Abstract
Background
Radiogenetic therapy is a novel approach in the treatment of cancer, which employs genetic modification to alter the sensitivity of tumor cells to the effect of applied radiation.
Aim
To select a potent radiation inducible promoter in the context of brain tumors and to investigate if CArG radio responsive motifs or other elements in the promoter nucleotide sequences can correlate to its response to radiation.
Methods
To select initial candidates for promoter inducible elements, the levels of mRNA expression of six different promoters were assessed using Quantitative RTPCR in D54 MG cells before and after radiation exposure. Recombinant Ad/reporter genes driven by five different promoters; CMV, VEGF, FLT-1, DR5 and survivin were constructed. Glioma cell lines were infected with different multiplicity of infection of the (promoter) Ad or CMV Ad. Cells were then exposed to a range of radiation (0–12 Gy) at single fraction. Fluorescent microscopy, Luc assay and X-gal staining was used to detect the level of expression of related genes. Different glioma cell lines and normal astrocytes were infected with Ad survivin and exposed to radiation. The promoters were analyzed for presence of CArG radio-responsive motifs and CCAAT box consensus using NCBI blast bioinformatics software.
Results
Radiotherapy increases the expression of gene expression by 1.25–2.5 fold in different promoters other than survivin after 2 h of radiation. RNA analysis was done and has shown an increase in copy number of tenfold for survivin. Most importantly cells treated with RT and Ad Luc driven by survivin promoter showed a fivefold increase in expression after 2 Gy of radiation in comparison to non-irradiated cells. Presence or absence of CArG motifs did not correlate with promoter response to radiation. Survivin with the best response to radiation had the lowest number of CCAAT box.
Conclusion
Survivin is a selective potent radiation inducible promoter for glioblastoma viral gene therapy and this response to radiation could be independent of CArG motifs.
Keywords: Survivin, Promoter, Gene therapy, CArG motifs, Glioblastoma multiforme, Transcription regulation
Background
Brain tumor gliomas are among the most aggressive of human malignancies. Patients with histopathologic subtype, glioblastoma multiform (GBM) have the worst prognosis; despite aggressive surgery, radiation, and chemotherapy [1, 2]. Radiation remains an integral part of the conventional treatment of brain malignancies [3, 4]. However, many gliomas are resistant to radiotherapy [5]. Additionally higher doses of radiotherapy are intolerable to normal brain tissue bringing with it complications that can lead to the deterioration of a patient’s general health. Therefore it is crucial to employ multidisciplinary approaches to overcome such obstacles and offer new solutions on a molecular level.
In this regard, the molecular and genetic basis underlying pathogenesis and treatment resistance of these tumors is under active investigation and is becoming better understood [6]. An important mediator of both tumorigenesis and resistance to treatment involves inhibition of apoptosis [7]. Recently, survivin has been characterized as an important member of the inhibitor of apoptosis family, with a very complex biology that has not been fully understood yet [8]. Survivin expression has been found to be undetectable in normal adult tissues [9]. However, it has been found to be abundantly expressed in a wide variety of human malignancies, including brain tumors [9].
Cancer radiogenetic therapy is a new approach that utilizes a multifunctional platform for tumor imaging, targeting, and gene delivery [10–13]. It employs using genetic modification to alter the sensitivity of malignant or normal tissue to the effect of radiation [14–17]. In this approach many vectors capable of delivering any payload to the tumor cells are designed. Recently, after the success of oncolytic virus in clinical trials and the recent FDA approval for its use in the treatment of melanoma locally [18], it is concluded that viral vectors exhibit great advantages due to their natural capability of efficient cell attachment, entry and high level of transgene expression as part of the viral replication cycle [19–22]. Human Ad-based vectors are now considered a major tool for gene therapy with more than 100 various adenoviral vectors developed for glioma targeting. Additionally, advances in viral based therapy have led to the use of condition replicative adenovirus controlling the replication of the virus mainly through transcription regulation by using tumor specific radioinducible promoters [23, 24].
One of the radiation-inducible promoters described is the RecA promoter that was used to increase tumor necrosis factor-α (TNF-α) production in Clostridium sp. [25]. The Egr-1 promoter has also been used as a radioinducible promoter to deliver TNF-α to tumor cells [26–28]. Also, it was studied in the context of radioprotective effect of FLT-3 in severe combined immunodeficient mice [27] for in vitro studies on gene activation [29] and for gene expression in the context of hypoxia inducible promoters [30]. One of the proposed mechanisms of the radiation mediated transcription regulation is the presence of CArG box in the nucleotide sequence of different promoters regions including radiosensitive EGR-1 [31]. Unfortunately, these genes are neither up-regulated in gliomas nor specifically expressed in tumor cells. Nowadays, advances in bioinformatics provide a powerful tool for elucidating the functional features of genes or their promoters, and also prediction tools to identify specific elements within promoters sequence directly with no detectable sequence similarity [32].
