Abstract
Aim:
Sex-based differences in response to adverse prenatal environments and infant outcomes have been observed, yet the underlying mechanisms for this are unclear. The placental epigenome may be a driver of these differences.
Methods:
Placental DNA methylation was assessed at more than 480,000 CpG sites from male and female infants enrolled in the extremely low gestational age newborns cohort (ELGAN) and validated in a separate US-based cohort. The impact of gestational age on placental DNA methylation was further examined using the New Hampshire Birth Cohort Study for a total of n = 467 placentas.
Results:
A total of n = 2745 CpG sites, representing n = 587 genes, were identified as differentially methylated (p < 1 × 10-7). The majority (n = 582 or 99%) of these were conserved among the New Hampshire Birth Cohort. The identified genes encode proteins related to immune function, growth/transcription factor signaling and transport across cell membranes.
Conclusion:
These data highlight sex-dependent epigenetic patterning in the placenta and provide insight into differences in infant outcomes and responses to the perinatal environment.
Keywords: : CpG DNA methylation, epigenetics, placenta, sexual dimorphism
The placenta serves as the primary organ responsible for regulation of the prenatal environment critical for optimal fetal development. It is a nutrient transporter and produces vital hormones to maintain pregnancy and support the fetus [1]. In contrast to these essential functions, the placenta also serves as a source of exposure for toxic substances, dysregulated hormone signaling and immune-related proteins [2]. Such exposures including maternal stress, excess hormones, cytokines or environmental toxicants impact fetal health and influence later-life health outcomes [3–11].
An increasing body of literature supports sex-specific birth and later-life outcomes in relation to adverse in utero environments [12]. For example, sex-specific perinatal and later-life outcomes have been linked to toxic substance exposure [5,6,13–16], maternal stress [7] and maternal immune status [9,17,18]. We hypothesize that sex-dependent health outcomes in infants may be associated with sexual dimorphism of the placenta. In support of this hypothesis, physiologic differences between male and female placentas have been observed [19]. For example, placentas derived from pregnancies of males and females display variation in the abundance and type of glucocorticoid transporter proteins, the expression of hormones and the production of immune-related proteins including cytokines [12,19–21]. In addition, sex-based differences in response to prenatal stressors have been observed at the levels of the placental proteome, transcriptome and epigenome [3,7,14,20,22–27]. Thus, the sexually dimorphic nature of the placenta could influence variation in toxicant transport and accumulation, hormone levels and immune response experienced by the fetus. While key physiological differences have been observed in male and female placentas, the underlying mechanisms are not well established.
Epigenetic regulation may underlie the physiologic differences observed between male and female placentas [12]. For instance, key differences in chromatin structure have been observed between male and female placentas, suggesting that there is a role for epigenetic regulation in placental sexual dimorphism [12,21]. Understanding epigenetic regulation in the placenta is important given the modifiability of its epigenome in response to prenatal stressors, and its role as a mediator of the developmental origins of health and disease [2,20,21,28]. Additionally, because certain epigenomic markers are stable over time [29], it is possible that changes to the fetal placenta methylome explain the sex-based differences in later-life disease risks.
To our knowledge, this study is among the first to address the gap in knowledge related to sexual dimorphism of the placental DNA methylome. Here, we investigated whether DNA (CpG) methylation patterns in the placenta differed based on the sex of the fetus. Sex-based differences within the placenta were identified in a subset of subjects from the extremely low gestational age newborns (ELGAN) cohort [30–35], and validated in a replication cohort comprised of women recruited at the University of North Carolina (UNC) hospitals [36]. To confirm that gestational age was not a major driver of the differences in DNA methylation, all data were subsequently compared with CpG methylation assessed in the New Hampshire Birth Cohort. The goal of these analyses is to provide key insights into whether sexual dimorphism of the placental methylome may explain differential susceptibility to adverse prenatal environmental conditions.
Methods
ELGAN study subject recruitment & sample collection
ELGAN study recruitment has been previously described in detail [37]. In short, from 2002 to 2004, women giving birth at one of the 14 participating sites before 28 weeks gestation were asked to enroll in the ELGAN study. Their consent was provided either upon hospital admission prior to or shortly after delivery. All procedures were approved by the Institutional Review Board at each of the 14 participating study sites.
A total of 1506 infants and 1249 mothers enrolled in the ELGAN study. A subcohort of 84 mother-infant pairs, representative of the larger cohort, were selected for this analysis. As part of participation in ELGAN, women were asked to contribute their placentas. A description of the methodology that was used for placental collection is as follows: delivered placentas were placed in a sterile exam basin and transported to a sampling room, where they were biopsied under sterile conditions. At the midpoint of the longest distance between the cord insertion and the edge of the placental disk, the amnion was pulled back using sterile technique to expose the chorion. Traction was applied to the chorion and the underlying trophoblast tissue and a piece of tissue was removed by cutting at the base of the section. The tissue was placed into a cryo vial and immediately immersed in the liquid nitrogen. Specimens were stored at -80°C until laboratory processing [38].
The replication cohort included 40 subjects of women receiving obstetric care at UNC hospitals who consented to collection of placental samples at the time of birth. Subjects gave written informed consent prior to enrollment and participation in this study. A full-thickness placental biopsy was obtained after delivery, avoiding the periphery and areas of obvious infarction. Samples were flash frozen in liquid nitrogen, and stored at -70°C until analysis. This research was approved by the Institutional Review Board at the University of North Carolina (#11–2054).
Data from a third cohort comprising 343 placenta samples obtained from women and children involved in the New Hampshire Birth Cohort, an ongoing prospective pregnancy and birth cohort, were utilized as a contrast and validation of the results in a cohort with a more generalizable spread of gestational age. This cohort and the placental DNA methylation array data have been previously described [39]. Briefly, pregnant women whose primary source of residential water is a private well and who obtained their prenatal care at clinics in New Hampshire, were recruited into the study. Eligibility criteria were that women were currently pregnant; 18–45 years old; received routine prenatal care at one of the study clinics; used a private well that serves <15 households or 25 individuals at their residence; resided in the sample place since their last menstrual period; and did not plan to move prior to delivery. Like the replication cohort, a full-thickness placental biopsy was obtained after delivery, avoiding the periphery and areas of obvious infarction, and placed immediately in RNA later for at least 48 h, then removed from the fixative and stored at -80°C until laboratory processing.
