Abstract
Skeletal muscle possesses remarkable regenerative potential due to satellite cells, an injury-responsive stem cell population located beneath the muscle basal lamina that expresses Pax7. By lineage tracing of progenitor cells expressing the Twist2 (Tw2) transcription factor in mice, we discovered a myogenic lineage that resides outside the basal lamina of adult skeletal muscle. Tw2+ progenitors are molecularly and anatomically distinct from satellite cells, are highly myogenic in vitro, and can fuse with themselves and with satellite cells. Tw2+ progenitors contribute specifically to type IIb/x myofibers during adulthood and muscle regeneration, and their genetic ablation causes wasting of type IIb myofibers. We show that Tw2 expression maintains progenitor cells in an undifferentiated state that is poised to initiate myogenesis in response to appropriate cues that extinguish Tw2 expression. Tw2-expressing myogenic progenitors represent a previously unrecognized, fiber-type specific stem cell involved in post-natal muscle growth and regeneration.
Introduction
Skeletal muscle is among the most regenerative adult tissues. Its remarkable regenerative capacity originates from a population of resident stem cells, termed satellite cells (SCs), located beneath the muscle basal lamina 1. SCs are marked by expression of Pax7, a transcription factor critical for muscle regeneration 1. In response to injury and disease, SCs become activated and undergo self-renewal and differentiation to form new myofibers 1-3. While SCs are essential for muscle regeneration, their their genetic ablation in adult mice does not accelerate sarcopenia 4-6. Thus, additional mechanisms or cell types might contribute to maintenance of muscle mass during aging.
Skeletal muscle is composed of heterogeneous myofiber types that differ in contractile and metabolic properties and expression of distinctive myosin isoforms. Four major fiber types are present in rodent muscles: one type of slow-twitch fiber (type I) and 3 types of fast-twitch fibers (type IIa, IIx/d, and IIb). While type I and type IIa fibers exhibit oxidative metabolism and high endurance; type IIx and IIb fibers are glycolytic and display low endurance 7. Slow and fast twitch fibers also differ in their responses to hypertrophic or atrophic stimuli. For example, type IIb and IIx myofibers are more susceptible than slow twitch fibers to a variety of atrophic signals such as denervation, nutrient deprivation, cancer cachexia, and chronic heart failure 8-10. While SCs can fuse into all myofiber types in injured muscle 11, it remains unknown whether fiber-type specific myogenic progenitors might also exist.
The Drosophila basic helix-loop-helix transcription factor Twist is expressed in muscle progenitors during embryogenesis and is essential for the formation of mesoderm and muscle 12-14. Within the adult musculature of Drosophila, Twist expression is restricted to muscle precursors that are normally quiescent but are activated by extracellular cues to regenerate the adult musculature during metamorphosis 15-17. Two mammalian Twist genes, Twist1 (Tw1) and Twist2 (Tw2), are expressed in various mesenchymal cell types, but not in differentiated myofibers18, 19. Tw1 and Tw2 have been shown to block myogenesis in vitro 19-22,23, but their potential roles in muscle formation or regeneration in mammals have not been explored.
Here, we traced the fate of Tw2-dependent cell lineages in mice and discovered that Tw2 expression marks a previously unrecognized interstitial myogenic progenitor cell that forms type IIb/x myofibers in adult muscle. Tw2-expressing progenitors represent a population of myogenic progenitor cells that contributes to specific fiber types during muscle homeostasis and regeneration, highlighting the ancestral functions of Twist as a regulator of muscle formation.
Results
Twist Expression in Interstitial Cells Within Adult Skeletal Muscle
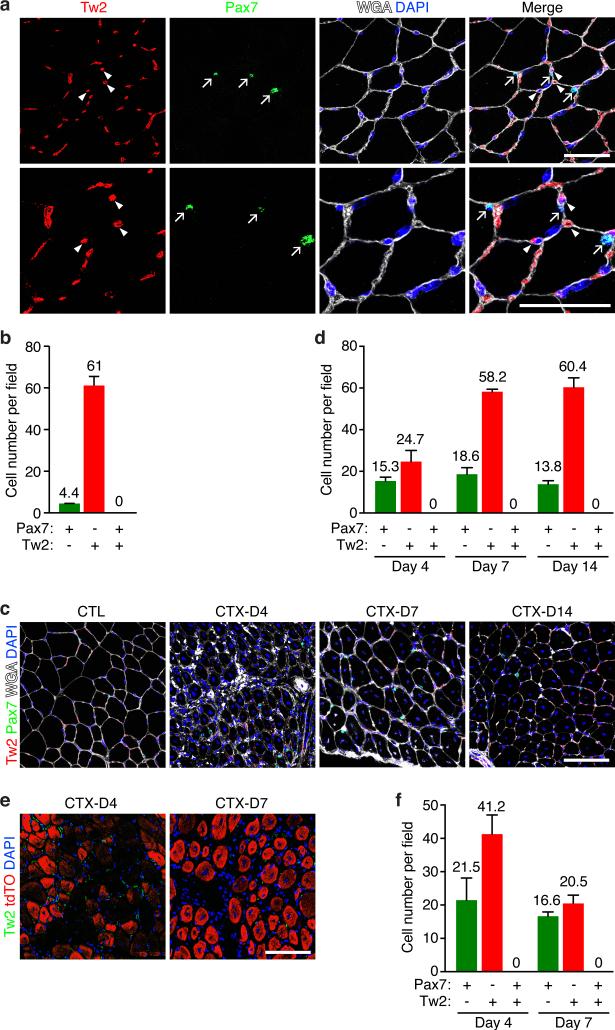
In adult muscle, Tw2 transcript is barely detectable in whole G/P muscle at 1, 2 and 4 months of age by RNA-seq analysis, in contrast to MyoD and Myh4 that are readily detected (Supplementary Fig. 1a). Real-time RT-PCR revealed that Tw2 was highly enriched in mononuclear non-myofiber cells compared to whole quadriceps muscle (Supplementary Fig. 1b). Immunostaining of transverse sections of gastrocnemius muscle from 3 months old wild-type (WT) mice revealed Tw2 protein in interstitial cells outside of the myofibers, but not within myofibers (Fig. 1a). Furthermore, Tw2 protein was not co-localized with Pax7, which was restricted to SCs beneath the basal lamina (Fig. 1a and b). Similar mutual exclusivity of expression of Tw2 and Pax7 was observed in muscles of 12 month-old mice (Supplementary Fig. 1c). We conclude that Tw2 is expressed in the myofiber interstitium and not in mature myofibers or SCs in adult muscle.
Figure 1. Tw2-expressing and Pax7-expressing cells are distinct cell types in skeletal muscle.
(a) Immunostaining of Tw2 (red) and Pax7 (green) on transverse sections of gastrocnemius muscle of 3-month old C57Bl6 WT mice. Myofibers were co-stained with wheat germ agglutinin (WGA) (white) and DAPI (blue). Arrows indicate Pax7+ cells and arrowheads indicate Tw2+ cells. Scale bar: 50 um.
(b) Quantification of the number of Pax7+, Tw2+, and Pax7+/Tw2+ double positive cells per field in 3-month old C57Bl6 WT mice. Quantification was performed on the top panel images in Fig. 1a. For each muscle section, at least 6 different fields were quantified and averaged. Data are mean ± S.E.M. N=6 mice.
(c) TA muscle of C57Bl6 WT mice was injured by CTX injection and harvested on Days 4, 7 and 10 post injury. Muscle sections were stained for Pax7 (green), Tw2 (red), DAPI (blue) and WGA (white). CTL: contralateral TA muscle. Scale bar: 100 um.
(d) Quantification of the number of Pax7+, Tw2+, and Pax7+/Tw2+ double positive cells per field in C57Bl6 WT mice on days 4, 7 and 14 post-CTX injury. For each muscle section, at least 6 different fields were quantified and averaged. Data are mean ± S.E.M. N=3 mice.
(e) Tw2 expression in TA muscle of Pax7-CreERT2; R26-tdTO mice after CTX injury. Adult Pax7-CreERT2; R26-tdTO mice were treated with TMX for 3 alternating days. One week after the first dose of TMX, CTX injury was performed on TA muscle. Muscles were harvested at Days 4 and 7 post injury and immunostained for Tw2 (green) and DAPI (blue). Scale bar: 100 um.
