Abstract
MicroRNA-21 (miR-21) is consistently up-regulated in various neurological disorders, including epilepsy. Here, we show that the biogenesis of miR-21 is altered following pilocarpine status epilepticus (SE) with an increase in precursor miR-21 (pre-miR-21) in rats. We demonstrate that pre-miR-21 has an energetically favorable site overlapping with the miR-21 binding site and competes with mature miR-21 for binding in the 3′UTR of TGFBR2 mRNA, but not NT-3 mRNA in vitro. This binding competition influences miR-21-mediated repression in vitro and correlates with the increase in TGFBR2 and decrease in NT-3 following SE. Polysome profiling reveals co-localization of pre-miR-21 in the ribosome fraction with translating mRNAs in U-87 cells. The current work suggests that pre-miR-21 may post-transcriptionally counteract miR-21-mediated suppression following SE and could potentially lead to prolonged TGF-β receptor expression impacting epileptogenesis. The study further supports that the ratio of the pre to mature miRNA may be important in determining the regulatory effects of a miRNA gene.
Keywords: Status epilepticus, pilocarpine, pre-miR-21, regulation of mRNA, TGFβ receptor 2
INTRODUCTION
MicroRNAs (miRNAs) are short non-coding RNA molecules (~20–25 nucleotides) that regulate gene expression of target messenger RNAs (mRNAs) post-transcriptionally in diverse cellular pathways (Kim, 2005). MiRNAs bind to mRNAs blocking translation and/or enhancing degradation, allowing for a large number of proteins to be concurrently regulated. To date, there are hundreds of miRNA members identified in higher eukaryotes targeting more than 60% of protein-encoding genes (Bartel, 2004; Friedman et al., 2009b; Kim, 2005). In miRNA biogenesis, primary miRNAs (pri-miRNAs) are first transcribed by RNA polymerase II and cleaved into precursor miRNAs (pre-miRNAs) by the RNase III Drosha in the nucleus; pre-miRNAs are then exported into the cytoplasm where they are processed by another RNase III Dicer to produce a short miRNA duplex consisting of a guide strand (mature miRNA) and passenger strand (miRNA*). The mature miRNA and Argonaute (Ago2) form a RNA-induced silencing complex to target and silence mRNA, whereas the miRNA* is often rapidly degraded (Ha and Kim, 2014; Kosik, 2006). The miRNA intermediates were merely considered as byproducts with no biological function until recent in vitro studies, Trujillo et al. described function of pri-miRNAs in target recognition and repression while Okamura et al. showed miRNA-type repression of specific miRNA stem loops (Okamura et al., 2013; Roy-Chaudhuri et al., 2014; Trujillo et al., 2010). Our previous study demonstrated competition between pre- and mature miRNAs for binding to the 3′UTR of their target mRNAs (Roy-Chaudhuri et al., 2014). However, the physiological consequences of these additional layers of translational regulation by miRNA intermediates have not been explored in epilepsy or other disease models.
Alterations in miRNA expression have been implicated in a wide variety of neurological disorders, including epilepsy (Bian and Sun, 2011; Bot et al., 2013; Jimenez-Mateos and Henshall, 2013; Kosik, 2006). Epilepsy is one of the most common diseases of the nervous system with a lifetime incidence of 3% and is associated with comorbid learning and memory problems (de Boer et al., 2008; Hesdorffer et al., 2011; Jimenez-Mateos et al., 2012; Pohlmann-Eden et al., 2006). Development of epilepsy after a neurologic insult has been associated with a large number of cellular and molecular changes, such as neuronal loss, gliosis, neurogenesis, synaptic organization, and alternation of gene expression including miRNAs (Pitkanen and Lukasiuk, 2009). Indeed, miRNA dysregulation is observed in a variety of epilepsy models and in tissue from patients undergoing surgery for medically refractory epilepsy (Dogini et al., 2013; Henshall, 2014). Altered miRNA regulation has been implicated in apoptosis, neurogenesis, synaptic functions, inflammation, and gliosis (Dogini et al., 2013; Jimenez-Mateos and Henshall, 2013). However, further studies are needed to understand how miRNAs regulate epileptogenesis and if they provide a novel therapy to treat epilepsy.
Studies on epileptogensis use models that have high rates of epilepsy following an initial insult. Treating rodents with pilocarpine, a chemoconvulsant, is a widely used model to induce status epilepticus (SE, a prolonged seizure) from which the animals initially recover but go on to develop spontaneous seizures or epilepsy (Curia et al., 2008). Animals present behavioral, electrographic, and neuropathologic features in the limbic structures similar to those observed in patients with temporal lobe epilepsy. MicroRNA-21 (miR-21), an evolutionarily conserved miRNA, increases following a variety of brain insults including head trauma, stroke, and SE that are associated with increased risk of developing epilepsy (Buller et al., 2010; Gorter et al., 2014; Peng et al., 2013).
Here, we observed differential expression of miR-21 precursors and mature miR-21 in the hippocampus following pilocarpine SE. To understand the functional significance of differential levels of precursor and mature miR-21 following SE, we carried out bioinformatics and biochemical approaches to show that pre-miR-21 can compete with mature miR-21 for regulating mRNA levels of Transforming Growth Factor β Receptor 2 (TGFBR2), but not Neurotrophin-3 (NT-3), both of which are targets of mature miR-21. TGF-β is a master anti-inflammation cytokine controlling neurological and immune functions following a neurologic insult and binds to TGF-β receptor 1 and receptor 2 (TGFBR1 and TGFBR2, respectively) resulting in a cascade of molecular changes implicated in epileptogenesis (Cacheaux et al., 2009; Friedman et al., 2009a; Okamoto et al., 2010). Together, our data suggest that pre-miR-21 may compete with mature miR-21 and block miRNA-mediated mRNA degradation and/or translational repression in the hippocampus following SE. This possibly contributes to the prolonged activation of TGF-β signaling with implications for epileptogenesis.
RESULTS
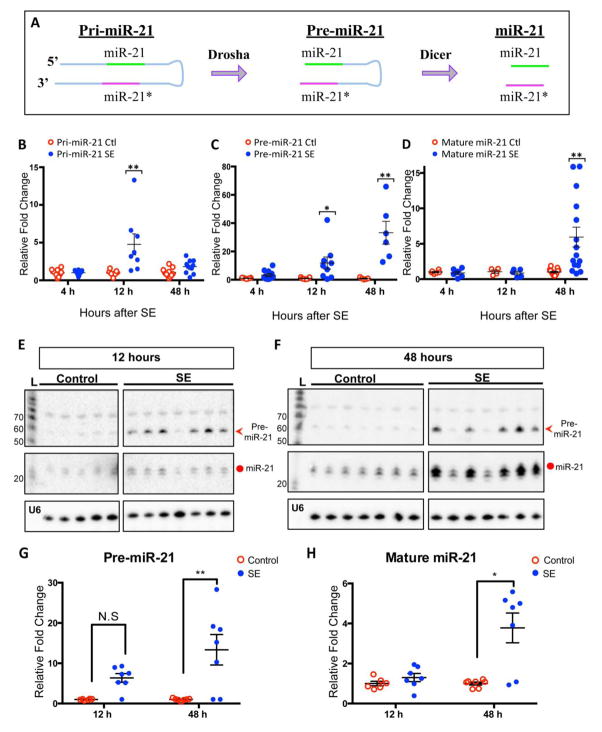
1. Differential changes in expression of pri-miR-21, pre-miR-21, and mature miR-21 in the hippocampus following status epilepticus
Pri-miR-21, pre-miR-21 and mature miR-21 levels in the rat hippocampi were assayed using qRT-PCR and northern blot analysis 4, 12, and 48 h after the animals had experienced a pilocarpine-induced Racine stage V seizure (Racine, 1972) (Supplementary Fig 1). The SE animals exhibited substantial increases in each transcript compared to controls, but each transcript displayed a unique time course (Fig 1). The qRT-PCR data showed that pri-miR-21 levels did not increase at 4 h post SE but increased by ~ 4.8-fold at 12 h post SE and then decreased at 48 h post SE (Fig 1B). Pre-miR-21 levels increased at each time point studied: 3.3-fold at 4 h, 11.8-fold at 12 h and 33.2-fold at 48 h post SE (Fig 1C). However, the change in mature miR-21 levels was substantially delayed reaching a ~ 5.9-fold increase at 48 h after SE (Fig 1D). Small RNA northern blot analysis confirmed the differential changes of pre- and mature miR-21 identified by qRT-PCR (Fig 1E & 1F; see quantification in Fig 1G &H). Pri-miR-21 and mature miR-21 increased after SE but the pre-miR-21 transcript increased as early as 4 hours after SE and to a greater extent than pri-miR-21 suggesting that processing of pre-miR21 is altered following SE.
