Abstract
Sustaining a balanced intestinal microbial community is critical for maintaining intestinal health and preventing chronic inflammation. The gut is a highly dynamic environment, subject to periodic waves of peristaltic activity. We hypothesized that this dynamic environment is a prerequisite for a balanced microbial community and that the enteric nervous system (ENS), a chief regulator of physiological processes within the gut, profoundly influences gut microbiota composition. We found that zebrafish lacking an ENS due to a mutation in the Hirschsprung disease gene, sox10, develop microbiota-dependent inflammation that is transmissible between hosts. Profiling microbial communities across a spectrum of inflammatory phenotypes revealed that increased levels of inflammation were linked to an overabundance of pro-inflammatory bacterial lineages and a lack of anti-inflammatory bacterial lineages. Moreover, either administering a representative anti-inflammatory strain or restoring ENS function corrected the pathology. Thus, we demonstrate that the ENS modulates gut microbiota community membership to maintain intestinal health.
Author summary
Intestinal health depends on maintaining a balanced microbial community within the highly dynamic environment of the intestine. Every few minutes, this environment is rocked by peristaltic waves of muscular contraction and relaxation through a process regulated by the enteric nervous system (ENS). We hypothesized that normal, healthy intestinal microbial communities are adapted to this dynamic environment, and that their composition would become perturbed without a functional ENS. To test this idea, we used a model organism, the zebrafish, with a genetic mutation that prevents formation of the ENS. We found that some mutant individuals without an ENS develop high levels of inflammation, whereas other mutants have normal intestines. We profiled the intestinal bacteria of inflamed and healthy mutants and found that the intestines of inflamed individuals have an overabundance of pro-inflammatory bacterial lineages, lack anti-inflammatory bacterial lineages, and are able to transmit inflammation to individuals with a normally functioning ENS. Conversely, we were able to prevent inflammation in the ENS mutants by either administering a representative anti-inflammatory bacterial strain or restoring ENS function. From these experiments, we conclude that the ENS modulates intestinal microbiota community membership to maintain intestinal health.
Introduction
The intestinal tract serves to harvest nutrients and energy, protect against harmful toxins and pathogens, and clear out waste. These functions can be modulated by both the enteric nervous system (ENS) and the trillions of symbiotic bacteria that reside within the gut [1–3]. Importantly, the influence of microbiota on intestinal functions and health depends on the constituent microbes. Alterations in microbial composition from those observed in “healthy” subjects are often defined as “dysbiotic,” which refers to communities that become perturbed in their composition such that they acquire pathogenic properties [4–6]. Given that the composition of the microbiota is critical for host health, it is significant that the intestinal microbial community is generally stable despite the highly dynamic internal environment of the intestinal tract [7,8], which experiences disruptions such as influxes of ingested matter, host secretion and epithelial cell turnover, and coordinated outward flow of material. How microbial community stability is achieved amid these constant perturbations is unknown. Hosts with impaired intestinal motility can develop dysbiosis and intestinal pathology [9,10], which suggests a profound role for the ENS in constraining microbiota composition. Here, we explore how the ENS shapes the ecology of the intestine, and we address key questions about the assembly of dysbiotic microbial communities, their functional properties, and strategies for their treatment—three aspects of dysbiosis that have been challenging to address from observational studies in humans. Our analysis reveals how, without ENS constraint, imbalances in pro- and anti-inflammatory members of the microbiota can drive intestinal pathology.
The most severe example of ENS dysfunction in humans is Hirschsprung disease (HSCR), an enteric neuropathy that results from a failure of neural crest–derived cells to form the distal ENS [3]. Approximately 30% of HSCR patients develop a severe form of intestinal dysbiosis, known as Hirschsprung-associated enterocolitis (HAEC) [9–11], which is distinguished by diarrhea, distension, fever, and, in extreme cases, sepsis and death [12]. Studies suggest that the etiology of HAEC has a microbial component, as both pathogenic bacteria [13] and alterations in commensal communities [9,10] have been linked to HAEC. Interestingly, patients with a broad range of human diseases, such as inflammatory bowel disease (IBD), cystic fibrosis [14], diabetes [15], malnutrition [16], and myotonic muscular dystrophy [17,18], also experience debilitating gastrointestinal (GI) symptoms. Although cause and effect are difficult to determine, these diseases are associated with both small intestinal bacterial overgrowth, a clinical syndrome often seen with impaired intestinal motility, and an altered microbiota, suggesting that impaired ENS function could be a driver of dysbiosis.
To explore how the ENS may prevent dysbiosis by constraining microbial populations, we turned to a zebrafish model of HSCR. Multiple well-described zebrafish lines carry mutations in HSCR loci [19–22]. The most extreme ENS loss is seen in mutants homozygous for a null mutation in the HSCR gene sox10 [23,24]; these mutants entirely lack an ENS [24]. The mutant allele t3 (sox10t3) homozygotes have diminished rhythmic peristaltic activity [21], making this an ideal model for dissecting the role of the ENS in host–microbe interactions. Zebrafish are well suited for examining ENS contributions to microbiota composition because we can monitor ENS development, absolute bacterial abundance, and disease phenotypes, such as neutrophil accumulation, across the entire intestine of individual larvae. Thus, we can assess system-level functional readouts that describe properties of the associated microbiota. Furthermore, the high fecundity and ease of working with zebrafish provide us with large sample sizes to increase the power of our experiments such that we can monitor how natural microbiota variation at the species level drives phenotypic variation.
In this study, we demonstrate that the ENS constrains the abundance and composition of the microbiota. We find that loss of the ENS in sox10t3 mutants results in assembly of a dysbiotic community leading to a microbe-driven intestinal inflammation that varies among individuals and resembles HAEC. Microbiota profiling across the spectrum of inflammatory states revealed that extreme intestinal inflammation is linked to an outgrowth of pro-inflammatory bacterial lineages and a reduction of anti-inflammatory bacterial lineages. Moreover, administering representative anti-inflammatory bacterial strains or transplanting wild-type (WT) ENS precursors to restore a WT ENS corrects the pathology in sox10t3 mutant hosts. Our analysis reveals that ENS function is a key feature of intestinal health that constrains the composition of the resident microbiota and prevents overgrowth of bacterial lineages that can drive disease.
Results
Loss of sox10 results in intestinal bacterial overgrowth
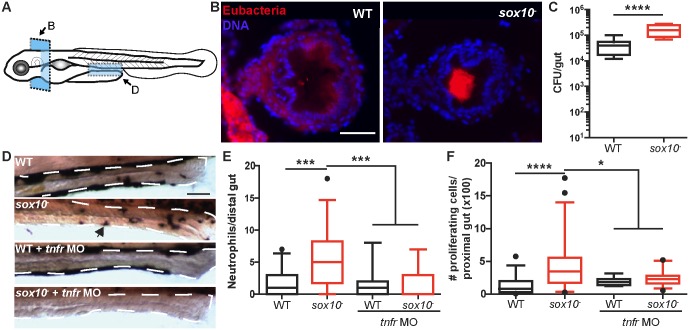
The complete loss of ENS in sox10t3 mutants (S1 Fig) results in defective intestinal motility [21]. Given the connection between altered intestinal motility and small intestinal bacterial overgrowth, we hypothesized that functional consequences of these mutants would include changes to intestinal ecology and alterations in resident microbial populations. To visualize the abundance and distribution of bacteria along the length of the intestine, we used fluorescent in situ hybridization (FISH). In sox10t3 mutants, we noted large populations of bacteria throughout the intestine, with marked accumulations of bacteria at the esophageal-intestinal junction (Fig 1A and 1B), a location not typically heavily colonized with bacteria. We also quantified the total number of colony-forming units (CFU) per intestine and found that sox10t3 mutants had a significantly higher bacterial load (Fig 1C). These results suggest that sox10t3 mutants experience bacterial overgrowth, which is consistent with defective intestinal transit. Defective intestinal transit has been observed in mutants in another allele, sox10m241, which have intestinal peristalsis but do not clear ingested fluorescent beads as well as WTs [25]. To demonstrate delayed intestinal transit in sox10t3 mutants, we adapted a previous single color assay [26] into a two-color intestinal transit assay (S1 Fig). The delayed transit and impaired clearance we observed in sox10 mutants likely contribute to bacterial overgrowth within their intestines. For the work described in this manuscript, we use sox10t3 mutants, hereafter referred to as sox10 mutant or sox10-.
Fig 1. sox10 mutants experience bacterial overgrowth and physiological indications of dysbiosis.
(A) Schematic representation of the location and orientation of images in B and D. (B) Representative images of the panbacterial population by FISH on the esophageal-intestinal junction of WT (left) and sox10- (right) fish. Blue, DNA; red, eubacteria. (C) Quantification of bacterial colonization level in sox10 mutants and WT siblings. (D) Representative images of WT, sox10 mutant, and tumor necrosis factor receptor (tnfr) morpholino (MO) injected larvae of both genotypes. Arrowhead indicates neutrophil. (E) Quantification of intestinal neutrophil number per 140 μm of distal intestine. (F) Total numbers of proliferating cells over 30 serial sections beginning at the esophageal-intestinal junction and proceeding into the bulb in 6-d-post-fertilization (dpf) fish. Box plots represent the median and interquartile range; whiskers represent the 5–95 percentile. n > 15 per group, *p < 0.05, ***p < 0.001, ****p < 0.0001, ANOVA with Tukey’s range test. Also see S1 Fig. Scale bars = 50 μm.