To that end, In this study we tried a combined technique of radiation plus transcription regulation to prove the principle of regulating gene expression. The aim of this study is to investigate and select the best radiation inducible promoter in the context of viral brain tumors therapy. We also aim to investigate, using bioinformatics, if the presence of CArG radio responsive motifs or other elements in a promoter’s nucleotide sequences is related to our selection.
Methods
Cell culture
The human glioblastoma cell lines D54 MG, U251 MG and human astrocytes (from Dr. Yancey Gillespie, university of Alabama at Birmingham, Birmingham, AL) were maintained in Dulbecco’s modified Eagle’s medium/F12, supplemented with 10% fetal calf serum, l-glutamine (200 μg/mL), 100 U/mL penicillin and 100 μg/mL streptomycin, at 37 °C in a 100% humidified 5% CO2 atmosphere.
Initial screening for mRNA copies in response to radiation
Six different human promoters (FLT-1, VEGF, Cox2, INOS, DR5 and survivin) were assessed for expression of mRNA in D54 MG cells 2 h after exposure to 2 Gy radiation using quantitative RT-PCR. Radiation provided using a 60Co therapy unit (Picker, Cleveland, OH).
Construction of adenovirus with proposed radiation inducible promoter
We constructed recombinant CMV Ad and Ad/Luc (encoding the luciferase reporter gene, a kind gift of Robert D. Gerard university of Leuven, Belgium) Ad/GFP (encoding the reporter green fluorescent protein) or Ad/LacZ (encoding the Escherichia coli b galactosidase gene, provided by De-chu Tang, university of Alabama at Birmingham, Birmingham, AL) driven by different promoters; 0.51 kb of CMV [33], 0.26 kb of survivin [34], 2.6 kb of VEGF promoter region [35], 1.2 kb of DR5 [36], flt-1 as described before [37]. Reporter genes replaced the E1A region in these vectors, under control of human promoters [36, 38]. These replication-deficient adenoviral vectors were constructed based on homologous recombination between pCMV, pVEGF, pDR5, FLt-1 promoter, or pSurvivin shuttle vectors and pVK500 adenoviral backbone that contain the entire adenoviral genome with E1A region deleted from it. Viruses were propagated in 293 cells, purified by centrifugation in CsCl gradients, and plaque titered in 293 cells following standard protocols [39].
Validation of radiation inducible adenovirus vector and selection of potent radiation inducible promoter in context of adenoviral vector
D54 MG cells were plated per well in 6-well plates at 2 × 105. Then, cells were infected with different multiplicity of infection (MOI) of different (promoter) Ad or CMV Ad then cells were exposed to a range of radiation (0–12 Gy) at single fraction. Control cells were left without radiation. Fluorescent microscopy, luciferase assay, and X-gal staining of the corresponding genes were used to detect the level of expression of related genes to control cells and radiation treated cells after 2 h of radiation and 24 h of radiation to select a potent radiation inducible promoter in context of adenoviral vector.
Reporter gene assays
X-gal assay
Cells were seeded in 24-well plates in quadruplets at a density of 5 × 104 cells/well. The following day, the cells were infected with AdDR5-LacZ at an m.o.i. of 100 in DMEM with 2% FBS for 1 h and then maintained in complete medium. For staining, wells were washed with 1 mM MgCl2 in PBS, and cells were fixed by 0.5% (w/v) glutaraldehyde at room temperature, then stained using X-gal reaction solution, and incubated at 37 °C until a color change was obtained. The end absorbance was then measured at 420 nm using a V Max plate reader (Molecular Devices Corp., Sunnyvale, CA, USA). LacZ activities were normalized for differences in incubation times.
EGFP expression assay
Cellular EGFP expression was quantitatively examined by FACS analysis and visualized using fluorescent microscopy. Cells were collected 48 h after Ad/VEGF-EGFP infection and approximately 10,000 cells were illuminated at 488 nm and fluorescence was detected in the FITC (525/20 nm) channel. Nonspecific fluorescence was detected using a 575/30 nm emission filter in the PI channel. EGFP fluorescence is expressed as the mean fluorescence signal in EGFP-positive cells in relative units [18] after subtraction of background fluorescence. An Olympus IX70 inverted microscope system (Olympus America, Melville, NY) was used for the screening of EGFP expression in cell monolayers.
Quantitative reverse transcription-PCR and PCR
Total RNA (5 μg) was isolated by RNeasy kit (Qiagen) and used for cDNA synthesis by oligo [18] and SuperScript II RNase H reverse transcriptase (Invitrogen). Primers used in the reverse transcription-PCR (RT-PCR) assays were described before [40, 41]. Quantitative real-time PCR using SYBR Green PCR Master Mix (Applied Biosystems) was performed with (Bio-Rad) systems according to the manufacturer’s protocol.
Detection of luciferase expression
Standard single luciferase assay (Promega, Madison, WI) was used to measure firefly luciferase enzyme activity as per the manufacturer’s instruction. For compensation of the differences in cell numbers in different samples, Bradford protein assay (Bio-Rad, Hercules, CA) was used to measure the protein concentration of the lysates. Luciferase is presented as normalized to cellular protein concentration.