(f) Quantification of the number of Pax7+ (tdTO+), Tw2+, and Pax7+/Tw2+ double positive cells per field in Pax7-CreERT2; R26-tdTO on days 4 and 7 post-CTX injury. Pax7+ cells were detected by tdTO+ signal. For each muscle section, at least 6 different fields were quantified and averaged. Data are mean ± S.E.M. N=3 mice.
Statistic source data for b,d,f are provided in Supplementary Table 3.
To analyze Tw2 expression during muscle regeneration, we performed cardiotoxin (CTX) injury on tibialis anterior (TA) muscle of WT mice, and harvested muscles on 4, 7, and 14 days post-CTX injury. Tw2+ and Pax7+ cells were detected by immunostaining. We did not observe cells that expressed both Pax7 and Tw2 proteins at any time point examined (Fig. 1c and d). Tw2+ cells declined immediately following CTX injury but the number of Tw2+ cells was rapidly restored by day 7 post-CTX.
To further compare the contributions of Tw2+ and Pax7+ cells to adult muscle, we performed lineage tracing for Pax7 by breeding Pax7-CreERT2 mice 24 with ROSA26-loxp-stop-loxp-tdTomato (R26-tdTO) reporter mice. At 3 months of age, Pax7-CreERT2; R26-tdTO mice were treated with tamoxifen (TMX) on 3 alternate days, and TA muscles were injected with CTX 1 week later. Immunostaining revealed that upon CTX injury, tdTO+ labeling (representing the Pax7+ lineage) marked regenerating myofibers but did not co-localize with Tw2+ cells (Fig. 1e and f). There was also no overlap between tdTO signal and Tw2 protein expression in uninjured contralateral muscle (Supplementary Fig. 1d). Together, these results confirm that Tw2+ cells and Pax7+ SCs represent distinct cell lineages.
Tw2+ Cells Contribute to a Subset of Type II Myofibers
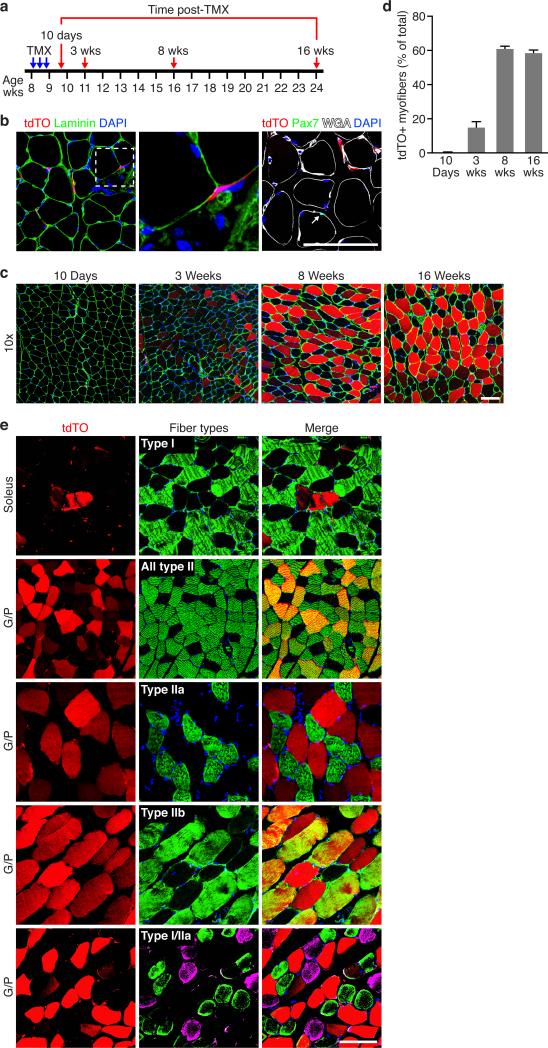
To identify and trace the fates of Tw2-expressing cells in vivo, we introduced a TMX-inducible Cre expression cassette (CreERT2) into the mouse T w2 gene by homologous recombination and bred mice harboring this allele to the R26-tdTO mice (Supplementary Fig. 2a, b). Following treatment of Tw2-CreERT2; R26-tdTO mice with TMX on 3 alternate days at 8 weeks of age, we monitored tdTO expression as a marker for the Tw2 lineage in skeletal muscle (Fig. 2a). Ten days after TMX treatment, we observed tdTO labeling of individual cells within the muscle interstitium, outside of the basal lamina, but no labeling of myofibers (Fig. 2b, left and middle panels). The anatomical location of the tdTO+ interstitial cells differed from that of Pax7+ SCs, which reside beneath the basal lamina. Immunostaining confirmed that tdTO+ cells do not express Pax7 protein (Fig. 2b, right panel). The tdTO+ interstitial cells often exhibited a distinctive spindle-shaped appearance with elongated processes that was also clearly distinct from the morphology of SCs (Fig. 2b, middle panel).
Figure 2. Progressive contribution of Tw2+ cells to adult skeletal muscle.
(a) Schematic of TMX treatment. Tw2-CreERT2; R26-tdTO mice on a mixed genetic background were injected with 1 mg TMX at 8 weeks of age on 3 alternating days. Mice were analyzed at various time points following initial TMX injection.
(b) At 10 days post-TMX, Tw2+ cells marked by tdTO+ (red) were located outside of the basal lamina (green) (left panel). Middle panel is an enlarged image of the left panel showing the morphology of Tw2+ cells. In contrast, Pax7+ cells (stained with Pax7 antibody, green) were beneath the basal lamina. Arrow indicates a Pax7+ cell. Scale bar: 100 um.
(c) Progressive tdTO labeling of myofibers of G/P muscle at the indicated times following TMX treatment. Transverse-sections of G/P muscle were co-stained with laminin (green) and DAPI (blue). Scale bar: 100 um.
(d) Quantification of percentage of tdTO+ myofibers among all myofibers in each field in G/P muscle. Data are mean ± S.E.M. For each time point, N=3 mice were analyzed. For each mouse, between 1000 and 2000 myofibers of G/P muscle were quantified.
(e) Transverse sections of different muscle groups obtained from Tw2-CreERT2; R26-tdTO mice at 4 months post-TMX were stained with various fiber type-specific antibodies (green as indicated). Scale bar: 100 um.
Statistic source data for d are provided in Supplementary Table 3.
By 3 weeks following TMX treatment, strong tdTO staining was observed in the G/P muscles, and the number of tdTO+ myofibers continued to increase up to 8 weeks post-TMX treatment, at which time 58% of myofibers were labeled by tdTO (Fig. 2c, d). No tdTO+ myofibers were observed in the absence of TMX (Supplementary Fig. 2c), validating the specificity of the lineage tracing method. Strong tdTO labeling was observed in all hindlimb muscles, diaphragm, and masseter muscles by 5 months post-TMX with the exception of the tongue (Supplementary Fig. 2d, e). Instead, interstitial cells within the tongue and adjacent epithelial cells along the surface of the tongue were labeled with tdTO (Supplementary Fig. 2e). Longitudinal sections of G/P muscle at 4 months post-TMX also revealed tdTO expression throughout the entire length of myofibers (Supplementary Fig. 2f). Tw2+ cells did not give rise to other cell types such as endothelial cells (marked by CD31) or fibroblasts (marked by vimentin) in adult muscle (Supplementary Fig. 2g).
To determine whether the Tw2+ lineage contributed to specific myofiber types, we analyzed tdTO labeling in G/P and soleus muscles, which are comprised of different proportions of type I and type II myofibers. There was no overlap between type I myofibers and tdTO+ myofibers in the soleus, but a subset of type II fibers within G/P that also expressed tdTO (Fig. 2e). Further staining revealed tdTO labeling of type IIb but not type IIa myofibers in G/P muscle (Fig. 2e). Some tdTO+ fibers were negative for IIb staining (Fig. 2e). By exclusion, we interpret these to be type IIx fibers. Therefore, we conclude that Tw2 cells specifically label type IIb/x fibers. The fiber type-specific tdTO labeling remained even 18 months after the TMX pulse (Supplementary Fig. 2h,i), To determine whether the fiber-type specificity also remained in aged mice, we performed lineage tracing on 7-month old Tw2-CreERT2; R26-tdTO mice, and observed robust contribution of tdTO+ cells to myofibers and the same type II fiber-specific labeling at 3 months post-TMX (Supplementary Fig. 2j,k).