The sox10- intestine exhibits increased neutrophil response and epithelial cell proliferation
We next asked whether the bacterial overgrowth phenotype in sox10- resulted in signs of intestinal inflammation. Thus, we quantified intestinal neutrophil populations, a marker of inflammation, in cohoused WTs and sox10 mutants by staining for the neutrophil-specific enzyme myeloid peroxidase. At 6 d post fertilization (dpf), intestinal neutrophil accumulation in sox10 mutants was significantly increased compared to WTs (Fig 1D and 1E). Notably, sox10 mutants exhibited a much greater variation in intestinal neutrophil accumulation (0–18; n = 30) compared to WT siblings (0–7; n = 31); some sox10 mutants had intestinal neutrophil levels similar to WTs, whereas others had significantly elevated neutrophil populations. Intestinal neutrophil accumulation under homeostatic conditions in WT fish requires the pro-inflammatory tumor necrosis factor (TNF) pathway [27,28]. The increased neutrophil response in sox10 mutants also depends on this pathway, as inhibiting expression of the TNF receptor using an antisense morpholino [27,28] abolished the increased neutrophil response (Fig 1D and 1E). Another indicator of intestinal pathology is epithelial cell proliferation. At 6 dpf, sox10 mutants had markedly increased intestinal cell proliferation relative to cohoused WT animals. Unlike the normal intestinal epithelial cell proliferation response to microbiota, which is TNF independent [29], we found that elevated cell proliferation in the sox10 mutant intestine was TNF dependent (Fig 1F), suggesting that this was an inflammation-dependent pathological response.
The intestinal microbiota of sox10 mutants is necessary and sufficient to induce a hyper-inflammatory state
To determine whether the intestinal microbiota of sox10- hosts is necessary to induce the increased intestinal neutrophil response, we derived sox10 mutants and their WT siblings germ free (GF). We found that GF sox10 mutants have a low neutrophil population, indistinguishable from their WT siblings (Fig 2A). To determine if the microbial community established in sox10 mutants is sufficient to induce inflammation, we performed an experiment in which we transferred microbiota from sox10 mutants into WTs. As donors, we used microbial communities from conventionally raised (CV) WT, sox10 mutant, or WT intestinal alkaline phosphatase morpholino (iap MO)-injected larvae. iap MO-injected fish are hypersensitive to lipopolysaccharide and thus develop elevated intestinal inflammation without evidence of dysbiosis [27]. These fish serve as control for the possibility that nonbacterial factors such as host pro-inflammatory cytokines rather than microbial derived factors cause transmissible intestinal inflammation (Fig 2B) [30]. At 6 dpf, for each separate group (WT, sox10-, and iap MO), we dissected, pooled, and homogenized the donor intestines. As a negative control, we included transplantation from homogenized intestines of GF fish. The homogenate from each group was inoculated into flasks housing GF 4 dpf WT fish (Fig 2C). We found that inoculation with microbes from sox10 mutants was sufficient to induce elevated intestinal inflammation in WTs as compared to inocula from GF, CV WT, or CV iap MO fish, none of which induced intestinal inflammation (Fig 2D). To test whether the capacity of sox10 mutant microbiota to induce elevated neutrophils was due to increased bacterial load, we transplanted 5× CV WT microbes, which corresponded to the bacterial load of sox10 mutant transplants. This larger inoculum did not induce more intestinal inflammation (S2 Fig), which indicates that the microbial community assembled in sox10- hosts is functionally distinct from WT microbiota and is sufficient to induce inflammation in fish with a normal, functional ENS.
Fig 2. Intestinal microbiota are necessary and sufficient to induce increased intestinal neutrophil accumulation in sox10 mutants.

(A) Quantification of intestinal neutrophil number per 140 μm of distal intestine. Neutrophil accumulation was inhibited when sox10 mutants were raised GF compared to CV controls. n > 21 per condition. (B) Schematic of fish used as donors in the transmission experiment. Intensity of red indicates level of intestinal inflammation. (C) Schematic of the experimental protocol. Intestines of GF, CV WT, sox10 mutants, or iap MO were dissected for use as inoculum for 4 dpf GF WT recipients. Recipient fish were colonized for 2 d before examination of intestinal neutrophil number. (D) Transfer of intestinal microbes from inflamed intestines of sox10 mutants causes increased intestinal neutrophil number in WTs. n ≥ 10, *p < 0.05, ***p < 0.001, ****p < 0.0001, ANOVA with Tukey’s range test. See also S2 Fig.
Bacterial overgrowth does not explain increased intestinal neutrophil response
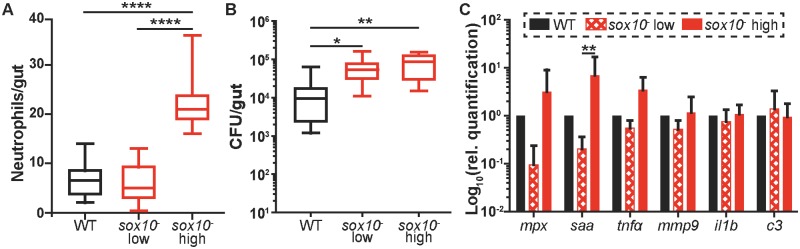
sox10 mutants exhibit a wide range of intestinal neutrophil populations (Figs 1D and 2A) as well as variation in bacterial load (Fig 1B). Therefore, we asked whether intestinal neutrophil abundance corresponded to increased bacterial abundance. We used transgenic sox10 mutant hosts expressing green fluorescent protein (GFP) under control of the neutrophil-specific mpx promoter to quantify both neutrophil population and intestinal bacterial load in individual fish (Fig 3A). When we compared sox10 mutants that fell in the bottom half of neutrophil response (“sox10- low”) or in the top half of neutrophil response (“sox10- high”) to WTs, we found that all sox10 mutants, regardless of neutrophil level, carried significantly higher bacterial loads than WTs (Fig 3B). Thus, impaired intestinal clearance (S1 Fig) leads to an increased bacterial load; however, the bacterial overgrowth per se in sox10- does not drive an increased intestinal neutrophil response. We further characterized the pro-inflammatory signature of the sox10- high- and low-neutrophil subsets by monitoring expression of a panel of immune genes in the intestine (Fig 3C). These results aligned with our observations of the neutrophil population, as the sox10- high-neutrophil subset had elevated levels of mpx, saa, and tnfα expression compared to WT and the sox10- low-neutrophil subset (Fig 3C); however, the increase in saa transcription was the only one to reach statistical significance. Consistent with the significantly elevated intestinal neutrophil response in these samples, saa is known to mediate intestinal neutrophil behavior stimulated by microbes [31]. Collectively, our results suggest that a pro-inflammatory compositional change occurs in the microbial community of a subset of sox10 mutants.
Fig 3. Increased bacterial colonization level does not drive increased intestinal neutrophil accumulation or pro-inflammatory gene expression.
Quantification of intestinal neutrophil number (A) and bacterial colonization level (B) in the sox10-, Tg(mpx:GFP) line. sox10- fish were split into two groups, “sox10- low” (bottom half) and “sox10- high” (top half) based on intestinal neutrophil number. Ten representative fish from each group were plated to determine total CFU/intestine. n ≥ 9 per group. *p < 0.05, **p < 0.01, ****p < 0.0001, ANOVA with Tukey’s range test. (C) Relative expression calculated by the 2-ΔΔCt method of immune genes from dissected intestines. For mpx, saa, il1b, and c3, n = 5 pools of 5 dissected intestines; for tnfα and mmp9, n = 3 pools of 18 dissected intestines. Graph displays average ± standard deviation (SD); **p < 0.01, t test corrected for multiple comparisons using Holm–Šidák method.
Changes in the abundance of two dominant microbial genera drive intestinal neutrophil accumulation
To address the possibility of a pro-inflammatory compositional change in the sox10- microbiota, we profiled microbial communities by performing 16S rRNA gene sequencing on intestinal communities isolated from cohoused WT and sox10 mutant individuals. We collected samples across three independent experiments. To uncover differences in microbiota composition that explain the variable severity of neutrophil accumulation in sox10 mutants, we collected intestinal neutrophil response data for the same individuals from which we isolated microbial DNA and grouped samples as “WT,” “sox10- low” (intestinal neutrophil response 0–8), or “sox10- high” (intestinal neutrophil response of greater than or equal to 22); these groups include the top 26% and the bottom 29%, respectively (Fig 4A). By standard metrics of community variability (non-metric multidimensional scaling of Canberra distances, richness, Faith’s Phylogenetic Diversity, unweighted UniFrac), these three groups were not significantly different (S3 Fig), which indicates that these communities are largely made up of the same microbes, and community differences driving neutrophil differences are perhaps due to changes in minor members [28].