DNA extraction & assessment of DNA methylation
DNA extraction and assessment of DNA methylation was performed as follows. A 0.2 g subsection of placental tissue was cut from each frozen biopsy on dry ice, washed briefly in sterile 1× phosphate-buffered saline (PBS) to remove any residual blood, and homogenized in Buffer RLT™ with β-mercaptoethanol (Qiagen, CA, USA). DNA and RNA sequences >18 nucleotides in length were collected using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, CA, USA), according to manufacturer’s instructions. CpG methylation was assessed using the Illumina HumanMethylation450 BeadChip© array (Illumina, Inc., CA, USA). This platform assesses the DNA methylation levels of 486,428 individual probes at single nucleotide resolution. Isolated DNA was first bisulfite converted using the EZ DNA methylation kit (Zymo Research, CA, USA) and converted DNA was then hybridized onto the array. The DNA methylation data were collected at Expression Analysis, Inc. (NC, USA; www.expressionanalysis.com). Methylation levels were calculated and expressed as β values (β = intensity of the methylated allele [M]/intensity of the unmethylated allele [U] + intensity of the methylated allele [M] + 100).
Statistical analysis
The array signal data were processed using R software (version 3.0.3). For each set of samples, batch effect was evaluated using principle component analysis and was determined to not be a significant source of variation. Any methylation β-values with a detection p-value above 0.01 were considered unreliable and were removed from analysis. β-mixture quantile normalization was performed using the WateRmelon package (version 1.11.0) in R [40]. Following normalization, probes containing a single nucleotide polymorphism in the assayed CpG dinucleotide, as well as those for which two or more single nucleotide polymorphisms were located in the probe sequence were removed. Last, probes on the Y chromosome were removed from this study as they were not comparable between male and female placental samples [41,42]. A total of n = 377,673 probes, representing 20,442 genes, remained for analysis. β-differences were calculated for differentially methylated probes by subtracting the median female β-value from the median male β-value. To identify covariates, a directed acyclic graph approach was used. No factors were identified that influenced both the sex of the infant and the methylome. For this reason, p-values were calculated from t-statistics. Multiple testing correction of the p-values was performed using the Bonferroni correction (p < 1 × 10-7) as well as a q-value correction, using the Partek Genomic Suite (MO, USA).
To replicate these findings, CpG methylation data were analyzed in a separate cohort based at UNC hospitals using the HumanMethylation450 BeadChip© (Illumina, CA, USA). Data were processed and analyzed according to the protocol described above, however data for only those CpG locations which were statistically significant in the ELGAN dataset were tested (n = 4371). This increased the Bonferroni corrected p-value threshold to 1×10- 5. In addition to the validation performed, t-statistics and a multiple test-corrected q-value were calculated for all probes.
Given the known impact of gestational age on the placental DNA methylome [43,44], and that the two previously described cohorts utilized in this study were of lower gestational age, data from the New Hampshire birth cohort [39] were also assessed (n = 343 placentas). In addition, we also performed a second analysis where linear regression modeling was used to control for gestational age in each of the cohorts.
Transcription factor binding site analysis of validated gene sets
To characterize transcription factor binding site patterns, two separate analyses were conducted. First, enriched transcription factor binding site motifs were identified using Gene Set Enrichment Analysis (GSEA) [45]. This analysis relies on a Kolmogorov–Smirnov like statistic of an enrichment score to calculate p-values. Subsequently, the p-values are false discovery rate corrected by calculating a normalized enrichment score. Second, identified transcription actors were validated with the Genomatix Common Transcription Factor Binding Site module using methods as previously described [46].
Gene set-based analysis
In addition to transcription factor binding site analysis, we analyzed whether the differentially methylated probes in the placenta were enriched for specific biological functions. Four gene sets were analyzed where gene content was established using www.uniprot.org. Specifically, four gene sets were tested: transport-related genes (n = 3704), immune-related genes (n = 1622), inflammation-related genes (n = 410) and growth/transcription-related genes (n = 2030). A χ2 test was used to identify enrichment of key functions. This test compares the relative abundance of genes associated with a key biological function among those found to be differentially methylated to the relative abundance across the genome. The p-value represented the probability of identifying a similar number of genes from a random sample of the same size from the genome. In addition to the comparison to these known functional groups, the identified differentially methylated sexually dimorphic genes were also compared with 142 genes that have been previously identified as having sexually dimorphic gene expression in human placentas [47]. These data were collected from a previous meta-analysis that identified genes differentially expressed in male and female placentas [47]. For these analyses, relaxed statistical significance was used to define differentially methylated probes (p < 0.05; q < 0.1) [47], to enable comparability to the previously published work.
Results
Study subject characteristics
In the ELGAN cohort subset, 58 (69%) were male and 26 (31%) were female. In the UNC cohort, 19 (48%) were male and 21 (53%) were female (Table 1). For both cohorts, none of the demographic variables differed between males and females. These variables included average maternal age, gestational age, parity, smoking status and race. Demographic characteristics for the New Hampshire Birth Cohort Study have been previously published [39].
Table 1. . Demographics.
| Sex |
ELGAN |
UNC |
||||
|---|---|---|---|---|---|---|
| Overall N (%), (range) | Male (n = 58) N (%), (range) | Female (n = 26) N (%), (range) | Overall N (%), (range) | Male (n = 19) N (%), (range) | Female (n = 21) N (%), (range) | |
| M |
58 (69%) |
|
|
19 (48%) |
|
|
| F |
26 (31%) |
|
|
21 (53%) |
|
|
| Gestational age |
25.5 (23–27) |
25.3 (23–27) |
25.9 (24–27) |
35.6 (22–41) |
34.4 (22–41) |
36.7 (27–41) |
| Maternal age |
28.6 (16–41) |
28.3 (16–41) |
29.2 (18–39) |
28.3 (19–38) |
28.8 (20–35) |
27.7 (19–38) |
| Race |
|
|
|
|
|
|
| White |
38 (45%) |
24 (41%) |
14 (39%) |
13 (33%) |
6 (32%) |
5 (24%) |
| Nonwhite |
46 (55%) |
34 (49%) |
12 (61%) |
27 (67%) |
13 (68%) |
16 (76%) |
| Smoking status |
|
|
|
|
|
|
| Yes |
12 (14%) |
7 (13%) |
5 (19%) |
3 (8%) |
1 (5%) |
2 (10%) |
| No |
70 (84%) |
49 (84%) |
21 (81%) |
37 (92%) |
18 (95%) |
19 (90%) |
| Not reported | 2 (2%) | 2 (3%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
ELGAN: Extremely low gestational age newborn; UNC: University of North Carolina.