To further assess the type IIb/x myofiber specificity of the Tw2 lineage, we compared the contribution of SCs to adult myofibers by treating Pax7-CreERT2;R26-tdTO mice with TMX at 8 weeks of age. Pax7+ SCs contributed to myofibers in all muscles including tongue at 8 weeks post-TMX (Supplementary Fig. 2l), consistent with prior studies 11,5. SCs also labeled all types of myofibers (Supplementary Fig. 2m). This finding is consistent with a recent study showing that Pax7+ SCs fuse into myofibers in uninjured skeletal muscle and that a fiber-type preference does not exist for Pax7+ SCs during fusion 11. Thus, in contrast to the broad muscular contributions of SCs, the Tw2-expressing lineage contributes specifically to adult type IIb/x myofibers
To determine whether Tw2+ cells contribute to embryonic myogenesis during development, we performed lineage tracing of Tw2+ cells by breeding mice harboring a constitutively active Cre expression cassette inserted in the Tw2 locus 25, 26 with R26-tdTO mice. At embryonic day (E) 10.5, Tw2+/tdTO+ cells were present in regions surrounding the somites, but not within somitic muscle (Supplementary Fig. 2o). At E15.5 and P1, tdTO was expressed in interstitial cells within various muscle groups but not within myofibers (Supplementary Fig. 2o). Together, these findings demonstrate that Tw2+ cells do not contribute to primary or secondary myogenesis in embryos, but are dedicated to the formation of muscle postnatally.
Ablation of the Tw2+ Lineage Causes Atrophy of Type IIb Myofibers
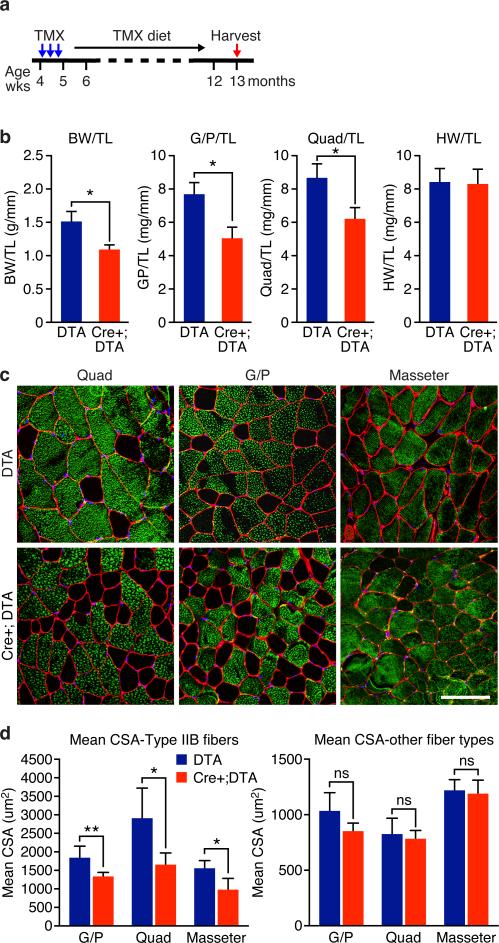
We genetically ablated Tw2+ cells in adult mice by breeding the Tw2-CreERT2 mice to mice harboring a diphtheria toxin (DTA) expression cassette in the Rosa26 locus (R26-DTA mice)27. Tw2-CreERT2; R26-DTA/+ (named Cre+;DTA) mice and control R26-DTA/+ (named DTA) mice were injected with TMX and maintained on TMX-containing diet beginning at 4 weeks of age (Fig. 3a) and analyzed at 13 months of age. G/P and quadriceps muscles were significantly smaller in Cre+;DTA mice compared to DTA control mice, whereas other tissues such as heart, kidney and liver were approximately comparable in size (Fig. 3b and Supplementary Fig. 3). Histological analysis of liver, WAT and BAT also revealed no significant difference between DTA and Cre+; DTA mice (Supplementary Fig. 3c). Intriguingly, in Cre+; DTA mice, we observed a significant decrease in the mean cross-sectional area (CSA) of type IIb fibers in G/P, quadriceps and masseter muscles (Fig. 3c,d). The mean CSA of other fiber types remained unchanged (Fig. 3d). The number of type IIb fibers per field as well as the total fiber numbers were also increased in quadriceps and masseter but not G/P muscles (Supplementary Fig. 3d). No centralized nuclei were observed in Cre+ DTA mice. These results indicate that ablation of Tw2+ cells caused type IIb fiber-specific atrophy, supporting the notion that Tw2+ cells are important for maintenance of type IIb myofiber size during adulthood.
Figure 3. Ablation of the Tw2+ lineage causes type IIb myofiber atrophy.
(a) Schematic of TMX treatment. Tw2-CreERT2; R26-DTA/+ (Cre+;DTA) and R26-DTA/+ (DTA) mice on a mixed genetic background were injected with 1 mg TMX at 4 weeks of age on 3 alternating days. Mice were kept on TMX-containing diet until the time of analysis.
(b) Measurement of body weight (BW), heart weight (HW) and muscle mass normalized to tibia length (TL) of mice. Data are mean ± S.E.M; two sample t-test; *: P < 0.05. N =5 male mice for each genotype.
(c) Type IIb (green) and laminin (red) immunostaining of transverse sections of quad, G/P, and masseter muscles of Cre+;DTA and DTA mice. Scale bar: 100 um.
(d) Measurement of mean myofiber cross-sectional area (CSA) of type IIb fibers (left) and other fibers (right). Data are mean ± S.E.M; two sample t-test; *: P < 0.05. **: P < 0.01. ns: not significant. N =5 mice for each genotype. For each muscle, more than 300 fibers were measured and averaged.
Statistic source data for b,d are provided in Supplementary Table 3.
The Tw2 Lineage Contributes to Muscle Regeneration
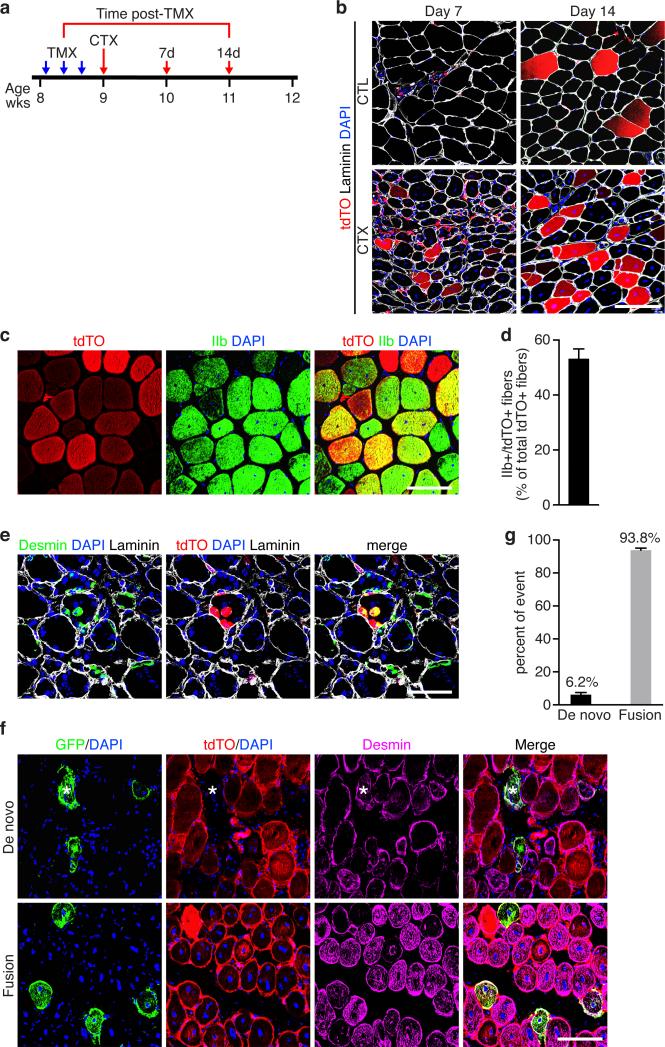
To determine whether the Tw2 lineage forms new myofibers in response to injury, we performed CTX injury on TA muscle (Fig. 4a). Seven days after CTX injection, we observed tdTO labeling of newly regenerated myofibers, marked by centralized nuclei, and the labeling became more robust by day 14 post-CTX when tdTO+ myofibers represented ~27.4% of newly regenerated myofibers (Fig. 4b). Myosin staining revealed that 55% of the regenerated tdTO+ myofibers were type IIb myofibers (Fig. 4c, d and Supplementary Fig. 4a).