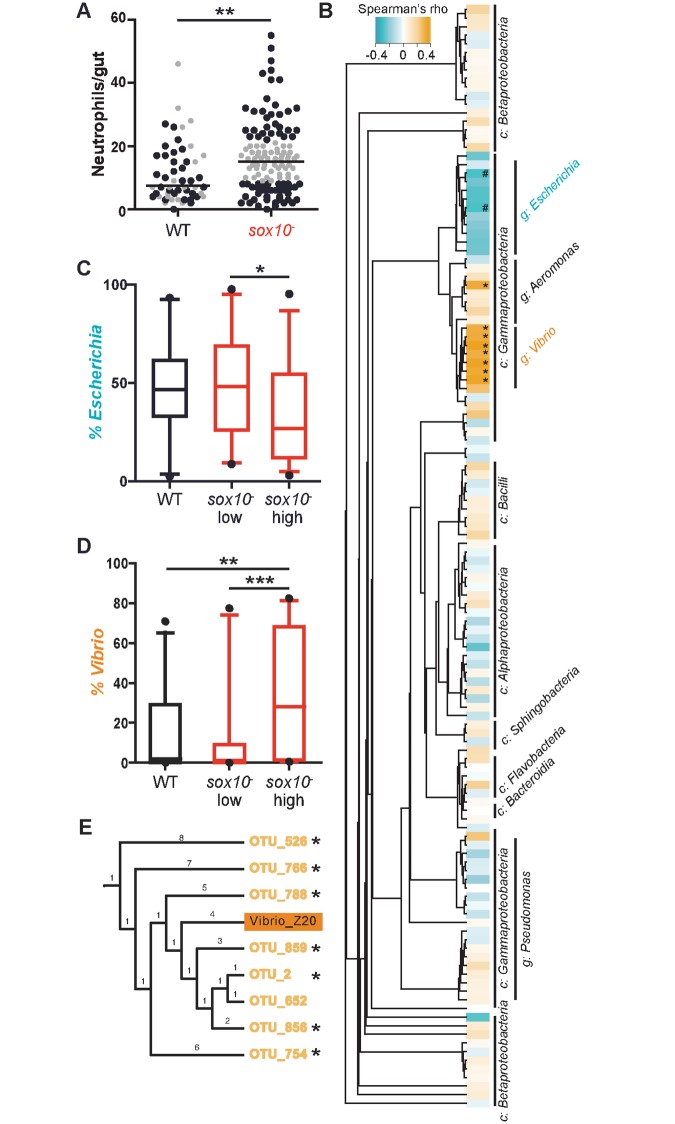
Fig 4. Changes in microbial lineages define low and high neutrophil accumulation.
(A) Intestinal neutrophil accumulation from samples used for 16S rRNA gene sequencing. Each dot represents an individual fish; black circles indicate sequenced samples, WT, n = 32; sox10- high, n = 30; sox10- low, n = 31. Gray circles indicate samples that were not sequenced. Line indicates median. **p < 0.01 Student’s t test. (B) Spearman’s rank correlation between intestinal neutrophil number and each operational taxonomic unit (OTU) present in at least 20 samples. After false discovery rate correction, two genera, Escherichia and Vibrio, stand out with correlations to neutrophil number. Asterisks represent significance of Spearman correlation, *p < 0.05, #p = 0.08, c: class, g: genus. The percent abundance of Escherichia (C) and Vibrio (D) across genotypes and intestinal neutrophil levels. (E) Phylogenetic tree of OTUs from the Vibrio genus and our Vibrio zebrafish isolate (Vibrio Z20). For D, E: *p < 0.05, **p < 0.01, ***p < 0.001, ANOVA. See also S3 Fig.
We next asked whether the relative abundance of any bacterial operational taxonomic units (OTUs) correlated with intestinal neutrophil number across all individuals surveyed in the study. Of 129 OTUs present in at least 20 individuals, we found a small subset whose percent abundance was associated with neutrophil number, as measured by Spearman’s correlations. Strikingly, these neutrophil-associated OTUs were tightly clustered in only two genera found in this population of fish intestines (Fig 4B). The Escherichia/Shigella genus (hereafter referred to as Escherichia) had ten OTUs that negatively correlated with neutrophil abundance, although only two had borderline significance after false discovery rate correction. All OTUs of the Vibrio genus had significant positive correlations with neutrophil abundance (Fig 4B). Examination of OTU abundances revealed not only that the two most abundant genera were Vibrio and Escherichia (S3 Fig) but also that they were significantly decreased and increased, respectively, in the “sox10- high” group relative to the WT and “sox10- low” groups (Fig 4C and 4D).
The observation of pro-inflammatory activity associated with Vibrio is consistent with our previous analysis of a zebrafish-derived Vibrio strain (ZWU0020) [32] that is phylogenetically closely related to the Vibrio OTUs in the current experiment (Fig 4E). Previously, we showed that Vibrio strain ZWU0020 (hereafter referred to as Vibrio Z20) promotes intestinal neutrophil accumulation in a concentration-dependent manner in gnotobiotic zebrafish [28]. Similarly, in the current study, the log10(relative abundance) of Vibrio was significantly positively correlated with neutrophil number (Fig 5A), and we also observed that the log10(relative abundance) of Escherichia was negatively correlated with intestinal neutrophil accumulation, although the amount of variation explained was low (Fig 5A). This relationship mirrors the relationship we previously observed in simple microbial communities in gnotobiotic zebrafish between the abundance of Shewanella strain ZOR0012 (hereafter referred to as Shewanella Z12) and a proportional decrease in neutrophil number [28]. Of note, two OTUs from the Shewanella genus included in our analysis in this study did not have a significant correlation with neutrophil number. Combining the loss of Escherichia and the gain of Vibrio does not increase the amount of variation in neutrophil number explained by the gain of Vibrio alone (Table 1, Fig 5B). We used Akaike’s Information Criterion (AIC) [33] to test the relative quality of each of these models; the model that accounts only for Vibrio reports the lowest AIC value, which identifies Vibrio as the best microbial predictor of intestinal neutrophil number variability (Table 1). These analyses suggest that a balance of the Vibrio and Escherichia lineages may be important for maintaining intestinal homeostasis, with Vibrio abundance being a major determinant of intestinal inflammation.
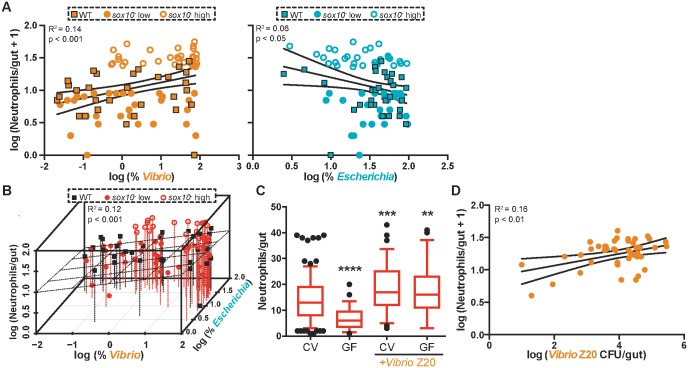
Fig 5. An increase in Vibrio contributes dominantly to intestinal neutrophil number.
(A) Correlation between the log percent Vibrio abundance (left) and log percent Escherichia abundance (right) with log10(intestinal neutrophil number +1). Linear regression analysis with 95% confidence intervals. Each point represents an individual fish: WT, squares, n = 32; sox10- high, open circles, n = 30; sox10- low, closed circles, n = 31. (B) Correlation between both log10(percent Escherichia) (z-axis) and log10(percent Vibrio) (x-axis) abundance with log10(intestinal neutrophil number +1) (y-axis). Planar regression analysis, n = 93. WT, black squares; sox10- low, red closed circles; sox10- high, red open circles. (C) Addition of Vibrio Z20 increased intestinal neutrophil accumulation in both CV and GF sox10 mutants. n ≥ 49, from at least three independent experiments. **p < 0.01, ***p < 0.001, ****p < 0.0001, indicates difference from CV, ANOVA. (D) Correlation between the absolute abundance of Vibrio Z20 in CV sox10 mutants and log10(intestinal neutrophil number + 1) in experiments with exogenously added Vibrio Z20. Linear regression analysis with 95% confidence intervals. For C-D, data from four to six independent experiments, n > 35. See also S4 Fig.
