Abstract
In this paper we describe an ontological scheme for representing anatomical entities undergoing morphological transformation and changes in phenotype during prenatal development. This is a proposed component of the Anatomical Transformation Abstraction (ATA) of the Foundational Model of Anatomy (FMA) Ontology that was created to provide an ontological framework for capturing knowledge about human development from the zygote to postnatal life. It is designed to initially describe the structural properties of the anatomical entities that participate in human development and then enhance their description with developmental properties, such as temporal attributes and developmental processes. This approach facilitates the correlation and integration of the classical but static representation of embryology with the evolving novel concepts of developmental biology, which primarily deals with the experimental data on the mechanisms of embryogenesis and organogenesis. This is important for describing and understanding the underlying processes involved in structural malformations. In this study we focused on the development of the lips and the palate in conjunction with our work on the pathogenesis and classification of cleft lip and palate (CL/P) in the FaceBase program. Our aim here is to create the Craniofacial Human Development Ontology (CHDO) to support the Ontology of Craniofacial Development and Malformation (OCDM), which provides the infrastructure for integrating multiple and disparate craniofacial data generated by FaceBase researchers.
1 INTRODUCTION
The domain of descriptive embryology [1] has not been comprehensively formalized in a semantically sound system. In fact, many of the embryological terms for concepts and relationships have been used ambiguously since von Baer [2] proposed the germ layer theory in 1828. This has remained problematic especially for correlating embryology with the rapidly evolving advancement of developmental biology, which primarily deals with experimental data on the mechanisms of embryogenesis and organogenesis. It is therefore necessary to establish a formal representation of knowledge in embryology that can be used to organize, manage and correlate discoveries in developmental biology. The Anatomical Transformation Abstraction (ATA) of the FMA [3] was proposed as the mechanism that would represent the knowledge of descriptive embryology in a formal, computer-processable way. The ATA component of the FMA was not built as part of the original ontology, but a need for it has come up in our current work on the design and implementation of the Ontology of Craniofacial Development and Malformation (OCDM), which we are building as part of our NIDCR-sponsored project for the FaceBase Consortium (https://www.facebase.org/) [4], which deals with craniofacial abnormalities.
Where applicable we leverage existing sources of developmental anatomy and incorporate any related representation. The Edinburgh Mouse Atlas Project or e-Mouse Atlas Project (EMAP) [5] is such a resource of relevance to our work. This project established a system for comparing the spatiotemporal expression of genes in staged embryos. EMAP deals with individual embryos and databases but our approach is to capture development information from a knowledge representation perspective. Another project designed to mesh with EMAP is the Human Developmental Anatomy (EHDA) [6], which is a structured controlled vocabulary that is complementary to our efforts. But unlike EHDA we extend ontological representation to spatio-structural and processual relationships between the developing structures at different stages of development.
Craniofacial development consists of a complex set of embryological events that are affected and controlled by both genetic and environmental factors. Any disturbance at any time in the development can result in structural malformations, such as facial clefts, and the most common types are cleft lip and/or cleft palate (CL/P) [7,8], which served as the driving use case for our study. For this purpose we therefore designed developmental properties and attributes relating to developing structures involved in the pathogenesis of CL/P (e.g. lips, alveolar ridge, incisor teeth and palate). The description is carried down to granularity levels necessary to identify where malformation can occur.
2 ADDING DEVELOPMENT TO THE FOUNDATIONAL MODEL OF ANATOMY ONTOLOGY
The structures upon which we have built the OCDM are derived from the Foundational Model of Anatomy Ontology (FMA), a reference ontology for the domain of anatomy, which is based on a disciplined ontological approach that declares foundational principles for representing anatomical entities and relationships. It has a high level scheme that specifies the domain and scope of the ontology as a four-tuple component: Anatomy Taxonomy (AT), Anatomical Structural Abstraction (ASA), Anatomical Transformation Abstraction (ATA) and Metaknowledge (Mk). AT, the backbone of the FMA, represents anatomical entities as classes (or types) in an Aristotelian inheritance hierarchy; ASA [9,10] elaborates on the spatio-structural relationships, such as parthood, connectivity and spatial association, between entities represented in AT; ATA describes the morphological and phenotypic transformation of anatomical entities during pre- and postnatal development; and Mk encompasses the principles and rules governing the relationships represented in the ontology’s other three component abstractions. The most recent account of these components is discussed in detail in [3]. Here in this paper we elaborate on the pre-natal component of the ATA on which our work is based. The anatomical entities are classes or types and the relations are the spatio-structural relationships that exist among them. In this paper, classes are represented in Courier New font and relations in bold italic.