Figure 1. Levels of miR-21 transcripts at different time points in rats after SE.
(A) A schematic diagram of miR-21 biogenesis. MiR-21 is transcribed as a long primary transcript (~3400 bp), pri-miR-21, from its own promoter. Drosha cleaves pri-miR-21 into a precursor transcript (~ 70 bp), pre-miR-21. Dicer further cleaves pre-mi-21 into short strands of ~22 nt, containing guide/mature miR-21 (green) and passenger miR-21 (miR-21*) (magenta). (B–C) qRT-PCR showing changes of pri-, pre-, and mature miR-21 levels at 4, 12, and 48 h after SE. Both pri- and mature miR-21 levels show significant increase only at 12 h and 48 h, respectively (B & D), whereas pre-miR-21 levels are elevated at all time points (C). Relative fold change is normalized to PPIA (cyclophilin) mRNA expression for pri-miR-21 and to 4.5S (ribonuclear small RNA) expression for pre-miR-21 and for mature miR-21. Shown are mean±SEM. Kruskal-Wallis test, *p<0.05, **p<0.01, *** p<0.001. (E & F) Small RNA northern blots detecting pre-miR-21 and mature miR-21 at 12 and 48 h post SE. The blots used LNA probe that detected both pre-miR-21 (red arrowhead) and mature miR-21 (circle). At 12 h time point, the intensity of the pre-miR-21 bands in the SE groups was stronger than that in the control group, but the intensities of the mature miR-21 bands were comparable between the SE and control groups (E). At 48 h time point, the levels of pre-miR-21 and mature miR-21 in the SE group were higher than that in the control group (F). (G & H) Quantification for northern blots. L= ladder for RNA sizes in nucleotides. U6 serves as a loading control. For pri-miR-21: 4h (Control N=10, SE N=12), 12h (Control N=7, SE N=8), 48h (Control N =11, SE N=10). For pre-miR-21: 4h (Control N= 7, SE N=11), 12h (Control N=7, SE N=9), 48h (Control N=5, SE N=6). For mature miR-21: 4h (Control N= 6, SE N=6), 12h (Control N=4, SE N=5), 48h (Control N=6, SE N=6).
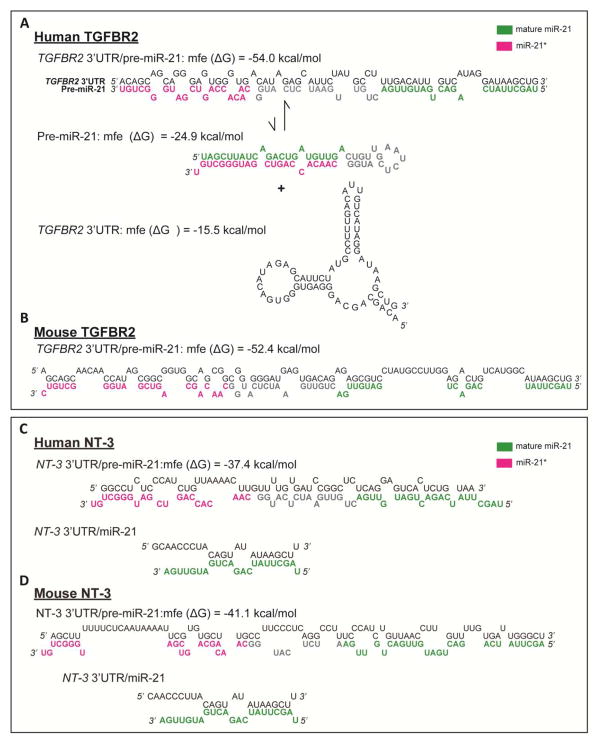
2. Pre-miR-21 potentially binds TGFBR2 mRNA 3′UTR to regulate its translation
We next asked if there was a biological significance of the changes in pri-, pre-, and mature miR-21 levels over time in the pilocarpine-induced seizure model. Our recent study (Roy-Chaudhuri et al., 2014) has uncovered a novel regulatory role of pre-miRNAs in miRNA-mediated target suppression. Previously, we showed that pre-miR-151 and pre-miR-124 can compete with their mature counterparts in vitro for binding to the miRNA-response elements (MREs) in the 3′ UTRs of selected target mRNAs to regulate their expression using biochemical analyses and bioinformatics (Roy-Chaudhuri et al., 2014). Furthermore, the same pre-miRNA can have differential effects on the expression of different target genes owing to its ability to compete with its mature counterpart to bind to one target but not to others (Roy-Chaudhuri et al., 2014).
Many miR-21 targets have been identified, including TGFBR2 mRNA and NT-3 mRNA (Kim et al., 2009; Mishra et al., 2014; Risbud et al., 2011; Yu et al., 2012). TGFBR2 and NT-3 are part of the TGF-β signaling and neurotrophin signaling pathways both of which have been implicated in epileptogenesis (Cacheaux et al., 2009; Elmer et al., 1997; Weissberg et al., 2015; Xu et al., 2002). Intriguingly, we found that the mature miR-21 binding region in TGFBR2 3′UTR as shown previously (Kim et al., 2009) and as predicted by TargetScan overlaps with the most favorable binding region of pre-miR-21 to TGFBR2 3′UTR. Furthermore, thermodynamic consideration using RNAhybrid (Rehmsmeier et al., 2004) favors the inter-molecular binding of pre-miR-21 to the TGFBR2 3′UTR region (minimum free energy or ΔG = −54.0 kcal/mole in humans and −52.4 kcal/mole in mice) over the combined intra-molecular folding energy of the pre-miR-21 and the target region in TGFBR2 3′UTR (Fig 2A & 2B). In contrast, the minimum ΔG for pre-miR-21 to 3′ UTR of NT-3 are −37.4 kcal/mol and −41.1 kcal/mol in human and mouse, respectively (Fig 2C & 2D). Based on our previous work (Roy-Chaudhuri et al., 2014), miRNA transcript (precursor or mature form) with ΔG of −50 kcal/mol or lower (more negative) exhibits favorable binding to its corresponding 3′UTR of a target mRNA. Thus, the 3′UTR of NT-3 does not contain a thermodynamically favorable region for pre-miR-21 binding. Together, it is likely that an increase in the pre-miR-21 concentration in SE would favor de-repression of TGFBR2 mRNA, and not NT-3 mRNA.
Figure 2. Putative binding site and thermodynamics of pre-miR-21 to TGFBR2 mRNA 3′UTR and to NT-3 mRNA 3′ UTR.
(A & B) A schematic shows the energetically favorable binding sties (in green for mature miR-21 and magenta for miR-21*) of pre-miR-21 to TGFBR2 mRNA for human (A) and for mouse (B). The minimum free energy change (ΔG) of human and mouse pre-miR-21 binding to TGFBR2 3′UTR are −54.0 kcal/mol and −52.4 kcal/mol, respectively. (C & D) A schematic shows the energetically favorable binding sties (in green for mature miR-21 and magenta for miR-21*) of pre-miR-21 to NT-3 mRNA for human (C) and in mouse (D). The ΔG of human and mouse pre-miR-21 binding to NT-3 3′UTR are −37.4 kcal/mol and −41.1 kcal/mol, respectively.