Identification & replication of differentially methylated probes in ELGAN
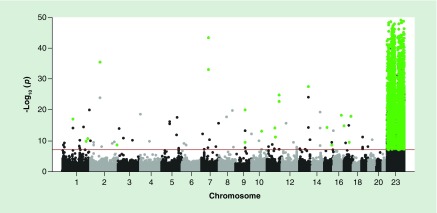
In the ELGAN cohort, we analyzed CpG methylation differences between male and female infants for 377,673 CpG probes representing 20,442 genes across 84 placentas. For this analysis, statistical significance was set at the Bonferroni-corrected p-value threshold of p < 1 × 10-7. A total of 4371 probes, representing 714 genes were found to be differentially methylated between male and female placentas (Figure 1, Supplementary Table 1). The vast majority (97.9%) of these probes (n = 4280, 666 genes) were located on the X chromosome (Table 2). Only 91 differentially methylated probes (2.1%), representing 48 genes, were located in autosomal regions (Table 2). When comparing probes that were differentially methylated between males and females, the greatest absolute β-difference displayed was 44.3% and the smallest was 2.5% (Figure 1, Supplementary Table 1). The majority of probes (99.2%) were greater than 5%, a level that has been previously used as a threshold while identifying sex-based differences [41]. Interestingly, and in contrast to our a priori hypothesis, 52.5% (n = 2296) probes were hypermethylated in males as compared with females (Table 2). Hypermethylation of probes in males relative to females occurred on both autosomal chromosomes (n = 73, 82.4%) and the X chromosome (n = 2223, 51.9%) (Table 2).
Figure 1. . Manhattan plot displaying all CpG probes tested for the analysis of sex-dependent DNA methylation in the placenta.
Probes are organized according to chromosomal positions. Probes in black and gray represent probes tested. Probes above the red line were those that were statistically significant. Probes in green are those that were validated in the replication cohort.
Table 2. . Comparison of methylation levels among sex-dependent differentially methylated probes.
| Differentially methylated probes (DMPs) |
Overall |
X |
Autosomal |
|||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
|
DMPs identified in ELGAN | ||||||
| All probes |
4371 |
- |
4280 |
- |
91 |
- |
| Hypermethylated probes (βMales > βFemales) |
2296 |
52.5 |
2223 |
51.9 |
73 |
80.2 |
| Hypomethylated probes (βMales < βFemales) |
2075 |
47.5 |
2057 |
48.1 |
18 |
19.8 |
|
DMPs validated in the replication cohort | ||||||
| All probes |
2745 |
- |
2724 |
- |
21 |
- |
| Hypermethylated probes (βMales > βFemales) |
1407 |
51.3 |
1394 |
51.2 |
13 |
61.9 |
| Hypomethylated probes(βMales < βFemales) | 1338 | 48.7 | 1330 | 48.8 | 8 | 38.1 |
DMP: Differentially methylated probe; ELGAN: Extremely low gestational age newborn.
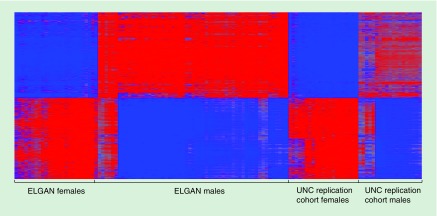
To establish whether CpG methylation levels in the ELGAN subjects replicated in a separate cohort, we analyzed placental CpG methylation data from a cohort of 40 pregnant women from the UNC hospital. A total of 2745 probes, representing 587 genes, replicated across both cohorts as differentially methylated between male and female placentas (Figure 2, Figure 1, Supplementary Table 1).
Figure 2. . Heatmap displaying the 2745 validated sexually dimorphic probes.
Red represents increased methylation in males relative to females and blue represents decreased methylation in males relative to females.
ELGAN: Extremely low gestational age newborn; UNC: University of North Carolina.
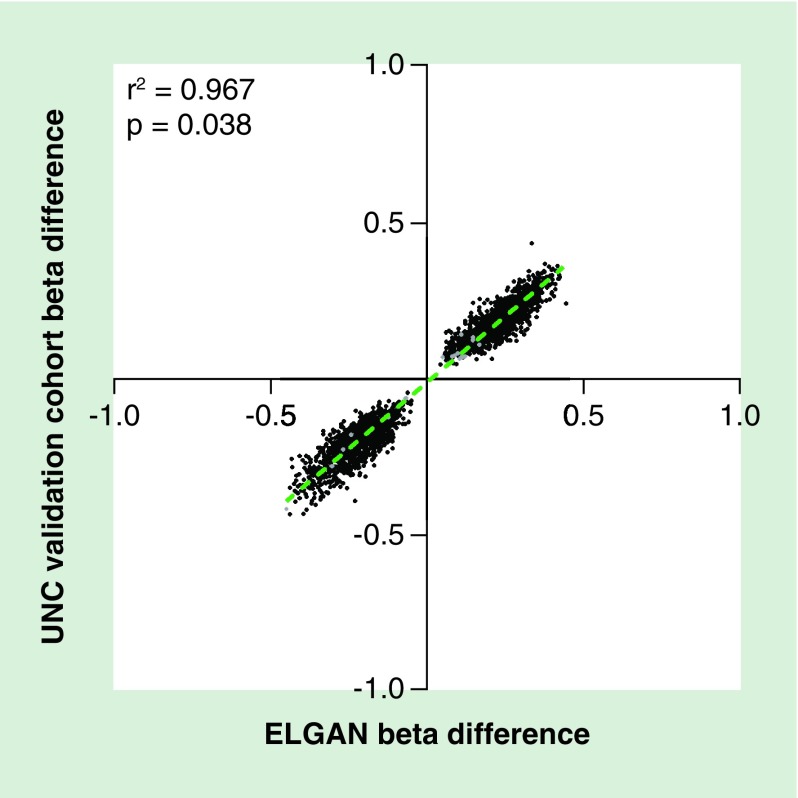
As noted within the ELGAN subcohort, the vast majority of probes 99.2% (n = 2724, 574 genes) were located on the X chromosome, and only 21 probes (13 genes) occurred on autosomal chromosomes (Table 2, Table 3). Again we observed similar patterns of hypermethylation where 51.3% (n = 1407) probes were hypermethylated in males as compared with female placentas, and this pattern was true for probes on the X chromosome (n = 1394, 51.2%) (Table 2). Additionally, across cohorts changes in the magnitude and direction of methylation were highly consistent as represented by the strong correlation between β-values (r2 = 0.95; p < 0.0001) (Figure 3).
