Abstract
In patients with colorectal cancer (CRC) that metastasizes to the liver, there are several key goals for improving outcomes including early detection, effective prognostic indicators of treatment response, and accurate identification of patients at high risk for recurrence. Although new therapeutic regimens developed over the past decade have increased survival, there is substantial room for improvement in selecting targeted treatment regimens for the patients who will derive the most benefit. Recently, there have been exciting developments in identifying high-risk patient cohorts, refinements in the understanding of systemic vs localized drug delivery to metastatic niches, liquid biomarker development, and dramatic advances in tumor immune therapy, all of which promise new and innovative approaches to tackling the problem of detecting and treating the metastatic spread of CRC to the liver. Our multidisciplinary group held a state-of-the-science symposium this past year to review advances in this rapidly evolving field. Herein, we present a discussion around the issues facing treatment of patients with CRC liver metastases, including the relationship of discrete gene signatures with prognosis. We also discuss the latest advances to maximize regional and systemic therapies aimed at decreasing intrahepatic recurrence, review recent insights into the tumor microenvironment, and summarize advances in noninvasive multimodal biomarkers for early detection of primary and recurrent disease. As we continue to advance clinically and technologically in the field of colorectal tumor biology, our goal should be continued refinement of predictive and prognostic studies to decrease recurrence after curative resection and minimize treatment toxicity to patients through a tailored multidisciplinary approach to cancer care.
Keywords: Colorectal Cancer Liver Metastasis, Biomarkers, Hepatic Arterial Infusion, High-Risk Colorectal Cancer, Recurrence
Abbreviations used in this paper: CDX2, caudal-type homeobox transcription factor 2; CEA, carcinoembryonic antigen; cfDNA, cell-free DNA; CK, cytokeratin; CRC, colorectal cancer; CRLM, colorectal cancer liver metastasis; CTC, circulating tumor cells; DFS, disease-free survival; dMMR, deficient mismatch repair; EGFR, epidermal growth factor receptor; EpCAM, epithelial cell adhesion molecule; 5-FU, fluorouracil; HAI, hepatic arterial infusion; IL, interleukin; LV, leucovorin; miRNA, microRNA; MSI, microsatellite instability; OS, overall survival; PD, programmed death; TH, T-helper
Summary.
Colorectal cancer ranks as the second leading cause of cancer-related deaths, with metastatic disease to the liver a common cause. Here we discuss exciting developments in the field that promise innovative approaches for early detection and treatment of metastatic colorectal cancer in the liver.
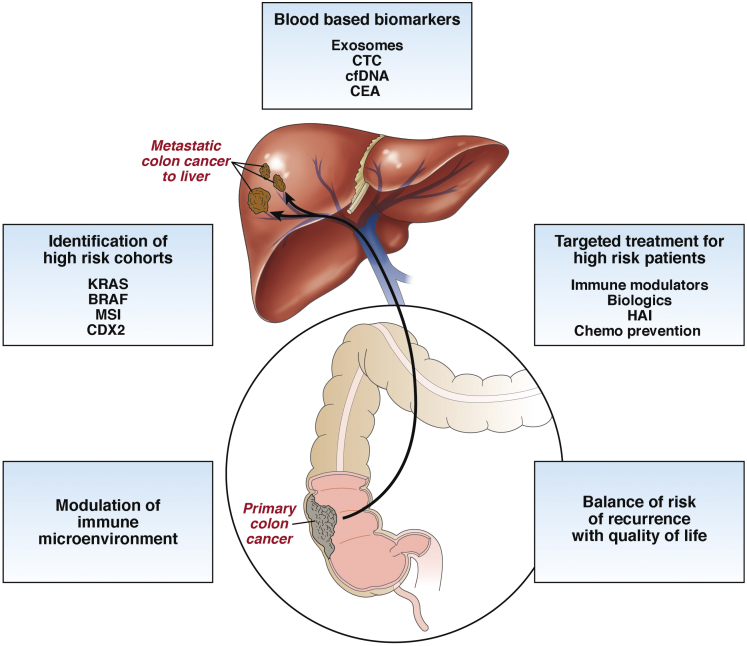
Colorectal cancer (CRC) is the third most common cancer worldwide, ranking as high as the second leading cause of cancer-related deaths in developed countries.1, 2, 3 The liver is recognized as the most common site of CRC metastasis because the majority of the intestinal mesenteric drainage enters the hepatic portal venous system. More than 50% of patients with CRC will develop metastatic disease to their liver over the course of their life, which ultimately results in death for more than two thirds of these patients.4, 5 Currently, hepatic resection of colorectal cancer liver metastasis (CRLM) in patients with isolated liver metastasis remains the only option for potential cure. However, even when resection is combined with modern adjuvant systemic regimens, it is curative in only 20% of patients,4, 5, 6 with 70% developing recurrence, primarily in the liver.4 Efforts to prevent recurrence are limited by the cumulative side effects of systemic therapy, development of chemoresistant cancer clones, and the inability to detect progression of radiographically occult micrometastatic disease. In an updated analysis of a large randomized controlled trial that examined the role of perioperative systemic therapy in patients with resectable CRLM before and after curative hepatic resection, there was no improvement in 5-year overall survival (OS) compared with patients treated with hepatic resection alone (51% vs 48%; P = .34).7, 8 Although perioperative systemic therapy remains the standard of care for patients with resected CRLM, there is significant opportunity to identify patients more accurately with a molecular high-risk signature who will benefit from adjuvant treatment aimed to decrease intrahepatic recurrence.9 In addition, for patients with liver-only metastatic CRC treated with curative intent surgery, detecting disease recurrence at the earliest stage and monitoring response to treatment are paramount to moving the field forward. In this report, we review modern approaches for treating patients with CRLM and ongoing work to optimize molecular risk stratification to direct systemic treatment and to monitor for intrahepatic recurrence (Figure 1).
Figure 1.
Treatment and management of metastatic colon cancer to the liver will require a multidisciplinary approach that includes identification of high-risk cohorts, understanding how to modulate the immune microenvironment, and identification of novel effective blood-based biomarkers to design targeted and real-time treatment for high-risk patients. Ultimately, successful management of these patients will acknowledge a balance of risk recurrence with their quality of life. Chemo, chemotherapy.
