Abstract
Homeodomain-leucine zipper (HD-Zip) gene family plays important roles in various abiotic stresses and hormone signaling in plants. However, no information is currently available regarding this family in cassava (Manihot esculenta), an important drought-tolerant crop in tropical and sub-tropical areas. Here, 57 HD-Zip genes (MeHDZ01-57) were identified in the cassava genome, and they were classified into four subfamilies based on phylogenetic analysis, which was further supported by their gene structure and conserved motif characteristics. Of which five gene pairs were involved in segmental duplication but none for tandem duplication, suggesting that segmental duplication was the main cause for the expansion of MeHDZ gene family in cassava. Global expression profiles revealed that MeHDZ genes were constitutively expressed, or not expressed, or tissue-specific expressed in examined tissues in both cultivated and wild subspecies. Transcriptomic analysis of three genotypes showed that most of MeHDZ genes responded differently to drought and polyethylene glycol treatments. Subsequently, quantitative RT-PCR analysis revealed comprehensive responses of twelve selected MeHDZ genes to various stimuli including cold, salt, and ABA treatments. These findings will increase our understanding of HD-Zip gene family involved in abiotic stresses and signaling transduction, and will provide a solid base for further functional characterization of MeHDZ genes in cassava.
Introduction
Homeodomain-leucine zipper (HD-Zip) proteins are one of key transcription factors in plants. Based on sequence analysis, HD-Zip proteins were characterized by the presence of two functional domains: a homeodomain (HD) responsible for specific binding to DNA and a leucine zipper (LZ) motif which closely linked to HD and acted as a dimerization motif [1,2]. To date, many members of HD-Zip proteins have been found in a large number of plant species, including Arabidopsis [3], rice [4], maize [5], soybean [6], legume [7], banana [8], and so on. Based on the structures, HD-Zip proteins were mainly categorized into four groups: HD-Zip I to HD-Zip IV [2]. Members of HD-Zip I and II contained the conserved HD and LZ domains and bound to the similar sequence CAAT-N-ATTG [9]. In addition, HD-Zip II members contained other motifs, such as CPSCE [2]. Besides the HD and LZ domains, members of both HD-Zip III and IV also contained a START domain with putative lipid binding capability [10]. However, a conserved C-terminal MEKHLA motif, which was probably involved in oxygen redox and light signaling, was present in the members of HD-Zip III but absent in the HD-Zip IV subfamily [11].
HD-Zip proteins have been demonstrated to participate in many biological processes related to plant growth and development [1,2]. Members of HD-Zip I subfamily mainly participated in abiotic stresses such as drought, extreme temperatures, osmotic and light stresses [2,3]. For example, AtHB5, -6, -7 and -12, which belonged to Arabidopsis HD-Zip I subfamily, were up-regulated or down-regulated by drought stress or externally applied abscisic acid (ABA) [12–15]. Other members of this subfamily, such as AtHB52 and AtHB53, were up-regulated in dark condition [3]. HD-Zip II proteins were mainly involved in shade avoidance and auxin response [16]. For examples, AtHB2 in this subfamily was specifically and reversibly regulated by red to far-red ratio, and it was participated in the regulation of Arabidopsis shade avoidance response [17]. HAT2, another member of HD-Zip II subfamily, was isolated as an auxin-inducible gene through DNA microarray screening [18]. HD-Zip III members mainly participated in the developmental regulation of apical meristem, vascular bundles, auxin transport and lateral organ initiation [2]. For examples, AtHB8 and AtHB15 were involved in vascular development [19,20]. Several studies have demonstrated that REV, PHB and PHV, along with KANADI, were involved in controlling the abaxial-adaxial patterning of lateral organs [21]. HD-Zip IV proteins generally played important roles in anthocyanin accumulation, epidermal cell differentiation, trichome formation and root development [1,2]. For examples, atml1 pdf2 double mutants resulted in severe defects in shoot epidermal cell differentiation and failed to survive after germination [22]. ANL2 affected anthocyanin accumulation in subepidermal tissues [23], while other members in this subfamily, such as HDG11, HDG12 and GL2, were involved in trichome development [2,24]. Recently, many studies have demonstrated that HD-Zip III and IV subfamilies were also involved in abiotic stresses such as salt, drought and ABA treatment [6,8,25].
Cassava (Manihot esculenta) is one of the most important root crops in tropical and sub-tropical areas and provides staple food for over 700 million people around the world [26]. Due to its good performance under adverse conditions such as drought and low fertilization, cassava has been considered as an important food security crop worldwide. However, as a representative tropical crop, cassava is very sensitive to cold stress [27]. To date, the mechanisms underlying its abiotic stresses remain elusive. With the availability of the cassava genome sequences [28,29], it provides an excellent opportunity for genome-wide annotation, classification and comparative genome research. Although HD-Zip genes have been extensively characterized in many species, a systemic study of HD-Zip gene family (especially for the stress-responsive members) has not been performed in cassava. In this study, a total of 57 HD-Zip genes were identified and characterized in the cassava genome. Besides their basic characteristics including phylogenetic relationship, gene structure, conserved protein motif and genome location, the expression profiles of these HD-Zip genes were investigated in different organs of various genotypes and several stress conditions, including drought, polyethylene glycol (PEG), cold, salt, and ABA treatments through RNA-seq or quantitative RT-PCR (qRT-PCR). These findings will provide a useful foundation for further investigations into the molecular biological functions of the HD-Zip genes in cassava.
Materials and methods
Plant materials and stress treatments
To reveal the expression of HD-Zip genes either in different tissues or in response to drought stress in cassava, several previously generated RNA-seq data [30,31] were used. In total four cassava accessions, including one wild subspecies (W14, M. esculenta subsp. flabellifolia) and three cultivated varieties (Arg7, Ku50 and SC124), were used in this study. Of which, Arg7 is adapted to high-latitude geographical region of Argentina and of moderate drought-tolerance; SC124 is a widely planted cultivar in China and can survive in severe drought condition, while Ku50 is a representative cultivar with high root yield and high starch content in root tubers and can grow under unfavorable environmental conditions [29,32,33].
In cassava growing season, Arg7, Ku50 and W14 were planted in the farm of Chinese Academy of Tropical Agricultural Sciences (Haikou, China) in the summer of 2013. To detect the changes of gene expression in different cassava tissues, samples from leaf (90 days after planting), stem (90 days after planting), early storage root (ESR, 90 days after planting), middle storage root (MSR, 150 days after planting) and last storage root (LSR, 210 days after planting) were collected for RNA-seq analysis.
To investigate the changes of gene expression in response to drought stress, stems of about 15 cm in length and two to three buds were selected and planted vertically in pots (sand: vermiculite = 1: 1; height × upper diameter × bottom diameter = 18.8 × 18.5 × 14.8 cm), as previously described [34]. About 45 days later, two experiments were conducted: (1) seedlings of Arg7, SC124 and W14 were withheld water for 0 day (control) and 12 days, and their leaves and roots were collected for RNA-seq, respectively; (2) seedlings of Ku50 were watered by 20% polyethylene glycol (PEG) 6000 solution, then the leaves, including folded leaf (FL), full expanded leaf (FEL) and bottom leaf (BL), as well as root (RT) were collected at 0, 3, 24 h after the treatment for RNA-seq, respectively. Each sample was pooled from 15 plants.
To examine the response of cassava to different abiotic stresses, including cold, salt, and ABA treatment, 2-month-old cassava seedlings of Arg7 were used and their leaves were collected for qRT-PCR. For salt stress, cassava seedlings were watered with 300 mM NaCl solution for 24 days. For ABA treatment, cassava seedlings were sprayed with 100 μM ABA for 72 h. For cold stress, cassava seedlings were exposed to 4°C for 48 h, and then returned to normal growth conditions for 14 days of recovery.