Table 3. . Autosomal sex-dependent differentially methylated probes (n = 21 probes, 13 gene).
| Probe ID | Gene | Position | CHR |
ELGAN |
Replication |
||||
|---|---|---|---|---|---|---|---|---|---|
|
Median β |
Median β |
||||||||
| p-value | Male | Female | p-value | Male | Female | ||||
| cg26827413 |
|
|
1 |
1.26 × 10-17 |
0.873 |
0.765 |
3.21 × 10-06 |
0.808 |
0.739 |
| cg07170752 |
DNAH14 |
Body |
1 |
2.94 × 10-11 |
0.830 |
0.782 |
5.03 × 10-06 |
0.748 |
0.678 |
| cg15817705 |
|
|
1 |
1.84 × 10-10 |
0.712 |
0.632 |
7.89 × 10-06 |
0.668 |
0.595 |
| cg11955727 |
|
|
2 |
3.44 × 10-36 |
0.593 |
0.898 |
1.92 × 10-10 |
0.532 |
0.811 |
| cg19484299 |
MSL3L2 |
Body |
2 |
2.53 × 10-09 |
0.877 |
0.711 |
1.46 × 10-06 |
0.761 |
0.651 |
| cg04462931 |
|
|
7 |
4.49 × 10-44 |
0.430 |
0.879 |
2.48 × 10-12 |
0.367 |
0.783 |
| cg12949927 |
|
|
7 |
8.67 × 10-34 |
0.623 |
0.891 |
4.61 × 10-09 |
0.551 |
0.777 |
| cg20926353 |
TLE1 |
5′UTR |
9 |
1.52 × 10-20 |
0.085 |
0.327 |
3.90 × 10-09 |
0.128 |
0.306 |
| cg14095100 |
TLE1 |
5′UTR |
9 |
3.47 × 10-10 |
0.053 |
0.124 |
5.23 × 10-06 |
0.098 |
0.159 |
| cg00167275 |
FAM35A |
TSS1500 |
10 |
9.37 × 10-14 |
0.058 |
0.181 |
1.82 × 10-07 |
0.089 |
0.210 |
| cg04455759 |
TIMM8B |
Body |
11 |
1.88 × 10-23 |
0.922 |
0.813 |
3.21 × 10-09 |
0.845 |
0.701 |
| cg04981952 |
PRKRIR |
3′UTR |
11 |
7.36 × 10-12 |
0.668 |
0.571 |
4.84 × 10-09 |
0.626 |
0.544 |
| cg10402023 |
SHANK2 |
Body |
11 |
8.39 × 10-15 |
0.819 |
0.945 |
2.45 × 10-08 |
0.735 |
0.856 |
| cg15756407 |
TIMM8B |
Body |
11 |
1.82 × 10-25 |
0.751 |
0.628 |
3.23 × 10-06 |
0.675 |
0.581 |
| cg24056269 |
STK24 |
Body |
13 |
3.83 × 10-28 |
0.804 |
0.692 |
1.49 × 10-07 |
0.711 |
0.640 |
| cg24400806 |
|
|
15 |
2.38 × 10-09 |
0.842 |
0.909 |
6.08 × 10-06 |
0.746 |
0.807 |
| cg02015053 |
EIF3J |
3′UTR |
15 |
5.98 × 10-15 |
0.674 |
0.571 |
9.07 × 10-06 |
0.608 |
0.543 |
| cg03704667 |
CHTF8 |
3′UTR |
16 |
6.56 × 10-19 |
0.767 |
0.646 |
6.01 × 10-06 |
0.700 |
0.628 |
| cg02798874 |
TRPV1 |
3′UTR |
17 |
2.16 × 10-15 |
0.655 |
0.509 |
3.99 × 10-08 |
0.603 |
0.478 |
| cg13192180 |
NOL11 |
Body |
17 |
1.20 × 10-18 |
0.869 |
0.723 |
4.41 × 10-07 |
0.787 |
0.652 |
| cg11851257 | MYST2 | Body | 17 | 4.66 × 10-10 | 0.820 | 0.733 | 1.93 × 10-06 | 0.745 | 0.669 |
CHR: Chromosome; ELGAN: Extremely low gestational age newborns.
Figure 3. . Comparison of sex-dependent beta differences between the extremely low gestational age newborns (X-axis) and University of North Carolina replication cohort (Y-axis) for all 2745 validated probes.
Autosomal probes are highlighted in gray, while those found on the X-chromosome are represented in black.
ELGAN: Extremely low gestational age newborn; UNC: University of North Carolina.
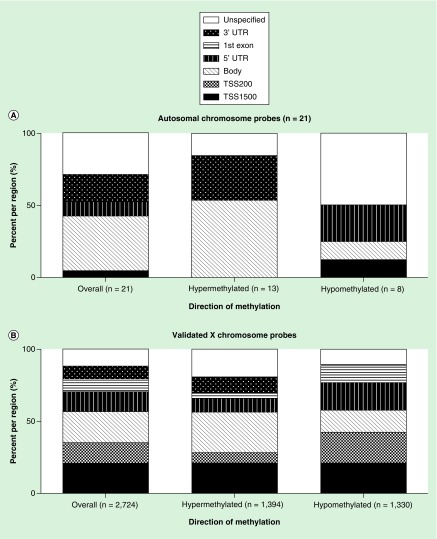
Among the replicated sexually dimorphic probes in all cohorts (n = 2745 probes, 587 genes), the majority of autosomal probes were located in the gene body (38.1%) and were hypermethylated. Interestingly, no sexually dimorphic probes were found within the TSS200 or 1st Exon (Figure 4). The majority of sexually dimorphic probes identified in the gene body and the 3′UTR were hypermethylated, with the TSS1500 and unspecified region showing higher proportions of hypomethylation (Figure 4). Of the sexually dimorphic probes on the X chromosome, the highest proportion were identified in the gene body (21.9%), and the majority of these were hypermethylated in males relative to females (Figure 4). Additionally, only the TSS200, 5′UTR and 1st Exon displayed higher proportions of hypomethylation (Figure 4).