Confirmation of selectivity of survivin as radiation inducible promoter in glioma
In order to detect the optimum dose of radiation for further applications, D54 MG glioma cell lines were plated in six-well plated. The cells were then infected with either AdSurvivin-Luc or AdCMV-Luc (1000 viral particles per cell). Virus-containing medium was replaced with fresh growth medium, after 1-h adsorption. After 24 h, cells were irradiated with different doses of 0, 2, 4, 8, and 12 Gy using a 60Co therapy unit (Picker, Cleveland, OH) at a dose of 80 cGy/min. Twenty-four hours later, cells were analyzed by luciferase assay to select the optimum radiation dose. To ensure the effect of this selected dose in different glioma cell lines and to detect the difference of survivin expression levels between tumor and normal cells the same steps were repeated on D54 MG, U251 MG and human astrocytes cell lines. Cells were irradiated with the convention selected dose of 2 GY using a 60Co therapy unit (Picker, Cleveland, OH) and analyzed after 24 h by luciferase assay.
All the assays were carried out in quadruplets and several independent experiments were performed to verify the reproducibility of the results.
Bioinformatics analysis to detect radiation inducible elements
We used the NCBI Nucleotide blast database (https://www.ncbi.nlm.nih.gov/nucleotide/), in order to identify the nucleotide sequence of mRNA transcripts of different available in-house promoters. These included: 1-survivin (NCBI Reference Sequence: NM_001012271.1), 2-FLT-1 (NCBI Reference Sequence: XM_017020485.1), 3-COX-2 (NCBI Reference Sequence: Gene Bank: U20548.1), 4-DR-5(NCBI Reference Sequence: XM_017013944.1), 5-VEGFA (NCBI Reference Sequence: NM_003376.5), 6-iNOS (NCBI Reference Sequence: NM_001204218.1).
The CArG motifs were previously described as motifs which respond to radiation and are responsible for radiation mediated transcription regulation [31]. Also, it was described that CCAAT box (a cis acting element) are significantly present with lower frequencies in radio responsive genes in comparison to regular genes [42].
Therefore we used NCBI FASTA sequence finder to detect the Number of CAAG motifs and the 10-nucleotide motif of consensus sequence in the CArG [CC (A+T rich) 6GG or serum response element] in these promoters. We also screened the promoters using the same software for CCAAT box consensus sequence in attempts to determine a cis acting elements responsible for survivin response to radiation.
Statistical analysis
Results are expressed as mean ± SD. Student’s t test was used according to the distribution of experimental values. P values of <0.05 were accepted as significant differences between groups.
Results
Evaluation of mRNA accumulation as an indicator of radiation mediated transcription regulation
Six different human promoters (FLT-1, VEGF, Cox2, INOS, DR5 and survivin) were assessed for expression of mRNA in D54 MG cells 2 h after exposure to 2 Gy radiation using quantitative RT-PCR. Levels of expression of FLT-1, DR5, VEGF, INOS and Cox2 increased twofold to threefold, while the survivin gene was upregulated to ~10 folds as depicted in (Fig. 1), indicating a strong radiation-inducible promoter. Columns, mean; P < 0.05, compared with non-radiated control. However, additional elevation of survivin RNA copy number was reported after 24 h of radiation.
Fig. 1.

Effect of radiation on mRNA expression of different genes promoters
Radiotherapy increases level of gene expression in the context of adenovirus with radiation relevant promoter in brain tumor
Adenoviral constructs containing VEGF, DR5, FLT-1 and survivin promoter driving lacZ, GFP and luciferase reporter genes were used for infection of D54 MG cells. Fluorescent microscopy was used to detect the effect of radiation on VEGF promoter in Ad/GFP as depicted in (Fig. 2a). X-gal staining used to detect the effect of radiation on DR5 promoter in Ad/LacZ as depicted in (Fig. 2b). Total luciferase activity was measured and values were normalized to amounts of total protein. Experiments were performed in quadruplets using replication-deficient adenovirus. CMV promoters a strong constitutive gene promoter, producing high basal levels of reporter gene expression but shows a negligible increase on irradiation [43]. Therefore, results are shown before and after 24 h of radiation, as a percentage of CMV promoter-induced luciferase expression. We have found that Ad/VEGF-Luc and AdDR5-Luc AdFLT1-LUC showed an increase luciferase expression in response to radiation by 1.25-fold in VEGF, 2.5 fold in DR5 and twofold in FLT-1, as depicted in (Fig. 2c). Ad/survivin-Luc however showed a fivefold increase in response to radiation suggesting also strong radiation-inducible promoter. Columns, mean; P < 0.05, compared with non-radiated control. For quantification, maximum projections and total fluorescence measurements were performed with Image J and the corrected total cell fluorescence (CTCF) was calculated using the formula CTCF = Integrated Density − (Area of selected cell × Mean fluorescence of background readings) data not shown.
Fig. 2.