Scope of the Clinical Problem for Patients With Colorectal Cancer Liver Metastasis
Detecting primary CRC and CRLM at an early stage results in better outcomes.10 At a molecular level, CRC consists of a heterogeneous group of diseases with molecularly, as well as clinically, distinct tumors based on the primary site of origin (eg, colon vs rectal, and right-sided vs left-sided). Chromosomal instability, deficient mismatch repair (dMMR) with resultant microsatellite instability (MSI), aberrant DNA methylation,11 as well as altered molecular signaling pathways all have been described in the transformation from normal mucosa to adenocarcinoma.12, 13, 14, 15, 16 The role of biologics in the adjuvant treatment of resected primary CRC has been evaluated, including cetuximab for Kirsten rat sarcoma viral oncogene (KRAS) wild-type cancers and the vascular endothelial growth factor inhibitor bevacizumab; however, these targeted treatments have not shown the benefit seen in the metastatic or advanced setting.17, 18, 19 More recently, those altered pathways and mutations have been used for therapy modification and patient stratification in metastatic CRC based on the sidedness of the primary tumor, supporting the use of different biologic agents for distinct primary biology underlying the disease.17, 18, 19 Chromosomal anomalies with demonstrated importance in tumorigenesis, including DNA gains or losses, result in changes in gene expressions that might lead to a differential response to chemotherapeutic agents. This recently was studied in an analysis of cell-free DNA (cfDNA) showing acquired resistance to anti–epidermal growth factor (EGFR) therapies,20 as well as recent investigations reporting a correlation between DNA copy number losses and an association with response to fluorouracil (5-FU), irinotecan, and capecitabine.21
Given the extensive molecular and clinical heterogeneity of CRC, it is essential to individualize therapy on the basis of molecular profiling to avoid treatment-related toxicities without a realized survival benefit. Some of the strongest data to support the need for identification of high-risk cohorts among patients with CRLM come from adjuvant trials for primary CRC. The 2004 adjuvant the Multicenter International Study of Oxaliplatin/5-Fluorouracil/Leucovorin in the Adjuvant Treatment of Colon Cancer (MOSAIC) trial22 assessed the impact of an oxaliplatin-containing systemic regimen (folinic acid, 5-FU, and oxaliplatin) for patients with resected primary CRC compared with 5-FU alone in patients with stage II and III disease. A significant survival benefit for patients with stage III disease was found and has been maintained in recently updated 10-year results.23 However, these benefits come with significant morbidity impacting patient quality of life. For patients with stage III CRC treated with folinic acid, 5-FU, and oxaliplatin, instead of 5-FU and leucovorin (LV), there is a consequent 4% decrease in mortality.23 However, to achieve this 4% reduction in mortality with oxaliplatin, 92% of those patients will suffer from treatment-associated peripheral neuropathy, with approximately 15% experiencing permanent neuropathy when followed up longitudinally for 2 years.24 It is clear that even among patients with stage III disease there is an underappreciated disease heterogeneity that at present is being treated with an often-homogenous systemic approach. These data in the primary CRC setting underscore the need for molecularly driven systemic treatment to avoid both the financial and quality-of-life costs to patients with liver-only metastatic CRC. Work is ongoing to identify molecular subsets of patients with CRLM to personalize targeted treatments to maximize therapeutic interventions. In this review, we describe the role of liquid biopsies (ie, analyses of tumor cells or tumor derived material that is circulating in the blood) along with novel cancer and immunologic cell populations to both surveil and assess treatment response in patients with CRLM. We also propose using this information to guide the design and development of therapeutic strategies for liver-directed treatments.
Treatment Challenges for Patients With Liver-Only Metastases
For patients with liver-only metastatic CRC, there is a pressing need for a more robust molecular characterization of the primary and metastatic lesions to direct perioperative management of patients at highest risk for disease recurrence.25 In the primary disease setting, the focus has been directed toward patients with high-risk stage II CRC—those patients with negative lymph nodes but other high-risk features such as T4 lesions, obstruction or perforation, cancers with lymphovascular invasion, and poorly differentiated histology. One of the early investigations on the impact of adjuvant treatment on stage II CRC was the 2007 QUick and Simple And Reliable (QUASAR) trial, in which patients with stage II CRC were randomized to treatment with adjuvant 5-FU/LV or observation after curative resection of their primary cancer.26 The results of this trial showed an approximate 3% improvement in outcome when 5-FU/LV was given in the adjuvant setting. In other words, 97% of patients were exposed to chemotherapy without any benefit. Because standard stage II patients do not benefit from adjuvant therapy as shown in the QUASAR, MOSAIC, and other trials, it is currently at the discretion of the treating clinician to decide if the high-risk features of the patient’s primary CRC support adjuvant treatment.22, 23, 26 According to the current National Comprehensive Cancer Network guidelines the “definition of high-risk stage II colon cancer is clearly inadequate, because many patients with high-risk features do not have a recurrence whereas some patients deemed to be average-risk do. Furthermore, no data point to features that are predictive of benefit from adjuvant chemotherapy, and no data correlate risk features and selection of chemotherapy in patients with high-risk stage II disease.”27 The challenges of selecting patients with high-risk stage II CRC are remarkably similar to and parallel the issues of directing perioperative treatment in patients with CRLM.
Aside from the pathologic risk factors of stage II CRC, several investigators have sought a correlation between discrete gene signatures and higher-risk patient populations to stratify patients molecularly and to better direct adjuvant therapy. Efforts to improve patient stratification have been ongoing through comprehensive molecular characterization of hypermutated genes, microsatellite instability, and hypermethylated genes to characterize colon cancer stages into subtypes to better predict outcomes as well as to further refine chemotherapy selection.28, 29, 30 Rodriguez et al31 reported that loss of corticotropin-releasing hormone receptor-2 expression in CRC specimens—a well-characterized neuropeptide that participates in the regulation of intestinal inflammation—promotes proinflammatory signals through interleukin (IL)6 and vimentin. Based on clinicopathologic data they were able to show that a decreased expression of corticotropin-releasing hormone receptor-2 in patients with CRC was associated with an increased risk of distant metastasis and a worse 5-year survival after initiation of treatment. In 2016, Dalerba et al32 reported on the expression of the caudal-type homeobox transcription factor 2 (CDX2), a critical regulator of intestinal development and oncogenesis, as a prognostic biomarker in patients with stage II CRC. Their work combined insights from basic science discoveries in normal colon stem cells and cancer stem cells, the availability of public databases of sequenced tumors (National Center for Biotechnology Information Gene Expression Omnibus and National Cancer Institute–Cancer Diagnosis Program), and the power of bioinformatics to query more than 2329 human samples. They identified 16 genes that were not present in colorectal epithelia that expressed high levels of the cancer stem cell marker, Activated Leukocyte Cell Adhesion Molecule (ALCAM or CD166). Of these genes, they focused on CDX2, a protein already identified for pathologic assessments of resected CRC specimens. In a discovery data set of patients with stage II CRC and a subsequent validation data set, the investigators went on to show that patients with CDX2-negative tumors had a worse 5-year disease-free survival (DFS) compared with patients with CDX2-positive cancers (49% among 15 patients with CDX2-negative tumors vs 87% among 191 patients with CDX2-positive tumors; P = .003). Furthermore, using a discovery data set of stage III CRC tumors, investigators identified an association of CDX2 expression and the treatment of adjuvant therapy with survival. Again, these findings were validated in a larger data set, in which the investigators found an increased 5-year DFS in patients treated with adjuvant chemotherapy in stage II CDX2-negative tumors vs individuals who did not undergo adjuvant chemotherapy (91% vs 56%; P = .006).32 Although the study population was small, these data show an identifiable profile in CRC patients who may achieve a survival benefit from adjuvant treatment that outweighs the treatment-associated morbidity. This work was updated most recently in the metastatic CRC population,33, 34 in which patients with CDX2-negative metastatic CRC were found to have a median OS of 8 vs 39 months in individuals with CDX2-positive metastatic CRC (hazard ratio, 4.04; 95% confidence interval, 2.49–6.54; P < .0001). CDX2-negative patients were more likely to have right-sided primary tumors, poorly differentiated cancers, distant lymphatic metastasis, and be women. Although the prevalence of CDX2-negative disease is low, these insights continue to stratify a subgroup of patients with advanced CRC who would derive a DFS benefit from adjuvant treatment after curative hepatic resection of their disease. The continued focus to elucidate the underlying biology driving disease recurrence in more diverse and larger subsets of patients will clarify the effective treatment for patients at all stages of disease.