Figure 4. . Distribution of sex-dependent differential methylation by region for all probes.
(A) displays probe locations for overall, hypermethylated and hypomethylated autosomal probes. (B) displays probe locations for overall, hypermethylated and hypomethylated X-chromosome probes.
X-inactivation has been previously cited as a potential mechanism which can account for many of the differences for the sexually dimorphic methylation changes observed on the X chromosome [26,42]. For this reason, the data were compared with genes that escape X-inactivation [49] and pseudo-autosomal genes [50]. Interestingly, a subset of the genes (n = 10) are known to escape X-inactivation (n = 10, 2%), and one XG blood group (one, namely XG, was a member of the human pseudoautosome (Figure 1, Supplementary Table 2).
Due to concerns about the potential for gestational age to impact CpG methylation, these sites were further tested in a cohort of 343 women recruited from the New Hampshire Birth cohort. In support of the primary data, we found replication of n = 2662 probes and n = 582 genes (99% of validated probes and genes). Importantly, replication occurred for all probes across the studies that passed p-value detection. Similar results were observed when gestational age was included in the model where n = 2561 probes and n = 568 genes (replication of 92% of probes and genes). These data were not considered during downstream analyses, but can be found in the Supplementary Materials (Figure 1, Supplementary Tables 1 & 2).
Transcription factor binding site analysis
In addition to characterization of the replicated probes/genes by region and methylation status, transcription factor binding site enrichment analysis was performed on the promoter regions of differentially methylated genes. An analysis of hypermethylated probes revealed an enrichment for binding sites for NFATC (GSEA p = 1.07 × 10- 26, Genomatix p = 2.23 × 10-4), and PAX4 (GSEA p = 1.39 × 10-17, Genomatix p = 9.62 × 10- 81). Among hypomethylated probes, binding sites for MAZ (GSEA p = 3.68 × 10-27, Genomatix p = 3.56 × 10-131) and FOXO4/MLLT7 (GSEA p = 4.54 × 10-20, Genomatix p = 2.41 ×10-37) were enriched. SP1 was significant in both the hypermethylated (GSEA p = 1.6 × 10-17, Genomatix p = 2.29 × 10-49) and hypomethylated (GSEA p = 2.29 × 10-30, Genomatix p = 7.82 × 10-48) gene sets.
Differentially methylated gene sets associated with response to the prenatal environment and sex-based differential gene expression
In addition to the transcription factor binding site analysis, we analyzed whether the differenitally methylated probes in the placenta were enriched for specific biological functions, namely transport, immune response, inflammation and growth/transcription factors. For this analysis, probes that were differentially methylated (p < 0.05 and q < 0.1) in both cohorts were considered (n = 4900 probes, n = 761 genes) (Supplementary Table 2). Using a Yates corrected χ2 test it was determined that immune proteins (n = 31, p = 0.0337), transporters (n = 119, p = 0.0233) and growth/transcription factors (n = 66, p = 0.0001) were enriched (Supplementary Table 2). Only inflammation proteins were not enriched (n = 12, p = 0.5623). Significantly enriched immune-related genes were involved in primary immunodeficiency and Toll-like receptor signaling, such as TLR7 and TLR8. The inflammatory-response genes included NOD-like receptor signaling pathways. Growth/transcription factors related genes were associated with neurotrophin and mTOR signaling pathways. Last, transporters tended to be involved in neuroactive ligand–receptor interaction and calcium signaling pathways, such as voltage-dependent anion-selective channel protein 1.
Following enrichment analysis, the differentially methylated genes identified in the present study were compared with a 142 gene set from a meta-analysis of sex-dependent differential gene expression within the placenta [47]. A total of 23 genes (n = 112 probes) that we identified as differentially methylated were also differentially expressed in the previous study [47] (Supplementary Table 2). This represents 16.2% of the 142 differentially expressed genes. Using a Yates corrected χ2 test it was determined that the overlap between studies was statistically significant (p < 0.0001). This suggests that DNA methylation may play a key role in the regulation of gene expression linked to sexual dimorphism.
Discussion
Fetal sex is known to influence susceptibility to the prenatal environment and can influence differential biological signaling within the placenta [3,5–7,12–15,23–25,27,41]. Therefore, we set out to identify sex-based differences in the placental DNA methylomes in three independent US-based birth cohorts. We identified a total of 587 genes (2745 probes), which were differentially methylated (p < 1 × 10-7) in male placentas as compared with female placentas in two cohorts. We further validated these probes in a third US cohort of normal gestational age and found that 99.2% of probes replicated. The majority of these probes were located on the X-chromosome, but a subset (n = 21 probes, 13 genes) were found on autosomal chromosomes. Many of the identified genes are involved in the transport and transcriptional control of the immune response. Interestingly, some of the genes identified here have been previously shown to be sexually dimorphic at the level of the placental transcriptome. Overall, our findings demonstrate that there is a sexual epigenetic dimorphism of the placental DNA methylome.
As previously demonstrated in other studies of fetal and adult tissues [22,26,41,42,48,51], the majority of differentially methylated genes were located on the X-chromosome. X-inactivation of one female X chromosome has been cited as potential explanation for sex-based differential methylation of the X-chromosome [26,42]. Interestingly, a subset of the genes identified in this study (n = 10) are known to escape X-inactivation [49]. One of these differentially methylated genes, XG, resides on the human pseudo-autosomal region of the X chromosome [50]. Our data highlight the intriguing finding that a very slight majority (51%) of the identified X-chromosome probes were hypermethylated in males relative to females. These data contradicted our a priori hypothesis that the majority of probes on the X-chromosome would have higher levels of methylation in females relative to males. Strong support for these data come from their reproducibility across three independent cohorts. In addition, other studies have identified some sites on the X chromosome that display higher levels of methylation in males relative to females [41,51]. While it may be intuitive that X-inactivation would likely present as a pattern of hypermethylation in females as compared with male placentas, prior research has shown that gene silencing on the X-chromosome is dependent on gene region and in some cases hypomethylation-associated silencing has been observed [49,52,53]. Taken together, these data support that the functional impact of methylation on X-chromosome inactivation exhibits positional dependencies, similar to those observed previously [54].
