Malignant populations constitute a mixture of multiple genetically distinct clones, which can often be dramatically altered with therapy[1]. Clonal evolution was recently shown to be frequent in chronic lymphocytic leukemia (CLL) by sequential whole-exome sequencing (WES) of matched sequential samples taken at treatment-initiation and first relapse following chemo(immuno)therapy from 59 individuals[2]. This finding raises important questions: Is evolution occurring as frequently during the preceding ‘watch and wait’ period, or is it primarily driven by therapeutic intervention? Are there clinical features associated with higher rates of evolution? Do the genetic variations that emerge over the course of evolution alter the clinical outcome?
To address these questions, we analysed serial samples from 103 individuals treated within the CLL8 trial, in which patients uniformly received fludarabine and cyclophosphamide as first-line treatment with/without additional rituximab (FC/FCR). Samples taken long before treatment-initiation, at treatment-initiation, and at relapse/progressive disease (PD) were included. Genomic analysis was performed by single nucleotide polymorphism (SNP) arrays thereby focussing on the evolution of somatic copy number alterations (CNAs). Comparing sequential samples prior to therapy (N=27), CNA evolution occurred in 19% of cases. In contrast, when comparing treatment-initiation and relapse genomic profiles (N=86), CNA evolution was seen twice as frequently. This suggested an association of FC(R) treatment with higher rate of CNA evolution. The only clinical feature significantly associated with CNA evolution was FCR therapy compared with FC (OR=2.806, p=0.024). As this might be related to narrower evolutionary bottlenecks imposed by the more effective FCR therapy, which could execute more selection pressure, we examined matched minimal residual disease (MRD) data and found CNA evolution more frequently in cases with CLL cell numbers rapidly receding in early phases of treatment. Finally, we observed frequent rises of TP53 mutant/deficient clones with therapy (in N=19 cases, 22%) that were associated with decreased overall survival (p=0.016).
Selection of serial samples from the CLL8 trial (German CLL Study Group, ClinicalTrials.gov number NCT00281918) was guided by availability: 216 samples were taken from 103 individuals. All treatment-initiation samples were tumor-cell enriched via CD19 MicroBeads (Miltenyi®), whereas pre-treatment and the majority of relapse samples were comprised of peripheral blood mononuclear cells (Sample characteristics in Supplementary Table S1).
Analysis for CNAs including copy-neutral losses of heterozygosities (CN-LOHs) as allelic CNAs was done by using 6.0 SNP-arrays (Affymetrix®; GEO-accession-number: GSE83566). The threshold for CNA detection was 10 to 15% affected cells for monoallelic lesions. Screening for TP53 and ATM mutations was performed by WES (N=108, median depths 96x) or by amplicon based targeted next generation sequencing (N=150, median depths 1332x; N=42 samples analyzed on both platforms). Details are provided in Supplemental Methods.
Statistical analyses were done in SPSS v22 using Fisher’s exact and Mann-Whitney U tests. Associations of CNA evolution with distinct variables were evaluated by binary logistic regression models and odds ratios (OR) are provided to describe the particular risks for CNA evolution. MRD levels are reported as fraction of CLL cells of all nucleated cells. MRD negativity was defined as a fraction <10−4. Progression-free survival (PFS) and overall survival (OS) were estimated by Kaplan-Meier methodology and survival curves were compared via logrank test. All tests were two-sided.
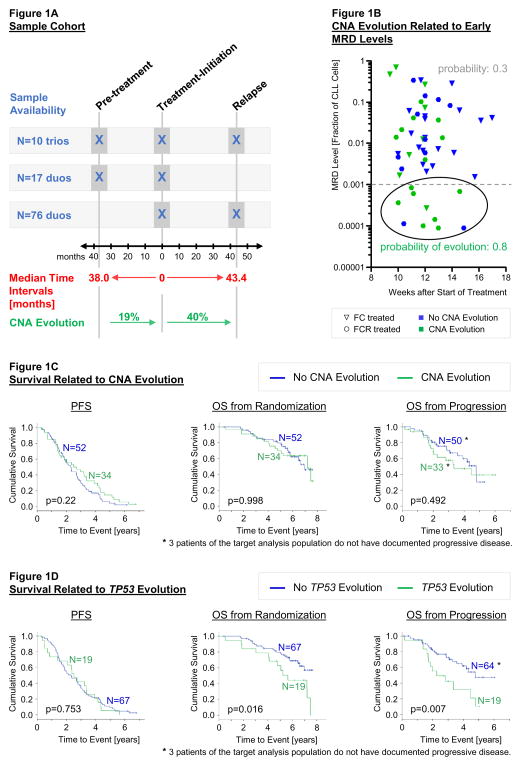
The available samples allowed 27 comparisons of matched pairs going back in time from treatment-initiation (a median of 3.2 years apart) and 86 comparisons going forward to relapse/PD (a median of 3.6 years apart). Among those, ten individuals had three serial samples each. These sample trios enabled an intra-individual comparison of pre- and post-treatment CNA evolution (Figure 1A).