2.1 Anatomical Transformation Abstraction (ATA)
The Anatomical Transformation Abstraction (ATA) of the FMA was designed to provide an ontological framework for capturing knowledge about human development from the zygote to postnatal life. It is designed to initially describe the spatio-structural properties of the anatomical entities that participate in human development and then enhance their description with developmental properties, such as temporal attributes and developmental processes.
This approach facilitates the correlation and integration of the classical but static representation of embryology with the evolving novel concepts of developmental biology. In this work we applied the high level structure of canonical adult anatomy to developmental anatomy and utilized the “principle of organizational units”, where entities are classified according to the salient structural units they express at increasing levels of granularity [3]. As shown in Figure 1 the same approach was carried out using the same anatomical structure classes in the adult version: Cell, Tissue, Organ part, Organ, Organ system, Cardinal body part and Body.
Figure 1.
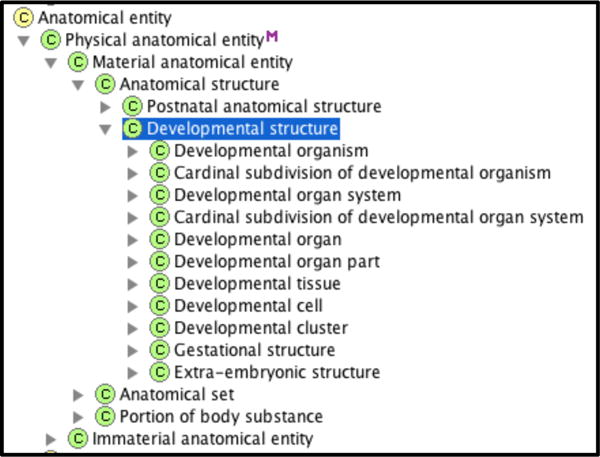
Protégé screen capture showing class inheritance hierarchy of developmental structure.
For example, Neural tube is_a Embryonic organ, Rhombomere is_a Embryonic organ part, and Endoderm is_a Embryonic tissue. In contrast to the adult anatomy, developmental entities undergo significant phenotypic changes within a very short period of time and therefore we represent them at different stages of their development. Here, developmental organisms are identified based not only on their structural make-up but on their age as well (Figure 2).
Figure 2.

Developmental organism at different stages of development.
In humans, an embryo is a developmental organism from 4 weeks to 8 weeks of gestation and a fetus is a developmental organism from 8 weeks of gestation to birth. We also specified time-dependent properties using the Carnegie staging system [11], the gestational age of the entities and the post-ovulation date.
We then elaborated on the spatio-structural properties and attributes of the developmental structures using textbook knowledge, such as parthood relation ( Bilaminar disc part_of Blastocyst and Bilaminar disc has_part Epiblast and Hypoblast), adjacency ( Cytotrophoblast surrounds Extra-embryonic mesoblast), and connectivity ( Midgut continuous_with Hindgut). We also recorded the phenotypic changes from one stage to the next using processual relations, such as tranforms and derives, as defined in the OBO Relation ontology [12]. The relation transforms_into (inverse transformation_of) entails no change in the identity of the structure, e.g. Embryo transforms_into Fetus. On the other hand in the relation derives_from, the identity of the structure is not preserved. This process results in the creation of a new distinct structure as in the case of a chondrocyte being derived from the chondroblast. Other developmental processes, such as fuses_with and fusion_of, are prevalent during development and are therefore instantiated in the ontology as well. These time-dependent processual relationships will support the modeling of genetic and molecular networks and mechanisms that regulate pre-natal development.