Identification of HD-Zip family genes in cassava
The whole genome and protein sequences of cassava were downloaded from phytozome database v9.0 (https://phytozome.jgi.doe.gov/). Hidden Markov Model (HMM) profiles of homeobox (HD, PF00046) and homeobox associated leucine zipper (HALZ, PF02183) were downloaded from PFam (http://pfam.sanger.ac.uk/) and were used as queries to identify putative cassava HD-Zip genes by local HMM-based searches setting E-values < 0.01 using HMMER3 [35]. Additionally, protein sequences of HD-Zip genes from Arabidopsis and rice [36] were used in a BLAST search using BLASTP against cassava proteins to explore more HD-Zips which might be missed by HMM profile searching. After removing redundant sequences, each HD-Zip candidate was further confirmed by investigating conserved domains using database of CDD (http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) in automatic mode (threshold = 0.01, maximum hits = 500) and PFam setting E-value equal to 1.0, respectively. The molecular weight and isoelectric points of each HD-Zip protein were calculated using ExPASy tools (http://www.expasy.org/tools/).
Phylogenetic and sequence analysis
Besides HD-Zips from Arabidopsis and rice, protein sequences of published HD-Zip genes from Vitis vinifera, Medicago truncatula, Populus trichocarpa, and maize [5,36] were also used. Protein sequences of HD-Zip genes were aligned using MUSCLE [37]. Phylogenetic tree was conducted in MEGA6 [38] using Maximum-Likelihood method with Jones-Taylor-Thornton (JTT) amino acid substitution model. Bootstrap analysis was performed using 1000 replicates with the partial deletion model for gaps/missing data.
HD-Zip exon-intron structure was performed with Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/) [39] by aligning cDNA to their corresponding genomic DNA sequences. Protein motifs were predicted by MEME software [40] with optimum motif width ranged from 11 to 50 and setting the maximum number of motifs equal to 15. The predicted motifs of HD-Zip proteins were annotated by searching against InterProScan database (http://www.ebi.ac.uk/Tools/pfa/iprscan/).
To demonstrate the possible regulatory mechanism of HD-Zip genes in various stress responses, 1500 bp promoter region upstream of the transcription start site (ATG) was used to analyze cis-elements using PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The overrepresentation analysis of cis-elements was performed by exact binominal test in each subfamily of HD-Zip genes.
Chromosomal location and gene duplication
Cassava HD-Zip (referred to MeHDZ) genes were mapped on cassava chromosomes according to their positions in the early genome version 6.1 from phytozome database (https://phytozome.jgi.doe.gov/). The schematic diagram of MeHDZ genes was drawn by MapInspect (http://www.plantbreeding.wur.nl/UK/software_mapinspect.html).
Tandem duplicates were defined as genes that occurred within a 50 kb region. Segmental duplicates were detected by BLASTN (score < 1e-5) using 100 kb (containing 50 kb upstream and 50 kb downstream) sequences flanking to the genes, and were further confirmed by chaining alignments (alignment length > 200 bp and sequence identity > 85%) [41].
Transcriptome and qRT-PCR analysis
Total RNA was extracted using RNA plant reagent kits (Tiangen Company, Beijing, China). Each RNA-Seq library was constructed as previously described in Yang et al [42]. Then, libraries were indexed, pooled and sequenced on Illumina HiSeq2000. Transcriptome analysis was performed as previously described [33,34]. For examples, sequence quality was examined using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), clean reads were mapped to cassava genome (version 4.1) using Tophat v2.0.13 [43], and raw count data were obtained and normalized by Cuffdiff embedded in Cufflinks pipeline v2.1.1 [44]. The transcriptome data were submitted to NCBI database and their accessions were shown in S1 Table.
Expression of MeHDZ genes in response to various stimuli, including cold, salt, and ABA treatments, was examined by qRT-PCR method using SYBR-green (TaKaRa Biotechnology Co. Ltd, Dalian, China) and Stratagene Mx3005P system (Stratagene, CA, USA) as previously described [34]. The cassava actin gene [45] was used as an internal control. The specific primers of target MeHDZ genes were designed by Primer 5.0 software (S2 Table). Subsequently, the specificity of each primer pair was examined by qRT-PCR melting curve analysis, agarose gel electrophoresis, and PCR products sequencing [30]. For each sample, qRT-PCR reaction was performed with three independent biological replicates and the relative mRNA expression level was calculated by 2-ΔΔCt as before [42]. Statistical difference was examined by Duncan’s multiple range test (n = 3) and expression values with different letters were significant at P < 0.05.
Results
Identification of HD-Zip genes in cassava
In total, 125 HD-Zip candidate sequences were found through searching against cassava genome using HMM and BLASTP methods. After removing redundant sequences and confirming the presence of both HD and LZ domains by CDD and PFam databases, 57 non-redundant HD-Zip genes (designated as MeHDZ01-57) were finally retained and used for further analysis. These proteins varied from 79 (MeHDZ57) to 853 (MeHDZ01) amino acid in length, and their molecular weight ranged from 9.12 (MeHDZ05) to 93.83 (MeHDZ13) kDa (S3 Table).
Phylogenetic and evolutionary analysis of HD-Zip genes
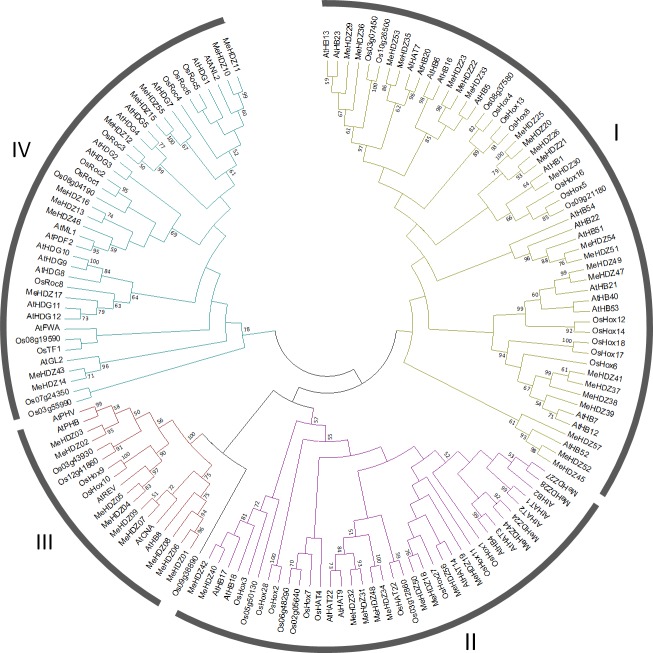
To study the evolutionary relationship of HD-Zip genes between cassava and other species, protein sequences of 57 MeHDZ genes, together with 48 from Arabidopsis and 44 from rice [36], were aligned and used for phylogenetic analysis (S4 Table and S1 File). As shown in Fig 1, phylogenetic tree well clustered these MeHDZ proteins into four groups (subfamily I-IV), together with known HD-Zip I to IV members of rice and Arabidopsis respectively in each clade, indicating that there were four major types of MeHDZ genes in cassava.