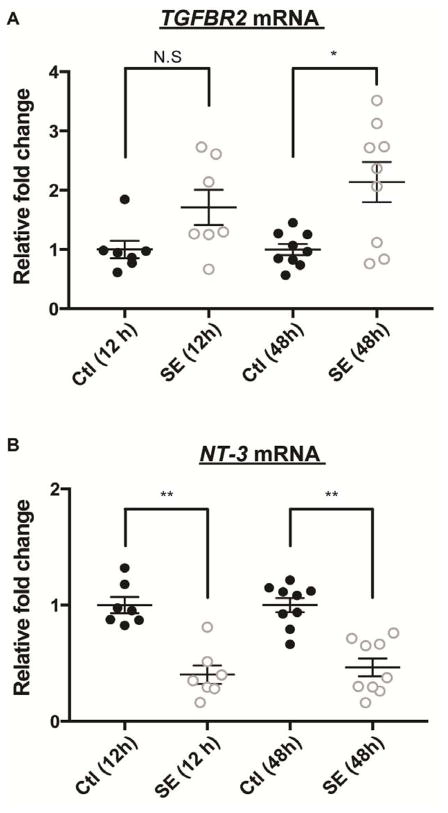
The bioinformatics analyses suggest the potential regulatory role of pre-miR-21 in TGFBR2 mRNA but not in NT-3 mRNA. To determine if the in vivo mRNA levels of TGFBR2 and NT-3 correlate with these bioinformatics predictions, we measured TGFBR2 mRNA and NT-3 mRNA levels at 12 h and 48 h after SE from the hippocampi of rats using qRT-PCR. There was an increase in the TGFBR2 mRNA level in the seizure groups at both time points (Fig 3A) with a statistically significant increase at 48 h (p-value =0.040). Conversely, we detected a comparable reduction of NT-3 mRNA at 12 h and 48 h after SE (Fig 3B), consistent with prior studies showing that NT-3 mRNA is a putative miR-21 target and is down-regulated following SE (Binder, 2007; Mudo et al., 1996; Risbud et al., 2011). The results suggest that in vivo pre-miR-21 might compete with mature miR-21 to prevent TGFBR2 mRNA repression by mature miR-21 after SE.
Figure 3. TGFBR2 mRNA and NT-3 mRNA levels at 12 and 48 h post SE.
Quantitative RT-PCR of TGFBR2 mRNA and NT-3 mRNA from the rat hippocampal lysate at 12 and 48 h after SE. (A) The TGFBR2 mRNA level in SE group was elevated by 1.7-fold at 12 h and 2.14-fold at 48 h compared to the control group. The 48 h SE group showed statistically significant increase. (B) The NT-3 mRNA level was significantly reduced at both 12 h (0.4±0.089) and 48 h (0.46±0.076) post SE compared to the control group. Relative fold change was normalized to housekeeping PPIA (cyclophilin) mRNA expression. Shown are mean±SEM. Control N= 7 samples and SE N= 9 samples. One-way ANOVA, Kruskal-Wallis test. *p<0.05, ** p<0.01, N.S.= not significant.
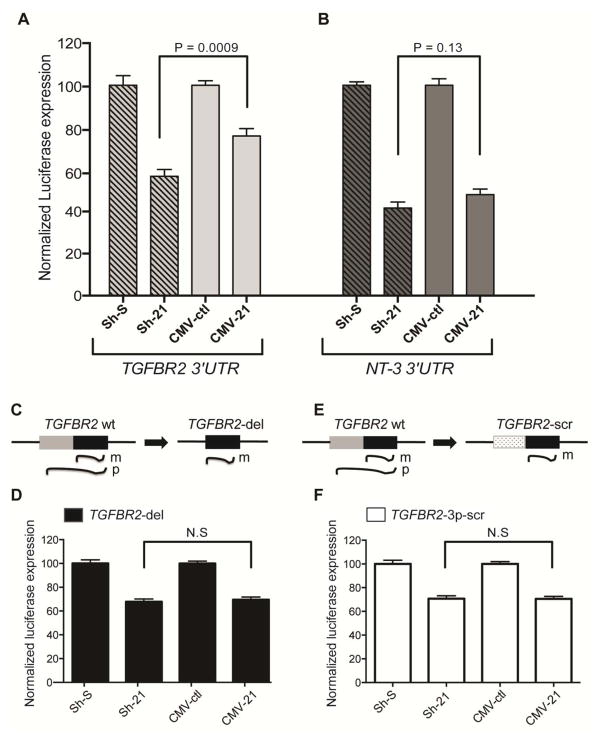
3. Pre-miR-21 competes with mature miR-21 to regulate TGFBR2 mRNA at the post-transcription level in vitro
To determine whether pre-miR-21 can bind and compete with mature miR-21 for the overlapping binding sites within the identified TGFBR2 3′UTR region, we performed a dual-luciferase reporter assay in HEK 293 cells that have negligible miR-21 expression (Supplementary Fig 2) (Li et al., 2009). Reporter construct (PsiCheck2) containing the human TGFBR2 3′UTR (about 1 kb) was co-transfected with a miR-21 overexpression construct. Ectopic expression of miR-21 was driven either by a polymerase (Pol) III-based small hairpin (Sh) system that expressed mature miR-21 (Sh-21) or a Pol II-based system that expressed both pre-miR-21 and mature miR-21 (CMV-21). A scrambled control (Sh-S) and vector expressing GFP (CMV-ctl) were the controls for Sh-21 and CMV-21, respectively. In the presence of overexpressed mature miR-21 (from Sh-21 construct), we observed an approximately 40% repression of TGFBR2 3′UTR-luciferase expression compared to Sh-S (Fig 4A; grey shaded). This repression, however, was significantly de-repressed in the presence of CMV-21 (Fig 4A; light grey) that expressed both the precursor and mature forms of miR-21. This suggests that the presence of pre-miR-21 binding to TGFBR2 3′UTR antagonized the binding and activity of mature miR-21 on TGFBR2 3′UTR-luciferase expression. To test the specificity of the competition between pre-miR-21 and mature miR-21, we also performed a similar reporter assay using human NT-3 3′UTR. The thermodynamic calculation predicted that the most favorable binding region of pre-miR-21 to NT-3 3′UTR (predicted by RNAhybrid) did not overlap with the putative binding site of mature miR-21 to NT-3 3′UTR. The human NT-3 3′UTR was also co-transfected with Sh-21 or CMV-21 expression plasmids in HEK 293 cells. In the luciferase assay, Sh-21 expression provided 60% suppression of NT3 3′UTR-luciferase expression, and CMV-21 showed the comparable suppressive activity on NT-3 (Fig 4B). To validate further the competition between pre- and mature miR-21 for binding to TGFBR2 3′UTR, we modified the TGFBR2 3′UTR putative target site such that the modified target region lost the ability to bind to pre-miR-21 but retained the ability to bind to mature miR-21. We hypothesized that in the presence of modified target, repression levels by reporter assay would be similar using Sh-21 or CMV-21. To create a modified TGFBR2 3′UTR target which might lose the ability to bind to pre-miR-21 without affecting mature miR-21 binding, we constructed the target region in two ways: i) by deleting the region in TGFBR2 3′UTR that was predicted to bind to miR-21* (TGFBR2-del) (see schematics in Fig 4C) and ii) by replacing the predicted miR-21* binding region with a scrambled sequence that has the identical length and GC content (TGFBR2-3p-scr) (see schematics in Fig 4E). The thermodynamic calculation showed an increased ΔG (less favorable) for the binding of pre-miR-21 to either of these modified TGFBR2 3′UTR targets, indicating no thermodynamic advantage for pre-miR-21 relative to the mature form for binding to the target (Supplementary Fig 3). In the presence of overexpressed mature miR-21 from the Sh-21 construct, the modified TGFBR2 reporter constructs exhibited 30–35% repression of luciferase expression compared to Sh-S (Fig 4D & Fig 4F; left two bars). Interestingly, we found a similar level of suppression in TGFBR2 expression in the presence of CMV-21 (Fig 4D & Fig 4F; right two bars). This suggests that in the absence of pre-miR-21 binding to TGFBR2 3′UTR, mature miR-21 resulted in a similar amount of repression. The luciferase assay data, taken together, demonstrated that pre-miR-21 can compete with mature miR-21 for binding to TGFBR2 3′UTR and thereby impede the repression activity of mature miR-21. The data suggests that the elevated pre-miR-21 after SE may have a biological function via regulation of TGFBR2 during epileptogenesis.
