Abstract
Using an elementary physical model for protein folding, of self-avoiding short copolymer chains on two-dimensional square lattices, we address two questions regarding the evolution and origins of globular proteins. (i) How will protein native structures and stabilities be affected by single-and double-site mutations? (ii) What is the probability that a randomly chosen sequence of amino acids will be compact and globular under folding conditions? For a large number of different sequences, we search the conformational space exhaustively to find unequivocally the "native" conformation(s), of global minimum free energy, for each sequence. We find that replacing nonpolar residues in the core by polar residues is generally destabilizing, that surface sites are less sensitive than core sites, that some mutations increase the degeneracy of native states, and that overall it is most probable that a mutation will be neutral, having no effect on the native structure. These results support a "Continuity Principle," that small changes in sequence seldom have large effects on structure or stability of the native state. The simulations also show that (i) the number of "convergent" sequences (different sequences coding for the same native structure) is extremely large and (ii) most sequences become quite dense under folding conditions. This implies that the probability of formation of a globular protein from a random sequence of amino acids by prebiotic or mutational methods is significantly greater than zero.
Full text
PDF
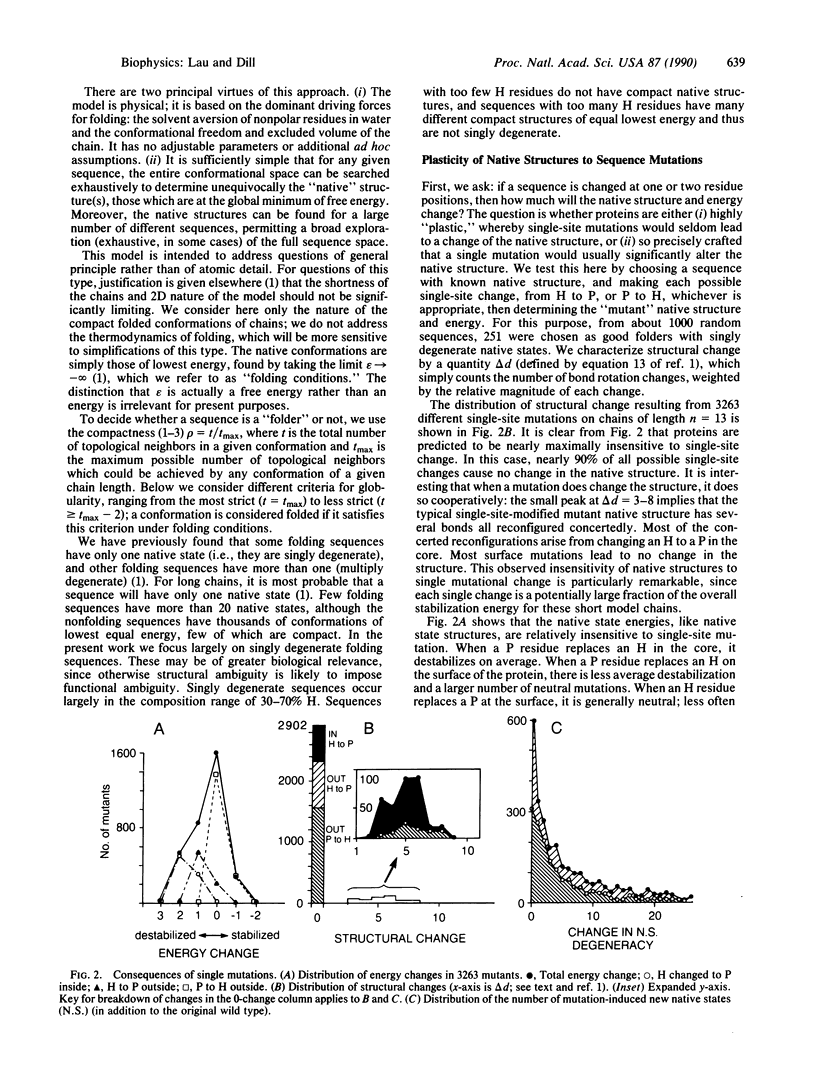
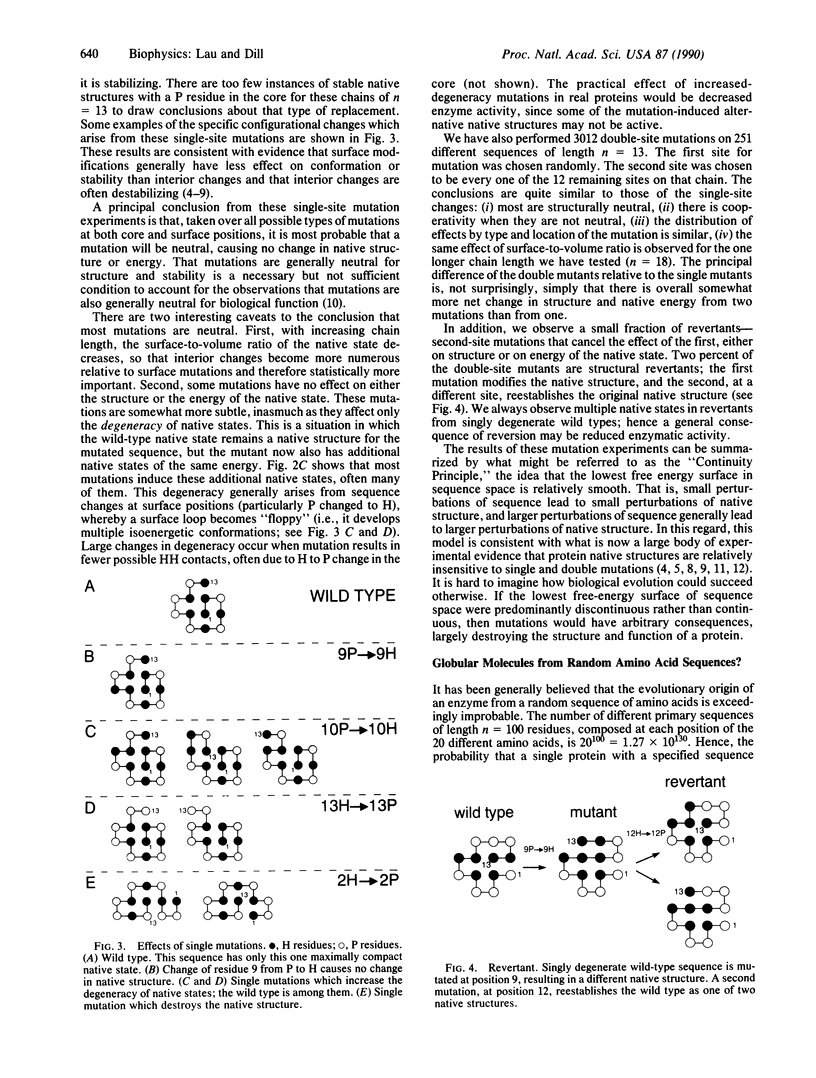
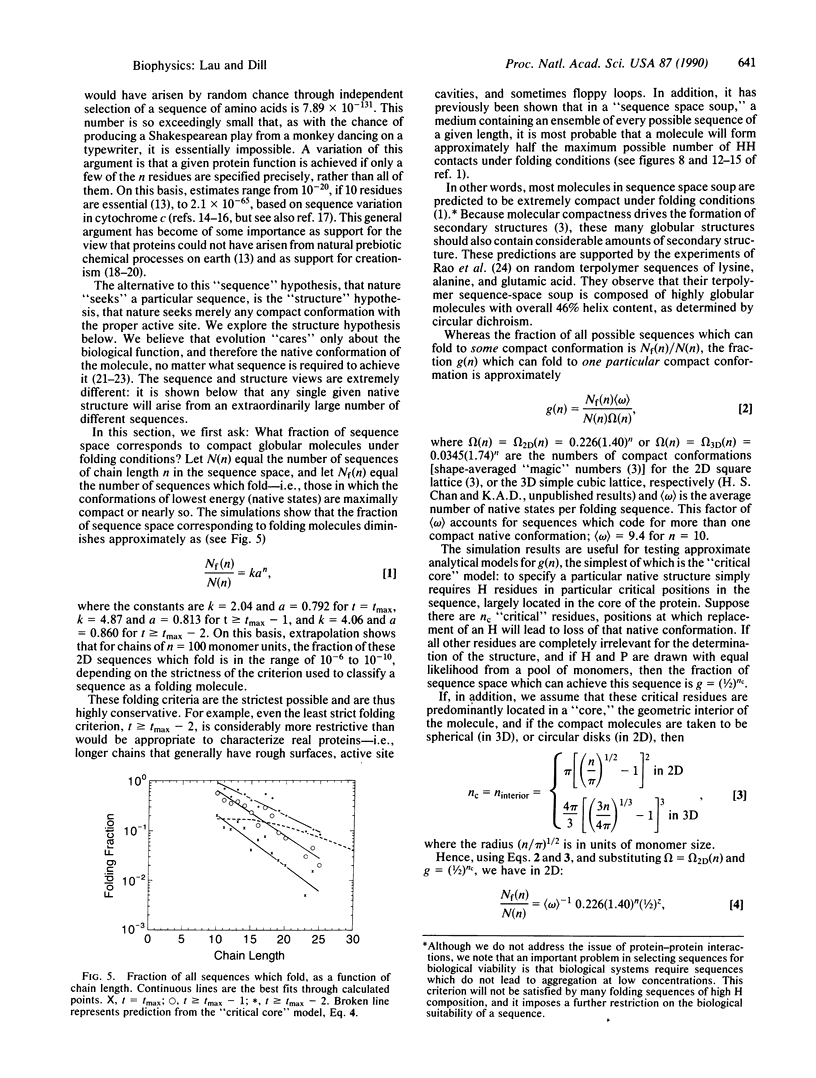

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Bell J. A., Sun D. P., Nicholson H., Wozniak J. A., Cook S., Matthews B. W. Replacements of Pro86 in phage T4 lysozyme extend an alpha-helix but do not alter protein stability. Science. 1988 Feb 5;239(4840):631–635. doi: 10.1126/science.3277275. [DOI] [PubMed] [Google Scholar]