Figure 4. Tw2+ cells contribute to skeletal muscle regeneration.
(a) Schematic of TMX and CTX treatment on adult Tw2-CreERT2; R26-tdTO mice on a mixed genetic background. CTX was injected into the TA muscle at 1 week after the 1st dose of TMX. Both contralateral (CTL) and CTX-injured TA muscles were harvested at 7 and 14 days post-CTX.
(b) Immunostaining of transverse sections of TA muscles revealed that Tw2+ cells are activated and contribute to regenerating myofibers (indicated by centralized nuclei) on days 7 and 14 after CTX injury. Sections were co-stained with laminin (white) and DAPI (blue). Scale bar: 100 um.
(c) Co-staining with IIb myosin (green) with newly regenerated tdTO+ myofibers on day 14 post-CTX. Scale bar: 100 um.
(d) Quantification of the percentage of tdTO+ myofibers that are type IIb-positive among all tdTO+ myofibers on day 14 post-CTX. Data are mean ± S.E.M; N=5 mice.
(e) Three days post-CTX, occasional small tdTO+ cells expressed desmin (green), indicating that these cells differentiated into desmin-positive myoblasts. Scale bar: 100 um.
(f) Transverse-sections of TA muscle on Day 7 post-CTX from Tw2-CreERT2; R26-mT/mG mice on a mixed genetic background at 4 months post-TMX. Asterisk represents a GFP+ regenerated new myofiber that lost mT (tdTO) expression. Scale bar: 100 um.
(g) Percent of de novo versus fusion events in Tw2-CreERT2; R26-mT/mG mice on Day 7 post-CTX. Data are mean ± S.E.M; N= 3 mice were analyzed.
Statistic source data for d,g are provided in Supplementary Table 3.
To determine whether activated Tw2+ cells differentiate autonomously or fuse with new myofibers, we immunostained muscle sections for desmin, which is highly expressed in immature muscle fibers during fetal life and regeneration28, 29 (Fig. 4e). Although most of the activated Tw2+ cells were negative for desmin expression (Supplementary Fig. 4b), we occasionally observed small desmin+/tdTO+ myofibers indicating that Tw2+ cells differentiated into desmin-expressing myofibers (Fig. 4e). Activated Tw2+ cells did not express Pax7, as demonstrated by immunostaining (Supplementary Fig. 4c, d).
To further determine whether Tw2 cells contribute to regeneration by initiating myogenesis autonomously or by fusing with existing myofibers, we bred Tw2-CreERT2 mice to R26-mT/mG mice, which constitutively express a membrane-targeted tdTO protein (mT) from the Rosa26 locus30 (Supplementary Fig. 4e). Upon Cre-activation, tdTO fluorescence is lost and membrane-targeted eGFP (mG) becomes expressed. If Tw2 cells fuse with existing myofibers, both signals will be present, but a de novo myofiber formed from Tw2+ cells will only be green. We analyzed muscles from Tw2-CreERT2; R26-mTmG/+ mice at 4 months post-TMX. In the absence of injury, almost all GFP+ myofibers retained tdTO expression (Supplementary Fig. 4f), indicating that Tw2+ cells contribute to myofibers via fusion with existing fibers during homeostasis, as reported for satellite cells5. Upon CTX-injury, although the majority (93.8%) of mG+ new myofibers also expressed mT, 6.2% of new myofibers expressed mG and lost mT expression (Fig. 4f, g). These findings demonstrate that Tw2+ cells can initiate myogenesis autonomously during regeneration, although the majority of these cells fuse with newly regenerated myofibers.
For comparison, we performed CTX injury on Pax7-Cre-ERT2; R26-tdTO mice following the same treatment regimen as in Fig. 4a. The Pax7+ lineage labeled all regenerating myofibers (Supplementary Fig. 3n). We conclude that following injury Tw2+ cells contribute to a subset of new type IIb myofibers that are also derived from the Pax7 lineage.
To test whether Tw2+ cells also possess the capacity to engraft into injured muscle, we isolated Tw2+ (tdTO+) cells by FACS sorting at 10 days post-TMX, and injected them into the TA muscles of mdx mice, which display extensive muscle degeneration and regeneration. Four weeks post-transplantation, tdTO+ myofibers were observed in the TA muscle of mdx mice, revealing the ability of exogenous Tw2+ cells to engraft and form new muscle (Supplementary Fig. 4g). However, the engraftment efficiency was limited with an average of only 5.5 myofibers per field (Supplementary Fig. 4h), whcih is lower than the engraftment capacity of SCs.
Freshly Isolated Tw2+ Cells Are Distinct From Pax7+ SCs
To further characterize Tw2+ cells, we examined expression of cell-surface markers on freshly isolated Tw2+ cells by FACS analysis from 8-week old Tw2-CreERT2; R26-tdTO/+ mice 10 days post-TMX (Supplementary Table 1, and Supplementary Fig. 5a,b). Only 0.46% of Tw2+ cells expressed the SC-specific marker α7-integrin. Similarly, other SC markers, including CD34, Vcam1 (CD106), and CXCR4 (CD184), were expressed in only a small fraction of Tw2+ cells. The majority of tdTO+ cells also did not express markers for endothelial cells (CD31) or hematopoietic cells (CD45) (Supplementary Table 1, and Supplementary Fig. 5a,b). Approximately 98.8% of tdTO+ cells were positive for CD29 (β1-integrin), a widely expressed marker of mesenchymal stem cells (MSCs), lymphocytes, hematopoietic stem cells (HSC), and other cell types. In addition, tdTO+ cells positive for Sca1 were further separated as Sca1-High (68.8%) and Sca1-medium (8.6%) populations. Other MSC markers, including CD73 and CD105, were expressed in only a small fraction of tdTO+ cells (Supplementary Table 1). Together, these findings suggest that the Tw2 lineage represents a population of muscle precursors clearly distinct from SCs and most closely resembling cells of a mesenchymal lineage. Expression of cell surface markers remained largely unchanged in Tw2+ cells upon CTX injury (Supplementary Table 2).