The sexually dimorphic genes were enriched for proteins involved in immune function, micronutrient transport and transcription/growth factors [12,20,21]. Specifically, immune-related TLR7 was shown to be hypermethylated in males relative to females and TLR8 was hypomethylated in males as compared with females. These immune-related differences between the male and female placental methylomes may contribute to differential susceptibility to environmental exposures between the sexes [8]. For example, an enhanced capability for transporting toxicants across the placenta in males during the prenatal period may be accounted for by differences in activation of the placental transporter voltage-dependent anion-selective channel protein 1, a major calcium transport channel [55] critical for fetal development. This protein, which normally enhances placental micronutrient delivery [56], can be hijacked by toxicants to cross cell membranes [57] during the prenatal period.
One possible mechanism underlying gene-specific genome-wide patterns of DNA methylation is the transcription factor occupancy theory [46]. Briefly, this theory posits that gene-specific methylation is influenced by transcription factor binding that either prevents or provides access to the DNA sequence for the DNA methylation machinery. Supporting this theory, we identified that binding sites for several transcription factors were significantly enriched in the hyper- and hypomethylated genes identified in this analysis. For instance, binding sites for NFATC were enriched in the promoter regions of the hypomethylated genes. Interestingly, in a separate study of placental sexually dimorphic gene expression, binding sites for NFATC were similarly enriched among the identified genes [47]. Additionally, binding sites for MAZ were identified to be enriched among the hypomethylated genes. MAZ is of interest as it has been previously identified as a key transcription factor associated with differential methylation patterns observed in response to a diverse suite of environmental contaminants [46]. Last, SP1, a transcription factor with enriched binding sites among both the hyper- and hypomethylated gene sets, is a known target of environmental contaminants, specifically toxic metals [58]. These transcription factors may play a role in impacting CpG methylation as well as responding to adverse conditions experienced during the prenatal environment.
Several factors should be considered in interpreting the results of this study. While we identified and validated a set of sexually dimorphic CpG sites across three cohorts, gene expression was not assessed in these samples. To address this, the sexually dimorphic placental methylome data were integrated with an existing genomic dataset in order to establish functional epigenetics. Future research should integrate data CpG methylation, as well as mRNA and protein expression. A potential confounder to these analyses is gestational age [43,44]. To address this, we integrated CpG methylation across three cohorts with varying gestational age, and found strong conservation. Last, future research should characterize X-inactive and X-active methylation patterns, through sequencing, to allow for male-female X-chromosome comparisons. These additional data would enhance the understanding of functional differences between methylation between males and females.
In conclusion, this study demonstrates sexual dimorphism at the level of the human placental DNA methylome. The epigenetic dimorphism observed could result in the sex-dependent transport of toxicants, nutrients and signaling molecules across the placenta, thereby resulting in a sex-dependent response of the fetus. Furthermore, it is possible that these differences could have consequences for both early and later-life health outcomes. The sexually dimorphic nature of the placental methylome may be a key factor to consider when examining sex-dependent outcomes in children following adverse exposures during the prenatal period.
Executive summary.
The placenta provides an interface between the maternal and fetal compartments transporting nutrients, signaling molecules and toxic substances between mother and fetus.
Differences in CpG methylation between male and female placentas may provide mechanistic understanding for sex-based differences observed, following adverse exposures during the prenatal period.
To investigate sex-based differences in the epigenome of the placenta, we analyzed DNA methylation in relation to fetal sex using genome-wide techniques comparing data from three separate US-based cohorts.
Methylation at 2745 probe (n = 587 genes) was identified and replicated, with enrichment of binding sites for transcription factors previously related to sexually dimorphic gene expression and environmental contaminants.
Genes with sex-based differential methylation in the placenta enrich for functions related to response to environmental exposures including: cellular transport, immune response and growth/transcription factors.
Within the placental DNA methylome, differences in the regulation of genes related to crucial biological functions may account for sex-specific effects of adverse in utero environments.
Supplementary Material
Footnotes
Financial & competing interests disclosure
This research was supported by grants from the NIH including the Environmental Influences on Child Health Outcomes (ECHO) award (1U2COD023375, UG33OD023348 and 1UG3OD023275), the National Institute of Environmental Health Sciences (P42-ES007126, T32-ES007018, P42-ES007373, P01-ES022832) and the National Institute of Neurological Disorders and Stroke (5U01NS040069 and 2R01NS040069). Further support was provided by the Wake Forest School of Medicine Innovation Pilot Grant, the Harold M and Mary Earnhardt Eagle Endowed Fund for Pediatric and Neonatal Research, and the EPA (RD83544201). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Godfrey KM. The role of the placenta in fetal programming-a review. Placenta. 2002;23(Suppl. A):S20–S27. doi: 10.1053/plac.2002.0773. [DOI] [PubMed] [Google Scholar]
- 2.Burton GJ, Fowden AL, Thornburg KL. Placental origins of chronic disease. Physiol. Rev. 2016;96(4):1509–1565. doi: 10.1152/physrev.00029.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broberg K, Ahmed S, Engstrom K, et al. Arsenic exposure in early pregnancy alters genome-wide DNA methylation in cord blood, particularly in boys. J. Dev. Orig. Health Dis. 2014;5(4):288–298. doi: 10.1017/S2040174414000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao J, Rebuli ME, Rogers J, et al. Prenatal bisphenol A exposure alters sex-specific estrogen receptor expression in the neonatal rat hypothalamus and amygdala. Toxicol. Sci. 2013;133(1):157–173. doi: 10.1093/toxsci/kft035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kippler M, Wagatsuma Y, Rahman A, et al. Environmental exposure to arsenic and cadmium during pregnancy and fetal size: a longitudinal study in rural Bangladesh. Reprod. Toxicol. 2012;34(4):504–511. doi: 10.1016/j.reprotox.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Llop S, Guxens M, Murcia M, et al. Prenatal exposure to mercury and infant neurodevelopment in a multicenter cohort in Spain: study of potential modifiers. Am. J. Epidemiol. 2012;175(5):451–465. doi: 10.1093/aje/kwr328. [DOI] [PubMed] [Google Scholar]