a Fluorescent microscopy detecting effect of radiation on GFP reporter gene driven by VEGF promoter. b X-gal staining detecting effect of radiation on LacZ reporter gene driven by DR5 promoter. c Effect of radiation on different promoters as detected by Luciferase assay
Radiation induces different levels of gene expression by adenovirus with survivin promoter at different doses
To detect the optimal dose of radiation, glioma cell lines (D54 MG) were infected with constructed adenoviral particles (1000 viral particles per cell) containing the luciferase gene under either control of survivin or CMV promoters. After viral infection, we conducted a dose–response assay exposing cells to different fractions of radiation as shown in (Fig. 3) and grown for an additional 24 h. Total luciferase activity was measured and values were normalized to amounts of total protein. Experiments were performed in quadruplets. The maximum response to radiation exposure was observed at dose 2 Gy (~3-fold) with higher doses, the response diminished to 1.5-fold, as depicted in Fig. 3.
Fig. 3.

Effect of different doses of radiation on gene expression driven by survivin promoter
We therefore considered 2 Gy the optimum dose for radiation. Columns, mean; bars, SD, P < 0.05, compared with non-radiated control.
Radiation induces high level of gene expression by adenovirus with survivin promoter in different glioma cell lines
Different glioma cell lines D54 MG, U251 MG and human astrocytes were infected with Ad-survivin and exposed to the selected dose of 2 Gy. The luciferase activity was assessed before radiation and 24 h after radiation. Survivin promoter showed an increase in expression from 30 to 99% in glioma cell lines after 2 Gy of radiation compared to non-irradiated cells as depicted in (Fig. 4). The difference in survivin expression between glioma cell lines and normal human astrocytes suggest that survivin is a tumor selective promoter. Columns, mean; bars, SD, P < 0.05, compared with non-radiated control.
Fig. 4.
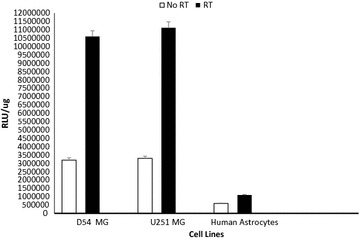
Selectivity of survivin promoter to Glioma cell lines
CARG Radiation inducible elements and other cis acting elements detection
After using NCBI nucleotide database to identify the nucleotide base sequence of the promoters used in this experiment, FASTA sequence finder was used to determine the presence of elements responding to radiation previously described. Variable number of CAAG elements was detected in the promoters independently from their response to radiation. Also, the CArG box which has been shown to be responsible for egr-1 increased transcription in response to radiation has not been detected in any of the promoters used in this experiment as depicted in Table 1. Therefore we concluded that radiation mediated transcription regulation is not dependent on CArG elements only
Table 1.
Number of CArG and CAAG elements in different promoters
| Survivin | FLT-1 | DR-5 | VEGF | COX-2 | iNOS | |
|---|---|---|---|---|---|---|
| CAAG elements | 21 | 83 | 58 | 36 | 17 | 167 |
| CArG box [CC (A+T rich) 6GG] | 0 | 0 | 0 | 0 | 0 | 0 |
Using NCBI nucleotide database and FASTA sequence finder we screened all the promoters for CCAAT box consensus. iNOS promoter with the lowest response to radiation had higher number of CCAAT (n = 14) as depicted in Fig. 5, while Survivin with the best response to radiation revealed two CCAAT boxes (n = 2) in its sequence as depicted in Fig. 6. The other promoters had numbers of this consensus ranging between these 2 values (data not shown).
Fig. 5.

Detetction of CCAAT box (a cis acting element) in iNOS promoter nucleotide sequence
Fig. 6.

Detetction of CCAAT box (a cis acting element) in Survivin promoter nucleotide sequence
Discussion
Treatment of brain tumors, especially GBM represents a great challenge [44, 45]. The standard GBM treatment includes surgical resection, radiation, and chemotherapy. Although, there has been an emergence of many innovations and advances in chemotherapy and radiation therapy, none of them have prevented recurrence [46, 47]. Rather they have only provided longer survival times. Radiogenetic therapy, is a novel approach that offers a combination model aiming to circumvent the obstacles faced by conventional treatments. It employs targeted viral vectors delivery of therapeutic transgenes under the control of radiation responsive promoters. Enhancing viral replication by using tumor selective promoters is likely to enhance virotherapy [48, 49]. This strategy would restrict therapeutic gene activation to tumor cells and/or irradiated tissues only, thereby providing selective expression for any tumor-located vector. Since gene activation would be controlled, damage to surrounding normal tissue would be reduced, thereby improving the therapeutic ratio replication such as targeting and transcription regulation. In this study we tried a combined technique of radiation plus transcription regulation to prove the principle of regulating gene expression.