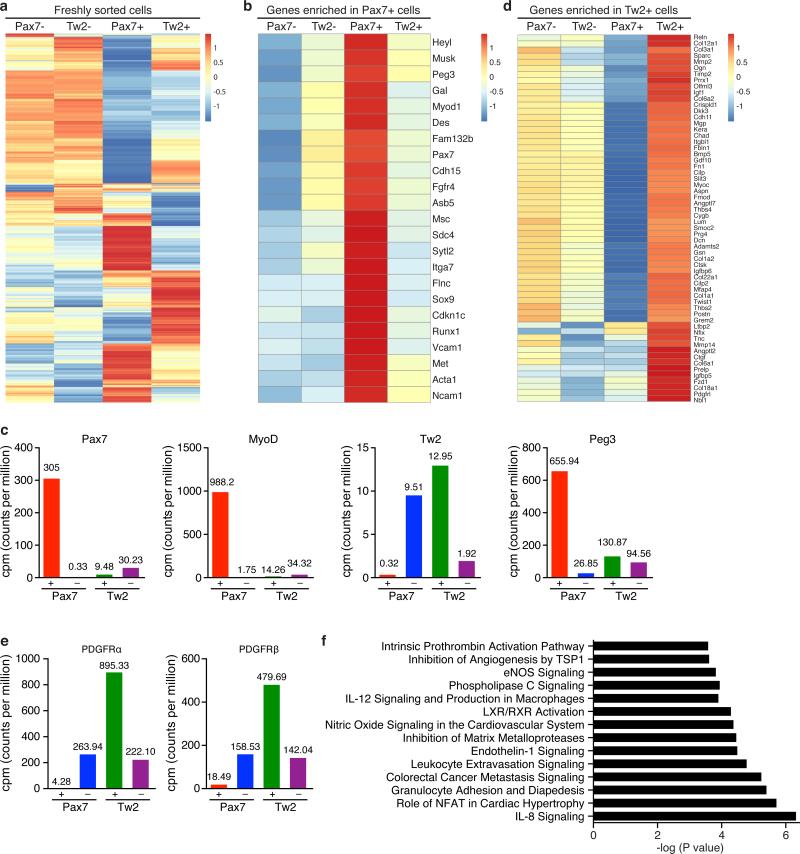
We performed RNA-seq analysis on freshly sorted Tw2+ and Tw2− cells from Tw2-CreERT2; R26-tdTO mice 10 days post-TMX treatment at 8 weeks of age. For comparison, freshly sorted Pax7+ and Pax7− cells from Pax7-CreERT2; R26-tdTO mice were also analyzed. Freshly isolated Tw2+ and Pax7+ cells showed distinct transcriptome signatures, whereas Tw2− and Pax7− cells displayed similar expression profiles (Fig. 5a). SC-enriched transcripts such as Pax7, Cadh15 (M-cad), Fgfr4, Sdc4, Met, Itga7 and Vcam1 were all strongly enriched in Pax7+ cells. Strikingly, none of these mRNAs were enriched in Tw2+ cells (Fig. 5b,c). MyoD transcript was barely detectable in freshly isolated Tw2+ cells in contrast to Pax7+ cells (Fig. 5b,c). Similarly, Peg3/PW1, a marker for PICs, was not enriched in the Tw2+ cells relative to either Tw2− cells or Pax7+ cells (Fig. 5b,c). On the contrary, genes highly enriched in Tw2+ cells (relative to Tw2− cells) were all repressed in Pax7+ cells, again confirming that Tw2+ and Pax7+ cells have distinct transcriptome signatures (Fig. 5d). Furthermore, platelet derived growth factor receptor alpha (PDGFRα), a marker for human and mouse MSCs, was highly enriched in Tw2+ cells but barely detected in Pax7+ cells (Fig. 5e). PDGFRβ, a marker for pericytes and vascular smooth muscle cells, was also highly enriched in Tw2+ but not Pax7+ cells (Fig. 5e). Ingenuity Pathway Analysis (IPA) revealed that mRNAs enriched in Tw2+ cells relative to Pax7+ cells are involved in cancer metastasis, interleukin/NFAT/endothelin signaling, and extra-cellular matrix (ECM) remodeling (Fig. 5f).
Figure 5. Molecular profiling of the Tw2+ lineage in skeletal muscle.
(a) Heat map of 4980 genes expressed in freshly sorted Tw2+, Tw2− Pax7+ and Pax7− cells identified by RNA-seq. Cells were isolated by FACS sorting from Tw2-CreERT2; R26-tdTO mice and Pax7-CreERT2; R26-tdTO mice at 10 days post-TMX, respectively.
(b) Heat map of the top 23 genes enriched in Pax7+ cells compared to Pax7− cells.
(c) CPM (counts per million) of Pax7, MyoD, Tw2, Peg3 expression by RNA-seq in Tw2+, Tw2−, Pax7+ and Pax7− cells.
(d) Heat map of top 60 genes enriched in Tw2+ cells relative to Tw2− cells.
(e) CPM (counts per million) of PDGFRα and PDGFRβ expression by RNA-seq in Tw2+, Tw2−, Pax7+ and Pax7− cells.
(f) Pathways enriched in freshly sorted Tw2+ cells relative to Tw2− cells identified by Ingenuity Pathway Analysis (IPA).
Tw2 Cells Transition Through a Pre-Myogenic Pax7+ State
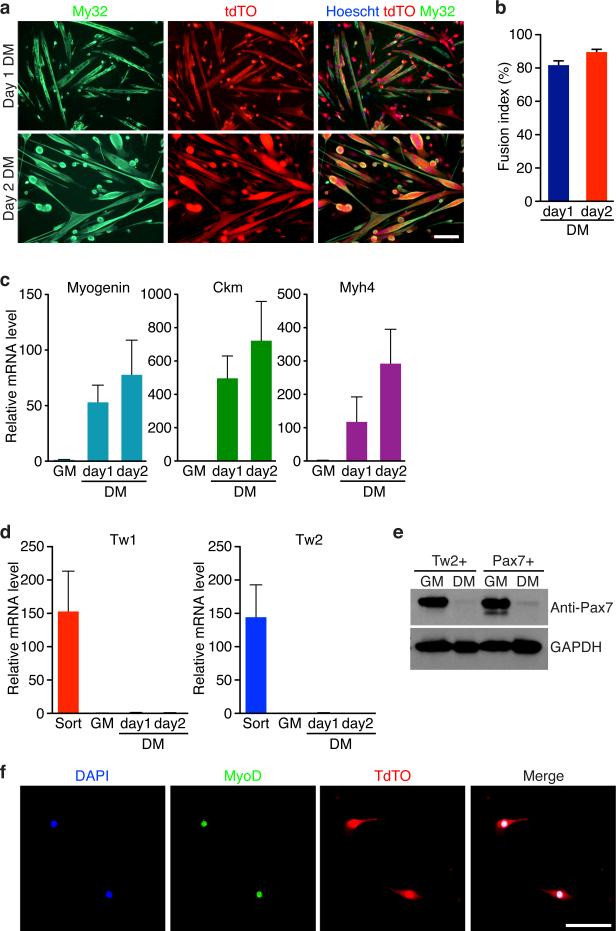
Freshly isolated Tw2+ cells proliferated rapidly in growth medium (GM) and remained tdTO-positive (Supplementary Fig. 6a). When switched to differentiation medium (DM), 81% of Tw2+ cells differentiated into multinucleated myosin-positive myotubes (Fig. 6a,b and Supplementary Fig. 6a). Muscle genes such as Myogenin, Ckm, and Myh4 were up-regulated in Tw2+ cells after 2 days in DM (Fig. 6c). The proliferative activity and efficiency of differentiation of Tw2+ cells are comparable to Pax7+ SCs and exceed those of other previously identified interstitial myogenic progenitors31-33.
Figure 6. Myogenic potential of Tw2+ cells in culture.
(a) Myosin staining (My32, green) revealed the majority of Tw2+ (tdTO+) cells expressed myosin and formed multinucleated myotubes starting at 1 and 2 days in DM. Scale bar: 20 um.
(b) Fusion index was calculated as the percentage of tdTO+ nuclei within multinucleated myotubes compared to the total number of tdTO+ nuclei in the culture. Data are mean ± S.E.M; N =3 independent experiments.
(c) Real-time RT-PCR revealed that muscle genes, such as Myogenin, Ckm and Myh4 are strongly activated when Tw2+ cells were cultured in DM. Values were normalized to those of the GM sample, which is set as 1. Data are mean ± SEM. N =4 independent experiments.
(d) Real-time RT-PCR revealed that Tw1 and Tw2 mRNA was enriched in freshly sorted Tw2+ cells (Sort). However, expression was extinguished once cells were plated in culture. Values were normalized to those of the GM sample, which is set as 1. Data are mean ± SEM. N =4 independent experiments.
(e) Western blot analysis for Pax7 protein in Tw2+ and Pax7+ cells. Pax7 protein is expressed in both Tw2+ and Pax7+ cells in GM, but not in cells cultured for 2 days in DM. GAPDH protein was detected as loading control. Unprocessed original scans of blots are shown in Supplementary Fig. 9.
(f) MyoD staining of Tw2+ (tdTO+) cells in GM revealed MyoD expression. Scale bar: 100 um.
Statistic source data for b,c,d are provided in Supplementary Table 3.
Although Tw1 and Tw2 transcripts were highly expressed in freshly isolated Tw2+ cells, expression was extinguished after maintaining the cells in culture (Fig. 6d and Supplementary Fig. 6b). In contrast, cultured Tw2+ cells quickly became Pax7-positive and MyoD-positive in GM (Fig. 6e,f), Thus, whereas Tw2+ cells do not express Pax7 in their native interstitial location in vivo, they transition through a Pax7+ state en route to differentiation. In addition, the full transcriptome profiles between Tw2+ cells and Pax7+ cells in GM and DM were largely similar (Supplementary Fig. 6c). Furthermore, when co-cultured at equal numbers in GM and exposed to DM, Tw2+ cells (tdTO+) and Pax7+ SCs (GFP+) fused with each other to form chimeric myotubes that were positive for both GFP and tdTO (Supplementary Fig. 6d,e). We conclude that although Tw2+ cells are distinct from Pax7+ SCs, when removed from their native environment and cultured in vitro, Tw2+ cells acquire a Pax7+ cell fate and display similar myogenic potential to Pax7+ cells.