Table 1. Coefficients of regression analysis of Escherichia, Vibrio, and intestinal neutrophil number.
| Estimate | Std. err. | t-value | p-value | MultipleR2 | p-value | A.I.C. | AIC relative likelihood | ||
|---|---|---|---|---|---|---|---|---|---|
| Escherichia | (Intercept) | 1.48 | 0.18 | 8.0 | <0.0001 | 0.06 | 0.02 | 93.5 | 0.04 |
| Escherichia | −12.5 | 3.7 | −3.3 | 0.001 | |||||
| Vibrio | (Intercept) | 0.99 | 0.043 | 23.5 | <0.0001 | 0.14 | 0.0003 | 87.1 | 1 |
| Vibrio | 0.12 | 0.035 | 3.5 | 0.0007 | |||||
| Escherichia and Vibrio | (Intercept) | 1.20 | 0.21 | 5.91 | <0.0001 | 0.13 | 0.0019 | 88.1 | 0.6 |
| Escherichia | −0.13 | 0.13 | −1.03 | 0.31 | |||||
| Vibrio | 0.11 | 0.04 | 2.73 | 0.0074 |
To confirm the functional contribution of Vibrio to the increased neutrophil responses in sox10-, we first added Vibrio Z20 to CV sox10 mutants at 4 dpf and assayed neutrophil numbers at 6 dpf. Exogenously added Vibrio Z20 induced a significant increase in neutrophil accumulation over the number seen in CV sox10 mutants (Fig 5C). Furthermore, the absolute abundance of Vibrio Z20 colonizing these fish was positively correlated with intestinal neutrophil number, similar to observations made in the 16S rRNA data set (Fig 5D). We noted that the extent of the increase in neutrophil accumulation upon addition of Vibrio depended on the level of intestinal neutrophils present in control hosts (S4 Fig), which we think reflects fluctuations in bacterial community composition in CV zebrafish between experiments and a limited ability to change the neutrophil-inducing capacity of an intestinal microbiota already dominated by Vibrio strains. We furthermore observed the inflammation-inducing capacity of Vibrio Z20 in monoassociation by adding Vibrio Z20 to GF sox10 mutants. In these conditions, Vibrio Z20 was still sufficient to induce high intestinal neutrophil influx (Fig 5C). In monoassociation the range of Vibrio colonization was too narrow (S4 Fig) to explore a correlative relationship. These experiments support our hypothesis, based on the microbiota profiling of these fish, that an overabundance of Vibrio species causes a dysbiotic and pro-inflammatory microbiota.
Intestinal hyper-inflammation is ameliorated by anti-inflammatory bacterial isolates or by transplantation of WT ENS precursors into sox10 mutants
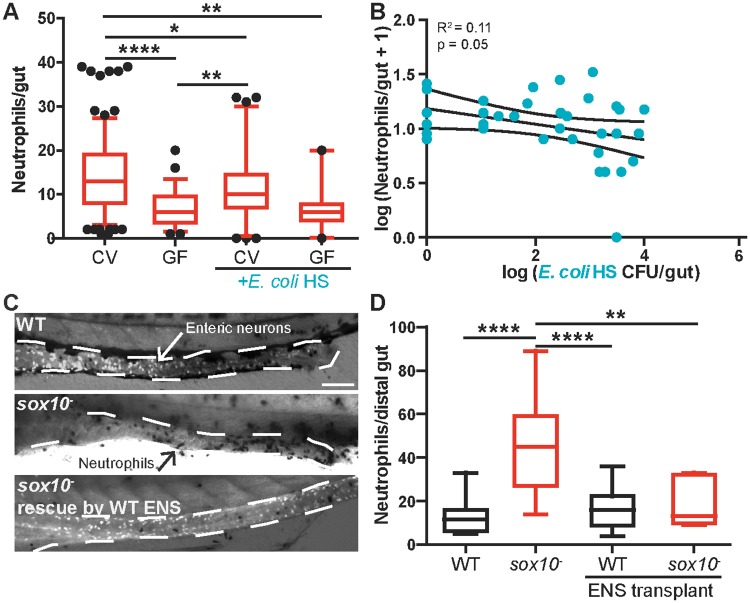
We hypothesized that the dysbiotic state of the sox10 mutant intestine could be corrected by balancing the pro-inflammatory activity of Vibrio species with the addition of anti-inflammatory isolates, such as Escherichia species or Shewanella Z12 [28]. Consistent with this prediction, we found that addition of Escherichia coli HS, a commensal Escherichia strain isolated from a healthy human adult [34] that is closely related to Escherichia OTUs in CV fish (S4 Fig), can colonize the zebrafish intestine (S4 Fig), reducing neutrophil numbers in CV sox10 mutants (Fig 6A) and maintaining GF levels of intestinal neutrophil accumulation in monoassociation (Fig 6A). Moreover, the absolute abundance of colonizing E. coli HS in CV sox10 mutants displayed a similar negative correlation with intestinal neutrophils (Fig 6B), as observed with the sequenced OTUs (Fig 5A). Shewanella Z12, another species that displays a negative correlation between abundance and intestinal neutrophil accumulation [28], also reduced intestinal neutrophils in sox10 mutants, which suggests this relationship may be a hallmark of anti-inflammatory bacterial strains (S4 Fig). Thus, sox10- dysbiosis can be corrected by adding anti-inflammatory bacteria to the community. Shewanella Z12 uses an unidentified secreted factor present in cell-free supernatant (CFS) to mediate its anti-inflammatory activity [28] (S4 Fig). However, E. coli HS CFS was insufficient to reduce sox10 mutant intestinal inflammation (S4 Fig), suggesting that these species use two distinct mechanisms to control the host innate immune response.
Fig 6. Inflamed intestines are rescued by anti-inflammatory bacterial isolates or transplantation of WT ENS into sox10 mutants.
(A) Addition of a representative Escherichia isolate, E. coli HS, to CV sox10 mutants reduces intestinal neutrophil accumulation. Monoassociation of sox10 mutants with E. coli HS does not increase neutrophil level over that observed in GF zebrafish. n > 20, from at least three independent experiments. (B) Correlation between absolute abundance of E. coli HS and log10(intestinal neutrophil number + 1) in experiments with added E. coli HS. Linear regression analysis with 95% confidence intervals. For A, B: n > 35, from three to six independent experiments. (C) Representative images of distal intestine from WT, sox10-, and sox10- rescued by WT ENS precursor transplantation. Anti-ElavI1–labeled enteric neurons are white (white arrow); neutrophils are black (black arrow). Scale bar = 100 μm. (D) Quantification of intestinal neutrophil number per 140 μm of distal intestine. n > 6 for all conditions, *p < 0.05, **p < 0.01, ****p < 0.0001, ANOVA with Tukey’s range test. See also S4 Fig.
As an alternative to manipulating the microbiota directly, we postulated that correcting the underlying deficit in the ENS would also alleviate the inflammation in sox10 mutants. To test this hypothesis, we performed a rescue experiment in which vagal neural crest cells, the ENS precursors, were transplanted from WT donors into sox10 mutant hosts. Our previous studies showed a correlation between the number of ENS neurons and gut motility [21]; thus, after the transplant, we assayed for formation of a normal-appearing ENS along with intestinal neutrophil accumulation. Following transplantation, the sox10 mutants that developed a normal-appearing ENS extending along the entire length of the intestine had WT levels of intestinal neutrophils (Fig 6C and 6D), demonstrating that the ENS is sufficient to prevent intestinal inflammation. Together, our results demonstrate that the ENS contributes to intestinal health by maintaining a balanced gut microbiota, revealing a previously unappreciated role for the ENS in host–microbe interactions.
Discussion
Many intestinal diseases, such as IBD, and many diseases with intestinal symptoms, such as cystic fibrosis [14], are associated with compositional changes in the intestinal microbiota, implying dysbiosis. However, it has been extremely challenging to establish whether such alterations are indeed dysbiotic and the underlying driver of disease. Overcoming this challenge will be an important step toward identifying new therapeutic targets and strategies for ameliorating dysbiosis-associated disease. To connect alterations in microbial communities with host pathology, three questions are crucial to address: (1) How do dysbiotic microbial communities assemble? (2) Which member(s) of dysbiotic communities contribute to disease? (3) How can dysbiosis-related disease be mitigated?
How do dysbiotic microbial communities assemble?
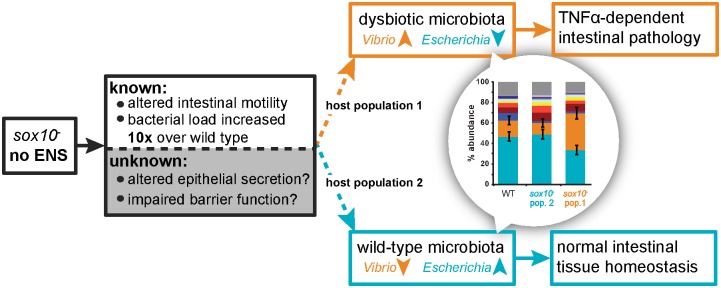
Microbiota are assembled through fundamental ecological processes, including dispersal, local diversification, ecological drift, and environmental selection [35]. We have previously shown that a portion of early larval zebrafish intestinal communities follow a neutral pattern of assembly [36]. This observation suggests that features of the gut environment constrain which microbes colonize and persist in the gut environment. We hypothesized that the ENS, which controls motility and aspects of intestinal homeostasis [3], may also directly or indirectly serve as a significant constraint on intestinal microbial community assembly, such that loss of the ENS constitutes a major ecological shift. Consistent with this hypothesis, we show that zebrafish lacking an ENS have an altered intestinal microbiota and deficits in clearing food from the gut, suggesting gut motility is a mechanism by which the ENS influences microbiota composition. This is further supported by the recent finding that GI transit time is one of the largest predictors of microbiota composition [37]. Moreover, intestinal motility profoundly influences the spatial organization of bacterial populations and has been found to promote competitive exclusion within resident communities [38]. This suggests that abnormal GI transit patterns can significantly reshape ecological interactions within the gut. The ENS also contributes to epithelial barrier function and secretion; however, whether and how these functions are altered in the sox10 mutant has not yet been described. Therefore, observed alterations to the microbial community may be the result of changes to any (or all) of these functions (Fig 7). Of all ENS mutants, the sox10 mutant has the most overlapping characteristics with the human disease HSCR; however, given that the ENS is interconnected with many other organ systems, our work reveals the need to investigate other model systems of ENS dysfunction. Currently, no other available mutant both entirely eliminates the ENS as seen in sox10 mutants and retains normal craniofacial structures [21]. For example, another severe mutant, ret, has a few residual ENS neurons and also exhibits severe craniofacial defects that may impair bacterial colonization [39]. A zebrafish sox10 cell ablation model exists [40] but requires treatment with the antibiotic metronidazole, which would alter the microbiota and confound our experiments. For future experiments, developing a new line in which it is possible to specifically ablate enteric neurons at specified developmental stages will be essential.