The majority of patients who have had an attempted curative hepatic resection of CRLM will have recurrence of their disease. Historically, several clinicopathologic factors (nodal status of the primary cancer, preoperative carcinoembryonic antigen [CEA] level, size of the largest liver lesion, and the number of hepatic metastases) have been shown to be independent predictors of both poor outcomes and intrahepatic recurrence of disease in patients with resected CRLM and collectively comprise the “clinical risk score.”6 Similar to the tumor characteristics in patients with clinically high-risk stage II CRC, these factors unfortunately provide a limited description of the disease. In addition to the prediction models, oncologists now are using mutational data in the EGFR pathways to select and treat patients who are most likely to respond to a given regimen (KRAS mutation status predicting poor response to anti-growth factor receptor therapies35, 36) and BRAF mutation status (conferring resistance to anti-EGFR therapy given beyond first-line treatment and associated with an increased risk of peritoneal disease).37, 38, 39, 40 Recent work has explored deriving cancer gene expression profiles as prognosticators of recurrence and survival for patients with CRLM. Balachandran et al9 reported a gene-expression classifier to correlate disease-specific survival as well as liver DFS in patients with resected CRLM. By using gene expression microarray on resected CRLM the investigators were able to identify and validate 20 genes that were associated with OS. Importantly, this so-called molecular risk score was shown to be an independent prognosticator of DFS, unlike the traditional clinical risk score. These findings suggest methods for identifying patients with high-risk primary CRC and resected CRLM who are at risk of recurrence and may benefit from directed and potentially prolonged adjuvant treatment. Further identification of patients with molecular subsets of CRLM that underlie discrete tumor biology, and subsequently predict treatment response and improve OS, are essential to realize the benefit of perioperative treatment with both biologic and cytotoxic therapy.
Maximizing Regional Treatment of Colorectal Cancer Liver Metastasis to Decrease Intrahepatic Recurrence
In patients with CRLM who undergo a hepatic resection with curative intent, it is estimated that approximately 75% of all recurrences—both intrahepatic and extrahepatic—occur within the first 2 years after surgery.41 Efforts over the past decades have sought to address the risk of recurrence, which is possibly the result of treatment-resistant micrometastatic disease. One avenue to obliterate micrometastatic disease in the liver focuses on maximizing locoregional therapy by exploiting basic tumor biology. Cancer cells from gastrointestinal malignancies, especially CRC, hematogenously spread via the portal circulation, often making the liver the first site of metastasis. Once hepatic metastases grow to more than 2 mm in size, they derive their blood supply from the hepatic artery, while normal hepatocytes are perfused mostly from the portal circulation.42 Understanding this biologic difference has led to treating select patients with CRLM using hepatic arterial infusion (HAI) therapy. This intense locoregional treatment is based on the extraction of chemotherapy from the hepatic arterial circulation, resulting in high local drug concentrations with the goal of minimizing systemic toxicity. The ideal agent should have a high dose-response curve, high extraction, and rapid total body clearance once the infusion is discontinued. Of the various agents studied, HAI-delivered floxuridine approximates this ideal with a short half-life (<10 min) and more than 90% hepatic extraction, resulting in a 16-fold higher concentration in hepatic tumors compared with venous administration.42, 43 By using floxuridine in combination with dexamethasone, patients with CRLM can have their liver disease maximally treated with modest side effects compared with standard systemic treatment.44 Several prospective trials45, 46, 47, 48 have investigated using HAI alone to circumvent the toxicity associated with systemic treatment of CRLM, to maximize hepatic response in an effort to improve both OS and progression-free survival, and potentially improve the patient’s quality of life.49, 50
The role of hepatic arterial infusion for patients with resected CRLM initially was tested without concurrent systemic therapy, which at the time of the initial trials did not include modern systemic agents such as oxaliplatin and irinotecan. To date, there have been no prospective randomized controlled trials comparing adjuvant HAI with modern systemic therapy vs modern systemic therapy alone in patients with resected CRLM. In 2016, Kemeny et al51 reported on an analysis of 4 consecutive HAI adjuvant trials for patients with resected CRLM from 1991 to 2009 (N = 287). The patients were divided into 2 groups: those treated before and after 2003, corresponding to the incorporation of modern systemic oxaliplatin or irinotecan-containing regimens. With a median follow-up period of 11 years, the investigators reported that patients treated after 2003 had a 5- and 10-year OS of 78% and 61%, respectively, with the median survival not being reached. Patients treated before 2003 had a 3- and 5-year DFS of 42% and 41%, respectively.51 Taken together, these data support that properly selected patients with CRLM can have hepatic resection of their disease followed by adjuvant systemic therapy plus HAI and achieve a 5-year survival as high as 78%. However, similar to toxicity associated with systemic therapy, treatment with HAI has risks, including biliary sclerosis in less than 5% of patients, that needs to be balanced with the anticipated benefit of treatment.51
For treatments such as HAI that seek to maximally treat the liver, it is imperative to begin integrating preoperative prognostic indicators that are predictive of a patient’s risk of intrahepatic recurrence after hepatic resection to select patients for intensive treatment regimens. Ultimately, we must develop a noninvasive test to determine the risk for both local and distant recurrence of disease with monitoring the response to systemic therapy as well as an early signal for intrahepatic recurrence. In addition to discrete gene expression profiles that identify patients at high risk of recurrence, blood-based biomarkers for noninvasive monitoring of early detection of recurrent disease actively in development may serve as a more reliable marker to monitor response to treatment, and ultimately to monitor patients for recurrence of disease after curative treatment.