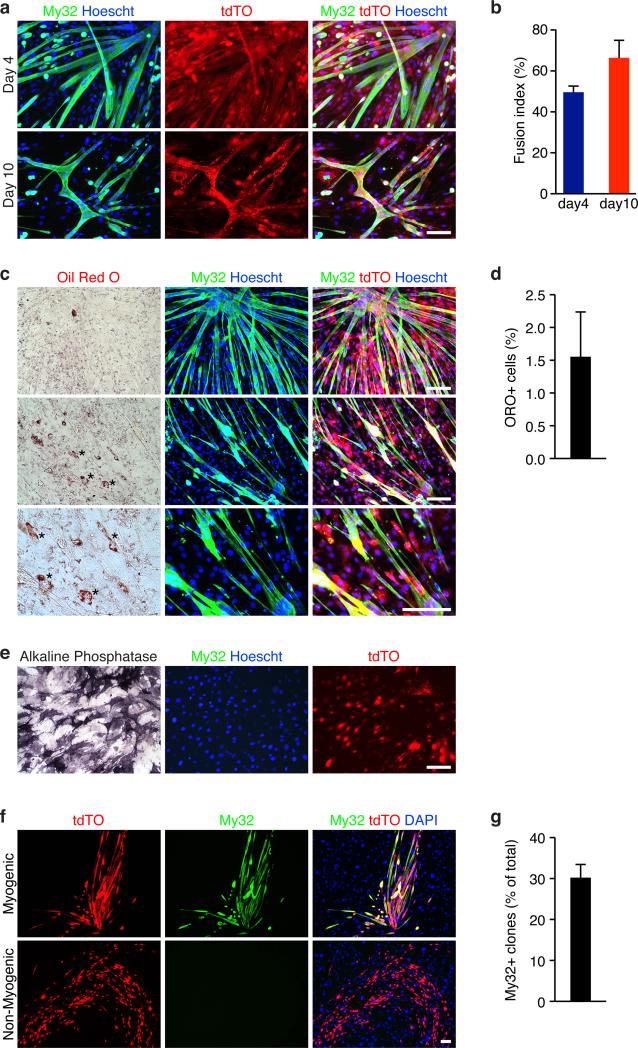
FACS analysis revealed that a small fraction (2.6%) of Tw2+ cells (tdTO+) expressed CD34, a marker commonly used to isolate SCs (Supplementary Table 1). To exclude the possibility that this small fraction of CD34+ cells might be responsible for the myogenic capacity of Tw2+ cells in vitro, we isolated the tdTO+/CD34- population by FACS. Freshly sorted tdTO+/CD34− cells proliferated rapidly within 48 hours post isolation (Supplementary Fig. 7a). After 4 days in DM, 50% of tdTO+/CD34− cells formed multi-nucleated myotubes, and after 10 days in DM, the fusion index increased to 66%, with many myotubes contracting spontaneously (Fig. 7a,b). These results exclude the possibility that the myogenic potential of Tw2+ cells comes from a rare population of tdTO+/CD34+ cells.
Figure 7. Myogenic and osteogenic potential of tdTO+/CD34- cells in vitro.
(a) Myosin staining (My32, green) revealed tdTO+/CD34− cells expressed myosin and formed multinucleated myotubes after 4 and 10 days in DM. Freshly sorted tdTO+/CD34− cells were cultured in GM for 48 hrs before induction of myogenesis. Scale bar: 20 um.
(b) Fusion index was calculated as the percentage of tdTO+ nuclei within multinucleated myotubes compared to the total number of tdTO+ nuclei in the culture. Data are mean ± SEM. N =3 independent experiments.
(c) Oil Red O staining revealed that only very few tdTO+ cells differentiated into adipocytes. Freshly sorted tdTO+/CD34- cells were cultured in GM for 48 hrs before induction of adipogenesis for 10 days. Cells were first stained with My32 and Hoechst before oil Red O staining. Scale bar: 20 um.
(d) Percentage of oil Red O positive cells per field. Data are mean ± SEM. N =3 independent experiments. For each experiment, more than 500 nuclei per field and total of 3 fields were counted and averaged.
(e) Alkaline phosphatase staining revealed tdTO+/CD34- cells can form osteoblasts when exposed to osteogenic medium. These cells did not express myosin. Scale bar: 20 um.
(f) Clonal analysis of tdTO+/CD34 cells for myogenic potential. Five 96-well plates were seeded by FACS. Single tdTO+/CD34 clones were grown on inactivated MEF feeder layers and induced for myogenesis. Myogenic clones were identified by My32 staining. DAPI staining identified both Tw2+ cells (tdTO+/CD34-) and MEFs. Scale bar: 100 um.
(g) Percent of myogenic clones (My32+) of the total surviving clones in the clonal analysis. Data are mean ± SEM. N=5 independent experiments.
Statistic source data for b,d,g are provided in Supplementary Table 3.
To examine whether the tdTO+/CD34− cells can form other cell lineages in addition to muscle cells, we exposed them to different conditions that support the formation of adipocytes and osteoblasts, respectively. After 10 days in adipogenic medium, the majority of tdTO+/CD34− cells formed multinucleated myotubes and only 1.5% of the cells differentiated into adipocytes (positive for Oil Red O staining), among more than 5,000 nuclei analyzed (Fig. 7c,d). Thus tdTO+/CD34− cells are not adipogenic in culture. Under osteogenic conditions, the majority of tdTO+/CD34− cells died within the first 24 hrs and after 10 days in culture, the few surviving cells proliferated and differentiated into osteoblasts, which stained for alkaline phosphatase (Fig 7e). No My32-positive myotubes were observed under osteoblastogenic conditions. Together, these results indicate that the Tw2 cells that did not express CD34 are myogenic and osteogenic in vitro.
To further confirm the myogenic capacity of Tw2+ cells, we performed clonal analysis on tdTO+/CD34− cells. Briefly, single tdTO+/CD34− clones were seeded by FACS sorting onto each well of 96-well plates that were pre-coated with inactivated mouse embryonic fibroblasts (MEFs) as feeder cells. Single clones were grown in GM with bFGF for 1 week before being switched to DM. After 1 week in DM, 29.8% of surviving clones formed My32+ myotubes, while the others did not express My32 and had different morphologies (Fig. 7f,g). These results confirmed the intrinsic myogenic capacity of Tw2+ cells.
Tw2 Blocks Myogenesis in vitro
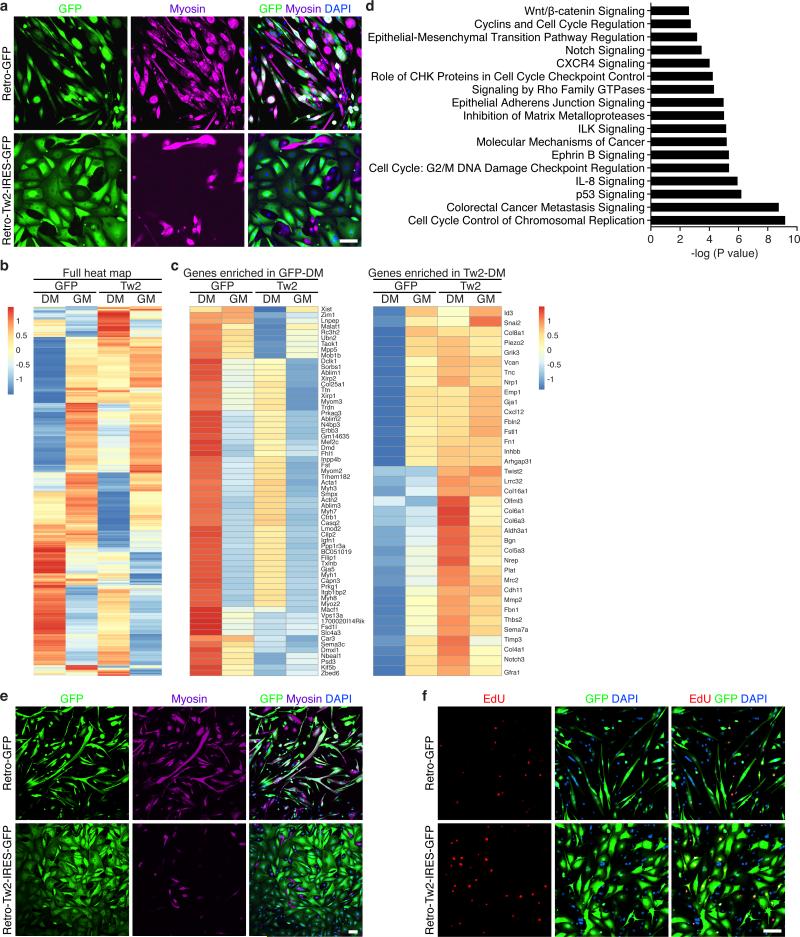
The finding that Tw2+ cells do not express Pax7 or MyoD in vivo, but quickly lose Tw2 gene expression and up-regulate Pax7 and MyoD expression in vitro suggested that Tw2 maintains cells in a progenitor cell state, preventing them from entering the muscle differentiation pathway. To test this hypothesis, we over-expressed Tw2 by retrovirus infection in Tw2+ cells and analyzed myotube formation. While control GFP retrovirus-infected cells formed multinucleated myotubes, Tw2-infected cells in DM underwent a dramatic change in morphology from spindle-shaped myoblasts to more flattened fibroblast-like shapes and did not form myotubes or express myosin (Fig. 8a). The few myosin-positive cells observed in Tw2-infected cultures were not infected by Tw2 retrovirus.