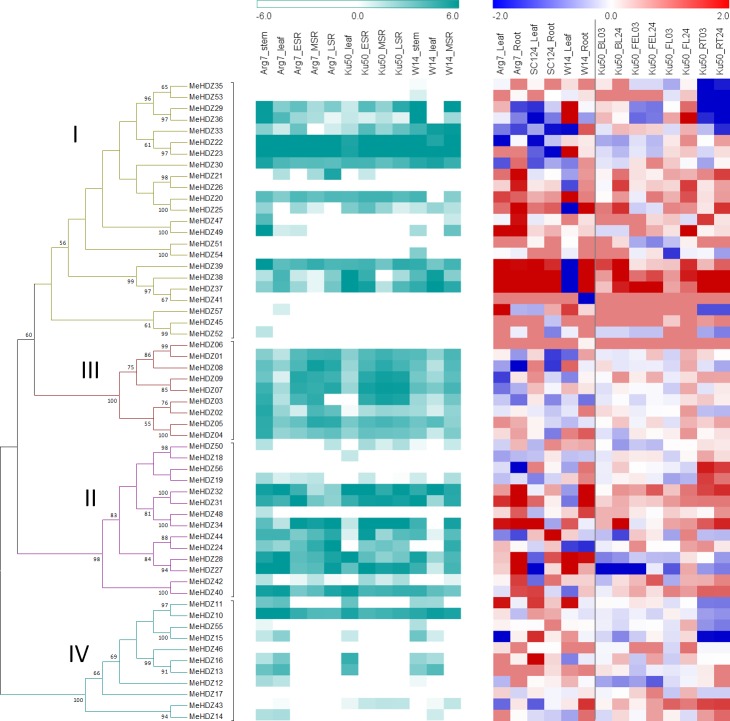
Fig 1. Phylogenetic clustering of HD-Zip proteins from Arabidopsis, rice and cassava.
The unrooted phylogenetic tree was constructed by Maximum-Likelihood method with 1000 bootstrap replicates. The HD-Zip protein sequences corresponding to 44 from rice (prefixed with ‘Os’), 48 from Arabidopsis (prefixed with ‘At’), and 57 from cassava (prefixed with ‘Me’) were well separated into four major groups (I-IV).
In addition, HD-Zip proteins from seven species including five eudicots (Arabidopsis, cassava, Vitis vinifera, Medicago truncatula, and Populus trichocarpa) and two monocots (rice and maize) were also aligned and further divided into subclasses (S1 Fig and S2 File), based on which four different evolutionary scenarios were revealed: (1) HD-Zip genes that were generated before the divergence of eudicots and monocots and they were highly conserved, as they were presented in all of the seven species although the number of HD-Zip genes might be varied. Moreover, this phenomenon was observed in each subfamily, e.g., subclass a, f, i in subfamily I, subclass m, p in subfamily II, subclass t in subfamily III, and subclass D in subfamily IV; (2) HD-Zip genes that were generated before the divergence of eudicots and monocots but their homologs in some speices might be lost during the evolution, because they were presented in both eudicots and monocots but not in all of examined species, e.g., subclass j, k in subfamily II, subclass q, r in subfamily III, and subclass u, w in subfamily IV; (3) HD-Zip genes that were generated after the divergence of eudicots and monocots, as they were exclusively presented in eudicots. They were either highly conserved (e.g, subclass c, h, n, z, A, and C containing HD-Zip genes presented in all of five examined eudicots) or not very conserved (e.g, subclass b, d, s, and y containing HD-Zip genes only presented in partial of examined eudicots); (4) HD-Zip genes that were generated after the divergence of eudicots and monocots, but they were exclusively presented in monocots. For examples, subclass g, l, o, x, B, and F only contained HD-Zip genes from rice and maize in this study. Taken together, these results indicated a complicated evolutionary process of HD-Zip genes in plants.
Gene structure and conserved motif analysis
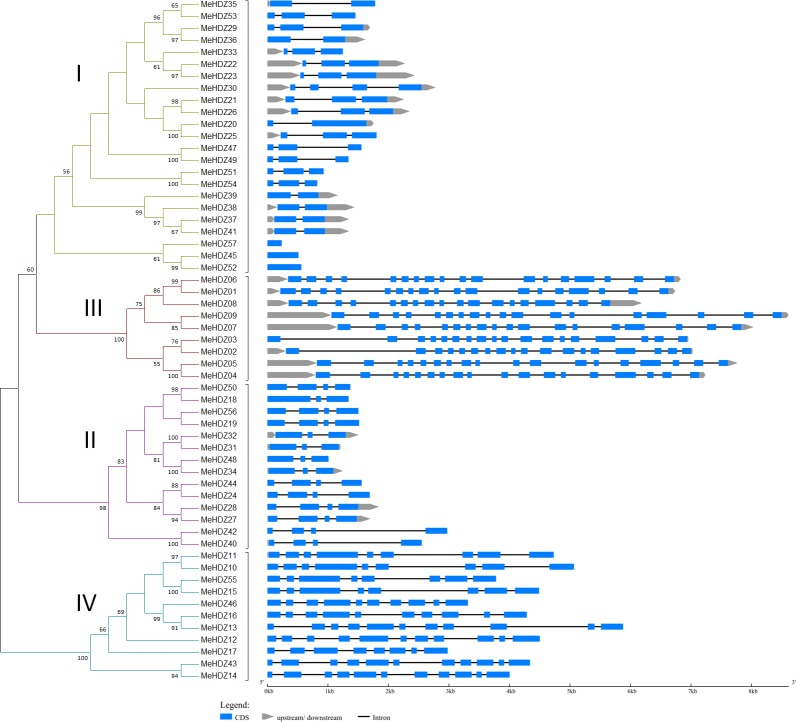
To gain information regarding the gene structure of MeHDZ genes, exon-intron structure analysis was performed for each MeHDZ gene individually (Fig 2). Overall, most closely related members within the same subfamily shared similar exon-intron structure and intron numbers. There were 23 members clustered in MeHDZ I subfamily and most of them contained one to three exons. There were 14 members in MeHDZ II subfamily, and they contained three to four exons. There were 9 and 11 members in MeHDZ III and IV subfamilies, respectively. Compared with MeHDZ I and II subfamilies, much more exons were found in MeHDZ III (18 in each) and IV (9.6 on average) subfamilies, respectively.
Fig 2. Exon-intron structure of 57 MeHDZ genes in cassava.
The phylogenetic tree was constructed by Maximum-Likelihood method with 1000 bootstrap replicates. The MeHDZ genes were clustered into four groups, I to IV. Exon-intron analysis was performed using GSDS (http://gsds.cbi.pku.edu.cn/). Length of exons and introns of each MeHDZ gene was displayed proportionally. Upstream was defined as the sequences before the transcription start site ‘ATG’, while downstream was defined as the sequences after the stop codon (e.g., ‘TAA’).