- 7.Ostlund BD, Conradt E, Crowell SE, Tyrka AR, Marsit CJ, Lester BM. Prenatal stress, fearfulness, and the epigenome: exploratory analysis of sex differences in dna methylation of the glucocorticoid receptor gene. Front. Behav. Neurosci. 2016;10:147. doi: 10.3389/fnbeh.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winans B, Humble MC, Lawrence BP. Environmental toxicants and the developing immune system: a missing link in the global battle against infectious disease? Reprod. Toxicol. 2011;31(3):327–336. doi: 10.1016/j.reprotox.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker-Athill EC, Tan J. Maternal immune activation and autism spectrum disorder: interleukin-6 signaling as a key mechanistic pathway. Neurosignals. 2010;18(2):113–128. doi: 10.1159/000319828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veru F, Laplante DP, Luheshi G, King S. Prenatal maternal stress exposure and immune function in the offspring. Stress. 2014;17(2):133–148. doi: 10.3109/10253890.2013.876404. [DOI] [PubMed] [Google Scholar]
- 11.De Escobar GM, Obregon MJ, Del Rey FE. Maternal thyroid hormones early in pregnancy and fetal brain development. Best Pract. Res. Clin. Endocrinol. Metab. 2004;18(2):225–248. doi: 10.1016/j.beem.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Gabory A, Roseboom TJ, Moore T, Moore LG, Junien C. Placental contribution to the origins of sexual dimorphism in health and diseases: sex chromosomes and epigenetics. Biol. Sex Differ. 2013;4(1):5. doi: 10.1186/2042-6410-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert-Diamond D, Emond JA, Baker ER, Korrick SA, Karagas MR. Relation between in utero arsenic exposure and birth outcomes in a cohort of mothers and their newborns from New Hampshire. Environ. Health Perspect. 2016;124(8):1299–1307. doi: 10.1289/ehp.1510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laine JE, Bailey KA, Rubio-Andrade M, et al. Maternal arsenic exposure, arsenic methylation efficiency, and birth outcomes in the Biomarkers of Exposure to ARsenic (BEAR) pregnancy cohort in Mexico. Environ. Health Perspect. 2015;123(2):186–192. doi: 10.1289/ehp.1307476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu L, Yokoyama K, Tian Y, et al. Decrease in birth weight and gestational age by arsenic among the newborn in Shanghai, China. Nihon Koshu Eisei Zasshi. 2011;58(2):89–95. [PubMed] [Google Scholar]
- 16.Tan J, Loganath A, Chong YS, Obbard JP. Exposure to persistent organic pollutants in utero and related maternal characteristics on birth outcomes: a multivariate data analysis approach. Chemosphere. 2009;74(3):428–433. doi: 10.1016/j.chemosphere.2008.09.045. [DOI] [PubMed] [Google Scholar]
- 17.Hodyl NA, Stark MJ, Osei-Kumah A, Clifton VL. Prenatal programming of the innate immune response following in utero exposure to inflammation: a sexually dimorphic process? Expert Rev. Clin. Immunol. 2011;7(5):579–592. doi: 10.1586/eci.11.51. [DOI] [PubMed] [Google Scholar]
- 18.Wang XY, Hagberg H, Nie CX, Zhu CL, Ikeda T, Mallard C. Dual role of intrauterine immune challenge on neonatal and adult brain vulnerability to hypoxia-ischemia. J. Neuropathol. Exp. Neurol. 2007;66(6):552–561. doi: 10.1097/01.jnen.0000263870.91811.6f. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld CS. Sex-specific placental responses in fetal development. Endocrinology. 2015;156(10):3422–3434. doi: 10.1210/en.2015-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Januar V, Desoye G, Novakovic B, Cvitic S, Saffery R. Epigenetic regulation of human placental function and pregnancy outcome: considerations for causal inference. Am. J. Obstet. Gynecol. 2015;213(Suppl. 4):S182–S196. doi: 10.1016/j.ajog.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Tarrade A, Panchenko P, Junien C, Gabory A. Placental contribution to nutritional programming of health and diseases: epigenetics and sexual dimorphism. J. Exp. Biol. 2015;218(Pt 1):50–58. doi: 10.1242/jeb.110320. [DOI] [PubMed] [Google Scholar]
- 22.De Coster S, Van Leeuwen DM, Jennen DG, et al. Gender-specific transcriptomic response to environmental exposure in Flemish adults. Environ. Mol. Mutagen. 2013;54(7):574–588. doi: 10.1002/em.21774. [DOI] [PubMed] [Google Scholar]
- 23.Filis P, Nagrath N, Fraser M, et al. Maternal smoking dysregulates protein expression in second trimester human fetal livers in a sex-specific manner. J. Clin. Endocrinol. Metab. 2015;100(6):E861–E870. doi: 10.1210/jc.2014-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drake AJ, O’shaughnessy PJ, Bhattacharya S, et al. In utero exposure to cigarette chemicals induces sex-specific disruption of one-carbon metabolism and DNA methylation in the human fetal liver. BMC Med. 2015;13:18. doi: 10.1186/s12916-014-0251-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen S, Strom M, Olsen SF, et al. Prenatal exposure to persistent organic pollutants and offspring allergic sensitization and lung function at 20 years of age. Clin. Exp. Allergy. 2016;46(2):329–336. doi: 10.1111/cea.12631. [DOI] [PubMed] [Google Scholar]
- 26.Mccarthy NS, Melton PE, Cadby G, et al. Meta-analysis of human methylation data for evidence of sex-specific autosomal patterns. BMC Genomics. 2014;15:981. doi: 10.1186/1471-2164-15-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Virani S, Rentschler KM, Nishijo M, et al. DNA methylation is differentially associated with environmental cadmium exposure based on sex and smoking status. Chemosphere. 2016;145:284–290. doi: 10.1016/j.chemosphere.2015.10.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bronson SL, Bale TL. The placenta as a mediator of stress effects on neurodevelopmental reprogramming. Neuropsychopharmacology. 2016;41(1):207–218. doi: 10.1038/npp.2015.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics. 2011;6(7):838–842. doi: 10.4161/epi.6.7.16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leviton A, Allred EN, Fichorova RN, Kuban KC, Michael O’shea T, Dammann O. Systemic inflammation on postnatal days 21 and 28 and indicators of brain dysfunction 2years later among children born before the 28th week of gestation. Early Hum. Dev. 2016;93:25–32. doi: 10.1016/j.earlhumdev.