Figure 4. Binding competition between pre-miR-21 and mature miR-21 to TGFBR2 3′ UTR.
(A & B) Dual-luciferase assay in HEK 293 cells in the presence of mature miR-21 (Sh-21) or both pre-miR-21 and mature miR-21 (CMV-21) to measure binding competition between pre-miR-21 and mature miR-21 to target mRNAs. (A) For binding to TGFBR2 3′UTR, mature miR-21 showed 40% suppression of TGFBR2 3′UTR-luciferase expression, and this suppression was alleviated to 20% in the presence of pre-miR-21 (p-value=0.0009). (B) For binding to NT3 3′UTR, mature miR-21 showed 60% suppression of NT3 3′UTR-luciferase expression, and this suppression was comparable in the presence of pre-miR-21 (p-value=0.19, not significant). Normalization was done with respect to a scramble control (Sh-S) for Sh-21 and to a CMV-driven GFP control (CMV-ctl) for CMV-21. Two-tailed Student’s t-test, and values are mean±SEM. N = 8; data points from two independent experiments measured in quadruplicate. (C & E) Schematics of the predicted loss of competition between pre-miR-21 and mature miR-21 for binding to TGFBR2 3′ UTR due to the deletion of the miR-21* binding site (TGFBR2-del) or the replacement of the miR-21* binding site by a scramble sequence (TGFB2R-3p-scr). (D & F) Dual-luciferase assay of the modified psiCHECK TGFBR2 constructs (TGFBR2-del and TGFB2R-3p-scr) in presence of Sh-21 and CMV-21. m= mature miR-21 and p= pre-miR-21.
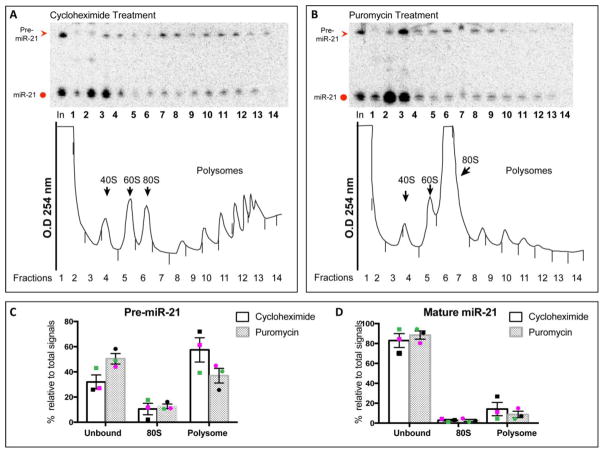
4. Polysome profiling revealed that pre-miR21 binds to translating mRNAs
Previous studies have shown that miRNAs associate with actively translating mRNAs (Maroney et al., 2006a; Maroney et al., 2006b). Various miRNAs including miR-21 have been shown to co-purify with polysomes generated by velocity sedimentation in sucrose gradient (Kim et al., 2004; Maroney et al., 2006a; Molotski and Soen, 2012; Nottrott et al., 2006). Our luciferase assay data suggests that pre-miR-21 base-pairs with the TGFBR2 3′UTR, which overlaps with mature miR-21 binding (miR-21-5p). Hence, we asked if, along with miR-21-5p, pre-miR-21 also co-localizes with translating mRNAs in polysomes. Northern analysis showed that pre-miR-21 accumulated to high levels in human glioblastoma cell line, U-87, compared to other cell lines (Supplementary Fig 2). Lysates from U-87 cells treated with cycloheximide, which freezes all ribosomes on translating mRNAs, were subjected to sucrose gradient fractionation to generate polysome profiles with distinct peaks representing 40S subunits, 60S subunits, 80S monosomes and polysomes (Fig 5A, bottom panel; black arrows). As expected, we found co-localization of mature miR-21 in the polysome fraction as well as in the ribonucleoprotein (RNP) faction of the profile by northern analysis. The presence of mature miRNAs in the RNP fractions is not uncommon (Tat et al., 2016; Kim et al., 2004; Nottrott et al., 2006) because mature miRNAs are not always associated with actively translating mRNAs. Interestingly, pre-miR-21 and mature miR-21 also co-localized in the polysome fractions (Fig 5A, top panel; red arrowhead; see quantification in Fig 5C & Fig 5D). To rule out the possibility of non-specific localization of pre- and mature miR-21 in the polysomes, we treated U-87 cells with puromycin, a molecule that mimics acyl-tRNA and terminates polypeptide chain elongation. In cells treated with puromycin, we observed loss of polysomes and an accumulation of the 80S monosomes (Fig 5B, bottom panel; black arrowhead). Northern analysis and quantification showed that localization of both pre- and mature miR-21 were dissociated from polysomes after puromycin treatment (Fig 5B, top panel; see quantification in Fig 5C & 5D), confirming that the sedimentation of pre-miR-21 in the polysome fraction in cycloheximide-treated cells was indeed a bona-fide interaction with translating mRNAs.
Figure 5. Pre-miR-21 is associated with mRNAs engaged in active translation.
(A & B) Representative Northern blots of ribosome profiles generated from lysate of U-87 cells that were treated with either cycloheximide (A) or puromycin (B) as described in Methods. (A) Top: Northern analysis to detect pre-miR-21 (red arrowhead) and mature miR-21 (red circle) in each of the fractions across the profile. Bottom: Tracing of the profile at 254 nm shows distinct peaks for ribosome subunits, monosome and polysome as indicated (arrows). (B) Top: Northern analysis to detect pre-miR-21 and mature miR-21 (red arrowhead) and mature miR-21 (red circle) in each of the fractions across the profile. Bottom: Tracing of the profile at 254 nm shows loss of polysomes and an accumulation of the 80S monosomes (arrowhead). Quantification of the levels of pre-miR-21 (C) and mature miR-21 (D) in three independent polysome profiles. Unbound= fractions 1–5, 80S= fraction 6, polysome= fractions 7–14. Percentage calculated is relative to the sum of fractions 1–14. Black, magenta, and green data points represent three independent polysome profiling experiments.
DISCUSSION
To our knowledge, this is the first study focusing on precursor miRNA expression and its novel regulation of miRNA activity in status epilepticus. In our prior study we showed that complementary sequences exist for both the miR-151-5p and -3p sequences within the 3′UTR of the transcription factor E2f6 gene (Roy-Chaudhuri et al., 2014). Pre-miR-151 binds to and competes with mature miR-151 (miR-151-5p) binding to the overlapping 3′UTR region of E2f6. In contrast, only mature miR-151 is capable of regulating an endogenous target present in the 3′UTR of RhoGDIA, a tumor suppressor gene, demonstrating how one miRNA can differentially regulate different mRNA targets. However, to date, such regulatory role of pre-miRNA in vivo or in a disease model has not been demonstrated.
We previously published a 4-fold increase of expression in miR-21* compared to mature miR-21 in the rat hippocampus following pilocarpine-induced SE, using a commercially available qRT-PCR assay that utilizes a hairpin primer (Risbud and Porter, 2013). While not confirmed the commercial assay for miR-21* may also recognize pre-miR21 as they share the same primer sequence used for the initial PCR reaction possibly explaining the divergent finding. Here using both a novel pre-miR-21 qRT-PCR assay and northern blot analysis, we show that the pre-miR-21 increase is more significant and is earlier than the mature miR-21 increase following pilocarpine-induced SE. Initially of unclear functional significance, pre-miRNAs may have a role in de-repressing mature miRNA repression of mRNAs. The degree of target mRNA translation repression can thus be altered by the pre/mature miRNA ratio. This mode of gene regulation provides a mechanism by which a single miRNA can differentially regulate unrelated mRNAs within a cell to increase target specificity (Roy-Chaudhuri et al., 2014).