- Alber T., Sun D. P., Nye J. A., Muchmore D. C., Matthews B. W. Temperature-sensitive mutations of bacteriophage T4 lysozyme occur at sites with low mobility and low solvent accessibility in the folded protein. Biochemistry. 1987 Jun 30;26(13):3754–3758. doi: 10.1021/bi00387a002. [DOI] [PubMed] [Google Scholar]
- Alber T., Sun D. P., Wilson K., Wozniak J. A., Cook S. P., Matthews B. W. Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme. Nature. 1987 Nov 5;330(6143):41–46. doi: 10.1038/330041a0. [DOI] [PubMed] [Google Scholar]
- Almassy R. J., Dickerson R. E. Pseudomonas cytochrome c551 at 2.0 A resolution: enlargement of the cytochrome c family. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2674–2678. doi: 10.1073/pnas.75.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grütter M. G., Weaver L. H., Matthews B. W. Goose lysozyme structure: an evolutionary link between hen and bacteriophage lysozymes? Nature. 1983 Jun 30;303(5920):828–831. doi: 10.1038/303828a0. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Sturtevant J. M., Sauer R. T. Effect of single amino acid replacements on the thermal stability of the NH2-terminal domain of phage lambda repressor. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5685–5689. doi: 10.1073/pnas.81.18.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R. W. Theoretical considerations on probabilities of biopolymers and life's origin. Radiat Environ Biophys. 1974 Mar 29;11(1):31–40. doi: 10.1007/BF01323098. [DOI] [PubMed] [Google Scholar]
- Lesk A. M., Chothia C. How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J Mol Biol. 1980 Jan 25;136(3):225–270. doi: 10.1016/0022-2836(80)90373-3. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Sauer R. T. Alternative packing arrangements in the hydrophobic core of lambda repressor. Nature. 1989 May 4;339(6219):31–36. doi: 10.1038/339031a0. [DOI] [PubMed] [Google Scholar]