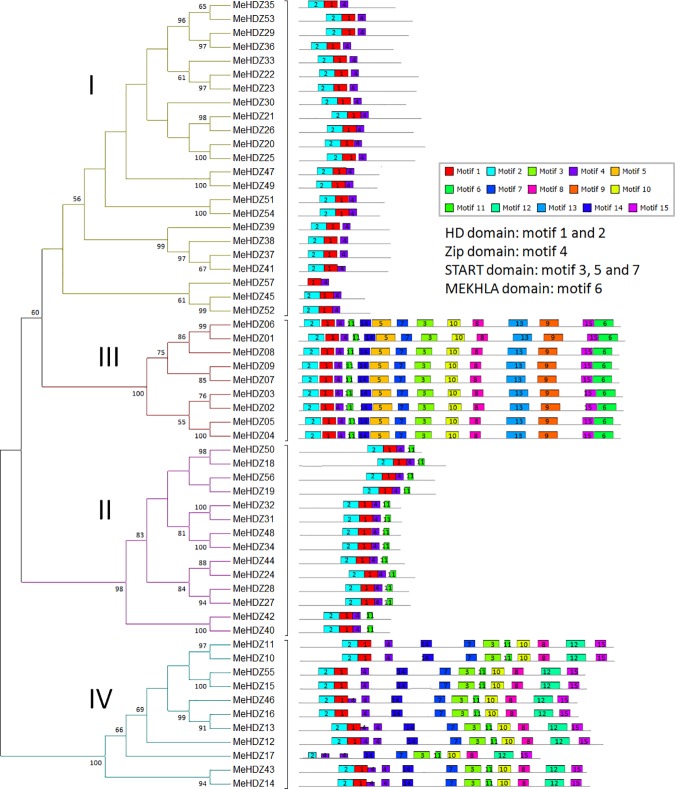
In total, 15 conserved motifs were captured by MEME software and subsequently annotated with InterProScan database (Fig 3, S2 Fig and S5 Table). As expected, HD domain (motif 1 and 2) and leucine zipper domain (motif 4) were found in all identified MeHDZ members. Some motifs that presented only in certain MeHDZ subfamilies were observed. For examples, START domain, which was represented by motif 3, 5 and 7, was not found in members of MeHDZ I and II subfamilies but presented in all members of MeHDZ III and IV subfamilies; MEKHLA domain, represented by motif 6, was presented only in members of MeHDZ III subfamily. Besides the well annotated, some unknown motifs (e.g., motif 9) were also found specifically in some subfamilies, indicating that these motifs might be key elements for the subfamily-specific functions.
Fig 3. Conserved motifs of MeHDZ proteins corresponding to their phylogenetic relationships.
The conserved motifs were identified by MEME software. Motifs were indicated by different colored boxes with the motif number, while non-conserved sequences were represented by grey lines. Length of motifs was exhibited proportionally.
Chromosomal location and gene duplication of MeHDZ genes
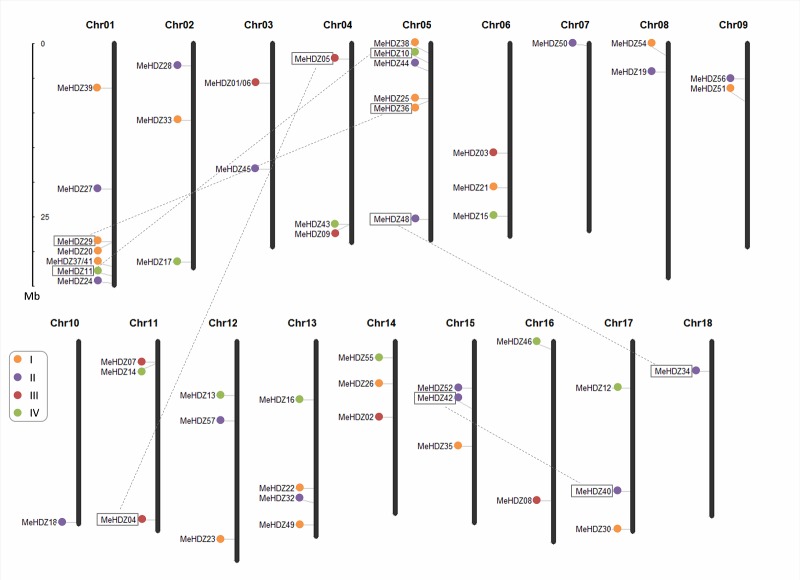
Genomic distribution of MeHDZ genes was determined by their chromosomal positions. To the best of our knowledge, this is the first report concerning the genomic distribution of gene family in cassava. Except for three members of MeHDZ31, -47 and -53 that were located on scaffolds, the remaining 54 MeHDZ genes were mapped on 18 cassava chromosomes (Fig 4; S6 Table). MeHDZ01 and MeHDZ06, together with MeHDZ37 and MeHDZ41, which may result from different alternative splicing, were mapped to the same loci on chromosome 3 and 1, respectively. Overall, an unequal distribution of MeHDZ genes was revealed in cassava. For example, a maximum of eight MeHDZ genes were presented on chromosome 1, in contrast, only one MeHDZ gene was respectively located on chromosome 7, 10 and 18, which contained the minimum MeHDZ gene in cassava (Fig 4).
Fig 4. Physical locations of MeHDZ genes on cassava chromosomes.
Chromosome numbers were indicated at the top of each chromosome. MeHDZ genes from different subfamilies were indicated by different colors. Five pairs of MeHDZ genes connected by dash lines were resulted from segmental duplication.
Segmental duplication and tandem duplication, which played important roles in expanding new members during the evolution of a gene family, were investigated to elucidate the potential evolution mechanism of MeHDZ genes in cassava. Totally, ten MeHDZ genes, forming five pairs, were identified as segmental duplications (Fig 4; S3 Fig). Of which, two pairs were from MeHDZ II subfamily, while the other three pairs were found in the remaining three subfamilies, respectively. However, none of the MeHDZ genes seemed to be generated from tandem duplications in our analysis. These results implicated that segmental duplication events were the main cause for the expansion of MeHDZ genes in cassava.
Cis-element identification of MeHDZ genes
To demonstrate the possible regulatory mechanism of MeHDZ genes, 1.5 kb promoter sequences upstream of the transcription start site (ATG) were extracted and used for cis-element prediction. As expected, many stress-related elements, such as MBS for drought, HSE for heat, LTR for low-temperature, and WUN-motif for wound-response, were identified in the promoter region of MeHDZ genes (S7 Table). Additionally, many hormone-related cis-elements, including ABRE for ABA, TGA-element for auxin, ERE for ethylene, P-box and GARE-motif for gibberellic acid (GA), CGTCA-motif and TGACG-motif for methyl jasmonate (MeJA), and TCA-element for salicylic acid, were also found (S7 Table). Moreover, other cis-elements such as circadian for circadian control, ARE for anaerobic induction, and as much as 20 cis-elements for light response were also observed.
To explore the particular roles of MeHDZ genes in each subfamily, overrepresentation analysis was conducted respectively for these cis-elements, and subfamily-specific overrepresented manners were revealed (S7 Table). Several cis-elements related to abiotic stress, e.g., ABRE, HSE, and TC-rich repeats, were significantly overrepresented in MeHDZ I and II subfamilies, while GARE-motif and MBS were overrepresented in MeHDZ III and IV subfamilies, respectively. Further, light responsive elements (e.g., Sp1 and G-box) also exhibited subfamily-specific overrepresented manners. Together, these results supported that subfamily-specific roles of HD-Zip subfamilies might be related to the regulatory elements in the promoter region.