The differential expression of pre- and mature miR-21 in a temporal fashion observed in our status epilepticus model prompted us to determine if pre-miR-21 could regulate the activity of mature miR-21 in a target specific manner. Using bioinformatics we identified two miR-21 targets that showed differential responses to the presence of pre-miR-21. NT-3 mRNA has a minimum free energy change (ΔG) considerably greater (less favorable) than that of TGFBR2 for binding of pre-miR21. We were able to corroborate the bioinformatic results with in vitro studies showing that pre-miR-21 can directly compete with mature miR-21 in the 3′UTR of TGFBR2 but not the 3′UTR of NT-3. Mutations in the putative pre-miR-21 binding site of TGFBR2 3′UTR were able to abolish its ability to compete with mature miR-21, strongly suggesting that pre-miR-21 can suppress degradation or silencing of an mRNA targeted by its mature counterpart or possibly other miRNAs that share nearby binding sites.
Consistent with several prior studies, we found that NT-3 mRNA is down-regulated in the hippocampus following SE (Kim et al., 1998; Mudo et al., 1996; Risbud et al., 2011). Conversely, TGFBR2 mRNA was increased following SE in spite of being a known miR-21 target (Kim et al., 2009; Mishra et al., 2014) and an eventual observed increase in miR-21. Here we demonstrated similar changes in mRNA levels of NT-3 and TGFBR2 in the hippocampus of our pilocarpine-induced SE rats. To date in an epilepsy model there are no protein measurements reported of NT-3 and TGFBR2 that will allow for a direct comparison of the protein, mRNA, miRNA and precursor miRNA concentrations. While technically challenging, protein measurements will be important to solidify our understanding the role that pre-miRNA-mediated regulation might play in epilepsy. While we do not have direct evidence that pre-miR-21 is competing with miR-21 in vivo, the reporter assays suggest a competition between pre- and mature miR-21 for binding to overlapping region in TGFBR2 3′UTR. Moreover, we found pre-miR-21 co-localizing with translating mRNAs in polysomes. We conjecture that after SE the elevated pre-miR-21 competes with mature miR-21 for binding to 3′UTR of TGFBR2 mRNA and thereby preventing degradation or silencing of TGFBR2 mRNA. This could explain the higher level of TGFBR2 mRNA we observed after SE, though in vivo expression studies are needed to confirm causality. It is also possible that pre-miR-21 may have other 3′UTR binding sites that compete with miR-21 and prevent those mRNAs from degradation or silencing during epileptogenesis. Future studies are needed to understand the extent of in vivo competition between pre- and mature miR-21 in epilepsy model. Additionally, it would be important to determine if there are other pre-miRNAs, including miR-146a (Aronica et al., 2010; Omran et al., 2012) and miR-132 (Nudelman et al., 2010) that are elevated following SE and their putative targets to understand the breadth of this novel form of mRNA regulation as these miRNAs are shown to play a role in epileptogenesis.
Markedly elevated expression of pre-miR-21 independent of pri- and mature miR-21 levels suggest altered rates of miR-21 processing or stabilization of pre-miR-21 following SE in our pilocarpine model. To date, miRNA processing has not been investigated sufficiently in epilepsy to offer an explanation to the observed high levels of pre-miR-21. On the other hand, the differential expression profile of pre- and mature miR-21 could in part be explained by a non-canonical processing of miRNA mediated by TGF-β1 signaling (Davis et al., 2008; Davis et al., 2010). TGF-β1 is a pleiotropic cytokine universally up-regulated in response to brain injury injuries, including SE. TGF-β1 expression is elevated in the hippocampal astrocytes of SE-experienced rats (Aronica et al., 2000) and in the neurons of amygdala-kindled rats (Plata-Salaman et al., 2000). Upon activation TGF-β1 binds TGFBR2 to recruit TGFBR1 for direct phosphorylation and nuclear translocation of the transcription factors Smad2 and Smad3 (Smad2/3) to regulate target gene expression (Massague and Wotton, 2000; Shi and Massague, 2003). Davis and colleagues demonstrated that TGF-β1 increased a subset of pre- and mature miRNAs, including miR-21 through a post-transcription step of processing pri-miRNAs. Specifically, Smad2/3 bind to R-SBE, a consensus sequence within the stem region of the pri- and pre-miRNA transcripts, to promote miRNA processing by Drosha (Davis et al., 2008; Davis et al., 2010). It is possible that during the early phase of epileptogenesis, activated TGF-β1 signaling results in Smad2/3 binding to pri-miR-21 for increased production and processing of pre-miR-21.
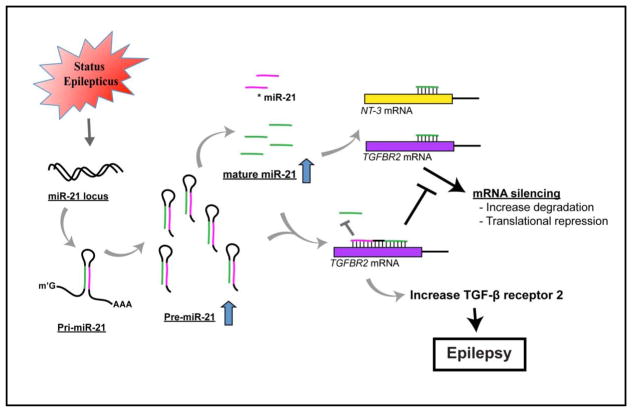
A number of miRNA expression profiling studies using different epilepsy models employed bioinformatics to predict the target mRNAs of the identified miRNAs (Bot et al., 2013; Gorter et al., 2014; Hu et al., 2011; McKiernan et al., 2012); however, few studies have validated these predicted epilepsy-associated miRNA targets (Jimenez-Mateos et al., 2012; Sano et al., 2012). To date no seizure studies have looked at pre-miRNA levels and their possible role in epileptogenesis. MiR-21 is an evolutionarily conserved miRNA (Krichevsky and Gabriely, 2009) and is consistently up-regulated in epilepsy samples and following other neurological injuries leading to epilepsy (Bot et al., 2013; Gorter et al., 2014; Risbud and Porter, 2013). The possibility of competitive binding of pre-and mature miRNAs for mRNA targets suggests that pre- and mature miRNAs need to be measured and the thermodynamics of each target mRNA may be needed to understand transcriptional regulation in epilepsy. MiR-21 has been implicated in neuronal survival following an insult and has many experimentally validated mRNA targets; however, none have been shown in an in vivo epilepsy model to be regulated by miR-21. Prolonged TGF-β signaling via infusion of TGF-β into the brain has been shown to be sufficient to induce epilepsy (Cacheaux et al., 2009). We speculate that the elevation in pre-miR-21 may allow for elevation of TGFBR2 mRNA levels, potentially leading to increased TGF-β signaling and epileptogenesis (Fig 6).
Figure 6.
A schematic model depicts the novel regulation of increased pre-miR-21 through completing with mature miR-21 to bind to 3′UTR of TGFBR2 mRNA and to regulate its expression following SE.
In summary, we propose that for a subset of mRNAs (TGFBR2 and E2f6) translation suppression and/or degradation is regulated by the ratio of precursor to mature miRNA. To fully understand miRNA regulation of mRNA translation, analysis requires quantification of pre- and mature miRNA. To date miR-151, miR-124 and miR-21 have been implicated in the precursor/mature competition mechanism of gene regulation but more studies are needed to understand the scope of this process in epilepsy and other disease models.
MATERIALS AND METHODS
Animals and Induction of Status Epilepticus
Adult male Spargue Dawley rats (Charles River Laboratories International, CA) between 60–90 days of age were provided with unrestricted access to food and water in temperature and humidity controlled housing with a 12 hour light- 12 hour dark cycle (lights on 7:00 AM). The rats were injected with 1mg/kg methyl-scopolamine (Sigma-Aldrich, MO) intraperitoneally (IP) to block peripheral cholinergic effects 30 minutes prior to pilocarpine-induced status epilpepticus (SE). 385mg/kg pilocarpine hydrochloride (Sigma-Aldrich Corp., MO) was administrated IP to the SE rats, and control rats were given 1/10 th dose (38.5mg/kg) of pilocaprine. Rats were monitored for appearance of stage V Racine seizures and received 6mg/kg diazapam (Hospira Inc., IL) one hour after the induction of SE. The seizure group received an additional half dose (3 mg/kg) every two hours after the initial diazepam injection until all behavioral seizures resolved. The Stanford Institutional Animal Care and Use Committee approved all the protocols used.