Understanding the molecular bases and mechanisms responsible for radiation mediated transcription regulation is crucial for better understanding of this approach and for classifying and selecting the best radioinducible gene promoter. Elements regulating transcriptional process are composed of both cis-acting and trans-acting elements [50, 51]. The cis-acting elements comprise promoters as well as enhancer regions (ex: CArG box, TATA box and CCAAT) that regulate expression of a distinct gene [52]. The trans-acting elements are related to a group of transcription factors that bind to specific sites within promoter and/or enhancer regions [53, 54]. Controlling the transcription of therapeutic gene expression [55] by using promoters that are radiation-responsive offers an attractive platform for the combination of radiation therapy and gene therapy [56].
We and others showed in this study, and other studies [57–59], that the survivin promoter in comparison to other promoters may represent an optimal radiation inducible promoter and provide compelling data to suggest that radiation, a mainstay of glioma therapy, further improves survivin-mediated adenoviral gene expression in targeted cells, which is a finding that might have significant implications for patients who may one day be treated with this virus and subsequently receive radiotherapy. Also, the difference in survivin expression levels between glioma cells and normal astrocytes suggest its tumor selectivity and potential safety in sparing normal cells from aggressive therapies.
In attempts to explain the radiosensitivity of survivin promoter, we screened all the promoters used in this experiment for the presence of CArG radio responsive motifs first described in EGR-1 promoter. The egr-1 was the first promoter described more than 2 decades ago to be responsive to radiation [27, 60, 61]. Functional analysis of Egr-1 revealed a number of DNA sequence motifs including GCGGGGGCG that can modulate the radiation-mediated response [31]. Ionizing radiation can readily induce the Egr-1 promoter via a 10-nucleotide motif of consensus sequence in the CArG (CC (A+T rich) 6GG or serum response element) [31, 62–65]. In our study we have analyzed several radio responsive promoters for the CArG elements. None of these sequences were detected in any promoters. Adding to that the number of CAAG sequence in all promoters was independent from response to radiation.
Additionally, our team as well as others have shown that radiation increased the number of survivin RNA transcripts in cells [57, 58, 66]. Therefore we do believe that elements other than CArG may be responsible for survivin radiosensitivity. Of note, it was concluded before that CCAAT box play a gene-specific transcriptional role depending on the type of gene [67]. Since survivin promoter has no TATA box, these sequences are involved in gene expression and cell cycle regulation. Therefore, they might also be responsible for radiosensitivity of survivin. Interestingly, a study by Wu et al. [68] correlated low frequency presence of CCAAT motifs to radio responsive genes. Following this concept we screened the promoters used in these study for this motif consensus. Our results came in accordance to this correlation where survivin promoter, with the best response to radiation, had the lowest number of CCAAT box motifs in comparison to other promoters tested. These preliminary results require future mutational analysis of different promoters in order to explore the mechanism behind radiation mediated transcription regulation and larger sample size to correlate between the different numbers of cis acting element in a given promoter and its response to radiation.
To this end we conclude that survivin is a selective potent radiation inducible promoter for glioma and CArG motifs are not the only elements involved in radiation mediated transcription regulation.
Authors’ contributions
All authors contributed equally to this study. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank Baher Ibrahim, MD, MA, MS (King's College, London), Maheen Sayeed, MS (Georgetown University, USA), Abanoub Gad ( Children's hospital of Philadelpia, USA), Mostafa Samir (Alexandria Comprehensive Cancer Center, Egypt) for their administrative and logistic help during manuscript preparation.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Most of this experiment was carried out in University of Alabama at Birmingham (UAB) laboratories. The bioinformatics analysis was carried out in the Alexandria Comprehensive Cancer Center, Alexandria, Egypt.
Contributor Information
George E. Naoum, Email: georgenaoum3@gmail.com
Zeng B. Zhu, Email: zeng.zhu@ccc.uab.edu
Donald J. Buchsbaum, Email: djb@uab.edu
David T. Curiel, Email: dcuriel@radonc.wustl.edu