Fig 7. Proposed model of sox10 mutant intestinal pathology.
sox10 mutants have altered intestinal motility and an increased bacterial load. Given the role of the ENS in intestinal function, sox10 mutants likely also experience alterations in epithelial secretion and permeability, although these phenotypes are yet to be examined. sox10 mutants can assemble a microbiota that mirrors WT intestinal microbiota (host population 2) or is dysbiotic (host population 1), characterized by an expansion of the Vibrio lineage and reduction of the Escherichia lineage. We do not yet know what determines which bacterial community assembles in sox10 mutants (dashed lines) but hypothesize that it could be due to the timing or order of exposure to bacterial strains, differences in epithelial permeability or secretion, or differences in other host compensatory mechanisms.
Mounting evidence suggests that ENS defects of HSCR patients, as well as those of HSCR animal models, are not restricted to the aganglionic region of the intestine but rather extend to more proximal intestinal regions; thus, these defects are poised to precipitate dysbiosis associated with HAEC [9,41,42]. Patients with chronic IBD can also have functional and structural abnormalities in the ENS that disrupt motility [43,44]. Although these pathologies are generally thought to be secondary to inflammation [45], our data raise the possibility that, regardless of the origin of ENS defects, they have the potential to disrupt the microbial community and thus contribute to a feedback loop that prevents a healthy microbiota from establishing after an inflammatory episode. Such a cycle could explain the variable outcomes of treating IBD patients with probiotics [46]. However, in healthy hosts, this feedback loop may instead enforce stability and homeostasis within the system.
Which member(s) of dysbiotic communities contribute to disease?
The intimate connection between human health and microbiota suggests that health is an effect of services provided by the microbial ecosystem [35], and thus to manage health through the microbiota, we need to identify the taxa that provide specific ecosystem services. One strategy to identify bacterial species that specifically influence host health or disease phenotypes is to define a dose–response relationship, or correlation, between a phenotype of interest and a microbial isolate. For example, we used gnotobiotic methods to identify Vibrio Z20 as pro-inflammatory in zebrafish by its positive correlation between abundance and intestinal neutrophil number [28], defining a dose–response relationship for this isolate. In this study, we have now expanded this approach to complex communities and discovered that Vibrio species fulfill this pro-inflammatory role in the highly inflamed sox10 mutant gut—finding a positive relationship between relative abundance of Vibrio and number of intestinal neutrophils. Correlations between OTUs and host phenotypes have been important in the identification of “indicator” species of interest in chronic obstructive pulmonary disease [47], ulcerative colitis [48], and asthma [49]. Perez-Losada and colleagues [49] expanded this concept by comparing the host and bacterial transcriptomes of asthmatics and healthy controls. They revealed positive correlations between both bacterial phyla (Proteobacteria) and functions (adhesion) with the pro-inflammatory cytokine IL1A [49]. Similarly, a recent large-scale study used correlations to identify microbial drivers of cytokine expression in healthy humans [50]. These studies highlight the potential for using correlation to identify bacterial species, or properties of bacterial species, that have functional consequences for the host in health and disease.
The zebrafish is an especially good model for this type of analysis because we can manipulate host genetics and the environment to control microbial variability across samples. For human studies, the heterogeneity in microbial communities among subjects may be a limiting factor in performing this type of analysis. Furthermore, zebrafish microbial communities are less complex than those of humans, which allows us to probe the data at a higher resolution, with less data reduction [48], and to analyze host–microbe interactions at the OTU level. Our analysis at this resolution revealed conserved functions at the genera level. The phylogenetic conservation of certain bacterial traits suggests that interactions between zebrafish and their resident microbiota serve as a model for identifying bacterial lineages that influence phenotypes across many host species. For example, like the pro-inflammatory Vibrio identified in sox10 mutants, some Vibrios, such as Vibrio parahaemolyticus, induce inflammatory gastroenteritis in humans [51]. We also identified the Escherichia genus as anti-inflammatory in the zebrafish, and some Escherichia are used as probiotics in the treatment of inflammatory intestinal disorders like ulcerative colitis [52]. These data suggest that characterizing species correlated with host phenotypes in model organisms may help to identify individual members of complex communities that contribute to disease phenotypes.
How can dysbiosis-associated disease be mitigated?
Viewing host–microbiota interactions as an ecological system allowed us to identify two system components, the ENS and key bacterial species, which greatly influence ecosystem function, as measured by host intestinal inflammation. With this information, we can ask whether manipulation of these components provides us with control over ecosystem function. For example, an expanded population of Vibrio lineages combined with a decreased population of Escherichia lineages in sox10 mutants induces increased neutrophil influx. We manipulated this component by introducing the anti-inflammatory Escherichia or Shewanella Z12 [28] and thus ameliorated the disease phenotype. Notably, the most consistent microbial signature of IBD patients is the loss of an anti-inflammatory species, Faecalibacterium prausnitzii [53], the colonization level of which decreases in a step-wise manner from healthy subjects, to patients in remission, to patients with active colitis, to patients with infective colitis [54]. Furthermore, administration of F. prausnitzii reduces disease severity in mice with chemically induced colitis [55]. These results highlight the important immunomodulatory role played by specific bacterial species within the intestinal microbiota and the need to identify these species to devise therapies for reestablishing control of the intestinal environment and ameliorating dysbiosis.
Treatment of dysbiosis-associated diseases with probiotics is likely to require continual probiotic administration if there is an underlying disease mechanism leading to its depletion. A more fruitful approach would include a treatment for the underlying ecological perturbation along with the introduction of probiotic strains. For example, restoring ENS function via transplantation or drug administration are possible ways to treat ENS dysfunction. We demonstrated that transplantation of WT ENS precursors into sox10 mutant hosts restored a normal-appearing ENS and rescued the inflammatory gut phenotype. We think that the normal inflammatory response indicates restored ENS function. However, in future experiments, determining the functional capacity of the transplanted ENS to restore motility, secretion, and epithelial barrier function will help elucidate which specific ENS functions contribute to the constraint on the intestinal microbiota. Recently, there has been significant success in establishing a functional ENS in mouse by transplantation of induced pluripotent stem cells [56], which, together with our results, suggests that this strategy could contribute to a successful cure of disease in cases of HAEC or IBD.
Conclusion
Here, we have utilized the zebrafish as a powerful model to examine the complex relationship between the ENS, the immune system, and the microbiota. We demonstrated the critical role played by the ENS in shaping the ecology of the intestine by constraining the functional properties of the resident microbiota. Our analysis reveals how, without this constraint, imbalances in pro- and anti-inflammatory members of the microbiota can drive intestinal pathology. The imbalances we discovered could not be described by large changes in phylum level abundances or the acquisition of a single pathogenic lineage but rather by subtle differences in the abundances of key commensal species that have the potential to either protect against or promote inflammation. We note that this discovery reveals the reciprocal relationship between the microbes and the ENS, as ENS activity and development can be altered by microbiota; in fact, individual bacterial species can have distinct effects on ENS function [57]. Furthermore, immune cell responses influence ENS function both under healthy conditions [58] and in inflammatory states [59]. Therefore, intestinal homeostasis depends on a complex tri-directional conversation that occurs between the microbiota, the ENS, and the immune system, with proper functioning of each branch depending on signals from the other two branches. Uncovering new therapeutic strategies for chronic intestinal diseases will require a profound understanding not only of each branch of this system but the multifaceted interactions that connect them and how alterations made to one system ripple out to affect the function of the other two branches. Developing scalable and tractable model systems, such as the zebrafish, in which we can monitor all three branches of this system will be critical for addressing these complex questions.
Materials and methods
Ethics statement
All zebrafish experiments were done in accordance with protocols approved by the University of Oregon Institutional Animal Care and Use Committee (protocol numbers 15–15, 14-14RR, and 15-83A8) and conducted following standard protocols as described in [60].