Figure 8. Overexpression of Tw2 blocks myogenesis in vitro.
(a) Tw2+ cells were infected with retroviruses expressing either GFP (retro -GFP) or Tw2 (Retro-Tw2-IRES-GFP) for 24 hours before switching to DM. After 5 days in DM, My32 staining (Purple) was performed to detect myotubes. Scale bar: 100 um.
(b) Heat map of genes expressed in GFP− or Tw2-infected Tw2+ cells in GM and DM identified by RNA-Seq.
(c) Heat maps of top genes enriched and repressed in GFP-DM vs. Tw2-DM.
(d) Pathways of genes enriched in Tw2-DM relative to GFP-DM identified by IPA analysis.
(e) Over-expression of Tw2 inhibits myotube formation of Pax7+ cells. Pax7+ cells were infected with retro-GFP or Retro-Tw2-IRES-GFP for 24 hours before switching to DM. After 5 days in DM, My32 staining (purple) was performed to detect myotubes. Scale bar: 100 um.
(f) EdU labeling revealed increased DNA-synthesis in Tw2-infected Pax7 cells in DM. Infected Pax7 cells were treated with EdU (10 uM) for 24 hours in DM before harvesting and staining. Scale bar: 100 um.
The gene expression profiles in GM were very similar between GFP-infected and Tw2-infected cells (Fig. 8b). However, after 5 days in DM, muscle-specific genes that were up-regulated in GFP-DM cells were not up-regulated in Tw2-DM cells, and conversely, genes strongly up-regulated in Tw2-DM cells were all down-regulated in GFP-DM (Fig. 8c). IPA pathway analysis revealed that genes involved in cell-cycle regulation, cancer metastasis, and EMT were specifically up-regulated in Tw2-DM cells (Fig. 8d). These findings confirm that Tw2-overexpression in Tw2+ cells inhibits myogenesis and induces EMT-like programs. Similarly, over-expression of Tw2 in Pax7+ cells inhibited myotube formation and caused morphological changes in DM (Fig. 8e). EdU labeling showed that Tw2-infected Pax7+ cells continued to proliferate in DM compared to GFP-infected cells, further demonstrating the ability of Tw2 to maintain cells in an undifferentiated state (Fig. 8f). Tw2 thus functions as a repressor of the myogenic program and its down-regulation triggers the onset of myogenesis in Tw2 progenitors (Supplementary Fig. 8).
Discussion
Our results reveal a previously unrecognized population of skeletal muscle progenitor cells marked by expression of Tw2. Tw2+ cells display a unique cell-surface marker expression profile and contribute specifically to type IIb/x fibers. These cells are distinct from Pax7+ SCs and appear to represent a subset of MSCs. In recent years, various non-SC muscle progenitors have been identified, including bone marrow-derived circulating stem cells, pericytes, PW1+ interstitial cells (PICs), muscle side population (SP) cells muscle-derived stem cells and mesoangioblasts 31-37. Tw2+ cells appear to be distinct from these cell lineages because they display a unique cell-surface marker profile, which does not resemble that of any known non-SC progenitors 31, 32, 38, 39. Tw2+ cells share overlapping cell surface marker profiles (e.g., Sca1 and PDGFRa) with muscle-resident fibroadipogenic progenitors (FAPs), but appear functionally distinct with respect to their differentiation potential 38. In addition to their robust myogenic potential, Tw2+ cells can form osteoblasts under appropriate conditions.
SCs contribute to all myofiber types in adult mice5, 11, whereas Tw2+ cells contribute specifically to adult type IIb/x myofibers. This specificity remained even in mice that were pulse-chased for 1.5 years. Remarkably, even when Tw2+ cells were in direct contact with type I myofibers, they remained completely distinct throughout life. Genetic ablation experiments revealed that Tw2+ cells are required for the maintenance of Type IIb/x fiber size during homeostasis without affecting muscle regeneration. In contrast, it has been reported that ablation of Pax7+ SCs completely abolished muscle regeneration, but its effect on myofiber size was minimal during aging 4-6, 40-42. The distinct phenotypes resulting from depleting Pax7+ versus Tw2+ cells suggested that unlike Pax7+ cells, which are essential for regeneration, Tw2+ cells play more important roles in normal muscle growth. Our results demonstrating that Tw2+ cells also contribute to muscle regeneration might seem contradictory to the absence of regeneration in Pax7-depleted muscle. An interpretation of these results is that although Tw2+ cells can initiate de novo myogenesis in vivo, the majority of them likely fuse with Pax7+ cells to form IIb/x myofibers. In addition, ablation of Tw2+ cells does not affect muscle regeneration, suggesting that Tw2+ cells are not a reservoir for rapid repair. The Tw2+ cells appear important for skeletal muscle maintenance and are insufficient to mount a rapid and comprehensive response to an acute injury.
Our results show that within their native interstitial locations in adult skeletal muscle, Tw2+ cells are distinct from SCs. However, following isolation they rapidly down-regulate Tw1 and Tw2 expression and activate Pax7 and MyoD expression to enter the myogenic pathway. Expression of Tw2 maintains these progenitors in a stem-like state, and the rapid down-regulation of Tw2 in isolated cells allows the transition to a Pax7+ state and acquisition of a muscle phenotype. Furthermore, forced expression of Tw2 in either Tw2+ or Pax7+ progenitors in culture prevented differentiation and maintained the cells in a proliferative state characterized by expression of genes involved in cell signaling, EMT and oncogenesis. Interestingly, Twist proteins can maintain an immature “stem-like” state and repress terminal differentiation of human MSCs and cancer cells43-46. Such functions would be consistent with the role of Twist that we uncovered in skeletal muscle progenitors.
It is curious that Tw2+ cells are specifically excluded from the tongue musculature. Tongue and laryngeal muscles are derived from occipital somites, located at the boundary of the trunk and head, whereas limb, trunk and diaphragm muscles are derived from trunk somites47. The intrinsic difference in initiation of the myogenic program between tongue and limb muscles could offer a clue into why Tw2+ cells are excluded from the tongue musculature. It will be interesting to investigate the potential contributions of Tw2+ cells in different muscle lineages during development.
Tw2+ progenitors are, to our knowledge, the first example of a fiber type specific myogenic progenitor population. In this regard, type IIb fibers are the most abundant fibers and are most susceptible to injury and disease in mice48.. Therefore, it is essential to maintain the size and integrity of type IIb fibers during aging, and the contribution of Tw2+ cells to type IIb fibers may represent such a mechanism. Humans do not have type IIb fibers in skeletal muscle; instead the predominant fiber type is equivalent to mouse IIx/d fibers 49. It will be interesting to study whether Tw2+ cells exist in human skeletal muscle and whether they contribute to specific fiber types.