2.2 Craniofacial Human Development Ontology (CHDO)
The CHDO provides the human development component of the OCDM. CHDO accounts for the developmental entities and processes that are involved in malformations and in particular, the pathogenesis of CL/P. We augmented the craniofacial content with spatio-structural, processual and temporal properties that relate to structures involved in CL/P.
As an example, Figure 3 shows the different properties of the intermaxillary process. We specified the developmental stage of structures using the Carnegie stages. We represented Carnegie stages as classes and then associated with them properties that describe 1) what developmental entities (material and immaterial) can be observed at a particular stage, 2) size, and 3) number of post-ovulation days, as shown for Carnegie stage 19 in Figure 4.
Figure 3.
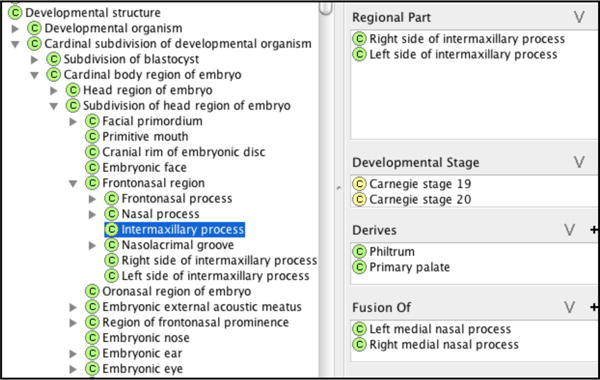
Properties of Intermaxillary process
Figure 4.
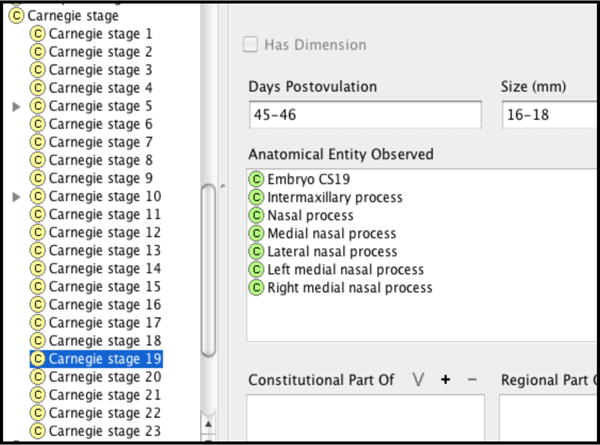
Properties of Carnegie stage 19.
We extended the description of entities and relations to granularity levels that would allow us to account for where and when any disturbance causing malformation, such as those affecting gene expressions, can occur anywhere in the pathway of normal development (Figure 5).
Figure 5.
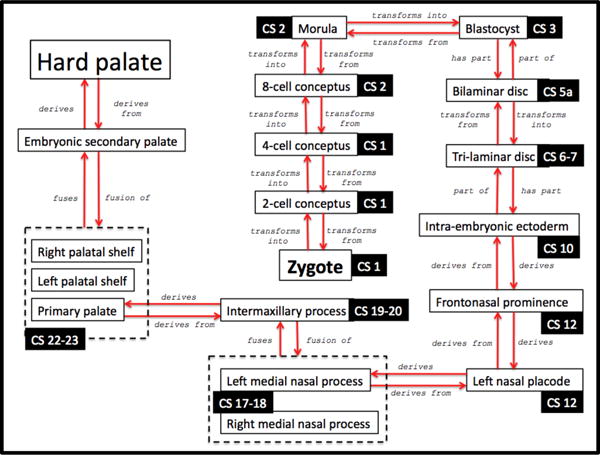
Schematic diagram of the development of hard palate from the zygote. Carnegie stages are represented as “CS”.
The ontology is currently implemented in Protégé frames but efforts are underway to translate the entire FMA, including the CHDO, into OWL 2 DL to allow better interoperability with other ontologies, such as those in the OBO Foundry. The OWL format would likewise facilitate the process of checking for errors and inconsistencies inadvertently entered in the ontology.