Tissue Collection and RNA Extraction
Both SE and control groups were sacrificed at 4, 12, and 48 h after the induction of SE. The animals were anesthetized with isoflurane, and the hippocampal tissue was dissected out, frozen on dry ice, and stored at −80°C until RNA extraction. For qRT-PCR and northern blots, total and small RNAs were extracted from the rat hippocampi using the mirVANA isolation kit according to the protocol (Thermo Fisher Scientific, MA). Total RNA was isolated from cell lines using Trizol (Thermo Fisher Scientific, MA) (48 h after transfection if transfected). RNA extraction of sucrose gradient fractions were carried out by phenol: chloroform: isoamylalcohol (Sigma-Aldrich, MO). The RNA concentration of extracted RNAs was measured and the A260/280 ratio was determined using Nanodrop spectrophotometer (Thermo Scientific NanoDrop, DE). The extracted RNAs were stored at −80°C until use.
Quantitative Real-time PCR
Total RNA and random hexamers were used for pri-miR-21, TGFBR2 mRNA, NT-3 mRNA; small RNA and specific stem loop primers were used for pre- and mature miR-21. All RNA concentrations were determined first, and reverse transcription was performed using TaqMan® Reverse Transcription Reagents (Thermo Fisher Scientific, MA). For pre-miR-21, small RNA and reverse transcription primers (designed by Thermo Fisher Scientific based on rat pre-miR-21 sequence) were incubated at a denaturing step (85°C for 5 min and 65°C for 5 min) before reverse transcription (25°C for 10 min, 48°C for 30 min, 95°C for 5 min). Real-time PCR reactions were carried out in 96-well OptiAmp fast plates and the StepOnePlus™ Real-Time PCR System (Stanford PAN Facility). The reaction mix in each well contained 10 ul of the TaqMan® Universal Master Mix II (Thermo Fisher Scientific, MA), 1 ul of the FAM-MGB probe/primer mix (20x) for gene of interest (purchased from Thermo Fisher Scientific, MA; pri-miR-21: Rn03464993_pri; TGFBR2: Rn00579682_m1; NT-3: Rn00579280; pre-miR-21: CS51000; mature miR-21: 000397), 9 ul of cDNA/ddH2O. For pri-miR-21, TGFBR2 mRNA, NT-3 mRNA and mature miR-21, 25 ng cDNA/well was used; for pre-miR-2, 125 ng cDNA/well was used. PPIA/cyclophilin (Rn00690933) (Kinjo et al., 2016; Risbud et al., 2011; Risbud and Porter, 2013) was used as housekeeping gene for pri-miR-21, TGFBR2 mRNA, NT-3 mRNA; 4.5S RNA(H) (001716) (Risbud et al., 2011; Risbud and Porter, 2013) was used as housekeeping gene for pre- and mature miR-21. The housekeeping genes did not vary across treatment groups for any of the time points. A relative standard curve was used to calculate the fold changes of pri-miR-21 in SE samples. Comparative CT Method (ΔΔCt) was used to calculate the fold changes of other genes of interest. Each sample was run in triplicates.
Small RNA Northern Blots
For small RNA northern blots, 3 ug of MirVana extracted small RNA from hippocampal tissues or 10 ug of Trizol extracted RNA from cell culture was electrophoresed on 15% (w/v) acrylamide/7M urea gel and transferred onto a Genescreen Plus (Perkin Elmer, MA) membrane. Hybridization was done overnight in PerfectHyb Plus (Sigma-Aldrich, MO) buffer at 65°C using 32P labeled hsa-miR-21-5p probe (miRCURY LNA™ Detection probe)(Exiqon, Vedbaek, Denmark) or at 35°C with U6 probe (IDT, CA) (Li et al., 2016; Roy-Chaudhuri et al., 2014).
Thermodynamic calculation
Minimum free energy of binding of pre-miR-21 to the target 3′UTRs (TGFBR2 and NT-3) and to the modified TGFBR2 3′UTRs was calculated using RNAhybrid. Minimum free energy of folding of the pre-miR-21 and TGFBR2 3′UTR was calculated using Mfold.
Cell culture and Plasmid constructs
HEK 293, U87, HeLa, and Neuro2A cell lines were grown in Dulbecco’s modified Eagle’s media (Thermo Fisher Scientific, MA) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% L-Glutamine, 1% Non-essential amino acids and 1% Pen-strep (Thermo Fisher Scientific, MA).
For cloning of the TGFBR2 reporter construct, the 3′UTR of TGFBR2 was PCR amplified (primers used: 5′-TAGGCGATCGCTCGAGCTCTTCTGGGGCAGGCTG -3′ and 5′-CCGCGTCGACACTAGTGCCAAACTGTGCTTGAGCAATC-3′) from human genomic DNA and cloned downstream of the Renilla luciferase gene in XhoI and SpeI digested psiCHECK-2 vector (Promega, WI) using Infusion Cloning HD kit (Clontech, CA). Similarly, the NT-3 reporter vector was constructed by PCR amplifying the 3′UTR of human NTF3 using primers (5′-TAGGCGATCGCTCGAGATTGGCATCTCTCCCCATAT-3′ and 5′-CCGCGTCGACACT AGTACAACAGTCATGGCCTTG AC-3′).
Site-directed mutagenesis using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, CA) was carried out on psiCheck-TGFBR2 construct to first generate the TGFBR2-del reporter construct and then on psiCheck-TGFBR2-del construct to generate the TGFBR2-3p-scr reporter construct. Primers for TGFBR2-del construct Primers for the TGFBR2-del construct were 5′-GCTGTGGGGATAAGCAGA AACAACATAGAGCATTCTATGCC-3′ and 5′-GGCATAGAATGCTCTATGTTGTT TCTGCTTATCCCCACAGC-3′. Primers for the TGFBR2-3p-scr construct were 5′GCTGTGGGGATAAGCAGAAACAGATGGGAGTAGAGC GGGCACACATAGAGCATTCTATGCCTTTG-3′ and 5′-CAAAGGCATAGAATGCTCTAT GTGTGCCCGCTCTACTCCCATCTGTTTCTGCTTATCCCCACAGC -3′.
To generate the pSh-21 construct for robust expression of mature hsa-miR-21, sense oligonucleotides (5′-GATCTCAACATCAGTCTGATAAGCTACTCCTGACCCAAGTAGCT TATCAGACTGATGTTGATTTTTGTAC-3′) and antisense oligonucleotides (5′AAAAATCAA CATCAGTCTGATAAGCTACTTGGGTCAGGAGTAGCTTATCAGACTGATGTTGA) were chemically synthesized (IDT, CA); both strands annealed and inserted between BglII and KpnI sites downstream of the U6 Pol III promoter. To generate the pCMV-21 construct overexpressing precursor and mature hsa-miR-21, a ~500 nt human genomic region harboring miR-21 locus was PCR amplified. Primers used were FW: 5′-GACAAGCTTGCGGCCGCCCTTTAGGAGCATT ATGAGCAT-3′ and RV: 5′-ATCCTCTAG AGTCGACGAAGGTCAAGTAACAGTCATAC-3′. The PCR amplicon was inserted into a digested CMV expression vector (Pol II promoter) using Infusion Cloning HD kit (Clontech, CA).
Reporter assays
Dual-luciferase assay (Promega, WI) was performed 24 hours after transfection according to manufacturer’s protocol and detected by a Modulas Microplate Luminometer (Turner Biosystems, CA). For transfection in a 24-well plate, 125 ng of psiCHECK reporter plasmid was co-transfected with 125 ng of miR-21 overexpression plasmids (pSh-21 or pCMV-21) in HEK 293 cells using Lipofectamine 2000 (Thermo Fisher Scientific, MA). Cell seeding was performed at a concentration of 5 × 104 cells for HEK 293 per well in a 24-well plate.
Ribosome profiles and Polysome Fractionation
U-87 cells (~ 6 × 107) grown in 15 cm dish for ~36 hours were treated with either cycloheximide (100 ug/ml) for 5 mins or puromycin (200 ug/ml) (Sigma-Aldrich, MO) for half hour before harvesting. Lysis was carried out in a buffer containing 150 mM KCl, 15 mM Tris pH 7.5, 5 mM MgCl2, 100 ug/ml cycloheximide, 1% Triton X-100 and 500 units/ml RNasin Plus (Promega, WI). Velocity sedimentation was carried out at 35,000 rpm for 2 hours 45 minutes in a 10–50% sucrose gradient. The absorbance profile was obtained at 254 nm by using a Foxy gradient fractionator (Brandel, MD).