Supplementary Material
Acknowledgements
We are grateful to UT Southwestern Flow Cytometry Core Facility, the Moody Foundation Flow Cytometry Facility, and UTSW Genomics and Microarray Core Facility for technical help and service. We thank Cheryl Nolen for technical assistance and Drs. Danel Garry, Michael Rudnicki, Margaret Buckingham and Hesham Sadek for constructive suggestions. This work was supported by grants from the NIH (HL-077439, AR-067294, HL-130253, DK-099653 and U01-HL-100401) and the Robert A. Welch Foundation (grant 1-0025 to E.N.O.). N.L. was supported by a Beginning-Grant-In-Aid (13BGIA17150004) from American Heart Association. G.G. is a research Fellow supported by Sarnoff Cardiovascular Research Foundation.
Footnotes
Author Contributions
Experiments were designed by N.L, G.A.G, and S.L. Experiments were performed by N.L, G.A.G, S.L, S.B, E.S, J.M.S, and P.J. Data were interpreted by N.L, G.A.G, S.L, B.C, R.B.D, and E.N.O. The paper was written by N.L and E.N.O.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Chang NC, Rudnicki MA. Satellite cells: the architects of skeletal muscle. Current topics in developmental biology. 2014;107:161–181. doi: 10.1016/B978-0-12-416022-4.00006-8. [DOI] [PubMed] [Google Scholar]
- 2.Brack AS, Rando TA. Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell stem cell. 2012;10:504–514. doi: 10.1016/j.stem.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fry CS, et al. Inducible depletion of satellite cells in adult, sedentary mice impairs muscle regenerative capacity without affecting sarcopenia. Nature medicine. 2015;21:76–80. doi: 10.1038/nm.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keefe AC, et al. Muscle stem cells contribute to myofibres in sedentary adult mice. Nature communications. 2015;6:7087. doi: 10.1038/ncomms8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy JJ, et al. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 2011;138:3657–3666. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiological reviews. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Pessin JE. Mechanisms for fiber-type specificity of skeletal muscle atrophy. Curr Opin Clin Nutr Metab Care. 2013;16:243–250. doi: 10.1097/MCO.0b013e328360272d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tonkin J, Villarroya F, Puri PL, Vinciguerra M. SIRT1 signaling as potential modulator of skeletal muscle diseases. Curr Opin Pharmacol. 2012;12:372–376. doi: 10.1016/j.coph.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Arany Z. PGC-1 coactivators and skeletal muscle adaptations in health and disease. Curr Opin Genet Dev. 2008;18:426–434. doi: 10.1016/j.gde.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pawlikowski B, Pulliam C, Betta ND, Kardon G, Olwin BB. Pervasive satellite cell contribution to uninjured adult muscle fibers. Skelet Muscle. 2015;5:42. doi: 10.1186/s13395-015-0067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes RM, Firulli AB. A twist of insight - the role of Twist-family bHLH factors in development. The International journal of developmental biology. 2009;53:909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baylies MK, Bate M. twist: a myogenic switch in Drosophila. Science. 1996;272:1481–1484. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]
- 14.Cripps RM, et al. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes & development. 1998;12:422–434. doi: 10.1101/gad.12.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bate M, Rushton E, Currie DA. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development. 1991;113:79–89. doi: 10.1242/dev.113.1.79. [DOI] [PubMed] [Google Scholar]
- 16.Cripps RM, Olson EN. Twist is required for muscle template splitting during adult Drosophila myogenesis. Developmental biology. 1998;203:106–115. doi: 10.1006/dbio.1998.9040. [DOI] [PubMed] [Google Scholar]
- 17.Currie DA, Bate M. The development of adult abdominal muscles in Drosophila: myoblasts express twist and are associated with nerves. Development. 1991;113:91–102. doi: 10.1242/dev.113.1.91. [DOI] [PubMed] [Google Scholar]
- 18.Fuchtbauer EM. Expression of M-twist during postimplantation development of the mouse. Developmental dynamics : an official publication of the American Association of Anatomists. 1995;204:316–322. doi: 10.1002/aja.1002040309. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Cserjesi P, Olson EN. Dermo-1: a novel twist-related bHLH protein expressed in the developing dermis. Developmental biology. 1995;172:280–292. doi: 10.1006/dbio.1995.0023. [DOI] [PubMed] [Google Scholar]
- 20.Rohwedel J, Horak V, Hebrok M, Fuchtbauer EM, Wobus AM. M-twist expression inhibits mouse embryonic stem cell-derived myogenic differentiation in vitro. Experimental cell research. 1995;220:92–100. doi: 10.1006/excr.1995.1295. [DOI] [PubMed] [Google Scholar]
- 21.Hebrok M, Wertz K, Fuchtbauer EM. M-twist is an inhibitor of muscle differentiation. Developmental biology. 1994;165:537–544. doi: 10.1006/dbio.1994.1273. [DOI] [PubMed] [Google Scholar]
- 22.Spicer DB, Rhee J, Cheung WL, Lassar AB. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- 23.Hjiantoniou E, et al. Twist induces reversal of myotube formation. Differentiation; research in biological diversity. 2008;76:182–192. doi: 10.1111/j.1432-0436.2007.00195.x. [DOI] [PubMed] [Google Scholar]
- 24.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sosic D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 26.Yu K, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 27.Voehringer D, Liang HE, Locksley RM. Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J Immunol. 2008;180:4742–4753. doi: 10.4049/jimmunol.180.7.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goebel HH. Desmin-related neuromuscular disorders. Muscle & nerve. 1995;18:1306–1320. doi: 10.1002/mus.880181114. [DOI] [PubMed] [Google Scholar]
- 29.Helliwell TR. Lectin binding and desmin staining during bupivicaine-induced necrosis and regeneration in rat skeletal muscle. The Journal of pathology. 1988;155:317–326. doi: 10.1002/path.1711550407. [DOI] [PubMed] [Google Scholar]
- 30.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell KJ, et al. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nature cell biology. 2010;12:257–266. doi: 10.1038/ncb2025. [DOI] [PubMed] [Google Scholar]
- 32.Dellavalle A, et al. Pericytes resident in postnatal skeletal muscle differentiate into muscle fibres and generate satellite cells. Nature communications. 2011;2:499. doi: 10.1038/ncomms1508. [DOI] [PubMed] [Google Scholar]
- 33.Doyle MJ, et al. Abcg2 labels multiple cell types in skeletal muscle and participates in muscle regeneration. The Journal of cell biology. 2011;195:147–163. doi: 10.1083/jcb.201103159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaBarge MA, Blau HM. Biological progression from adult bone marrow to mononucleate muscle stem cell to multinucleate muscle fiber in response to injury. Cell. 2002;111:589–601. doi: 10.1016/s0092-8674(02)01078-4. [DOI] [PubMed] [Google Scholar]
- 35.Minasi MG, et al. The meso-angioblast: a multipotent, self-renewing cell that originates from the dorsal aorta and differentiates into most mesodermal tissues. Development. 2002;129:2773–2783. doi: 10.1242/dev.129.11.2773. [DOI] [PubMed] [Google Scholar]
- 36.Qu-Petersen Z, et al. Identification of a novel population of muscle stem cells in mice: potential for muscle regeneration. The Journal of cell biology. 2002;157:851–864. doi: 10.1083/jcb.200108150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dellavalle A, et al. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nature cell biology. 2007;9:255–267. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- 38.Joe AW, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nature cell biology. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. The Journal of cell biology. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambasivan R, et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 43.Beck B, et al. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell stem cell. 2015;16:67–79. doi: 10.1016/j.stem.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Isenmann S, et al. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem cells. 2009;27:2457–2468. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt JM, et al. Stem-cell-like properties and epithelial plasticity arise as stable traits after transient Twist1 activation. Cell reports. 2015;10:131–139. doi: 10.1016/j.celrep.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 47.Michailovici I, Eigler T, Tzahor E. Craniofacial Muscle Development. Current topics in developmental biology. 2015;115:3–30. doi: 10.1016/bs.ctdb.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 48.Webster C, Silberstein L, Hays AP, Blau HM. Fast muscle fibers are preferentially affected in Duchenne muscular dystrophy. Cell. 1988;52:503–513. doi: 10.1016/0092-8674(88)90463-1. [DOI] [PubMed] [Google Scholar]
- 49.Scott W, Stevens J, Binder-Macleod SA. Human skeletal muscle fiber type classifications. Phys Ther. 2001;81:1810–1816. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.