Noninvasive Liquid Biomarkers for Early Detection of Primary and Recurrent Disease
Although newly identified gene signatures can identify at-risk patient populations for defined treatment regimens, a second approach for improving patient survival is in developing biomarkers with enhanced specificity and sensitivity for early detection of primary and recurrent disease. The goal of this approach is to identify recurrent or persistent disease at a point when traditional clinical indicators, such as radiographic signs, still are negative, and to treat or alter treatment of disease at the earliest time point—this almost certainly will improve overall disease control.
Genetic material sourced from blood-based material originating from primary and/or metastatic lesions can be used to inform noninvasive, blood-based biomarker discovery, and provide a nuanced view of the disease—specifically the temporal evolution of disease over treatment—to facilitate tailored therapy. The gold standard for a noninvasive early diagnosis test is the fecal occult blood test, but it has sensitivity limitations.52 Detection of the plasma-based factor CEA also widely is used to monitor disease status longitudinally in patients with treated CRC, but it also has limitations. To improve specificity and sensitivity, tumor biologists are pursuing a new generation of blood-based biomarkers that have correlative or biologic value, as well as providing tumor genomic information on which to alter treatment. Although still in development, these new factors, including new populations of circulating tumor cells (CTCs), cfDNA, micro-RNA (miRNA), and exosomes have the potential for the further development of critical assays to surveil patients with CRC and intervene at times that may improve disease control.53, 54
Conventionally isolated CTCs, defined by cell surface expression of epithelial cell adhesion molecule (EpCAM) or cytokeratin (CK), and the absence of the pan-leukocyte marker, CD45 expression, have been shown to correlate with progression-free survival and OS in patients with colorectal cancer,55 prostate cancer,56 and also with breast cancer.57 Although these data are predictive of prognosis, CTCs are rare entities in the circulation, and, more importantly, they have failed to provide biologic insights that may guide informed therapeutic treatment. Recent discoveries of novel CTC populations has re-energized the study of CTCs.
Standard CTC detection methods rely on the expression of specific epithelial markers, CK and/or EpCAM, and the exclusion of leukocyte-specific markers, typically CD45. CTCs also have been isolated based on size, density, charge, or various other properties that positively or negatively enrich a specific cell population.58 CellSearch (Janssen Diagnostics, Raritan, NJ) is the Food and Drug Administration–approved test to detect CTCs by magnetic separation of EpCAM+ cells followed by positive staining for CK and negative staining for CD45. These existing approaches bias the subsets of CTCs captured, and may be excluding biologically relevant subpopulations. For example, Zhang et al59 showed the high metastatic capability of an EpCAM- CTC population isolated from patients with breast cancer in a mouse xenograft assay. This EpCAM- CTC population may represent cancer cells that have undergone epithelial-to-mesenchymal transition, thereby losing expression of EpCAM-, and therefore represent a more migratory and invasive cell. Indeed, in CRC, CTCs that have lost EpCAM expression or gained N-cadherin, vimentin, or fibronectin expression are hypothesized to have undergone epithelial–mesenchymal transition and may be more stem cell–like.60 It reasons that to gain their full metastatic potential, tumor cells must cross several cellular barriers to travel through the circulation and seed metastatic sites. Recent mechanisms such as epithelial–mesenchymal transition have suggested enhanced motility and dissemination of these cells. Such cells appear to have adapted by loss of epithelial differentiation and acquisition of advantageous phenotypes to modulate and balance differentiation, self-renewal, and homeostasis in the selected environment.53, 54
An additional population of CTCs that have not been well studied are those expressing the leukocyte marker CD45. Peripheral blood cells from cancer patients, isolated by differential centrifugation and size exclusion, were found to harbor CTCs that expressed CK and CD45, yet conferred robust growth in culture.61 In addition, CD45+CK+ CTCs were identified in patients with metastatic pancreatic cancer from an EpCAM+-enriched population,62 and in metastatic breast cancer patients, even with partial CD45+ depletion with magnetic beads.63 Interestingly, the breast cancer CD45+ CTCs also expressed the macrophage marker CD68, indicating that CTC populations may acquire proteins typically expressed by macrophages, possibly through a cell fusion mechanism.64 To fully appreciate these CD45+ CTCs, direct visualization would rule cancer-immune cell clusters that could be construed as a CD45+ CTC by flow cytometry. If these cells arise from leukocyte-cancer fusion, this novel tumor biology may provide important insights that may guide therapy more effectively. Ongoing work is directed at investigating how these untapped and uninvestigated populations of CTCs potentially can contribute to our overall knowledge of disease and are being developed in parallel with the rapid advancements in other biomarker fields, such as that of cfDNA.
cfDNA is hypothesized to arise from cells that die, whether by necrosis, cell lysis, or apoptosis, releasing naked DNA into the circulation and creating a residual fingerprint. Although cfDNA was first detected in healthy individuals in the late 1940s, it was not until the 1970s–1980s that neoplastic characteristics were identified and that cfDNA was found to exist in higher concentrations in cancer patients relative to healthy controls.65, 66 Although quantification of cfDNA was useful in some disease states when used alongside classic blood tests (eg, CEA),67 cfDNA is being developed for the identification of gene mutations and microsatellite instability in early detection assays. Current technologic advancements in amplifying DNA and in sequencing supports the relevance of this biologic material. For example, de Kok et al68 showed concordance of KRAS point mutations between primary tumors and serum cfDNA amplified by polymerase chain reaction in 14 CRC patients. These data also supported that cfDNA could be derived from cancer cells.
In addition to point mutations, microsatellite abnormalities have been detected in patient blood from breast cancer, head and neck cancer, lung cancer, melanoma, and CRC.69, 70, 71, 72 Isolated cfDNA has many characteristics of tumor DNA, including the presence of oncogenes and other global molecular classifiers such as MSI, CpG island methylator phenotype,73 and chromosomal instability.74 El Messaoudi et al75 conducted a multiparametric analysis correlating cfDNA with OS in metastatic CRC patients (n = 97). Higher cfDNA levels were associated with a statistically significant decrease in OS (18.07 vs 28.5 mo; P = .0087). Furthermore, on multivariate analysis the investigators showed that a higher cfDNA level is an independent prognostic factor (P = .034) and that high levels of cfDNA fragmentation were associated with decreased OS in the mutant KRAS/BRAF population. Newer technologies under development such as the PlasmaSelect assay (Personal Genome Diagnostics, Baltimore, MD)76 allow for the identification of multiple mutations and genetic alterations resulting in a comprehensive genomic analysis of the tumor and the potential to track tumor evolution across treatment. Analyses of cfDNA along with evaluation of novel CTC populations have great potential to provide novel noninvasive approaches to diagnosis cancer, facilitate early detection of disease and recurrent disease, assessment of the evolving tumor biology, and provide a foundation for tailored treatment.