Zebrafish husbandry
CV-raised WT (AB x Tu strain), heterozygote sox10t3- (referred to as sox10-) [24], and Tg(BACmpx:GFP)i114 (referred to as mpx:GFP) [61] fish were maintained as described [60]. Homozygous sox10 mutants were obtained by mating heterozygotes and identified by lack of pigmentation [24]. The sox10t3 line was used for neutrophil experiments unless otherwise indicated. The mpx:GFP line [61] was crossed with sox10+/- adults to create a line that when in-crossed resulted in offspring that were sox10-/- and Tg(BACmpx:GFP)i114 (referred to as sox10, mpx:GFP). No defects were observed in heterozygous siblings, which have pigment, develop normally, and survive to adulthood, and thus they are grouped with homozygous WTs [21,23,24,38,62]. For all experiments, WT siblings and homozygous sox10 mutants were cohoused.
Food transit assay and fluorescent in situ hybridization
See S1 Text.
Morpholino injections
Splice-blocking MOs (Gene Tools, Corvallis, OR) were injected into embryos at the one cell stage. For knockdown of TNF, the tr1v1/tr1v2 MOs (1.2 moles and 6 moles, respectively) were used as previously described [27,28]. For knockdown of intestinal alkaline phosphatase, the iape212 MO (3 pmoles) was used as previously described [27].
Histology and quantification of neutrophils and proliferating cells
Zebrafish larvae were fixed in 4% paraformaldehyde (PFA) overnight. Whole larvae were stained with Myeloperoxidase kit (Sigma) following the manufacturer’s protocol and processed and analyzed as previously described [27]. For analysis of neutrophils in mpx:GFP fish, GFP+ cells in the intestine were quantified as previously described [28]. For proliferation, larvae were immersed in 100 μg/ml EdU (A10044, Invitrogen) for 16 h prior to PFA fixation. Subsequent processing and analysis were done as previously described [29]. See also Histology and neutrophil analysis in S1 Text.
Microbiota quantification
At 6 dpf, larvae were humanely killed with Tricaine (Western Chemical, Inc., Ferndale, WA), mounted in 4% methylcellulose (Fisher, Fair Lawn, NJ), and their intestines were dissected using sterile technique. Dissected zebrafish intestines were placed in 100-μl sterile EM, homogenized, diluted, and cultured on tryptic soy agar plates (TSA; BD, Sparks MD). After incubation at 32°C for 48 h for conventionally colonized fish or for 24 h for inoculated fish, colonies were counted.
Gnotobiotic fish husbandry
Zebrafish embryos were derived GF as previously described [63]. All manipulations to the GF flasks were performed under a class II A/B3 biological safety cabinet. Zebrafish inoculated with donor microbial populations were generated by inoculating flasks with 4 dpf GF zebrafish with 104 CFU/mL of donor microbes (1×). Donor microbes were collected by dissecting CV zebrafish and based on colonization data (Fig 1B) each fish was assumed to carry 105 total CFU/gut; a total of 25 dissected guts were pooled and homogenized to create the donor microbes. We inoculated CV fish with live Vibrio (106 bacterial cells/ml), E. coli HS (107 bacterial cells/ml), and Shewanella Z12 (106 bacterial cells/ml) as previously described [28]. For monoassociations, each strain was inoculated at 106 bacterial cells/ml. We isolated and concentrated CFS as previously described [28]. Flasks were kept at 28°C until analysis of myeloperoxidase positive cells on 6 dpf.
RNA isolation and qPCR
RNA isolation and cDNA preparation were performed as previously reported [27] except either five (for saa, mpx, and il1b primers) or 18 (for mmp9 and tnfα primers) dissected intestines were pooled. RNA was harvested by homogenizing and extracting with Trizol reagent (Invitrogen). Contaminating genomic DNA was eliminated using the Turbo DNA-free kit (Ambion) per manufacturer’s instructions. The RNA (100 ng for saa, mpx, and il1b primers; 320 ng for mmp9 and tnfα primers) was used as templates for generating cDNA with Superscript III Reverse Transcriptase and random primers (Invitrogen) following manufacturer’s instructions. The cDNA was measured in a qPCR reaction with SYBR Fast qPCR master mix (Kapa Biosystems). Assays were performed in triplicate using ABI StepOne Plue RealTime. Data were normalized to elfa and analyzed using ΔΔCt analysis. Sequences and annealing temperatures are presented in S1 Table.
Sample preparation and Illumina sequencing
Dissected intestines were placed in 2-mL screw cap tubes with 0.1 mm zirconia silica beads and 200-μL sterile lysis buffer (20 mM Tris-Cl; 2 mM EDTA; 2.5-mL 20% Tx-100) and frozen in liquid nitrogen. DNA was extracted using Qiamp DNA micro Kit (Qiagen) as detailed in S1 Text. The microbial communities of each sample were characterized by an Illumina HiSeq 2500 Rapid Run (San Diego, CA) sequencing the 16S rRNA gene amplicon by the University of Oregon Genomics and Cell Characterization Facility. The read length was paired-end 150 nucleotide, targeting the V4 region (primers listed in S2 Table). The 16S rRNA gene Illumina reads were clustered using USEARCH 8.1.1803 [64]. The final OTU table was rarefied to a depth of 100,000 (see S2 Data for metadata, S3 Data for OTU taxonomy, and S4 Data for OTU table). Measures of community diversity and similarity (OTU richness, phylogenetic distances, unweighted UniFrac) were calculated in R using vegan, picante, and GUniFrac (See S2 Code). Correlations were calculated in R, and false discovery rate was adjusted using the Benjamini & Hochberg correction in p.adjust (See S1 Code).
ENS transplantation
WT donor embryos were labeled by injection of 5% tetramethylrhodamine dextran (3000 MW) at the 1–2 cell stage and reared until the next manipulation in filter-sterilized EM. Embryos at the 12–14 somite stage were mounted in agar, a small hole dissected in the skin, and cells transplanted as previously described [65] and detailed in the S1 Text.
Statistics
Statistical analysis was performed using Prism (Graphpad software). Statistical significance was defined as p < 0.05. Data whose distributions were bounded by 0 were log transformed + 1 prior to statistical analysis. For correlations in Figs 5 and 6, log transformations of neutrophil number and percent OTU were performed so the data met the assumptions of normality and homoscedasticity for linear regression. We note that the relationships and result of multiple linear regression were the same if the data were not log transformed. Throughout, box plots represent the median and interquartile range; whiskers represent the 5–95 percentile. Data for all figures are available in S1 Data.
Supporting information
(A) Representative images of wild type (WT, left) and sox10- (middle and right images) distal intestine. Anti-ElavI labeled enteric neurons are green (green arrow). Scale bar 100 mm. There are no enteric neurons in the sox10- fish. (B) Schematic of the fluorescent food feeding schedule. Color indicates administration of fluorescent tracer; arrow indicates time of imaging for 8 dpf fish. (C) Representative images of 8 dpf wild types and sox10 mutants. Scale bar 100 mm. (D) The percent of fish with the indicated fluorescent food color in their intestines at 7 and 8 dpf (i.e. ‘eaters’). Each point represents the percentage of eaters from a separate dish of nine to 30 fish. In total, n > 200 fish per genotype per day. Bars represent mean ± SD. **** p < 0.0001, Student’s T-test.
(TIF)
Inoculating germ-free (GF) fish with a 5x concentrated donor inoculum from WTs does not increase intestinal neutrophil number over what is observed for a 1x concentration. Box plots represent the median and interquartile range, whiskers represent the 5–95 percentile; n ≥ 20; * p < 0.05, ANOVA.
(TIF)
(A) Bacterial communities in wild type (WT, black closed circles), sox10- low neutrophil (red closed circles), and sox10- high neutrophil (red open circles) do not differ based on Canberra distances, as shown in a NMDS analysis. (B-D) WT, sox10- low neutrophil, and sox10- high neutrophil intestinal microbiota are not different in the number of OTUs present in their communities (B); in phylogenic diversity based on Faith’s PD alpha diversity metric (C); and by pairwise comparisons of unweighted UniFrac distances (D). Box plots represent the median and interquartile range, whiskers represent the 5–95 percentile, n > 30 per group, collected from three independent experiments. (E) The average percent abundance of the top 11 representative genera. The ‘other’ group consists of all OTUs that made up less than 0.5% on average in all groups. Error bars represent SEM for the top two most abundant species.