Expression profiles of MeHDZ genes in different tissues of three genotypes
To reveal expression profiles of HD-Zip genes in different tissues, previous generated RNA-seq for the samples of stem, leaf, early storage root (ESR), middle storage root (MSR), and last storage root (LSR) in one wild subspecies (W14) and two cultivars (Arg7 and Ku50) was used. Expressed genes were arbitrarily defined as those with FPKM (Fragments Per Kilobase of exon per Million fragments mapped) > 1. Overall, different expression profiles were revealed within each subfamily of MeHDZ genes (Fig 5A; S8 Table). In MeHDZ I subfamily, most genes were expressed in all tested organs, of which MeHDZ22 and MeHDZ23 showed the highest expression level. Genes such as MeHDZ21, -47, -49 and -54 were expressed only in a few samples, while the others such as MeHDZ26, -41 and -51 were not expressed at all. Similar gene expression patterns were observed in MeHDZ II subfamily. For example, MeHDZ27, -28, -31, -32, -40 and -44 were expressed in all tested samples, while MeHDZ56 were not expressed and the remaining genes were expressed in partial samples. Compared with MeHDZ I and II subfamilies, most genes in MeHDZ III and IV subfamilies exhibited distinct tissue-specific expression patterns. For examples, eight out of nine genes (except MeHDZ06 that was not expressed in any samples) from MeHDZ III subfamily were lower expressed in leaves than in other tissues; and seven out of eleven genes (except MeHDZ10 that showed constitutive expression, MeHDZ17 and MeHDZ46 that were not expressed, and MeHDZ43 that expressed higher in root than in other samples) from MeHDZ IV subfamily were lower expressed in roots than in others (Fig 5A). Taken together, these results suggested that the functions of MeHDZ genes were diverged through constitutive expression, non-expression, or tissue-specific expression during cassava evolution processes.
Fig 5. Expression profiles of MeHDZ genes in cassava.
(A) Expression of MeHDZ genes in different tissues of three genotypes. Log2-transformed FPKM value was used to plot the heatmap. ESR: early storage root; MSR: middle storage root; LSR: last storage root. (B) Expression of MeHDZ genes in response to drought and PEG treatments. Log2 based fold change of treatment/control was used to plot the heatmap. FL: folded leaf; FEL: full expanded leaf; BL: bottom leaf; RT: root. The numbers attached behind samples represented the time points at which samples were collected: e.g., 03 and 24 represented 3 and 24 h, respectively.
Different expression patterns were also revealed in the same tissue among different genotypes, especially between the cultivated and the wild subspecies. Compared to W14, the expression of 22, 25 and 15 HD-Zip genes was greatly changed (absolute value of fold-change > 2) in stem, leaf and MSR in Arg7, while 15 and 17 HD-Zip genes were differentially expressed in leaf and MSR in Ku50, respectively (S8 Table).
Expression analysis of MeHDZ genes responding to drought in different genotypes
To demonstrate the responses of MeHDZ genes to drought stress, seedlings of one wild subspecies (W14) and two cultivated varieties (Arg7 and SC124) were subjected to 12-day water withholding treatment, and their leaf and root samples of control and drought-treated plants were collected for RNA-seq analysis, respectively. As shown in Fig 5B, the expression of 18 and 24 genes were greatly changed (absolute value of fold-change > 2) in leaf and root in Arg7, respectively. Comparable number of genes were observed in leaf in SC124 (19) and W14 (21), however, the number was decreased to about one-half in root, e.g., 13 genes in SC124 and 14 genes in W14, respectively (S9 Table). These results suggested that more MeHDZ genes were triggered by drought stress in the root of SC124 and W14 than that in Arg7, which might be one explanation for the good performance of SC124 and W14 relative to Arg7 under drought condition. It’s worthy to note that MeHDZ genes showed different response to drought between wild subspecies and cultivated varieties. For example, in leaf, MeHDZ25, -39, -38 and -37 from MeHDZ I subfamily were greatly depressed in W14 but extremely induced in Arg7 and SC124. On the contrary, the expression of MeHDZ36 and MeHDZ23 was greatly increased in leaf of W14 but decreased in that of SC124 (Fig 5B). The results implied that MeHDZ genes might play different roles in drought stress between wild subspecies and cultivated varieties. As expected, the expression of most members of MeHDZ IV subfamily was not changed in root, which was consistent with their root-specific non-expressed patterns (Fig 5A).
In addition, PEG solution, which is usually used to simulate drought stress, was applied to Ku50 seedlings to investigate the expression profiles of MeHDZ genes (S10 Table). Overall, the expression of most genes was more dramatically changed at 24 h than that at 3 h (Fig 5B), suggesting that longer periods of drought stress caused more serious influences on cassava. Consistent with water withholding treatment, genes such as MeHDZ20, -25, -21, -26, -39, -38, and -37 from MeHDZ I subfamily, and MeHDZ31, -32 and -34 from MeHDZ II subfamily, exhibited strong induction in the majority of tested samples. On the contrary, MeHDZ53, -35, -29 and -36 from MeHDZ I subfamily, and MeHDZ10, -11, -15 and -55 from MeHDZ IV subfamily were greatly depressed in root (Fig 5B). Taken together, these results indicated that all four MeHDZ subfamilies were responsive to drought stress and the expression analysis will provide useful clues for further characterizing their functions in cassava.
Expression profiles of MeHDZ genes in response to various treatments
Besides HD-Zip I members that were well demonstrated to associate with abiotic stress, it was of great interests to investigate whether genes from HD-Zip II to IV were involved in abiotic stress responses as previously proposed. Here twelve genes (including three from each MeHDZ subfamily), which exhibited similar expression patterns among different tissues within subfamily and differentially expressed under drought or PEG treatments based on RNA-seq data, were chosen to further examine their responses to cold, salt, and ABA treatment through qRT-PCR method.
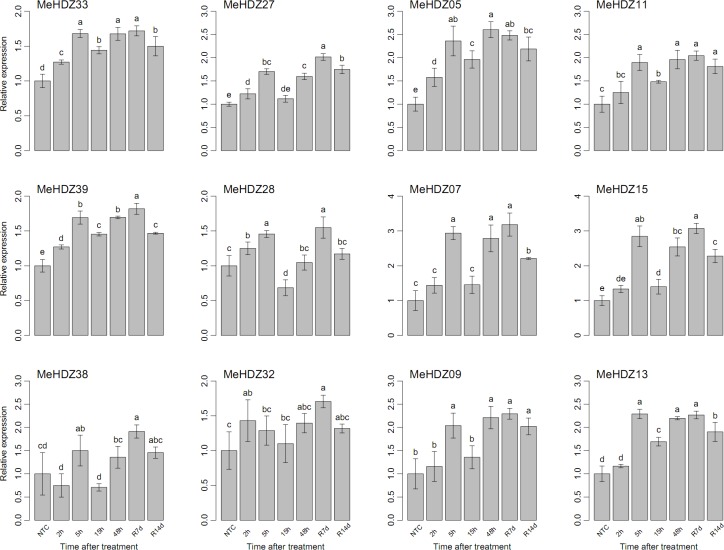
Under cold treatment following recovery, the majority of tested MeHDZ genes showed similar expression patterns, and they were significantly induced at 5h and depressed at 15h, and then increased from 48h to R7d and finally decreased at R14d (Fig 6).
Fig 6. Relative expression of MeHDZ genes in leaves under cold treatment.
NTC (no treatment control) was normalized as “1” at each graph. Data were shown as mean ± standard deviation derived from three biological replicates, and values with different letters were significant based on Duncan’s multiple range tests (P < 0.05, n = 3).