Figure 1.
1A: Description of sample cohort
Outlined is the distribution of available sequential samples over the three different time-points. The median time interval in-between sample collections was 38.0 months for the pre-treatment to treatment-initiation comparisons [range: 6 – 126 months] and 43.4 months for the treatment-initiation to relapse comparisons [range: 5 – 91 months]. Time intervals between the paired cases before and after treatment showed no statistically significant difference (p=0.473, Mann-Whitney U test calculated for 17 pre-treatment versus 86 post-treatment sample duos). The incidence of CNA evolution was increased after interval treatment (p=0.103, Fisher’s exact test calculated for 17 pre-treatment versus 86 post-treatment sample duos).
1B: CNA evolution related to early MRD levels.
Displayed are MRD levels measured by 4-color flow cytometry at interim response assessment (after 3 cycles of therapy) plotted against the time-point of measurement. The probability of CNA evolution was higher for cases with an MRD level <0.001 at this stage of treatment as calculated by binary logistic regression (p = 0.008).
1C: Survival related to CNA evolution.
Kaplan-Meier estimates for progression free survival (PFS) and overall survival (OS); according to the presence of CNA evolution. P-values are based on log-rank tests.
1D: Survival related to TP53 evolution.
Kaplan-Meier estimates for PFS and OS; according to the presence of TP53 mutant/deficient evolution. P-values are based on log-rank tests.
At treatment-initiation the cohort consisted of 37% of subjects with del(11q), 8% with trisomy 12, 59% with del(13q), 7% with del(17p), and 21% with normal karyotype, consistent with a more adverse risk profile of patients. In line with this, we note a relatively short median time from diagnosis to treatment-initiation (23.9 months) and from treatment-initiation to relapse (28.1 months) as well as a relatively low rate of complete remissions in our cohort compared with the entire CLL8 cohort (13% among FC-treated patients and 25% among FCR-treated patients compared to 22% and 44% previously reported in the entire CLL8 trial cohort[3]).
CNA evolution was defined as the appearance of at least one newly detected CNA after the considered time period. We first sought to determine the rate of CNA evolution during the ‘watch and wait’ period and found that in the 27 comparisons of pre-treatment and treatment-initiation samples, CNA evolution was infrequently observed (N=5; 19% of cases). In 3 of the 5 treatment-initiation samples with CNA evolution, the newly detected CNAs had log2-ratios that were only slightly above the detection limit. Three of the 5 cases with CNA evolution carried an ATM-deletion/mutation in their pre-treatment sample, whereas all 5 cases with TP53-alteration had no CNA evolution at treatment-initiation. The latter is in line with previously published data[4].
In the 86 comparisons of treatment-initiation and relapse samples, CNA evolution was observed twice as frequently (N=34, 40% of cases). We note that the time intervals between the matched pairs before and after treatment were not significantly different (p=0.473, calculated for 17 pre-treatment versus 86 post-treatment sample duos).
Though the formal comparison of frequencies for CNA evolution before and after treatment did not reach statistical significance (p=0.103, calculated for 17 pre-treatment versus 86 post-treatment sample duos), our results support the hypothesis that FC-based therapy accelerates clonal evolution as has been previously suggested[5]. The 10 cases with sample trios showed CNA evolution in 2 cases before treatment and in 5 cases after treatment.