Waleed O. Arafat, Phone: (+20)1223399328, Email: W.o.arafaat@gmail.com
References
- 1.Deb P, et al. Glioblastoma multiforme with long term survival. Neurol India. 2005;53(3):329–332. doi: 10.4103/0028-3886.16936. [DOI] [PubMed] [Google Scholar]
- 2.Ohgaki H. Epidemiology of brain tumors. Methods Mol Biol. 2009;472:323–342. doi: 10.1007/978-1-60327-492-0_14. [DOI] [PubMed] [Google Scholar]
- 3.Patel TR, Yu JB, Piepmeier JM. Role of neurosurgery and radiation therapy in the management of brain tumors. Hematol Oncol Clin North Am. 2012;26(4):757–777. doi: 10.1016/j.hoc.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Fiveash JB, et al. A phase 2 study of radiosurgery and temozolomide for patients with 1 to 4 brain metastases. Adv Radiat Oncol. 2016;1(2):83–88. doi: 10.1016/j.adro.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelley K, et al. Radioresistance of brain tumors. Cancers (Basel) 2016;8(4):42. doi: 10.3390/cancers8040042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn GP, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziegler DS, et al. Resistance of human glioblastoma multiforme cells to growth factor inhibitors is overcome by blockade of inhibitor of apoptosis proteins. J Clin Invest. 2008;118(9):3109–3122. doi: 10.1172/JCI34120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altieri DC. Survivin—the inconvenient IAP. Semin Cell Dev Biol. 2015;39:91–96. doi: 10.1016/j.semcdb.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guindalini RSC, Machado MCM, Garicochea B. Monitoring survivin expression in cancer: implications for prognosis and therapy. Mol Diagn Ther. 2013;17(6):331–342. doi: 10.1007/s40291-013-0048-1. [DOI] [PubMed] [Google Scholar]
- 10.Kwiatkowska A, et al. Strategies in gene therapy for glioblastoma. Cancers (Basel) 2013;5(4):1271–1305. doi: 10.3390/cancers5041271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Natsume A, Yoshida J. Gene therapy for high-grade glioma: current approaches and future directions. Cell Adhes Migr. 2008;2(3):186–191. doi: 10.4161/cam.2.3.6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonabend AM, Ulasov IV, Lesniak MS. Gene therapy trials for the treatment of high-grade gliomas. Gene Ther Mol Biol. 2007;11(A):79–92. [PMC free article] [PubMed] [Google Scholar]
- 13.Wollmann G, Ozduman K, van den Pol AN. Oncolytic virus therapy for glioblastoma multiforme: concepts and candidates. Cancer J. 2012;18(1):69–81. doi: 10.1097/PPO.0b013e31824671c9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao KC, et al. Molecular response of human glioblastoma multiforme cells to ionizing radiation: cell cycle arrest, modulation of the expression of cyclin-dependent kinase inhibitors, and autophagy. J Neurosurg. 2003;98(2):378–384. doi: 10.3171/jns.2003.98.2.0378. [DOI] [PubMed] [Google Scholar]
- 15.Arafat W, et al. Targeted radiotherapy potentiates the cytotoxicity of a novel anti-human DR5 monoclonal antibody and the adenovirus encoding soluble TRAIL in prostate cancer. J Egypt Natl Cancer Inst. 2015;27(4):205–215. doi: 10.1016/j.jnci.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Arafat WO, et al. An adenovirus encoding proapoptotic Bax synergistically radiosensitizes malignant glioma. Int J Radiat Oncol Biol Phys. 2003;55(4):1037–1050. doi: 10.1016/S0360-3016(02)04488-7. [DOI] [PubMed] [Google Scholar]
- 17.Arafat WO, et al. An adenovirus encoding proapoptotic Bax induces apoptosis and enhances the radiation effect in human ovarian cancer. Mol Ther. 2000;1(6):545–554. doi: 10.1006/mthe.2000.0071. [DOI] [PubMed] [Google Scholar]
- 18.Andtbacka RH, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33(25):2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 19.Asadi-Moghaddam K, Chiocca EA. Gene- and viral-based therapies for brain tumors. Neurotherapeutics. 2009;6(3):547–557. doi: 10.1016/j.nurt.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho DY, et al. Targeting cancer stem cells for treatment of glioblastoma multiforme. Cell Transpl. 2013;22(4):731–739. doi: 10.3727/096368912X655136. [DOI] [PubMed] [Google Scholar]
- 21.Ren H, et al. Immunogene therapy of recurrent glioblastoma multiforme with a liposomally encapsulated replication-incompetent Semliki forest virus vector carrying the human interleukin-12 gene—a phase I/II clinical protocol. J Neurooncol. 2003;64(1–2):147–154. doi: 10.1007/BF02700029. [DOI] [PubMed] [Google Scholar]
- 22.Naoum GE, et al. Role of nanotechnology and gene delivery systems in TRAIL-based therapies. Ecancermedicalscience. 2016;10:660. doi: 10.3332/ecancer.2016.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dey M, et al. Cancer stem cells: the final frontier for glioma virotherapy. Stem Cell Rev. 2011;7(1):119–129. doi: 10.1007/s12015-010-9132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanzawa T, et al. Current and future gene therapy for malignant gliomas. J Biomed Biotechnol. 2003;2003(1):25–34. doi: 10.1155/S1110724303209013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawakami Y, et al. Substitution of the adenovirus serotype 5 knob with a serotype 3 knob enhances multiple steps in virus replication. Cancer Res. 2003;63(6):1262–1269. [PubMed] [Google Scholar]