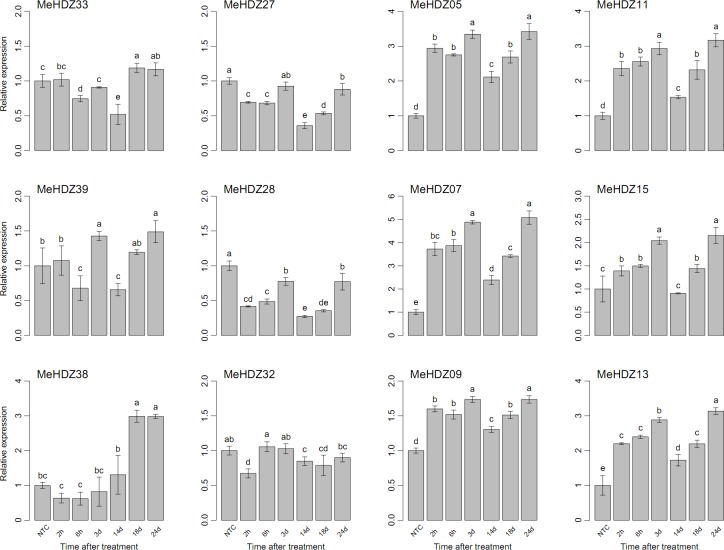
Under salt treatment, three members of MeHDZ I subfamily exhibited different expression trends, e.g., MeHDZ33 was significantly decreased from 6h to 14d but increased at 18d and 24d, MeHDZ39 was greatly induced at 3d and 24d, while MeHDZ38 was almost not changed before 14d but dramatically increased at 18d and 24d (Fig 7). MeHDZ27 and MeHDZ28 from MeHDZ II subfamily were significantly depressed, especially at 2-6h and 14-18d. Interestingly, members from MeHDZ III and IV subfamilies exhibited similar expression patterns and their expression was greatly induced from 2h to 3d, followed by a significant decrease at 14d and then increased at 18d and 24d (Fig 7).
Fig 7. Relative expression of MeHDZ genes in leaves under NaCl treatment.
NTC (no treatment control) was normalized as “1” at each graph. Data were shown as mean ± standard deviation derived from three biological replicates, and values with different letters were significant based on Duncan’s multiple range tests (P < 0.05, n = 3).
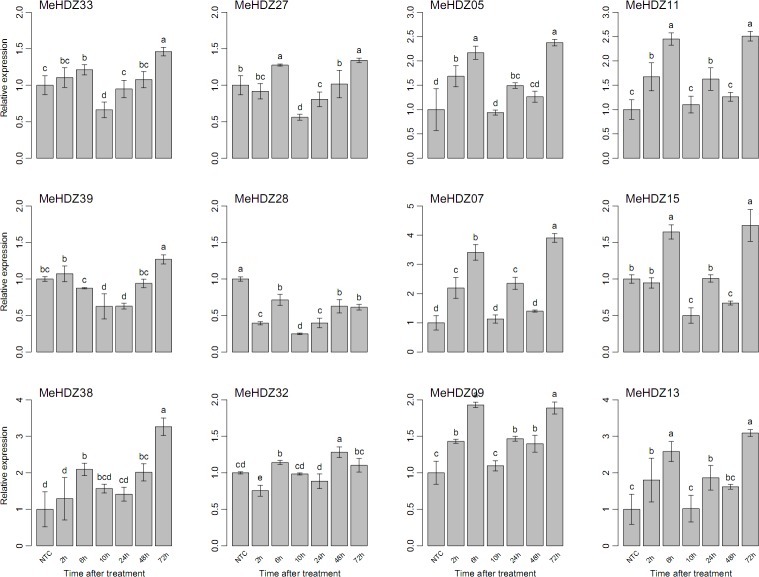
Under ABA treatment, the expression of MeHDZ33 and MeHDZ39 from MeHDZ I subfamily and MeHDZ27 from MeHDZ II subfamily was greatly depressed at 10h but induced at 72h. The expression of MeHDZ38 from MeHDZ I subfamily was not dramatically induced until at 72h, while the expression of MeHDZ28 from MeHDZ II subfamily was greatly depressed during the whole treatment (Fig 8). Interestingly, members from MeHDZ III and IV subfamilies showed very similar expression trends and their expression was greatly induced from 2h to 6h, followed by a significant decrease from 10h to 48h and then dramatically increased at 72h (Fig 8).
Fig 8. Relative expression of MeHDZ genes in leaves under ABA treatment.
NTC (no treatment control) was normalized as “1” at each graph. Data were shown as mean ± standard deviation derived from three biological replicates, and values with different letters were significant based on Duncan’s multiple range tests (P < 0.05, n = 3).
Together, these results indicated that similar to HD-Zip I, members from HD-Zip II and IV subfamilies were also involved in abiotic stresses (e.g., cold and salt) and ABA response in cassava.
Discussion
Cassava is an important crop for tropical and sub-tropical regions. It has good performance even grown under unfavorable environments (e.g., drought), however, the mechanisms underlying its abiotic stresses tolerance remain elusive. The ABA signaling pathway has well been demonstrated to play crucial roles in plant abiotic stresses, in which HD-Zips are important transcription factors involved in ABA signaling pathway. Many researches have further confirmed the involvement of HD-Zip I genes in abiotic stresses (e.g., drought, cold and salt) and ABA signaling response [2,3]. However, the identity, characterization and function of HD-Zip genes are still unknown in cassava.
In this study, a comprehensive analysis of HD-Zip genes was performed in cassava genome, and a total of 57 HD-Zip genes were identified. With the availability of complete genome sequences, it enabled detailed comparisons of HD-Zip genes among many species. Compared with cassava, comparable number of HD-Zip genes was found in maize (55) [5], although this number was higher in soybean (88) [6] but less in peach (33) [46], rice (44) [36] and Arabidopsis (48) [36]. Similar to Arabidopsis and rice, HD-Zip proteins were well classified into four subfamilies in cassava (Fig 1). Besides, the same number of HD-Zip subfamilies was also found in other monocot and dicot plants (e.g., maize, soybean and peach) [5,6,46]. This classification was well supported by gene structure analysis. For example, in cassava, the members of MeHDZ III subfamily had the highest numbers of exons (18), then followed by MeHDZ IV (8 to 11), I (1 to 4) and II (3 to 4) subfamilies (Fig 2). These findings were similar to that reported in maize, soybean and peach [5,6,46]. Conserved motif comparisons also supported the classification of HD-Zip genes. For example, in cassava, members of MeHDZ I and II subfamilies had less conserved motifs comparing to the members of MeHDZ III and IV subfamilies. Moreover, distinct motifs (e.g., START and MEKHLA domain) were exclusively found in the members of MeHDZ III and IV but not in MeHDZ I and II subfamilies (Fig 3). Similar conclusions were observed in many monocot and dicot plants such as Arabidopsis, rice, maize, soybean and peach [3–6,46]. Together, MeHDZ genes of the same subfamilies shared similar characteristics of gene structure and conserved motif, supporting the classification of MeHDZ subfamilies.
Gene duplication, through either segmental duplication or tandem duplication, played important roles in expansion of new members during the evolution of a gene family [47]. Duplicated genes usually have high levels of sequence similarities, however, over the course of evolution, the fates of duplicated genes are quite different: either lose its function (nonfunctionalization), or obtain new function (neofunctionalization), or keep the ancestral function (subfunctionalization) [48]. Comparison of expression profiles of cassava MeHDZ genes involved in tandem and segmental duplications would reveal similar, different or silenced gene expression relative to other members, signifying conservation, neofunctionalization and nonfunctionalization after the duplication events [48]. As expected, diverse expression patterns were revealed in different pairs of duplicated MeHDZ genes. For example, in MeHDZ I subfamily, MeHDZ29 and MeHDZ36 were duplicated genes and exhibited similar expression behavior, suggesting their conservation in the regulation of gene expression; in MeHDZ IV subfamily, MeHDZ10 and MeHDZ11 showed different expression patterns, indicating their divergence in gene expression and they acquired novel regulatory characteristic (e.g., function in different tissues); in MeHDZ II subfamily, MeHDZ34 exhibited a constitutive expression while MeHDZ48 was not expressed in almost all examined tissues (except for stem in Arg7) (Fig 5A), leading to nonfunctionalization after the duplication events. Similar results were also observed in HD-Zip genes from other plants [3,6].