CNA evolution consisted of many known recurrent CLL CNAs: del(17)(p13) [N=10], del(11)(q22.1–23.2) [N=9], del(13)(q14.3) [N=9], and +(2)(p16.1-p15) [N=4]. To distinguish between the CNA profiles at treatment-initiation and at relapse, we applied the genomic identification of significant targets in cancer (GISTIC) algorithm[6], which showed an increase in significance for 17p loss and new 8q gain including the c-MYC locus at 75% confidence (Supplementary Figure S1). The emergences of clones harbouring these lesions may reflect selective resistance to therapy afforded by the lesions[7,8,9] (SNP-array results in Supplementary Tables S2, S3). Two cases acquired losses in 9p21.3 and the minimally deleted region contained only the CDKN2A/CDKN2B gene loci (Supplementary Figure S2). Notably, this deletion was never identified at treatment-initiation[9], and was previously linked to disease progression and transformation into high-grade lymphoma.[10,11]
We next evaluated clinical parameters that may impact the likelihood of CNA evolution after therapy. Among all variables tested (Table 1), the only statistically significant predictor was the treatment type, with CNA evolution more frequently observed with FCR than FC (OR=2.806, 95% confidence interval (CI)=1.149–6.851, p=0.024). Both treatment groups had comparable risk profiles (Supplementary Table S4). The higher incidence of CNA evolution seen with FCR treatment may result from relatively focused selection pressure imposed specifically by rituximab exposure or from broader overall selection pressure by the more selective FCR treatment (compared to FC) leading to a narrower evolutionary bottleneck.
Table 1.
Odds ratios and probabilities for CNA evolution (CE) based on clinical parameters as calculated by binary logistic regression.
| Odds Ratio | p-Value | 95% Confidence Interval | Probability for CE if Characteristic present | Probability for CE if Characteristic absent | |
|---|---|---|---|---|---|
| IgHV | |||||
| IgHV unmutated (N = 63/84) | 1.142 | 0.797 | 0.414–3.146 | 0.413 | 0.381 |
| High-risk Genomic Abnormalities at Trial Enrollment | |||||
| TP53 Loss and/or Mutation (N = 14/86) | 1.667 | 0.384 | 0.527–5.270 | 0.500 | 0.375 |
| ATM Loss and/or Mutation (N = 34/86) | 1.12 | 0.801 | 0.463–2.707 | 0.412 | 0.385 |
| Treatment Arm | |||||
| FCR (N = 40/86) | 2.806 | 0.024 | 1.149–6.851 | 0.525 | 0.283 |
| Response | |||||
| Complete Response (N = 16/84) | 1.720 | 0.333 | 0.574–5.152 | 0.500 | 0.368 |
| Non-response (N = 10/84) | 0.629 | 0.525 | 0.150–2.626 | 0.300 | 0.405 |
To distinguish between these two possible explanations, we examined matched minimal residual disease data[12] (Supplementary Table S5). The frequency of CNA evolution was not significantly higher in patients achieving MRD negativity compared with those remaining MRD positive after therapy (46% versus 34%; p=0.436). However, early MRD measurements (after 3 cycles of FC/FCR) might provide an opportunity to identify patients whose tumors would encounter the narrowest bottleneck. Indeed, CNA evolution was more frequently observed in cases with low MRD levels at this time-point (in 8/10 cases when using 0.001 as cut-off, Figure 1B). With the caveat that almost only FCR-treated patients reached these low MRD levels during early treatment phases (9/10 cases), this observation is more likely supporting the hypothesis that clonal evolution is enhanced by a strong overall selection pressure imposed by chemo(immuno)therapy.
CNAs detected at treatment-initiation were rarely observed to recede below the detection limit at relapse (16 CNAs in 12/86 samples). Decline or loss of del(13q) [N=6] and del(11q) [N=4] in favour of a different, likely fitter del(13q)/del(11q) clone was commonly seen. Sample “CLL075_post treatment” lost evidence for chromothripsis [>10 switches between two copy-number states within 50 Mb] supporting the hypothesis that chromothripsis – though associated with inferior OS and PFS[9] – is not necessarily associated with relapsed/refractory tumor status.[13, 14].
Presence of CNA evolution itself was not associated with survival differences (Figure 1C, Supplementary Figure S3 for separate analyses of FC/FCR treated patients).
Built on our finding that TP53 deletion was specifically enriched at relapse, we examined the impact of TP53 driven evolution on outcome. Incidences for TP53 deletion as well as TP53 mutation were twice as high at relapse than at treatment-initiation (TP53 deletion: increase from 8% to 20%; TP53 mutation: from 15% to 31%). Emergence of clones harboring TP53 deletions (in 10/86 cases) and/or TP53 mutations (in 6/86 cases, Supplementary Table S6) was associated with a significantly shorter OS (Figure 1D) indicating that selection for resistant clones by therapeutic selective pressure is of important prognostic significance.
Collectively, these results suggest that clonal evolution is accelerated by therapy, and might be even more so by chemo(immuno)therapy capable of inducing profound remissions. TP53 disrupted clones emerge frequently upon relapse and carry a strong prognostic significance.