Aside from cfDNA and CTCs, there are active investigations into tumor-derived or tumor microenvironment-derived exosomal stable miRNAs that are released into the vasculature, glandular secretions, or waste excretions.77, 78, 79, 80 miRNAs are a small, recently discovered class of highly conserved noncoding RNAs that play key roles in the regulation of gene expression. In their transcriptional regulation, miRNAs possess differential short nucleotide length sequence complementarity to confer combinatorial diversity to affect hundreds of targets in similar gene networks. In this framework, a single miRNA exists in reciprocal inhibition with an entire network of functionally related genes. Thus, baseline activity or change in exosomal miRNAs provides a molecular snapshot of intracellular activity from their tissue of origin. Additional information can be derived because the presence of exosomal miRNAs dictates potential paracrine and endocrine roles that have been observed in a number of malignancies including CRC.81, 82, 83 Properties of exosomal miRNAs have been highlighted in recent clinical studies to ascertain their utility as novel minimally invasive biomarkers.84, 85, 86, 87 In CRC, Ogata-Kawata et al88 performed miRNA microarray analyses on serum exosomes derived from 88 CRC patients compared with healthy controls and identified a subset of 7 up-regulated diagnostic miRNAs that outperformed conventional CEA utility for surveillance of CRC recurrence. In CRLM, a similar study was performed in serum exosomes from both the liver metastases subgroup and the nonmetastatic group at different time points of treatment and resection.89 Microarray analyses showed a distinct subset of 6 tumor-derived exosomal micro RNAs (miRNA or miR), which were synchronized with liver metastasis development. Among this subset, exosomal miR-19a was found to be up-regulated compared with normal healthy volunteers, and the level of exosomal miR-19a was found to correlate with more aggressive disease including nodal involvement, liver metastases, and higher TNM stage.89 Taken together, these data provide a glimpse in the acquisition and analysis of exosomal miRNA diagnostic and predictive biomarkers in primary and metastatic CRC.
Immune Reprogramming as a Therapeutic Strategy
Over the past 5 years, there has been burgeoning interest and now demonstrated efficacy in exploiting the tumor immune microenvironment as a viable and effective treatment option for many cancers that previously had been recalcitrant to treatment. It has become increasingly clear that the tumor microenvironment plays a key role in tumor progression and response to therapies across many different cancer types.90 Immune check-point inhibitors such as ipilimumab, pembrolizumab, and nivolumab are being widely studied in prospective trials in a variety of cancers, including in patients with liver-only metastatic CRC. Several recent studies have highlighted the role of non-neoplastic cells, particularly stromal cells and immune cells, as prognostic markers in human CRC.91, 92, 93, 94 Immunologically, it has been shown that dMMR cancers harbor infiltrating tumor lymphocytes that actively are suppressed by immune-inhibitory signals such as the programmed death (PD) ligand-1 and PD-1 complexes.94 In 2015, Le et al reported the results of a phase II trial of patients with treatment-refractory progressive metastatic cancer treated with the anti–PD-1 antibody pembrolizumab.94 The results for 32 patients with metastatic CRC were stratified by dMMR CRC compared with patients with proficient MMR tumors. For patients with dMMR CRC, the immune-related objective response and immune-related progression-free survival were 40% and 78%, respectively, as compared with 0% and 11%, respectively, for patients with proficient MMR CRC. These findings currently are being evaluated in the KEYNOTE-177 trial in patients with MSI-high or dMMR metastatic CRC who have been randomized to treatment with pembrolizumab vs standard therapy.95
Specific features of the tumor microenvironment such as an abundance of T-helper (TH) 2 cytokines, proinflammatory molecules, pro-angiogenic molecules, and profibrotic molecules are immunosuppressive and considered protumorigenic. In contrast, an abundance of TH1 cytokines, angiostatic factors, and immunostimulatory molecules, along with the mobilization and reinvigoration of the CD8 T cells, all are characteristic of a robust antitumorigenic microenvironment. Therefore, understanding the recruitment and function of leukocytes in the cancer will enable the development of both targeted therapies and biomarkers that can predict emergence of treatment resistance and recurrence of cancer.96 Interestingly, there are several nuances that dictate the function of even the same leukocyte subsets in different cancers.97, 98 For example, protumorigenic macrophages are regulated by TH2-CD4+ T cells in mammary carcinomas and B cells in pancreatic adenocarcinomas and squamous cell carcinomas. Similarly, the soluble mediators and signaling pathways that regulate this cellular cross-talk also are different, with IL4/IL13 and colony stimulating factor-1 playing key roles in driving macrophage function in mammary carcinomas whereas Bruton tyrosine kinase and phosphoinositide 3′-kinase regulating the macrophage function in pancreatic and squamous cell carcinoma. As such, it is possible that the immune checkpoint pathways also are regulated differently in different tissues, necessitating the development of tailored approaches to immunotherapy across different cancers.99 In this context, we propose that a multimodal biomarker-based approach would be optimal for immune-mediated cancer control and assessing treatment response. Such an approach would rely on biomarkers that measure the protumorigenic and antitumorigenic factors elaborated earlier and provide multiple avenues to mobilize and reinvigorate the cytotoxic T-cell responses. This could include a combination of strategies such as neutralizing the TH2 responses, cytotoxic and targeted agents, immune checkpoint blockade, vaccines, and chimeric antigen-receptor T cells. This multimodal approach also will sample the microenvironment whenever the cancers escape and recalibrate the specific reprogramming strategy, specific immune checkpoints that can be targeted, and specific pathways that drive the microenvironment so cancer regression and control can be re-established. In summary, an approach that dynamically engages the immune system by constantly sampling the microenvironment to detect recurrence and relapse is essential to incorporate into the management of patients with advanced CRC and direct novel immunologic therapies.
