol J 12:203. doi: 10.1186/s12985-015-0433-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taddeo B, Roizman B. 2006. The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J Virol 80:9341–9345. doi: 10.1128/JVI.01008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zenner HL, Mauricio R, Banting G, Crump CM. 2013. Herpes simplex virus 1 counteracts tetherin restriction via its virion host shutoff activity. J Virol 87:13115–13123. doi: 10.1128/JVI.02167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Z, Su C, Zheng C. 28 September 2016. Herpes simplex virus 1 tegument protein UL41 counteracts IFIT3 antiviral innate immunity. J Virol doi: 10.1128/JVI.01672-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau L, Gray EE, Brunette RL, Stetson DB. 2015. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 350:568–571. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- 23.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B. 2015. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A 112:E4306–E4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esclatine A, Taddeo B, Evans L, Roizman B. 2004. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc Natl Acad Sci U S A 101:3603–3608. doi: 10.1073/pnas.0400354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkinson J, Everett RD. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol 74:10006–10017. doi: 10.1128/JVI.74.21.10006-10017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Wang K, Wang S, Zheng C. 2013. Herpes simplex virus 1 E3 ubiquitin ligase ICP0 protein inhibits tumor necrosis factor alpha-induced NF-kappaB activation by interacting with p65/RelA and p50/NF-kappaB1. J Virol 87:12935–12948. doi: 10.1128/JVI.01952-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orzalli MH, Knipe DM. 2014. Cellular sensing of viral DNA and viral evasion mechanisms. Annu Rev Microbiol 68:477–492. doi: 10.1146/annurev-micro-091313-103409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diner BA, Lum KK, Toettcher JE, Cristea IM. 2016. Viral DNA sensors IFI16 and cyclic GMP-AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during herpesvirus infection. mBio 7:e01553-16. doi: 10.1128/mBio.01553-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Wang K, Lin R, Zheng C. 2013. Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J Virol 87:12814–12827. doi: 10.1128/JVI.02355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xing J, Ni L, Wang S, Wang K, Lin R, Zheng C. 2013. Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-kappaB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J Virol 87:9788–9801. doi: 10.1128/JVI.01440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Wang S, Wang K, Zheng C. 2013. Herpes simplex virus 1 DNA polymerase processivity factor UL42 inhibits TNF-alpha-induced NF-kappaB activation by interacting with p65/RelA and p50/NF-kappaB1. Med Microbiol Immunol 202:313–325. doi: 10.1007/s00430-013-0295-0. [DOI] [PubMed] [Google Scholar]
- 35.Wang K, Ni L, Wang S, Zheng C. 2014. Herpes simplex virus 1 protein kinase US3 hyperphosphorylates p65/RelA and dampens NF-kappaB activation. J Virol 88:7941–7951. doi: 10.1128/JVI.03394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, Konrad A, Neipel F, Sturzl M, Whitby D, Li H, Zhu F. 2015. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18:333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D, Su C, Zheng C. 2016. Herpes simplex virus 1 serine protease VP24 blocks the DNA-sensing signal pathway by abrogating activation of interferon regulatory factor 3. J Virol 90:5824–5829. doi: 10.1128/JVI.00186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christensen MH, Jensen SB, Miettinen JJ, Luecke S, Prabakaran T, Reinert LS, Mettenleiter T, Chen ZJ, Knipe DM, Sandri-Goldin RM, Enquist LW, Hartmann R, Mogensen TH, Rice SA, Nyman TA, Matikainen S, Paludan SR. 2016. HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J 35:1385–1399. doi: 10.15252/embj.201593458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen CY, Shyu AB. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20:465–470. doi: 10.1016/S0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 40.Fallmann J, Sedlyarov V, Tanzer A, Kovarik P, Hofacker IL. 2016. AREsite2: an enhanced database for the comprehensive investigation of AU/GU/U-rich elements. Nucleic Acids Res 44:D90–D95. doi: 10.1093/nar/gkv1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xing J, Wang S, Lin R, Mossman KL, Zheng C. 2012. Herpes simplex virus 1 tegument protein US11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J Virol 86:3528–3540. doi: 10.1128/JVI.06713-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paz S, Vilasco M, Arguello M, Sun Q, Lacoste J, Nguyen TL, Zhao T, Shestakova EA, Zaari S, Bibeau-Poirier A, Servant MJ, Lin R, Meurs EF, Hiscott J. 2009. Ubiquitin-regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter. Mol Cell Biol 29:3401–3412. doi: 10.1128/MCB.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang TH, Liao CL, Lin YL. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-kappaB activation. Microbes Infect 8:157–171. doi: 10.1016/j.micinf.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 44.Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, Wilkinson P, Julkunen I, Vitour D, Meurs E, Hiscott J. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J Virol 80:6072–6083. doi: 10.1128/JVI.02495-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jordan M, Schallhorn A, Wurm FM. 1996. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res 24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu H, Zheng C, Xing J, Wang S, Li S, Lin R, Mossman KL. 2011. Varicella-zoster virus immediate-early protein ORF61 abrogates the IRF3-mediated innate immune response through degradation of activated IRF3. J Virol 85:11079–11089. doi: 10.1128/JVI.05098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]