- Perutz M. F. Species adaptation in a protein molecule. Adv Protein Chem. 1984;36:213–244. doi: 10.1016/s0065-3233(08)60298-3. [DOI] [PubMed] [Google Scholar]
- Rao S. P., Carlstrom D. E., Miller W. G. Collapsed structure polymers. A scattergun approach to amino acid copolymers. Biochemistry. 1974 Feb 26;13(5):943–952. doi: 10.1021/bi00702a019. [DOI] [PubMed] [Google Scholar]
- Rao S. T., Rossmann M. G. Comparison of super-secondary structures in proteins. J Mol Biol. 1973 May 15;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Reidhaar-Olson J. F., Sauer R. T. Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences. Science. 1988 Jul 1;241(4861):53–57. doi: 10.1126/science.3388019. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Viswanadhan V. N. On the prediction of functionally equivalent residues at given sites in proteins. J Theor Biol. 1987 Jan 7;124(1):123–125. doi: 10.1016/s0022-5193(87)80257-6. [DOI] [PubMed] [Google Scholar]
- Wright C. S., Alden R. A., Kraut J. Structure of subtilisin BPN' at 2.5 angström resolution. Nature. 1969 Jan 18;221(5177):235–242. doi: 10.1038/221235a0. [DOI] [PubMed] [Google Scholar]
- Yockey H. P. A calculation of the probability of spontaneous biogenesis by information theory. J Theor Biol. 1977 Aug 7;67(3):377–398. doi: 10.1016/0022-5193(77)90044-3. [DOI] [PubMed] [Google Scholar]
- Yockey H. P. A prescription which predicts functionally equivalent residues at given sites in protein sequences. J Theor Biol. 1977 Aug 7;67(3):337–343. doi: 10.1016/0022-5193(77)90042-x. [DOI] [PubMed] [Google Scholar]
- Yockey H. P. On the information content of cytochrome c. J Theor Biol. 1977 Aug 7;67(3):345–376. doi: 10.1016/0022-5193(77)90043-1. [DOI] [PubMed] [Google Scholar]