(TIF)
(A) Colonization level of E. coli HS or Vibrio Z20 monoassociated in sox10- mutants. n > 30. (B) The ability of exogenously added bacteria to alter the intestinal neutrophil response depended on the intestinal neutrophil response in the control, which is a result of the microbial community assembled. This graph represents four independent experiments and the difference in intestinal neutrophil influx between the control and the treated samples. The background colors indicate the approximate ranges of neutrophil numbers observed for wild-type germ free fish (WT GF), wild-type conventional fish (WT CV), and conventional sox10- fish. (C) Phylogenetic tree of Vibrio and Escherichia/Shigella OTUs including the zebrafish isolates Vibrio Z20 and Shewanella Z12 and E. coli HS, the representative of the Escherichia genus used in experiments. Tree based on 16S sequence. (D) A natural zebrafish isolate of Shewanella (Shw Z12) with a previously demonstrated negative correlation between colonization level and intestinal neutrophil accumulation reduces intestinal neutrophil number in sox10- mutants through a factor present in the cell free supernatant (CFS). E. coli HS CFS is not sufficient to reduce neutrophil number. *p < 0.05, ***p < 0.001, ANOVA. Box plots represent the median and interquartile range; whiskers represent the 5–95 percentile.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(CSV)
(CSV)
(CSV)
(R)
Acknowledgments
We thank Doug Turnbull and the University of Oregon Genomics and Cell Characterization Core Facility for expertise in Illumina sequencing and bioinformatics and Rose Sockol and the UO Zebrafish Facility staff for fish husbandry. We thank Adam Burns and Keaton Stagamann for assistance with R code.
Abbreviations
- AIC
Akaike’s Information Criterion
- CFS
cell-free supernatant
- CFU
colony-forming units
- CV
conventionally raised
- dpf
d post fertilization
- ENS
enteric nervous system
- FISH
fluorescent in situ hybridization
- GF
germ free
- GFP
green fluorescent protein
- GI
gastrointestinal
- HAEC
Hirschsprung-associated enterocolitis
- HSCR
Hirschsprung disease
- iapMO
intestinal alkaline phosphatase morpholino
- IBD
inflammatory bowel disease
- OTU
operational taxonomic unit
- TNF
tumor necrosis factor
- WT
wild-type
Data Availability
All relevant data are within the paper and its Supporting Information files except the 16S rRNA sequencing results, which are available in the NCBI SRA database, accession number SRP096662.
Funding Statement
NIGMS (grant number P50GM098911). Received by KJG. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. NIDDK (grant number 1F32DK098884-01A1). Received by ASR. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Broad Medical Research Program (grant number IBD-0209). Received by KJG. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. John Simon Guggenheim Memorial Foundation Fellowship. Received by JSE. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. NICHD (grant number P01HD22486). Received by JSE. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cummings JH, Macfarlane GT. Role of intestinal bacteria in nutrient metabolism. Clin Nutr. 1997;16: 3–11. [DOI] [PubMed] [Google Scholar]
- 2.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14: 685–690. 10.1038/ni.2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furness JB. The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol. 2012;9: 286–294. 10.1038/nrgastro.2012.32 [DOI] [PubMed] [Google Scholar]
- 4.Huttenhower C, Kostic AD, Xavier RJ. Inflammatory Bowel Disease as a Model for Translating the Microbiome. Immunity. Elsevier Inc.; 2014;40: 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Ley RE, Mahowald M a, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444: 1027–31. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 6.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455: 1109–13. 10.1038/nature07336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108: 4554–4561. 10.1073/pnas.1000087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozupone C a, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489: 220–30. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierre JF, Barlow-Anacker AJ, Erickson CS, Heneghan AF, Leverson GE, Dowd SE, et al. Intestinal Dysbiosis and Bacterial Enteroinvasion in a Murine Model of Hirschsprung’s Disease. J Pediatr Surg. 2014;49: 1242–1251. 10.1016/j.jpedsurg.2014.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward NL, Pieretti a., Dowd SE, Cox SB, Goldstein a. M. Intestinal aganglionosis is associated with early and sustained disruption of the colonic microbiome. Neurogastroenterol Motil. 2012;24: 874–885. 10.1111/j.1365-2982.2012.01937.x [DOI] [PubMed] [Google Scholar]
- 11.Rossi V, Avanzini S, Mosconi M, Mattioli G, Buffa P, Jasonni V, et al. Hirschsprung Associated Enterocolitis. J Gastroint Dig Syst. 2014;4: 2–5. [Google Scholar]
- 12.Austin KM. The pathogenesis of Hirschsprung’s disease-associated enterocolitis. Semin Pediatr Surg. Elsevier Inc.; 2012;21: 319–327. [DOI] [PubMed] [Google Scholar]
- 13.Bagwell CE, Langham MR, Mahaffey SM, Talbert JL, Shandling B. Pseudomembanous colitis following resection for Hirschsprung’s disease. J Pediatr Surg. 1992;27: 1261–1264. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Somerset S. Digestive system dysfunction in cystic fibrosis: Challenges for nutrition therapy. Dig Liver Dis. 2014;46: 865–874. 10.1016/j.dld.2014.06.011 [DOI] [PubMed] [Google Scholar]
- 15.Krishnan B, Babu S, Walker J, Walker A, Pappachan JM. Gastrointestinal complications of diabetes. World J Diabetes. 2013;4: 51–63. 10.4239/wjd.v4.i3.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown EM, Wlodarska M, Willing BP, Vonaesch P, Han J, Reynolds LA, et al. Diet and specific microbial exposure trigger features of environmental enteropathy in a novel murine model. Nat Commun. Nature Publishing Group; 2015;6: 7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellini M, Biagi S, Stasi C, Costa F, Mumolo MG, Ricchiuti A, et al. Gastrointestinal manifestations in myotonic muscular dystrophy. World J Gastroenterol. 2006;12: 1821–1828. 10.3748/wjg.v12.i12.1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarnopolsky MA, Pearce E, Matteliano A, James C, Armstrong D. Bacterial overgrowth syndrome in myotonic muscular dystrophy is potentially treatable. Muscle and Nerve. 2010;42: 853–855. 10.1002/mus.21787 [DOI] [PubMed] [Google Scholar]
- 19.Ganz J, Melancon E, Eisen JS. Zebrafish as a model for understanding enteric nervous system interactions in the developing intestinal tract. Methods in Cell Biology. Elsevier Ltd; 2016. [DOI] [PubMed] [Google Scholar]
- 20.Shepherd I, Eisen J. Development of the Zebrafish Enteric Nervous System. Methods Cell Biol. 2011;101: 143–160. 10.1016/B978-0-12-387036-0.00006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhlman J, Eisen JS. Genetic screen for mutations affecting development and function of the enteric nervous system. Dev Dyn. 2007;236: 118–127. 10.1002/dvdy.21033 [DOI] [PubMed] [Google Scholar]
- 22.Pietsch J, Delalande J-M, Jakaitis B, Stensby JD, Dohle S, Talbot WS, et al. Lessen Encodes a Zebrafish Trap100 Required for Enteric Nervous System Development. Development. 2006;133: 395–406. 10.1242/dev.02215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, et al. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128: 4113–4125. [DOI] [PubMed] [Google Scholar]
- 24.Kelsh RN, Eisen JS. The zebrafish colourless gene regulates development of non-ectomesenchymal neural crest derivatives. Development. 2000;127: 515–525. [DOI] [PubMed] [Google Scholar]
- 25.Davuluri G, Seiler C, Abrams J, Soriano a. J, Pack M. Differential effects of thin and thick filament disruption on zebrafish smooth muscle regulatory proteins. Neurogastroenterol Motil. 2010;22: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Field H a., Kelley K a., Martell L, Goldstein a. M, Serluca FC. Analysis of gastrointestinal physiology using a novel intestinal transit assay in zebrafish. Neurogastroenterol Motil. 2009;21: 304–312. 10.1111/j.1365-2982.2008.01234.x [DOI] [PubMed] [Google Scholar]
- 27.Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal Alkaline Phosphatase Detoxifies Lipopolysaccharide and Prevents Inflammation in Zebrafish in Response to the Gut Microbiota. Cell Host Microbe. 2007;2: 371–82. 10.1016/j.chom.2007.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolig AS, Parthasarathy R, Burns AR, Bohannan BJM, Guillemin K. Individual Members of the Microbiota Disproportionately Modulate Host Innate Immune Responses. Cell Host Microbe. Elsevier Inc.; 2015;18: 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci USA. 2011;108 Suppl: 4570–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bojanova DP, Bordenstein SR. Fecal Transplants: What Is Being Transferred? PLoS Biol. 2016;14: e1002503 10.1371/journal.pbio.1002503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanther M, Sun X, Mühlbauer M, Mackey LC, Flynn EJ, Bagnat M, et al. Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology. AGA Institute American Gastroenterological Association; 2011;141: 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephens W, Burns A, Stagamann K, S W, Rawls J, Guillemin K, et al. The composition of the zebrafish intestinal microbial community varies across development. ISME J. 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burnham KP. Multimodel Inference: Understanding AIC and BIC in Model Selection. Sociol Methods Res. 2004;33: 261–304. [Google Scholar]