Supplementary Material
Acknowledgments
Supported by Deutsche Forschungsgemeinschaft (ED 256/1-1, SFB 1074/B2 and Z1), Else Kröner-Fresenius-Stiftung (2012_A146), Virtual Helmholtz Institute (VH-VI-404, TP2), BMBF (CancerEpiSys, PRECISE), and F. Hoffmann-La Roche (genetic reference diagnostics of CLL8: CD19 enrichment, routine FISH analysis and IGHV-status).
The authors thank the patients who participated in the CLL8 trial; the investigators who treated patients and submitted samples; F. Hoffmann-La Roche (Basel, Switzerland) for support; and Sabrina Kless, Christina Galler, Sabrina Rau, Karin Lanz and Andreas Streicher for excellent assistance and support.
Footnotes
Disclosure of Conflicts of Interest:
The authors declare no competing financial interests.
Authorship Contributions:
J.E., E.T., D.A.L., S.B., C.J.W., H.D., and S.S. designed the research. J.E., E.T., and D.A.L. performed experiments. J.E., E.T., D.A.L., S.R., J.B., K.F., A.F., J.B., K.H., S.B., L.W., M.K., C.J.W., M.H., H.D., and S.S. collected and analyzed data. J.E., E.T., D.A.L., S.R., J.B., K.F., A.F., J.B., K.H., S.B., L.W., J.G.G., D.S.N., C.J.W., H.D. and S.S. interpreted the data. J.E., D.A.L., S.R., J.B., J.B., K.H., S.B., J.G.G., D.S.N., C.J.W., H.D. and S.S. contributed to write the manuscript.
References
- 1.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hallek M, Fischer K, Fingerle-Rowson G, Fink AM, Busch R, Mayer J, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010;376:1164–1174. doi: 10.1016/S0140-6736(10)61381-5. [DOI] [PubMed] [Google Scholar]
- 4.Ouillette P, Saiya-Cork K, Seymour E, Li C, Shedden K, Malek SN. Clonal evolution, genomic drivers, and effects of therapy in chronic lymphocytic leukemia. Clin Cancer Res. 2013;19:2893–2904. doi: 10.1158/1078-0432.CCR-13-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 8.Zenz T, Eichhorst B, Busch R, Denzel T, Häbe S, Winkler D, et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28:4473–4479. doi: 10.1200/JCO.2009.27.8762. [DOI] [PubMed] [Google Scholar]
- 9.Edelmann J, Holzmann K, Miller F, Winkler D, Bühler A, Zenz T, et al. High-resolution genomic profiling of chronic lymphocytic leukemia reveals new recurrent genomic alterations. Blood. 2012;120:4783–4794. doi: 10.1182/blood-2012-04-423517. [DOI] [PubMed] [Google Scholar]
- 10.Chigrinova E, Rinaldi A, Kwee I, Rossi D, Rancoita PM, Strefford JC, et al. Two main genetic pathways lead to the transformation of chronic lymphocytic leukemia to Richter syndrome. Blood. 2013;122:2673–2682. doi: 10.1182/blood-2013-03-489518. [DOI] [PubMed] [Google Scholar]
- 11.Fabbri G, Khiabanian H, Holmes AB, Wang J, Messina M, Mullighan CG, et al. Genetic lesions associated with chronic lymphocytic leukemia transformation to Richter syndrome. J Exp Med. 2013;210:2273–2288. doi: 10.1084/jem.20131448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Böttcher S, Ritgen M, Fischer K, Stilgenbauer S, Busch RM, Fingerle-Rowson G, et al. Minimal residual disease quantification is an independent predictor of progression-free and overall survival in chronic lymphocytic leukemia: a multivariate analysis from the randomized GCLLSG CLL8 trial. J Clin Oncol. 2012;30:980–988. doi: 10.1200/JCO.2011.36.9348. [DOI] [PubMed] [Google Scholar]
- 13.Bassaganyas L, Beà S, Escaramís G, Tornador C, Salaverria I, Zapata L, et al. Sporadic and reversible chromothripsis in chronic lymphocytic leukemia revealed by longitudinal genomic analysis. Leukemia. 2015;29:758. doi: 10.1038/leu.2014.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst A, Jones DT, Maass KK, Rode A, Deeg KI, Jebaraj BM, et al. Telomere dysfunction and chromothripsis. Int J Cancer. 2016;138:2905–2914. doi: 10.1002/ijc.30033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.