Summary, Novel Molecules, and Future Clinical Trial Directions
Ultimately, the biology of the tumor—both cell intrinsic and cell extrinsic—underlies the clinical outcome for patients with metastatic CRC. Indeed, the concept of liver-only metastatic disease by definition implies a different biologic subtype. This difference is readily clinically apparent and our attempts to exploit this biology are the basis of all liver-directed therapy. Future directions in treating patients with the CRC liver-only subtype therefore must revolve around better molecular characterization and the development of improved therapeutic approaches. As liver-directed therapy continues to evolve with the development of other local treatment modalities—including microwave ablation, irreversible electroporation, and transarterial radioembolization (eg, Y-90)—our ability to identify and understand this biology becomes paramount. Currently, clinicians use the biologic test of time to ascertain if a hepatic-only metastatic state can be maintained while first-line systemic agents are used—and hence provide the rationale to attempt intensive liver-directed approaches, including hepatic resection and HAI. Banking tissue from these patients for molecular and clinicopathologic analyses, and correlating molecular and cellular correlates with high-quality clinical data, is essential and has become an integral aspect of modern prospective clinical trials. Tissue samples collected and analyzed across the continuum of a patient’s treatment are essential to facilitate discovery-based approaches to improve therapy. Specimens collected at several time points (eg, pretreatment, after each chemotherapeutic cycle, after completion of systemic therapy, and after hepatic resection) along with tissue from the surrounding hepatic parenchyma can be analyzed for a number of genomic and cellular alterations, including mutational analysis, copy number variability, and circulating biomarkers that have the potential to shed insight into evolving tumor biology that may predict treatment response. Data support that properly selected patients with CRLM can have hepatic resection of their disease followed by adjuvant systemic therapy plus HAI and achieve a 5-year OS as high as 78% with a hepatic DFS of 62% at 5 years.51 The role of HAI in the adjuvant treatment of patients with resected CRLM additionally offers the unique opportunity to deliver novel agents in a liver-directed fashion. Although floxuridine has been used for decades and represents a pharmacokinetically ideal agent, our rapidly expanding knowledge of CRC tumor biology and microenvironment begs for the development and study of novel agents coupled with the rational design of clinical trials that can exploit this knowledge in a liver-targeted fashion. Given the extensive molecular and clinical heterogeneity of the liver-only metastatic CRC, it is of great importance to individualize targeted therapy on the basis of molecular profiling through logical implementation of biomarker assessment in liquid biopsies for early metastasis to monitor for intrahepatic recurrence. We are charged with moving beyond a blunt one-size-fits-all approach in treating patients with CRLM. Ultimately, we believe that molecularly driven treatments will lead to improved OS, a reduction in intrahepatic recurrence, and will decrease the toxicity of perioperative therapies.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding The State of the Science Symposium held on June 17, 2016, in Portland, Oregon, and this review was supported by a Team Building Pilot Project grant from the Knight Cancer Institute at Oregon Health & Science University and Oregon State University.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Sameer A.S. Colorectal cancer: molecular mutations and polymorphisms. Front Oncol. 2013;3:114. doi: 10.3389/fonc.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J Clin. 2016;2016(66):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Tomlinson J.S., Jarnagin W.R., DeMatteo R.P. Actual 10-year survival after resection of colorectal liver metastases defines cure. J Clin Oncol. 2007;25:4575–4580. doi: 10.1200/JCO.2007.11.0833. [DOI] [PubMed] [Google Scholar]
- 5.House M.G., Kemeny N.E., Gönen M. Comparison of adjuvant systemic chemotherapy with or without hepatic arterial infusional chemotherapy after hepatic resection for metastatic colorectal cancer. Ann Surg. 2011;254:851–856. doi: 10.1097/SLA.0b013e31822f4f88. [DOI] [PubMed] [Google Scholar]
- 6.Fong Y., Fortner J., Sun R.L. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–321. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordlinger B., Sorbye H., Glimelius B. Perioperative chemotherapy with FOLFOX4 and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC Intergroup trial 40983): a randomised controlled trial. Lancet. 2008;371:1007–1016. doi: 10.1016/S0140-6736(08)60455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nordlinger B., Sorbye H., Glimelius B. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14:1208–1215. doi: 10.1016/S1470-2045(13)70447-9. [DOI] [PubMed] [Google Scholar]
- 9.Balachandran V.P., Arora A., Gönen M. A validated prognostic multigene expression assay for overall survival in resected colorectal cancer liver metastases. Clin Cancer Res. 2016;22:2575–2582. doi: 10.1158/1078-0432.CCR-15-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.UK Cancer Research. Early diagnosis. Vol 2016. Available from: http://www.cancerresearchuk.org/support-us/campaign-for-us/cross-cancer-out/early-diagnosis. Accessed October 8, 2016.
- 11.Powell S.M., Zilz N., Beazer-Barclay Y. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 12.Boland C.R., Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087 e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goel A., Nagasaka T., Arnold C.N. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 14.Pino M.S., Chung D.C. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacDonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw R.J., Cantley L.C. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 17.Venook A., Niedzwiecki D., Innocenti F. Impact of primary tumor location on overall survival and progression-free survival in patients with metastatic colorectal cancer: analysis of CALGB/SWOG 80405 (Alliance) J Clin Oncol. 2016;34(Suppl) abstract 3504. [Google Scholar]
- 18.Von Einem J., Heinemann V., von Weikersthal L.F. Left-sided primary tumors are associated with favorable prognosis in patients with KRAS codon 12/13 wild-type metastatic colorectal cancer treated with cetuximab plus chemotherapy: an analysis of the AIO KRK-0104 trial. J Cancer Res Clin Oncol. 2014;140:1607–1614. doi: 10.1007/s00432-014-1678-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tejpar S., Stintzing S., Ciardiello F. Prognostic and predictive relevance of primary tumor location in patients with RAS wild-type metastatic colorectal cancer: retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2016.3797. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spindler K.-L.G., Pallisgaard N., Vogelius I. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res. 2012;18:1177–1185. doi: 10.1158/1078-0432.CCR-11-0564. [DOI] [PubMed] [Google Scholar]
- 21.Leon L.G., Giovannetti E., Smid K. DNA copy number profiles correlate with outcome in colorectal cancer patients treated with fluoropyrimidine/antifolate-based regimens. Curr Drug Metab. 2011;12:956–965. doi: 10.2174/138920011798062337. [DOI] [PubMed] [Google Scholar]