3 CONCLUSION
CHDO is a symbolic model of craniofacial human development that now begins to correlate the static representation of classical embryology with the dynamic domain of developmental biology. Towards this goal it can serve at least three practical purposes: 1) to provide standards for communication and annotations of development-related data, 2) to serve as a developmental ontology template for model organisms, and 3) to leverage this underlying ontological framework for promoting integration and interoperability, reuse of knowledge and data discovery among different applications. As one of the components of the OCDM, CHDO is open source and freely available at the FaceBase website https://www.facebase.org/content/ocdm. It is regularly updated with new contributions or edits from domain experts.
Acknowledgments
This work is supported by grant NIH/NIDCR grant 5U01DE020050-04 from the National Institute of Dental and Craniofacial Research. We thank Melissa Clarkson for reviewing the manuscript and for providing the web viewer that facilitated our review of the ontology content.
References
- 1.Carlson BM. Human Embryology and Developmental Biology. St Louis: Mosby; 1994. [Google Scholar]
- 2.von Baer KE. Ueber Entwicklungsgeschichte der Tierre. Borntrager Konigsburg 1828 [Google Scholar]
- 3.Rosse Cornelius, Mejino Jose LV. The Foundational Model of Anatomy Ontology. In: Burger A, Davidson D, Baldock R, editors. Anatomy Ontologies for Bioinformatics: Principles and Practice. Springer; 2007. pp. 59–117. [Google Scholar]
- 4.Hochheiser H, Aronow B, Artinger K, Beaty TH, Brinkley JF, Chai Y, Clouthier D, Cunningham ML, Dixon M, Donahue LR, Fraser S, Hallgrimsson B, Iwata J, Kelin O, Marazita ML, Murray JC, Murray S, de Villena FPM, Postlethwait J, Potter S, Shapiro L, Spritz R, Visel A, Weinberg S, Trainor P. The FaceBase Consortium: A Comprehensive Program to Facilitate Craniofacial Research. Developmental Biology. 2011;355(2):175–182. doi: 10.1016/j.ydbio.2011.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldock RA, Dubreuil C, Hill B, Duncan Davidson. The Edinburgh Mouse Atlas: Basic structure and informatics. In: Levotsky S, editor. bioinformatics databases and systems. Kluwer Academic Pres; 1999. pp. 102–115. [Google Scholar]
- 6.Bard J. A new ontology (structured hierarchy) of human developmental anatomy for the first 7 weeks (Carnegie stages 1-20) J Anat. 2012;221(5):406–16. doi: 10.1111/j.1469-7580.2012.01566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marazita ML. The evolution of human genetic studies of cleft lip and cleft palate. Annu Rev Genomics Hum Genet. 2012;13:263–83. doi: 10.1146/annurev-genom-090711-163729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marazita ML, Mooney MP. Current concepts in the embryology and genetics of cleft lip and cleft palate. Clin Plas Surg. 2004 Apr;31(2):125–40. doi: 10.1016/S0094-1298(03)00138-X. [DOI] [PubMed] [Google Scholar]
- 9.Mejino Jose LV, Rosse Cornelius. Symbolic modeling of structural relationships in the Foundational Model of Anatomy. Proceedings, First International Workshop on Formal Biomedical Knowledge Representation (KR-MED 2004); Whistler Mountain, Canada. 2004. [Google Scholar]
- 10.Mejino Jose LV, Agoncillo Augusto V, Rickard KL, Rosse Cornelius. Representing Complexity in Part-Whole Relationships within the Foundational Model of Anatomy. Proceedings, American Medical Informatics Association Fall Symposium; 2003. pp. 450–454. [PMC free article] [PubMed] [Google Scholar]
- 11.O’Rahilly, Müller F. Developmental stages in human embryos. Washington, D. C.: Carnegie Institution of Washington; 1987. [Google Scholar]
- 12.Smith Barry, Ceusters Werner, Klagges Bert, Köhler Jacob, Kumar Anand, Lomax Jane, Mungall Chris, Neuhaus Fabian, Rector Alan L, Rosse Cornelius. Relations in biomedical ontologies. Genome Biology. 6(5):R46.1–R46.15. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]