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laughon M, Bose C, Allred EN, et al. Patterns of blood protein concentrations of ELGANs classified by three patterns of respiratory disease in the first 2 postnatal weeks. Pediatr. Res. 2011;70(3):292–296. doi: 10.1203/PDR.0b013e3182274f35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’shea TM, Shah B, Allred EN, et al. Inflammation-initiating illnesses, inflammation-related proteins, and cognitive impairment in extremely preterm infants. Brain Behav. Immun. 2013;29:104–112. doi: 10.1016/j.bbi.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’shea TM, Allred EN, Kuban KC, et al. Elevated concentrations of inflammation-related proteins in postnatal blood predict severe developmental delay at 2 years of age in extremely preterm infants. J. Pediatr. 2012;160(3):395–401. doi: 10.1016/j.jpeds.2011.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hecht JL, Fichorova RN, Tang VF, Allred EN, Mcelrath TF, Leviton A. Relationship between neonatal blood protein concentrations and placenta histologic characteristics in extremely low GA newborns. Pediatr. Res. 2011;69(1):68–73. doi: 10.1203/PDR.0b013e3181fed334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mcelrath TF, Allred EN, Van Marter L, Fichorova RN, Leviton A. Perinatal systemic inflammatory responses of growth-restricted preterm newborns. Acta Paediatr. 2013;102(10):e439–e442. doi: 10.1111/apa.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin E, Ray PD, Smeester L, Grace MR, Boggess K, Fry RC. Epigenetics and preeclampsia: defining functional epimutations in the preeclamptic placenta related to the TGF-beta pathway. PLoS ONE. 2015;10(10)(e0141294) doi: 10.1371/journal.pone.0141294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’shea TM, Allred EN, Dammann O, et al. The ELGAN study of the brain and related disorders in extremely low gestational age newborns. Early Hum. Dev. 2009;85(11):719–725. doi: 10.1016/j.earlhumdev.2009.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onderdonk AB, Hecht JL, Mcelrath TF, Delaney ML, Allred EN, Leviton A. Colonization of second-trimester placenta parenchyma. Am. J. Obstet. Gynecol. 2008;199(1):52e51–52e10. doi: 10.1016/j.ajog.2007.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green BB, Karagas MR, Punshon T, et al. Epigenome-wide assessment of DNA methylation in the placenta and arsenic exposure in the New Hampshire Birth Cohort Study (USA) Environ. Health Perspect. 2016;124(8):1253–1260. doi: 10.1289/ehp.1510437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris TJ, Beck S. Analysis pipelines and packages for Infinium HumanMethylation450 BeadChip (450k) data. Methods. 2015;72:3–8. doi: 10.1016/j.ymeth.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall E, Volkov P, Dayeh T, et al. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol. 2014;15(12):522. doi: 10.1186/s13059-014-0522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yousefi P, Huen K, Dave V, Barcellos L, Eskenazi B, Holland N. Sex differences in DNA methylation assessed by 450 K BeadChip in newborns. BMC Genomics. 2015;16:911. doi: 10.1186/s12864-015-2034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Novakovic B, Yuen RK, Gordon L, et al. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics. 2011;12:529. doi: 10.1186/1471-2164-12-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hillman SL, Finer S, Smart MC, et al. Novel DNA methylation profiles associated with key gene regulation and transcription pathways in blood and placenta of growth-restricted neonates. Epigenetics. 2015;10(1):50–61. doi: 10.4161/15592294.2014.989741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin EM, Fry RC. A cross-study analysis of prenatal exposures to environmental contaminants and the epigenome: support for stress-responsive transcription factor occupancy as a mediator of gene-specific CpG methylation patterning. Environ. Epigenet. 2016;2(1):dvv011. doi: 10.1093/eep/dvv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buckberry S, Bianco-Miotto T, Bent SJ, Dekker GA, Roberts CT. Integrative transcriptome meta-analysis reveals widespread sex-biased gene expression at the human fetal-maternal interface. Mol. Hum. Reprod. 2014;20(8):810–819. doi: 10.1093/molehr/gau035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singmann P, Shem-Tov D, Wahl S, et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenet. Chromatin. 2015;8:43. doi: 10.1186/s13072-015-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cotton AM, Price EM, Jones MJ, Balaton BP, Kobor MS, Brown CJ. Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet. 2015;24(6):1528–1539. doi: 10.1093/hmg/ddu564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helena Mangs A, Morris BJ. The human pseudoautosomal region (PAR): origin, function and future. Curr. Genomics. 2007;8(2):129–136. doi: 10.2174/138920207780368141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J, Morgan M, Hutchison K, Calhoun VD. A study of the influence of sex on genome wide methylation. PLoS ONE. 2010;5(4):e10028. doi: 10.1371/journal.pone.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharp AJ, Stathaki E, Migliavacca E, et al. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011;21(10):1592–1600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yasukochi Y, Maruyama O, Mahajan MC, et al. X chromosome-wide analyses of genomic DNA methylation states and gene expression in male and female neutrophils. Proc. Natl Acad. Sci. USA. 2010;107(8):3704–3709. doi: 10.1073/pnas.0914812107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rojas D, Rager JE, Smeester L, et al. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol. Sci. 2015;143(1):97–106. doi: 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gincel D, Zaid H, Shoshan-Barmatz V. Calcium binding and translocation by the voltage-dependent anion channel: a possible regulatory mechanism in mitochondrial function. Biochem. J. 2001;358(Pt 1):147–155. doi: 10.1042/0264-6021:3580147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baczyk D, Kingdom JC, Uhlen P. Calcium signaling in placenta. Cell Calcium. 2011;49(5):350–356. doi: 10.1016/j.ceca.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 57.Atchison WD. Effects of toxic environmental contaminants on voltage-gated calcium channel function: from past to present. J. Bioenerg. Biomembr. 2003;35(6):507–532. doi: 10.1023/b:jobb.0000008023.11211.13. [DOI] [PubMed] [Google Scholar]
- 58.Zawia NH, Sharan R, Brydie M, Oyama T, Crumpton T. Sp1 as a target site for metal-induced perturbations of transcriptional regulation of developmental brain gene expression. Brain Res. Dev. Brain Res. 1998;107(2):291–298. doi: 10.1016/s0165-3806(98)00023-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.