- 26.Nuyts S, et al. Radio-responsive recA promoter significantly increases TNFalpha production in recombinant clostridia after 2 Gy irradiation. Gene Ther. 2001;8(15):1197–1201. doi: 10.1038/sj.gt.3301499. [DOI] [PubMed] [Google Scholar]
- 27.Weichselbaum RR, et al. Radiation-induced tumour necrosis factor-alpha expression: clinical application of transcriptional and physical targeting of gene therapy. Lancet Oncol. 2002;3(11):665–671. doi: 10.1016/S1470-2045(02)00900-2. [DOI] [PubMed] [Google Scholar]
- 28.Weichselbaum RR, et al. Molecular targeting of gene therapy and radiotherapy. Acta Oncol. 2001;40(6):735–738. doi: 10.1080/02841860152619151. [DOI] [PubMed] [Google Scholar]
- 29.Du N, et al. Radioprotective effect of FLT3 ligand expression regulated by Egr-1 regulated element on radiation injury of SCID mice. Exp Hematol. 2003;31(3):191–196. doi: 10.1016/S0301-472X(02)01082-2. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt M, et al. Inducible promoters for gene therapy of head and neck cancer: an in vitro study. Eur Arch Otorhinolaryngol. 2004;261(4):208–215. doi: 10.1007/s00405-003-0621-z. [DOI] [PubMed] [Google Scholar]
- 31.Datta R, et al. Ionizing radiation activates transcription of the EGR1 gene via CArG elements. Proc Natl Acad Sci USA. 1992;89(21):10149–10153. doi: 10.1073/pnas.89.21.10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werner T. Cluster analysis and promoter modelling as bioinformatics tools for the identification of target genes from expression array data. Pharmacogenomics. 2001;2(1):25–36. doi: 10.1517/14622416.2.1.25. [DOI] [PubMed] [Google Scholar]
- 33.Takayama K, et al. Vascular endothelial growth factor promoter-based conditionally replicative adenoviruses for pan-carcinoma application. Cancer Gene Ther. 2007;14(1):105–116. doi: 10.1038/sj.cgt.7700991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu ZB, et al. Targeting mesothelioma using an infectivity enhanced survivin-conditionally replicative adenoviruses. J Thorac Oncol. 2006;1(7):701–711. doi: 10.1097/01243894-200609000-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida T, et al. Promoter structure and transcription initiation sites of the human death receptor 5/TRAIL-R2 gene. FEBS Lett. 2001;507(3):381–385. doi: 10.1016/S0014-5793(01)02947-7. [DOI] [PubMed] [Google Scholar]
- 36.Zhu ZB, et al. Transcriptional targeting of tumors with a novel tumor-specific survivin promoter. Cancer Gene Ther. 2004;11(4):256–262. doi: 10.1038/sj.cgt.7700679. [DOI] [PubMed] [Google Scholar]
- 37.Kaliberov SA, et al. Adenovirus-mediated transfer of BAX driven by the vascular endothelial growth factor promoter induces apoptosis in lung cancer cells. Mol Ther. 2002;6(2):190–198. doi: 10.1006/mthe.2002.0648. [DOI] [PubMed] [Google Scholar]
- 38.Ulasov IV, et al. Survivin-driven and fiber-modified oncolytic adenovirus exhibits potent antitumor activity in established intracranial glioma. Hum Gene Ther. 2007;18(7):589–602. doi: 10.1089/hum.2007.002. [DOI] [PubMed] [Google Scholar]
- 39.Rosenfeld ME, et al. Adenoviral-mediated delivery of the herpes simplex virus thymidine kinase gene selectively sensitizes human ovarian carcinoma cells to ganciclovir. Clin Cancer Res. 1995;1(12):1571–1580. [PubMed] [Google Scholar]
- 40.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 41.Cooney R, et al. Adenoviral-mediated gene transfer of nitric oxide synthase isoforms and vascular cell proliferation. J Vasc Res. 2006;43(5):462–472. doi: 10.1159/000095163. [DOI] [PubMed] [Google Scholar]
- 42.Wu J, et al. The initiator motif is preferentially used as the core promoter element in ionizing radiation-responsive genes. Radiat Res. 2006;166(5):810–813. doi: 10.1667/RR0570.1. [DOI] [PubMed] [Google Scholar]
- 43.Scott SD, Joiner MC, Marples B. Optimizing radiation-responsive gene promoters for radiogenetic cancer therapy. Gene Ther. 2002;9(20):1396–1402. doi: 10.1038/sj.gt.3301822. [DOI] [PubMed] [Google Scholar]
- 44.Carlsson SK, Brothers SP, Wahlestedt C. Emerging treatment strategies for glioblastoma multiforme. EMBO Mol Med. 2014;6(11):1359–1370. doi: 10.15252/emmm.201302627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson TA, Karajannis MA, Harter DH. Glioblastoma multiforme: State of the art and future therapeutics. Surg Neurol Int. 2014;5:64. doi: 10.4103/2152-7806.137196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nieder C, et al. Treatment of unresectable glioblastoma multiforme. Anticancer Res. 2005;25(6c):4605–4610. [PubMed] [Google Scholar]