Statistics and Quantification
Statistical analysis was performed using two-tailed Student’s t-Test or one-way ANOVA with Kruskal-Wallis (Prism 6.0). A p-value of 0.05 or lower was considered statistically significant. The qRT-PCR assays were performed in triplicate. The luciferase assays were done independently at least twice with each experiment done in 4 replicates to generate individual data points that were all used to calculate SEM. In northern blots, the pre-miR-21 and mature miR-21 levels were normalized to U6 using ImageJ software. In polysome profiling, the fractions were grouped into unbound (fractions 1–5), 80S (fraction 6) and polysome (fractions 7–14). Quantification of pre-miR-21 or mature miR-21 in each group was presented as percentage of the total signals of pre-miR-21 or mature miR-21 (sum of pre-miR-21 or mature miR-21 signals in all fractions after background subtraction from northern blots using Bio-Rad Quantify One ®).
Supplementary Material
Pilocarpine is administrated to animals until they reach repetitive stage V seizures (Racine’s scale) in which generalized motor convulsions are exhibited. The seizure-exposed animals initially recover but most will go on to develop spontaneous seizures or epilepsy (Curia et al., 2008). The present study focuses on the molecular changes during the early period of epileptogenesis. The brains are collected at 4, 12, and 48 h after SE. Timeline is not drawn to scale.
Northern analysis shows expression level of pre-miR-21 (red arrowhead) and mature miR-21 (red circle) in Neuro 2A, HEK 293, HeLa, and U87 cell lines. U87 has the highest levels of pre- and mature miR-21 compared to other cell lines.
A schematic shows the putative binding sties and ΔG of pre-miR-21 to wild-type TGFBR2 mRNA 3′UTR (left), to TGFBR2-del mRNA 3′UTR (right top), and to TGFBR2-3p-scr mRNA 3′UTR (right bottom). ΔG for both modified TGFBR2 mRNA 3′UTR are calculated.
Highlights.
Pre-miR-21 markedly increases following status epilepticus (SE).
Pre-miR-21 localizes to polysomes with translating mRNAs.
Pre-miR-21 competes with mature miR-21 for binding in the 3′UTR of TGFBR2 mRNA, but not NT-3 mRNA, de-repressing miR-21-mediated gene suppression in vitro.
TGFBR2 increases and NT-3 decreases following SE, implicating pre-miR-21-mediated gene regulation.
Acknowledgments
We thank Deepti Dueby for review of this manuscript. This work was supported by National Institute of Neurological Disorders and Stroke (NINDS) (NSRO1 N5056222) (B.E.P) and RO1-DK078424 (M.A.K.). K.C. is supported by the Child Health Research Institute, Lucile Packard Foundation for Children’s Health and the Stanford CTSA (UL1 TR001085) from Stanford University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aronica E, Fluiter K, Iyer A, Zurolo E, Vreijling J, van Vliet EA, Baayen JC, Gorter JA. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. The European journal of neuroscience. 2010;31:1100–1107. doi: 10.1111/j.1460-9568.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- Aronica E, van Vliet EA, Mayboroda OA, Troost D, da Silva FH, Gorter JA. Upregulation of metabotropic glutamate receptor subtype mGluR3 and mGluR5 in reactive astrocytes in a rat model of mesial temporal lobe epilepsy. The European journal of neuroscience. 2000;12:2333–2344. doi: 10.1046/j.1460-9568.2000.00131.x. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bian S, Sun T. Functions of noncoding RNAs in neural development and neurological diseases. Molecular neurobiology. 2011;44:359–373. doi: 10.1007/s12035-011-8211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder DK. Neurotrophins in the dentate gyrus. Progress in brain research. 2007;163:371–397. doi: 10.1016/S0079-6123(07)63022-2. [DOI] [PubMed] [Google Scholar]
- Bot AM, Debski KJ, Lukasiuk K. Alterations in miRNA levels in the dentate gyrus in epileptic rats. PloS one. 2013;8:e76051. doi: 10.1371/journal.pone.0076051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller B, Liu X, Wang X, Zhang RL, Zhang L, Hozeska-Solgot A, Chopp M, Zhang ZG. MicroRNA-21 protects neurons from ischemic death. The FEBS journal. 2010;277:4299–4307. doi: 10.1111/j.1742-4658.2010.07818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacheaux LP, Ivens S, David Y, Lakhter AJ, Bar-Klein G, Shapira M, Heinemann U, Friedman A, Kaufer D. Transcriptome profiling reveals TGF-beta signaling involvement in epileptogenesis. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29:8927–8935. doi: 10.1523/JNEUROSCI.0430-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curia G, Longo D, Biagini G, Jones RS, Avoli M. The pilocarpine model of temporal lobe epilepsy. Journal of neuroscience methods. 2008;172:143–157. doi: 10.1016/j.jneumeth.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Molecular cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer HM, Mula M, Sander JW. The global burden and stigma of epilepsy. Epilepsy & behavior: E&B. 2008;12:540–546. doi: 10.1016/j.yebeh.2007.12.019. [DOI] [PubMed] [Google Scholar]
- Dogini DB, Avansini SH, Vieira AS, Lopes-Cendes I. MicroRNA regulation and dysregulation in epilepsy. Frontiers in cellular neuroscience. 2013;7:172. doi: 10.3389/fncel.2013.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer E, Kokaia M, Ernfors P, Ferencz I, Kokaia Z, Lindvall O. Suppressed kindling epileptogenesis and perturbed BDNF and TrkB gene regulation in NT-3 mutant mice. Experimental neurology. 1997;145:93–103. doi: 10.1006/exnr.1997.6478. [DOI] [PubMed] [Google Scholar]
- Friedman A, Kaufer D, Heinemann U. Blood-brain barrier breakdown-inducing astrocytic transformation: novel targets for the prevention of epilepsy. Epilepsy research. 2009a;85:142–149. doi: 10.1016/j.eplepsyres.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009b;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorter JA, Iyer A, White I, Colzi A, van Vliet EA, Sisodiya S, Aronica E. Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy. Neurobiology of disease. 2014;62:508–520. doi: 10.1016/j.nbd.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nature reviews Molecular cell biology. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Henshall DC. MicroRNA and epilepsy: profiling, functions and potential clinical applications. Current opinion in neurology. 2014;27:199–205. doi: 10.1097/WCO.0000000000000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesdorffer DC, Logroscino G, Benn EK, Katri N, Cascino G, Hauser WA. Estimating risk for developing epilepsy: a population-based study in Rochester, Minnesota. Neurology. 2011;76:23–27. doi: 10.1212/WNL.0b013e318204a36a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Zhang C, Long L, Long X, Feng L, Li Y, Xiao B. Expression profile of microRNAs in rat hippocampus following lithium-pilocarpine-induced status epilepticus. Neuroscience letters. 2011;488:252–257. doi: 10.1016/j.neulet.2010.11.040. [DOI] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, Sano T, O’Tuathaigh C, Waddington JL, Prenter S, Delanty N, Farrell MA, O’Brien DF, Conroy RM, Stallings RL, DeFelipe J, Henshall DC. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nature medicine. 2012;18:1087–1094. doi: 10.1038/nm.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Henshall DC. Epilepsy and microRNA. Neuroscience. 2013;238:218–229. doi: 10.1016/j.neuroscience.2013.02.027. [DOI] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Smith MA, Post RM, Rosen JB. Attenuation of kindling-induced decreases in NT-3 mRNA by thyroid hormone depletion. Epilepsy research. 1998;29:211–220. doi: 10.1016/s0920-1211(97)00087-9. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature reviews Molecular cell biology. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Hwang SJ, Bae YC, Jung JS. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem cells. 2009;27:3093–3102. doi: 10.1002/stem.235. [DOI] [PubMed] [Google Scholar]