- 34.Rasko DA, Rosovitz MJ, Myers GSA, Mongodin EF, Fricke WF, Gajer P, et al. The pangenome structure of Escherichia coli: Comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 2008;190: 6881–6893. 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman D a. The Application of Ecological Theory Toward an Understanding of the Human Microbiome. Science (80-). 2012;336: 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, et al. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. Nature Publishing Group; 2016;10: 655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falcony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science (80-). 2016;352: 560–564. [DOI] [PubMed] [Google Scholar]
- 38.Wiles T, Jemielita M, Baker R, Schlomann B, Logan S, Ganz J, et al. Host Gut Motility Promotes Competitive Exclusion within a Model Intestinal Microbiota. PLoS Biol. 2016;14: e1002517 10.1371/journal.pbio.1002517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knight RD, Mebus K, D’Angelo A, Yokoya K, Heanue T, Roehl H. Ret signalling integrates a craniofacial muscle module during development. Development. 2011;138: 2015–2024. 10.1242/dev.061002 [DOI] [PubMed] [Google Scholar]
- 40.Chung AY, Kim PS, Kim S, Kim E, Kim D, Jeong I, et al. Generation of demyelination models by targeted ablation of oligodendrocytes in the zebrafish CNS. Mol Cells. 2013;36: 82–87. 10.1007/s10059-013-0087-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gosain A, Barlow-Anacker AJ, Erickson CS, Pierre JF, Heneghan AF, Epstein ML, et al. Impaired Cellular Immunity in the Murine Neural Crest Conditional Deletion of Endothelin Receptor-B Model of Hirschsprung’s Disease. PLoS ONE. 2015;10: e0128822 10.1371/journal.pone.0128822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gosain A. Established and emerging concepts in Hirschsprung’s-associated enterocolitis. Pediatr Surg Int. Springer Berlin Heidelberg; 2016;32: 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villanacci V, Bassotti G, Nascimbeni R, Antonelli E, Cadei M, Fisogni S, et al. Enteric nervous system abnormalities in inflammatory bowel diseases. Neurogastroenterol Motil. 2008;20: 1009–1016. 10.1111/j.1365-2982.2008.01146.x [DOI] [PubMed] [Google Scholar]
- 44.Mawe GM. Colitis-induced neuroplasticity disrupts motility in the inflamed and post-inflamed colon. J Clin Invest. 2015;125: 949–955. 10.1172/JCI76306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lomax AE, Linden DR, Mawe GM, Sharkey KA. Effects of gastrointestinal inflammation on enteroendocrine cells and enteric neural reflex circuts. Auton Neurosci. 2006;126: 250–257. 10.1016/j.autneu.2006.02.015 [DOI] [PubMed] [Google Scholar]
- 46.Whelan K, Quigley EMM. Probiotics in the managements of irritable bowel syndrome and inflammatory bowel disease. Curr Opin Gastroenterol. 2013;29: 184–189. 10.1097/MOG.0b013e32835d7bba [DOI] [PubMed] [Google Scholar]
- 47.Sze MA, Dimitriu PA, Suzuki M, McDonogh JE, Campbell JD, Brothers JF, et al. Host Response to the Lung Microbiome in Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2015;192: 438–445. 10.1164/rccm.201502-0223OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgan XC, Kabakchiev B, Waldron L, Tyler AD, Tickle TL, Milgrom R, et al. Associations between host gene expression, the mucosal microbiome, and clinical outcome in the pelvic pouch of patients with inflammatory bowel disease. Genome Biol. 2015;16: 67 10.1186/s13059-015-0637-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Losada M, Castro-Nallar E, Bendall ML, Freishtat RJ, Crandall KA. Dual transcriptomic profiling of host and microbiota during health and disease in pediatric asthma. PLoS ONE. 2015;10: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 2016;167: 1125–1136.e8. 10.1016/j.cell.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritchie JM, Rui H, Zhou X, Iida T, Kodoma T, Ito S, et al. Inflammation and disintegration of intestinal villi in an experimental model for vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 2012;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kruis W, Fric P, Pokrotnieks J, Lukás M, Fixa B, Kascák M, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. 2004;53: 1617–23. 10.1136/gut.2003.037747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao Y, Shen J, Ran ZH. Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: A meta-analysis and systematic review of the literature. Gastroenterol Res Pract. 2014;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15: 1183–1189. 10.1002/ibd.20903 [DOI] [PubMed] [Google Scholar]
- 55.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105: 16731–6. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fattahi F, Steinbeck JA, Kriks S, Tchieu J, Zimmer B, Kishinevsky S, et al. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature. Nature Publishing Group; 2016;531: 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hyland NP, Cryan JF. Microbe-host interactions: Influence of the gut microbiota on the enteric nervous system. Dev Biol. Elsevier; 2016. [DOI] [PubMed] [Google Scholar]
- 58.Muller PA, Koscsó B, Rajani GM, Stevanovic K, Berres M-L, Hashimoto D, et al. Crosstalk between Muscularis Macrophages and Enteric Neurons Regulates Gastrointestinal Motility. Cell. 2014;158: 300–313. 10.1016/j.cell.2014.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Margolis KG, Gershon MD, Bogunovic M. Cellular Organization of Neuroimmune Interactions in the Gastrointestinal Tract. Trends Immunol. Elsevier Ltd; 2016;37: 487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westerfield M. The Zebrafish Book A Guide for the Laboratory Use of Zebrafish (Danio rerio). 4th ed Eugene: University of Oregon Press; 2000. [Google Scholar]
- 61.Renshaw SA, Loynes CA, Trushell DMI, Elworthy S, Ingham PW, Whyte MKB. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108: 3976–8. 10.1182/blood-2006-05-024075 [DOI] [PubMed] [Google Scholar]
- 62.Carney TJ, Dutton K a, Greenhill E, Delfino-Machín M, Dufourcq P, Blader P, et al. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;133: 4619–30. 10.1242/dev.02668 [DOI] [PubMed] [Google Scholar]
- 63.Milligan-Myhre K, Charette JR, Phennicie RT, Stephens WZ, Rawls JF, Guillemin K, et al. Study of Host-Microbe Interactions in Zebrafish. Third Edit. Methods in Cell Biology. Elsevier Inc.; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26: 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 65.Eisen JS. Determination of Primary Motoneuron Identity in Developing Zebrafish Embryos. Science (80-). 1991;252: 569–572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Representative images of wild type (WT, left) and sox10- (middle and right images) distal intestine. Anti-ElavI labeled enteric neurons are green (green arrow). Scale bar 100 mm. There are no enteric neurons in the sox10- fish. (B) Schematic of the fluorescent food feeding schedule. Color indicates administration of fluorescent tracer; arrow indicates time of imaging for 8 dpf fish. (C) Representative images of 8 dpf wild types and sox10 mutants. Scale bar 100 mm. (D) The percent of fish with the indicated fluorescent food color in their intestines at 7 and 8 dpf (i.e. ‘eaters’). Each point represents the percentage of eaters from a separate dish of nine to 30 fish. In total, n > 200 fish per genotype per day. Bars represent mean ± SD. **** p < 0.0001, Student’s T-test.
(TIF)
Inoculating germ-free (GF) fish with a 5x concentrated donor inoculum from WTs does not increase intestinal neutrophil number over what is observed for a 1x concentration. Box plots represent the median and interquartile range, whiskers represent the 5–95 percentile; n ≥ 20; * p < 0.05, ANOVA.
(TIF)
(A) Bacterial communities in wild type (WT, black closed circles), sox10- low neutrophil (red closed circles), and sox10- high neutrophil (red open circles) do not differ based on Canberra distances, as shown in a NMDS analysis. (B-D) WT, sox10- low neutrophil, and sox10- high neutrophil intestinal microbiota are not different in the number of OTUs present in their communities (B); in phylogenic diversity based on Faith’s PD alpha diversity metric (C); and by pairwise comparisons of unweighted UniFrac distances (D). Box plots represent the median and interquartile range, whiskers represent the 5–95 percentile, n > 30 per group, collected from three independent experiments. (E) The average percent abundance of the top 11 representative genera. The ‘other’ group consists of all OTUs that made up less than 0.5% on average in all groups. Error bars represent SEM for the top two most abundant species.
(TIF)
(A) Colonization level of E. coli HS or Vibrio Z20 monoassociated in sox10- mutants. n > 30. (B) The ability of exogenously added bacteria to alter the intestinal neutrophil response depended on the intestinal neutrophil response in the control, which is a result of the microbial community assembled. This graph represents four independent experiments and the difference in intestinal neutrophil influx between the control and the treated samples. The background colors indicate the approximate ranges of neutrophil numbers observed for wild-type germ free fish (WT GF), wild-type conventional fish (WT CV), and conventional sox10- fish. (C) Phylogenetic tree of Vibrio and Escherichia/Shigella OTUs including the zebrafish isolates Vibrio Z20 and Shewanella Z12 and E. coli HS, the representative of the Escherichia genus used in experiments. Tree based on 16S sequence. (D) A natural zebrafish isolate of Shewanella (Shw Z12) with a previously demonstrated negative correlation between colonization level and intestinal neutrophil accumulation reduces intestinal neutrophil number in sox10- mutants through a factor present in the cell free supernatant (CFS). E. coli HS CFS is not sufficient to reduce neutrophil number. *p < 0.05, ***p < 0.001, ANOVA. Box plots represent the median and interquartile range; whiskers represent the 5–95 percentile.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(CSV)
(CSV)
(CSV)
(R)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files except the 16S rRNA sequencing results, which are available in the NCBI SRA database, accession number SRP096662.