- 47.Pinsker M, Lumenta C. Experiences with reoperation on recurrent glioblastoma multiforme. Zentralbl Neurochir. 2001;62(2):43–47. doi: 10.1055/s-2002-19477. [DOI] [PubMed] [Google Scholar]
- 48.Dey M, Ulasov IV, Lesniak MS. Virotherapy against malignant glioma stem cells. Cancer Lett. 2010;289(1):1–10. doi: 10.1016/j.canlet.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy AM, Rabkin SD. Current status of gene therapy for brain tumors. Transl Res. 2013;161(4):339–354. doi: 10.1016/j.trsl.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shibata M, Gulden FO, Sestan N. From trans to cis: transcriptional regulatory networks in neocortical development. Trends Genet. 2015;31(2):77–87. doi: 10.1016/j.tig.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnston SJ, Carroll JS. Transcription factors and chromatin proteins as therapeutic targets in cancer. Biochim Biophys Acta. 2015;1855(2):183–192. doi: 10.1016/j.bbcan.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 52.Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2012;13(1):59–69. doi: 10.1038/nri3362. [DOI] [PubMed] [Google Scholar]
- 53.Nishizawa M, et al. Post-transcriptional inducible gene regulation by natural antisense RNA. Front Biosci (Landmark Ed) 2015;20:1–36. doi: 10.2741/4297. [DOI] [PubMed] [Google Scholar]
- 54.Yeh JE, Toniolo PA, Frank DA. Targeting transcription factors: promising new strategies for cancer therapy. Curr Opin Oncol. 2013;25(6):652–658. doi: 10.1097/01.cco.0000432528.88101.1a. [DOI] [PubMed] [Google Scholar]
- 55.Rad SM, et al. Transcription factor decoy: a pre-transcriptional approach for gene downregulation purpose in cancer. Tumour Biol. 2015;36(7):4871–4881. doi: 10.1007/s13277-015-3344-z. [DOI] [PubMed] [Google Scholar]
- 56.Kim TM, Park PJ. Advances in analysis of transcriptional regulatory networks. Wiley Interdiscip Rev Syst Biol Med. 2011;3(1):21–35. doi: 10.1002/wsbm.105. [DOI] [PubMed] [Google Scholar]
- 57.Arafat WO, et al. Survivin as a novel radiation/tissue specific promoter for adenovirus based gene therapy. Cancer Res. 2004;64(7 Supplement):959. [Google Scholar]
- 58.Arafat WO, et al. Survivin as a novel radiation/CIS platinum/tumor specific promoter for conditional oncolytic viral therapy. J Clin Oncol. 2004;22(14_suppl):1535. [Google Scholar]
- 59.Nandi S, et al. Low-dose radiation enhances survivin mediated virotherapy against malignant glioma stem cells. Cancer Res. 2008;68(14):5778–5784. doi: 10.1158/0008-5472.CAN-07-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ahmed MM. Regulation of radiation-induced apoptosis by early growth response-1 gene in solid tumors. Curr Cancer Drug Targets. 2004;4(1):43–52. doi: 10.2174/1568009043481704. [DOI] [PubMed] [Google Scholar]
- 61.Kufe D, Weichselbaum R. Radiation therapy: activation for gene transcription and the development of genetic radiotherapy-therapeutic strategies in oncology. Cancer Biol Ther. 2003;2(4):326–329. doi: 10.4161/cbt.2.4.495. [DOI] [PubMed] [Google Scholar]
- 62.Datta R, et al. Reactive oxygen intermediates target CC(A/T)6GG sequences to mediate activation of the early growth response 1 transcription factor gene by ionizing radiation. Proc Natl Acad Sci USA. 1993;90(6):2419–2422. doi: 10.1073/pnas.90.6.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsu H, et al. Combined radiation and cytochrome CYP4B1/4-ipomeanol gene therapy using the EGR1 promoter. Anticancer Res. 2003;23(3b):2723–2728. [PubMed] [Google Scholar]
- 64.Mezhir JJ, et al. Ionizing radiation: a genetic switch for cancer therapy. Cancer Gene Ther. 2006;13(1):1–6. doi: 10.1038/sj.cgt.7700879. [DOI] [PubMed] [Google Scholar]
- 65.Tsai-Morris CH, Cao XM, Sukhatme VP. 5′ Flanking sequence and genomic structure of Egr-1, a murine mitogen inducible zinc finger encoding gene. Nucleic Acids Res. 1988;16(18):8835–8846. doi: 10.1093/nar/16.18.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pennati M, Folini M, Zaffaroni N. Targeting survivin in cancer therapy: fulfilled promises and open questions. Carcinogenesis. 2007;28(6):1133–1139. doi: 10.1093/carcin/bgm047. [DOI] [PubMed] [Google Scholar]
- 67.Nenoi M, et al. Regulation of the catalase gene promoter by Sp1, CCAAT-recognizing factors, and a WT1/Egr-related factor in hydrogen peroxide-resistant HP100 cells. Cancer Res. 2001;61(15):5885–5894. [PubMed] [Google Scholar]
- 68.Wu J, et al. The initiator motif is preferentially used as the core promoter element in ionizing radiation-responsive genes. Radiat Res. 2006;166(5):810–813. doi: 10.1667/RR0570.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Most of this experiment was carried out in University of Alabama at Birmingham (UAB) laboratories. The bioinformatics analysis was carried out in the Alexandria Comprehensive Cancer Center, Alexandria, Egypt.