- Kinjo ER, Higa GS, Santos BA, de Sousa E, Damico MV, Walter LT, Morya E, Valle AC, Britto LR, Kihara AH. Pilocarpine-induced seizures trigger differential regulation of microRNA-stability related genes in rat hippocampal neurons. Scientific reports. 2016;6:20969. doi: 10.1038/srep20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nature reviews Neuroscience. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. Journal of cellular and molecular medicine. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Huang H, Sun L, Yang M, Pan C, Chen W, Wu D, Lin Z, Zeng C, Yao Y, Zhang P, Song E. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- Li Q, Hu B, Hu GW, Chen CY, Niu X, Liu J, Zhou SM, Zhang CQ, Wang Y, Deng ZF. tRNA-Derived Small Non-Coding RNAs in Response to Ischemia Inhibit Angiogenesis. Scientific reports. 2016;6:20850. doi: 10.1038/srep20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nature structural & molecular biology. 2006a;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Maroney PA, Yu Y, Nilsen TW. MicroRNAs, mRNAs, and translation. Cold Spring Harbor symposia on quantitative biology. 2006b;71:531–535. doi: 10.1101/sqb.2006.71.043. [DOI] [PubMed] [Google Scholar]
- Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. The EMBO journal. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKiernan RC, Jimenez-Mateos EM, Bray I, Engel T, Brennan GP, Sano T, Michalak Z, Moran C, Delanty N, Farrell M, O’Brien D, Meller R, Simon RP, Stallings RL, Henshall DC. Reduced mature microRNA levels in association with dicer loss in human temporal lobe epilepsy with hippocampal sclerosis. PloS one. 2012;7:e35921. doi: 10.1371/journal.pone.0035921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Deng JJ, Gowda PS, Rao MK, Lin CL, Chen CL, Huang T, Sun LZ. Androgen receptor and microRNA-21 axis downregulates transforming growth factor beta receptor II (TGFBR2) expression in prostate cancer. Oncogene. 2014;33:4097–4106. doi: 10.1038/onc.2013.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotski N, Soen Y. Differential association of microRNAs with polysomes reflects distinct strengths of interactions with their mRNA targets. Rna. 2012;18:1612–1623. doi: 10.1261/rna.033142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudo G, Jiang XH, Timmusk T, Bindoni M, Belluardo N. Change in neurotrophins and their receptor mRNAs in the rat forebrain after status epilepticus induced by pilocarpine. Epilepsia. 1996;37:198–207. doi: 10.1111/j.1528-1157.1996.tb00012.x. [DOI] [PubMed] [Google Scholar]
- Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nature structural & molecular biology. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM, Storm DR. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20:492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto OK, Janjoppi L, Bonone FM, Pansani AP, da Silva AV, Scorza FA, Cavalheiro EA. Whole transcriptome analysis of the hippocampus: toward a molecular portrait of epileptogenesis. BMC genomics. 2010;11:230. doi: 10.1186/1471-2164-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ladewig E, Zhou L, Lai EC. Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes & development. 2013;27:778–792. doi: 10.1101/gad.211698.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omran A, Peng J, Zhang C, Xiang QL, Xue J, Gan N, Kong H, Yin F. Interleukin-1beta and microRNA-146a in an immature rat model and children with mesial temporal lobe epilepsy. Epilepsia. 2012;53:1215–1224. doi: 10.1111/j.1528-1167.2012.03540.x. [DOI] [PubMed] [Google Scholar]
- Peng J, Omran A, Ashhab MU, Kong H, Gan N, He F, Yin F. Expression patterns of miR-124, miR-134, miR-132, and miR-21 in an immature rat model and children with mesial temporal lobe epilepsy. Journal of molecular neuroscience: MN. 2013;50:291–297. doi: 10.1007/s12031-013-9953-3. [DOI] [PubMed] [Google Scholar]
- Pitkanen A, Lukasiuk K. Molecular and cellular basis of epileptogenesis in symptomatic epilepsy. Epilepsy & behavior: E&B. 2009;14(Suppl 1):16–25. doi: 10.1016/j.yebeh.2008.09.023. [DOI] [PubMed] [Google Scholar]
- Plata-Salaman CR, Ilyin SE, Turrin NP, Gayle D, Flynn MC, Romanovitch AE, Kelly ME, Bureau Y, Anisman H, McIntyre DC. Kindling modulates the IL-1beta system, TNF-alpha, TGF-beta1, and neuropeptide mRNAs in specific brain regions. Brain research Molecular brain research. 2000;75:248–258. doi: 10.1016/s0169-328x(99)00306-x. [DOI] [PubMed] [Google Scholar]
- Pohlmann-Eden B, Beghi E, Camfield C, Camfield P. The first seizure and its management in adults and children. Bmj. 2006;332:339–342. doi: 10.1136/bmj.332.7537.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine RJ. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalography and clinical neurophysiology. 1972;32:281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud RM, Lee C, Porter BE. Neurotrophin-3 mRNA a putative target of miR21 following status epilepticus. Brain research. 2011;1424:53–59. doi: 10.1016/j.brainres.2011.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud RM, Porter BE. Changes in microRNA expression in the whole hippocampus and hippocampal synaptoneurosome fraction following pilocarpine induced status epilepticus. PloS one. 2013;8:e53464. doi: 10.1371/journal.pone.0053464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Chaudhuri B, Valdmanis PN, Zhang Y, Wang Q, Luo QJ, Kay MA. Regulation of microRNA-mediated gene silencing by microRNA precursors. Nature structural & molecular biology. 2014;21:825–832. doi: 10.1038/nsmb.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Reynolds JP, Jimenez-Mateos EM, Matsushima S, Taki W, Henshall DC. MicroRNA-34a upregulation during seizure-induced neuronal death. Cell death & disease. 2012;3:e287. doi: 10.1038/cddis.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Trujillo RD, Yue SB, Tang Y, O’Gorman WE, Chen CZ. The potential functions of primary microRNAs in target recognition and repression. The EMBO journal. 2010;29:3272–3285. doi: 10.1038/emboj.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissberg I, Wood L, Kamintsky L, Vazquez O, Milikovsky DZ, Alexander A, Oppenheim H, Ardizzone C, Becker A, Frigerio F, Vezzani A, Buckwalter MS, Huguenard JR, Friedman A, Kaufer D. Albumin induces excitatory synaptogenesis through astrocytic TGF-beta/ALK5 signaling in a model of acquired epilepsy following blood-brain barrier dysfunction. Neurobiology of disease. 2015;78:115–125. doi: 10.1016/j.nbd.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Michalski B, Racine RJ, Fahnestock M. Continuous infusion of neurotrophin-3 triggers sprouting, decreases the levels of TrkA and TrkC, and inhibits epileptogenesis and activity-dependent axonal growth in adult rats. Neuroscience. 2002;115:1295–1308. doi: 10.1016/s0306-4522(02)00384-6. [DOI] [PubMed] [Google Scholar]
- Yu Y, Kanwar SS, Patel BB, Oh PS, Nautiyal J, Sarkar FH, Majumdar AP. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFbetaR2) in colon cancer cells. Carcinogenesis. 2012;33:68–76. doi: 10.1093/carcin/bgr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pilocarpine is administrated to animals until they reach repetitive stage V seizures (Racine’s scale) in which generalized motor convulsions are exhibited. The seizure-exposed animals initially recover but most will go on to develop spontaneous seizures or epilepsy (Curia et al., 2008). The present study focuses on the molecular changes during the early period of epileptogenesis. The brains are collected at 4, 12, and 48 h after SE. Timeline is not drawn to scale.
Northern analysis shows expression level of pre-miR-21 (red arrowhead) and mature miR-21 (red circle) in Neuro 2A, HEK 293, HeLa, and U87 cell lines. U87 has the highest levels of pre- and mature miR-21 compared to other cell lines.
A schematic shows the putative binding sties and ΔG of pre-miR-21 to wild-type TGFBR2 mRNA 3′UTR (left), to TGFBR2-del mRNA 3′UTR (right top), and to TGFBR2-3p-scr mRNA 3′UTR (right bottom). ΔG for both modified TGFBR2 mRNA 3′UTR are calculated.