- 22.André T., Boni C., Mounedji-Boudiaf L. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350:2343–2351. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- 23.André T., De Gramont A., Vernerey D. Adjuvant fluorouracil, leucovorin, and oxaliplatin in stage II to III colon cancer: updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC study. J Clin Oncol. 2015;33:4176–4187. doi: 10.1200/JCO.2015.63.4238. [DOI] [PubMed] [Google Scholar]
- 24.André T., Boni C., Navarro M. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol. 2009;27:3109–3116. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- 25.Benson A.B., 3rd, Hamilton S.R. Path toward prognostication and prediction: an evolving matrix. J Clin Oncol. 2011;29:4599–4601. doi: 10.1200/JCO.2011.37.8646. [DOI] [PubMed] [Google Scholar]
- 26.Quasar Collaborative Group. Gray R., Barnwell J. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 27.National Comprehensive Cancer Network Clinical practice guidelines in oncology (NCCN guidelines) Colon cancer version. 2016;2 https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf Available from. Accessed October 8, 2016. [Google Scholar]
- 28.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinicrope F.A., Shi Q., Smyrk T.C. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology. 2015;148:88–99. doi: 10.1053/j.gastro.2014.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phipps A.I., Limburg P.J., Baron J.A. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148:77–87. doi: 10.1053/j.gastro.2014.09.038. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez J., Huerta-Yepez S., Law I. Diminished expression of CRHR2 in human colon cancer promotes tumor growth and EMT via persistent IL-6/Stat3 signaling. Cell Mol Gastroenterol Hepatol. 2015;1:610–630. doi: 10.1016/j.jcmgh.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalerba P., Sahoo D., Paik S. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N Engl J Med. 2016;374:211–222. doi: 10.1056/NEJMoa1506597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B.Y., Jones J.C., Briggler A.M. Lack of caudal-type homeobox transcription factor 2 expression as a prognostic biomarker in metastatic colorectal cancer. Clin Colorectal Cancer. 2016 doi: 10.1016/j.clcc.2016.09.003. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 34.Kemeny N.E., Chou J.F., Capanu M. KRAS mutation influences recurrence patterns in patients undergoing hepatic resection of colorectal metastases. Cancer. 2014;120:3965–3971. doi: 10.1002/cncr.28954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Krieken J., Jung A., Kirchner T. KRAS mutation testing for predicting response to anti-EGFR therapy for colorectal carcinoma: proposal for an European quality assurance program. Virchows Arch. 2008;453:417–431. doi: 10.1007/s00428-008-0665-y. [DOI] [PubMed] [Google Scholar]
- 36.Yaeger R., Cercek A., Chou J.F. BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer. 2014;120:2316–2324. doi: 10.1002/cncr.28729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Cutsem E., Köhne C.-H., Láng I. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2010;33:5091. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 38.Di Nicolantonio F., Martini M., Molinari F. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 39.Prahallad A., Sun C., Huang S. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 40.Fong Y., Cohen A.M., Fortner J.G. Liver resection for colorectal metastases. J Clin Oncol. 1997;15:938–946. doi: 10.1200/JCO.1997.15.3.938. [DOI] [PubMed] [Google Scholar]
- 41.Ensminger W.D., Gyves J.W. Clinical pharmacology of hepatic arterial chemotherapy. Semin Oncol. 1983;10:176–182. [PubMed] [Google Scholar]
- 42.Kelly R.J., Kemeny N.E., Leonard G.D. Current strategies using hepatic arterial infusion chemotherapy for the treatment of colorectal cancer. Clin Colorectal Cancer. 2005;5:166–174. doi: 10.3816/ccc.2005.n.027. [DOI] [PubMed] [Google Scholar]
- 43.Kemeny N., Seiter K., Niedzwiecki D. A randomized trial of intrahepatic infusion of fluorodeoxyuridine with dexamethasone versus fluorodeoxyuridine alone in the treatment of metastatic colorectal cancer. Cancer. 1992;69:327–334. doi: 10.1002/1097-0142(19920115)69:2<327::aid-cncr2820690209>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 44.Kemeny N., Daly J., Reichman B. Intrahepatic or systemic infusion of fluorodeoxyuridine in patients with liver metastases from colorectal carcinoma. A randomized trial. Ann Intern Med. 1987;107:459–465. doi: 10.7326/0003-4819-107-4-459. [DOI] [PubMed] [Google Scholar]
- 45.Rougier P., Laplanche A., Huguier M. Hepatic arterial infusion of floxuridine in patients with liver metastases from colorectal carcinoma: long-term results of a prospective randomized trial. J Clin Oncol. 1992;10:1112–1118. doi: 10.1200/JCO.1992.10.7.1112. [DOI] [PubMed] [Google Scholar]
- 46.Lorenz M., Muller H.H. Randomized, multicenter trial of fluorouracil plus leucovorin administered either via hepatic arterial or intravenous infusion versus fluorodeoxyuridine administered via hepatic arterial infusion in patients with nonresectable liver metastases from colorectal carcinoma. J Clin Oncol. 2000;18:243–254. doi: 10.1200/JCO.2000.18.2.243. [DOI] [PubMed] [Google Scholar]
- 47.Kerr D.J., McArdle C.S., Ledermann J. Intrahepatic arterial versus intravenous fluorouracil and folinic acid for colorectal cancer liver metastases: a multicentre randomised trial. Lancet. 2003;361:368–373. doi: 10.1016/S0140-6736(03)12388-4. [DOI] [PubMed] [Google Scholar]
- 48.Kemeny N.E., Niedzwiecki D., Hollis D.R. Hepatic arterial infusion versus systemic therapy for hepatic metastases from colorectal cancer: a randomized trial of efficacy, quality of life, and molecular markers (CALGB 9481) J Clin Oncol. 2006;24:1395–1403. doi: 10.1200/JCO.2005.03.8166. [DOI] [PubMed] [Google Scholar]
- 49.Allen-Mersh T.G., Earlam S., Fordy C. Quality of life and survival with continuous hepatic-artery floxuridine infusion for colorectal liver metastases. Lancet. 1994;344:1255–1260. doi: 10.1016/s0140-6736(94)90750-1. [DOI] [PubMed] [Google Scholar]
- 50.Kemeny N.E., Chou J.F., Boucher T.M. Updated long-term survival for patients with metastatic colorectal cancer treated with liver resection followed by hepatic arterial infusion and systemic chemotherapy. J Surg Oncol. 2016;113:477–484. doi: 10.1002/jso.24189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geiger T.M., Ricciardi R. Screening options and recommendations for colorectal cancer. Clin Colon Rectal Surg. 2009;22:209–217. doi: 10.1055/s-0029-1242460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dalerba P., Cho R.W., Clarke M.F. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 53.Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell. 2012;22:699–701. doi: 10.1016/j.ccr.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 54.Cohen S.J., Punt C.J.A., Iannotti N. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 55.de Bono J.S., Scher H.I., Montgomery R.B. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 56.Cristofanilli M., Budd G.T., Ellis M.J. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 57.Alix-Panabieres C., Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 58.Zhang L., Ridgway L.D., Wetzel M.D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5:180ra48. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hardingham J.E., Grover P., Winter M. Detection and clinical significance of circulating tumor cells in colorectal cancer—20 years of progress. Mol Med. 2015;21:S25–S31. doi: 10.2119/molmed.2015.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clawson G.A., Matters G.L., Xin P. Macrophage-tumor cell fusions from peripheral blood of melanoma patients. PLoS One. 2015;10:e0134320. doi: 10.1371/journal.pone.0134320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao Y., Zhu Y., Zhang Z. Clinical significance of pancreatic circulating tumor cells using combined negative enrichment and immunostaining-fluorescence in situ hybridization. J Exp Clin Cancer Res. 2016;35:66. doi: 10.1186/s13046-016-0340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lustberg M.B., Balasubramanian P., Miller B. Heterogeneous atypical cell populations are present in blood of metastatic breast cancer patients. Breast Cancer Res. 2014;16:1–15. doi: 10.1186/bcr3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Powell A.E., Anderson E.C., Davies P.S. Fusion between intestinal epithelial cells and macrophages in a cancer context results in nuclear reprogramming. Cancer Res. 2011;71:1497–1505. doi: 10.1158/0008-5472.CAN-10-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leon S.A., Shapiro B., Sklaroff D.M. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646. [PubMed] [Google Scholar]