Cis-elements played crucial roles in various aspects of biological processes through regulating gene expression. Several cis-elements related to abiotic stress (e.g., drought, low temperature and light) and response of hormones (e.g., ABA and auxin) were revealed in the promoter region of MeHDZ genes. This was coincident with the well characterized functions of HD-Zip genes in plants [2]. For example, members of HD-Zip I have been demonstrated to be involved in abiotic stresses such as drought/osmotic stresses [2,3]. Consistently, the expression of many members from MeHDZ I subfamily was significantly changed (either up-regulated or down-regulated) in three different genotypes under drought and PEG treatments in our study (Fig 5). Compared with HD-Zip I, the remaining three groups (II to IV) of HD-Zip genes were well characterized as they were involved in shade avoidance, auxin response, development and cell differentiation, rather than drought response, although a few evidences have showed that the expression of HD-Zip II to IV genes was up-regulated in response to water deficit [8,49,50]. In this study, the expression of many members from the MeHDZ II to IV subfamilies was dramatically increased or decreased under drought and PEG treatments, and additionally drought-inducible cis-element (e.g., MBS) was identified in these members, supporting possible roles of genes from MeHDZ II to IV subfamilies in response to drought stress in cassava.
Besides drought, lots of evidences have also revealed the involvement of HD-Zip I genes in other abiotic stresses (e.g., cold and salt) and related signal transduction pathways (e.g., ABA) [3]. However, no such information was available for the MeHDZ genes in response to various stimuli in cassava. Moreover, as previously proposed [51], it was of great interests to examine whether genes from HD-Zip II to IV were involved in abiotic stress responses. Thus, three genes of each MeHDZ subfamily were selected to further examine their expression in response to various treatments (Figs 6–8).
Two genes of MeHDZ I subfamily, MeHDZ38 and MeHDZ39 that were homologues of AtHB12 and AtHB7 respectively, were significantly induced under all tested treatments (Figs 6–8). This was consistent with previous reports that the expression of both AtHB12 and AtHB7 was greatly up-regulated in ABA, salt and cold treatments [3]. Similar results were also observed in Medicago truncatula where the homologues of AtHB7 and AtHB12 were induced under salt stress [52]. However, for another MeHDZ I subfamily gene, MeHDZ33, whose expression was also up-regulated in tested treatments, the expression of its Arabidopsis homologue (AtHB16) was decreased in salt and cold but not responded to ABA [3].
Overall, three examined genes of MeHDZ II subfamily were greatly induced by cold and depressed by salt stress, however, diverse responses were revealed under ABA treatment (Figs 6–8). In soybean, 14 and 16 out of 27 HD-Zip II genes significantly responded to salt and drought stresses, respectively [6]. Likely, HD-Zip II members of Craterostigma plantagineum were shown to be regulated by ABA and water availability [53]. In addition, the transcripts of HD-Zip II members from Arabidopsis and rice were dramatically changed upon drought exposure [4,49]. Together, these results suggested that HD-Zip II genes would play important roles in abiotic stresses.
Uniformly, each three tested members of MeHDZ III and IV subfamilies were all significantly induced in response to cold, salt and ABA treatments. Similar results were revealed in soybean that each six out of eight differentially expressed HD-Zip III genes were up-regulated by salt and drought, while eleven and nine out of eleven differentially expressed HD-Zip IV genes were up-regulated by salt and drought, respectively [6]. A recent study also revealed that three members (REV, PHB and PHV) of HD-Zip III subfamily in Arabidopsis were strongly decreased after three hours of ABA treatment [25]. These results indicated the involvement of HD-Zip III and IV genes in abiotic stresses and ABA response.
In conclusion, a total of 57 HD-Zip genes from cassava genome were identified, and their basic classification, gene structure, conserved protein motifs and evolutionary characteristics were revealed. Subsequently, the cis-elements in the promoter region were explored, and the expression profiles were investigated in different organs of three cassava genotypes, as well as in response to various stimuli through transcriptome or qRT-PCR analysis. These findings will provide a solid base for future studies on the functional characterization of HD-Zip genes, and will improve our understanding of MeHDZ genes involved abiotic stress and signaling transduction in cassava.
Supporting information
Each HD-Zip subfamily was further divided into subclasses (from a to F). The HD-Zip protein sequences were prefixed with ‘At’ for Arabidopsis, ‘Me’ for cassava, ‘Vv’ for Vitis vinifera, ‘Mt’ for Medicago truncatula, ‘Pt’ for Populus trichocarpa, ‘Os’ for rice, and ‘Zm’ for maize, respectively.
(TIF)
(TIF)
Red dots represented the relative positions of MeHDZ genes indicated at the bottom. Each 50 kb sequences upstream and downstream of the genes were selected to BLASTN against each other, and alignments with length > 200 bp and sequence identity > 85% were chained. Five pairs of MeHDZ genes identified as segmental duplications were marked with red boxes.
(TIF)
(TXT)
(TXT)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the International Science and Technology Cooperation Program of China (2010DFA62040, 2014DFA30680), the Natural Science Foundation of China (31471561), and the Natural Science Foundation of Hainan Province (20163120, 20153048).
References
- 1.Elhiti M, Stasolla C. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal. Behav. 2009; 4: 86–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007; 12: 419–426. 10.1016/j.tplants.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 3.Henriksson E, Olsson AS, Johannesson H, Johansson H, Hanson J, Engström P, et al. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 2005; 139: 509–518. 10.1104/pp.105.063461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agalou A, Purwantomo S, Övernäs E, Johannesson H, Zhu X, Estiati A, et al. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 2008; 66: 87–103. 10.1007/s11103-007-9255-7 [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, Zhou Y, Jiang H, Li X, Gan D, Peng X, et al. Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PLoS ONE 2011; 6: e28488 10.1371/journal.pone.0028488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Chen Z, Zhao H, Zhao Y, Cheng B, Xiang Y. Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments. PLoS ONE 2014; 9: e87156 10.1371/journal.pone.0087156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Jiang H, Zhou L, Deng L, Lin Y, Peng X, et al. Molecular evolution of the HD-ZIP I gene family in legume genomes. Gene 2014; 533: 218–228. 10.1016/j.gene.2013.09.084 [DOI] [PubMed] [Google Scholar]
- 8.Pandey A, Misra P, Alok A, Kaur N, Sharma S, Lakhwani D, et al. Genome-Wide Identification and Expression Analysis of Homeodomain Leucine Zipper Subfamily IV (HDZ IV) Gene Family from Musa accuminata. Front. Plant Sci. 2016; 7: 20 10.3389/fpls.2016.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palena C, Gonzalez D, Chan R. A monomer–dimer equilibrium modulates the interaction of the sunflower homeodomain leucine-zipper protein Hahb-4 with DNA. Biochem. J. 1999; 341: 81–87. [PMC free article] [PubMed] [Google Scholar]
- 10.Schrick K, Nguyen D, Karlowski WM, Mayer KF. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004; 5: R41 10.1186/gb-2004-5-6-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherjee K, Bürglin TR. MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol. 2006; 140: 1142–1150. 10.1104/pp.105.073833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soderman E, Mattsson J, Engström P. The Arabidopsis homeobox gene ATHB‐7 is induced by water deficit and by abscisic acid. The Plant Journal 1996; 10: 375–381. [DOI] [PubMed] [Google Scholar]
- 13.Soderman E, Hjellström M, Fahleson J, Engström P. The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves, roots and carpels and up-regulated by water deficit conditions. Plant Mol. Biol. 1999; 40: 1073–1083. [DOI] [PubMed] [Google Scholar]
- 14.Johannesson H, Wang Y, Hanson J, Engström P. The Arabidopsis thaliana homeobox gene ATHB5 is a potential regulator of abscisic acid responsiveness in developing seedlings. Plant Mol. Biol. 2003; 51: 719–729. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y-H, Oh H-S, Cheon C-I, Hwang I-T, Kim Y-J, Chun J-Y. Structure and expression of the Arabidopsis thaliana homeobox gene Athb-12. Biochem. Biophys. Res. Commun. 2001; 284: 133–141. 10.1006/bbrc.2001.4904 [DOI] [PubMed] [Google Scholar]
- 16.Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, et al. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 2005; 19: 2811–2815. 10.1101/gad.364005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, et al. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 1999; 126: 4235–4245. [DOI] [PubMed] [Google Scholar]
- 18.Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, et al. The HAT2 gene, a member of the HD‐Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. The Plant Journal 2002; 32: 1011–1022. [DOI] [PubMed] [Google Scholar]
- 19.Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, et al. The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 2001; 126: 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, et al. microRNA‐directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. The Plant Journal 2005; 42: 84–94. 10.1111/j.1365-313X.2005.02354.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilegems M, Douet V, Meylan-Bettex M, Uyttewaal M, Brand L, Bowman JL, et al. Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development 2010; 137: 975–984. 10.1242/dev.047662 [DOI] [PubMed] [Google Scholar]
- 22.Abe M, Katsumata H, Komeda Y, Takahashi T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 2003; 130: 635–643. [DOI] [PubMed] [Google Scholar]
- 23.Kubo H, Peeters AJ, Aarts MG, Pereira A, Koornneef M. ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. The Plant Cell 1999; 11: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, et al. Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol. 2006; 141: 1363–1375. 10.1104/pp.106.077388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandt R, Cabedo M, Xie Y, Wenkel S. Homeodomain leucine-zipper proteins and their role in synchronizing growth and development with the environment. J. Integr. Plant Biol. 2014; 56: 518–526. 10.1111/jipb.12185 [DOI] [PubMed] [Google Scholar]
- 26.Taylor N, Chavarriaga P, Raemakers K, Siritunga D, Zhang P. Development and application of transgenic technologies in cassava. Plant Mol. Biol. 2004; 56: 671–688. 10.1007/s11103-004-4872-x [DOI] [PubMed] [Google Scholar]
- 27.An D, Yang J, Zhang P. Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress. BMC Genomics 2012; 13: 64 10.1186/1471-2164-13-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Utsumi Y, Tanaka M, Morosawa T, Kurotani A, Yoshida T, Mochida K, et al. Transcriptome analysis using a high-density oligomicroarray under drought stress in various genotypes of cassava: an important tropical crop. DNA Res. 2012; 19: 335–345. 10.1093/dnares/dss016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, Feng B, Xiao J, Xia Z, Zhou X, Li P, et al. Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 2014; 5: 5110 10.1038/ncomms6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei Y, Shi H, Xia Z, Tie W, Ding Z, Yan Y, et al. Genome-Wide Identification and Expression Analysis of the WRKY Gene Family in Cassava. Front. Plant Sci. 2016; 7: 25 10.3389/fpls.2016.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei F, Hai M, Guo Y, Ding Z, Tie W, Ding X, et al. The ERF transcription factor family in cassava: genome-wide characterization and expression analyses against drought stress. Sci. Rep. 2016; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao P, Liu P, Shao J, Li C, Wang B, Guo X, et al. Analysis of different strategies adapted by two cassava cultivars in response to drought stress: ensuring survival or continuing growth. J. Exp. Bot. 2014; 66: 1477–1488. 10.1093/jxb/eru507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu W, Yang H, Yan Y, Wei Y, Tie W, Ding Z, et al. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 2016; 6: 22783 10.1038/srep22783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu L, Ding Z, Han B, Hu W, Li Y, Zhang J. Physiological Investigation and Transcriptome Analysis of Polyethylene Glycol (PEG)-Induced Dehydration Stress in Cassava. Int. J. Mol. Sci. 2016; 17: 283 10.3390/ijms17030283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eddy SR. Accelerated profile HMM searches. PLoS Comput. Biol. 2011; 7: e1002195 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu R, Chi X, Chai G, Kong Y, He G, Wang X, et al. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa). PLoS ONE 2012; 7: e31149 10.1371/journal.pone.0031149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 2014; 31: 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME Suite. Nucleic Acids Res. 2015; 43(W1): W39–49. 10.1093/nar/gkv416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khaja R, MacDonald JR, Zhang J, Scherer SW. Methods for identifying and mapping recent segmental and gene duplications in eukaryotic genomes. Methods Mol. Biol. 2006; 338: 9–20. 10.1385/1-59745-097-9:9 [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Ding Z, Ma F, Chauhan RD, Allen DK, Brutnell TP, et al. Transcriptional response to petiole heat girdling in cassava. Sci. Rep. 2015; 5: 8414 10.1038/srep08414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole Trapnell AR, Goff Loyal, Pertea Geo, Kim Daehwan, Kelley David R, Pimentel Harold, Salzberg Steven L, Rinn John L, Pachter Lior. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7: 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J, Duan X, Yang J, Beeching JR, Zhang P. Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots. Plant Physiol. 2013; 161: 1517–1528. 10.1104/pp.112.212803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang C, Ma R, Shen Z, Sun X, Korir N, Yu M. Genome-wide analysis of the homeodomain-leucine zipper (HD-ZIP) gene family in peach (Prunus persica). Gen. Mol. Res. 2014; 13: 2654–2668. [DOI] [PubMed] [Google Scholar]
- 47.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004; 4: 10 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science 2000; 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- 49.Huang D, Wu W, Abrams SR, Cutler AJ. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 2008; 59: 2991–3007. 10.1093/jxb/ern155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu H, Chen X, Hong Y-Y, Wang Y, Xu P, Ke S-D, et al. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. The Plant Cell 2008; 20: 1134–1151. 10.1105/tpc.108.058263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chew W, Hrmova M, Lopato S. Role of homeodomain leucine zipper (HD-Zip) IV transcription factors in plant development and plant protection from deleterious environmental factors. Int. J. Mol. Sci. 2013; 14: 8122–8147. 10.3390/ijms14048122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ariel F, Diet A, Verdenaud M, Gruber V, Frugier F, Chan R, et al. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. The Plant Cell 2010; 22: 2171–2183. 10.1105/tpc.110.074823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng X, Phillips J, Meijer AH, Salamini F, Bartels D. Characterization of five novel dehydration-responsive homeodomain leucine zipper genes from the resurrection plant Craterostigma plantagineum. Plant Mol. Biol. 2002; 49: 601–610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each HD-Zip subfamily was further divided into subclasses (from a to F). The HD-Zip protein sequences were prefixed with ‘At’ for Arabidopsis, ‘Me’ for cassava, ‘Vv’ for Vitis vinifera, ‘Mt’ for Medicago truncatula, ‘Pt’ for Populus trichocarpa, ‘Os’ for rice, and ‘Zm’ for maize, respectively.
(TIF)
(TIF)
Red dots represented the relative positions of MeHDZ genes indicated at the bottom. Each 50 kb sequences upstream and downstream of the genes were selected to BLASTN against each other, and alignments with length > 200 bp and sequence identity > 85% were chained. Five pairs of MeHDZ genes identified as segmental duplications were marked with red boxes.
(TIF)
(TXT)
(TXT)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.