- 65.Stroun M., Anker P., Maurice P. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46:318–322. doi: 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- 66.Shapiro B., Chakrabarty M., Cohn E.M. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51:2116–2120. doi: 10.1002/1097-0142(19830601)51:11<2116::aid-cncr2820511127>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 67.de Kok J.B., van Solinge W.W., Ruers T.J. Detection of tumour DNA in serum of colorectal cancer patients. Scand J Clin Lab Invest. 1997;57:601–604. doi: 10.3109/00365519709055283. [DOI] [PubMed] [Google Scholar]
- 68.Mayall F., Fairweather S., Wilkins R. Microsatellite abnormalities in plasma of patients with breast carcinoma: concordance with the primary tumour. J Clin Pathol. 1999;52:363–366. doi: 10.1136/jcp.52.5.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bruhn N., Beinert T., Oehm C. Detection of microsatellite alterations in the DNA isolated from tumor cells and from plasma DNA of patients with lung cancer. Ann N Y Acad Sci. 2000;906:72–82. doi: 10.1111/j.1749-6632.2000.tb06594.x. [DOI] [PubMed] [Google Scholar]
- 70.Nakayama T., Taback B., Nguyen D.H. Clinical significance of circulating DNA microsatellite markers in plasma of melanoma patients. Ann N Y Acad Sci. 2000;906:87–98. doi: 10.1111/j.1749-6632.2000.tb06596.x. [DOI] [PubMed] [Google Scholar]
- 71.Nawroz H., Koch W., Anker P. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med. 1996;2:1035–1037. doi: 10.1038/nm0996-1035. [DOI] [PubMed] [Google Scholar]
- 72.Kloten V., Rose M., Kaspar S. Epigenetic inactivation of the novel candidate tumor suppressor gene ITIH5 in colon cancer predicts unfavorable overall survival in the CpG island methylator phenotype. Epigenetics. 2014;9:1290–1301. doi: 10.4161/epi.32089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crowley E., Di Nicolantonio F., Loupakis F. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 74.El Messaoudi S., Mouliere F., Du Manoir S. Circulating DNA as a strong multimarker prognostic tool for metastatic colorectal cancer patient management care. Clin Cancer Res. 2016;22:3067–3077. doi: 10.1158/1078-0432.CCR-15-0297. [DOI] [PubMed] [Google Scholar]
- 75.Parpart-Li S., Angiuoli S., Chesnick B. Abstract P2-01-04: a method for comprehensive genomic analysis of cell free DNA. Cancer Res. 2016;76 P2-01-04. [Google Scholar]
- 76.Tran N. Cancer exosomes as miRNA factories. Trends Cancer. 2016;2:329–331. doi: 10.1016/j.trecan.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 77.Montani F., Marzi M.J., Dezi F. miR-Test: a blood test for lung cancer early detection. J Natl Cancer Inst. 2015;107:djv063. doi: 10.1093/jnci/djv063. [DOI] [PubMed] [Google Scholar]
- 78.Ge Q., Zhou Y., Lu J. miRNA in plasma exosome is stable under different storage conditions. Molecules. 2014;19:1568–1575. doi: 10.3390/molecules19021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balzano F., Deiana M., Dei Giudici S. miRNA stability in frozen plasma samples. Molecules. 2015;20:19030–19040. doi: 10.3390/molecules201019030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Felicetti F., De Feo A., Coscia C. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J Transl Med. 2016;14:56. doi: 10.1186/s12967-016-0811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh R., Pochampally R., Watabe K. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol Cancer. 2014;13:256. doi: 10.1186/1476-4598-13-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lugini L., Valtieri M., Federici C. Exosomes from human colorectal cancer induce a tumor-like behavior in colonic mesenchymal stromal cells. Oncotarget. 2016;7:50086–50098. doi: 10.18632/oncotarget.10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cazzoli R., Buttitta F., Di Nicola M. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8:1156–1162. doi: 10.1097/JTO.0b013e318299ac32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eichelser C., Stückrath I., Müller V. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5:9650–9663. doi: 10.18632/oncotarget.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thind A., Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292. doi: 10.3402/jev.v5.31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ye S.B., Li Z.L., Luo D.H. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5:5439–5452. doi: 10.18632/oncotarget.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ogata-Kawata H., Izumiya M., Kurioka D. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matsumura T., Sugimachi K., Iinuma H. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113:275–281. doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hanahan D., Coussens L.M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 90.Mlecnik B., Bindea G., Kirilovsky A. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Sci Transl Med. 2016;8:327ra26. doi: 10.1126/scitranslmed.aad6352. [DOI] [PubMed] [Google Scholar]
- 91.Mlecnik B., Bindea G., Angell H.K. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44:698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 92.Isella C., Terrasi A., Bellomo S.E. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47:312–319. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 93.Calon A., Lonardo E., Berenguer-Llergo A. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320–329. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 94.Le D.T., Uram J.N., Wang H. PD-1 blockade in tumors with mismatch-repair deficiency. NEJM. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Study of Pembrolizumab (MK-3475) vs. Standard Therapy in Participants with Miscrosatellite Instability-High or Mismatch Repair Deficient Stage IV Colorectal Carcinoma (MK-34-177/KEYNOTE-177). Available at: https://clinicaltrials.gov/ct2/show/NCT02563002. Accessed February 6, 2017.
- 96.Palucka A.K., Coussens L.M. The basis of oncoimmunology. Cell. 2016;164:1233–1247. doi: 10.1016/j.cell.2016.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ruffell B., Chang-Strachan D., Chan V. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26:623–637. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Affara N.I., Ruffell B., Medler T.R. B cells regulate macrophage phenotype and response to chemotherapy in squamous carcinomas. Cancer Cell. 2014;25:809–821. doi: 10.1016/j.ccr.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Topalian S.L., Taube J.M., Anders R.A. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]