Abstract
Purpose
Ovarian cancer is the leading cause of death from gynecologic malignancy in the United States, with high rates of recurrence and eventual resistance to cytotoxic chemotherapy. Model systems that allow for accurate and reproducible target discovery and validation are needed to support further drug development in this disease.
Experimental Design
Clinically-annotated patient-derived xenograft (PDX) models were generated from tumor cells isolated from the ascites or pleural fluid of patients undergoing clinical procedures. Models were characterized by immunohistochemistry and by molecular analyses. Each PDX was luciferized to allow for reproducible in vivo assessment of intraperitoneal tumor burden by bioluminescent imaging (BLI). Plasma assays for CA125 and human LINE-1 were developed as secondary tests of in vivo disease burden.
Results
14 clinically annotated and molecularly characterized luciferized ovarian PDX models were generated. Luciferized PDX models retain fidelity to both the non-luciferized PDX and the original patient tumor, as demonstrated by immunohistochemistry, array CGH, and targeted and whole-exome sequencing analyses. Models demonstrated diversity in specific genetic alterations and activation of PI3K signaling pathway members. Response of luciferized PDX models to standard of care therapy could be reproducibly monitored by BLI or plasma markers.
Conclusions
We describe the establishment of a collection of 14 clinically annotated and molecularly characterized luciferized ovarian PDX models in which orthotopic tumor burden in the intraperitoneal space can be followed by standard and reproducible methods. This collection is well-suited as a platform for proof-of-concept efficacy and biomarker studies and for validation of novel therapeutic strategies in ovarian cancer.
Introduction
Ovarian cancer is the leading cause of death from gynecologic cancer in the United States, with an estimated 21,290 cases and 14,180 deaths occurring in 2015 (1). Thus, development of new therapeutic strategies for ovarian cancer remains a critical need. While a large number of ovarian cancer cell lines exist to aid with pre-clinical investigation, characterization of these cell lines has demonstrated that many of the most commonly utilized cell lines do not exhibit molecular features consistent with the most common form of ovarian cancer, high grade serous ovarian cancer (HGSOC) (2). Furthermore, cell lines that appear most representative of HGSOC have limited utility as they frequently do not efficiently form tumors in vivo (3, 4).
Patient-derived xenografts (PDX) are emerging as an alternative pre-clinical model that may offer additional insights into the development of novel targeted therapies in a number of tumor lineages (reviewed in (5, 6)). Putative advantages of PDX models include preservation of histologic appearance of the cancer cells and increased molecular fidelity to the original tumor, both in terms of genomic characteristics and gene expression, and retention of intratumoral heterogeneity.
While PDX models of ovarian cancer have been described and demonstrate fidelity to the original cancer (7, 8), some of these models present challenges for pre-clinical modeling, especially with regard to tracking tumor growth or regression in an intraperitoneal environment. We therefore sought to establish a well-characterized collection of ovarian cancer PDX models whose growth kinetics can be readily assessed by either bioluminescent imaging or serum biomarker measurement, enabling robust pre-clinical evaluation of novel therapies in ovarian cancer.
Materials and Methods
Establishment of patient-derived tumor xenografts
Under IRB-approved protocols, tumor ascites or pleural effusions were collected from patients with suspected or established ovarian cancer at the Brigham and Women’s Hospital or the Dana-Farber Cancer Institute (DFCI). Tumor cells were isolated from samples after centrifugation and red blood cell lysis. Ovarian PDXs were established by implanting these cells intraperitoneally in irradiated nude mice (Taconic, Hudson, NY). Depending on the number of tumor cells isolated, one to three mice were implanted with cells from each collected sample. All animal studies were performed in accordance to DFCI institutional animal care and use committee guidelines per DFCI-approved animal protocols.
Mice were followed three times per week for abdominal distention or palpable tumor for assessment of tumor development. Mice were euthanized if they developed signs of morbidity or ascites, a bodyweight gain of ~40%, or if there was no evidence of tumor development after a period of 1 year. After euthanization, necropsy was performed and major organs were collected and FFPE blocks were prepared. In mice with evidence of ascites, ascites was collected and tumor cells were isolated following red blood cell lysis. A portion of ascitic tumor cells was suspended in PBS and transplanted into new irradiated nude mice for serial transplantation.
Development of luciferized PDX models
Lentiviral vector FUW-Luc-mCherry-puro lentivirus (FmC) used in this study, encoding Firefly luciferase and mCherry (from Dr. Andrew Kung, Columbia University) was packaged in 293T cells using a helper virus-free packaging system. Optimal conditions for successful luciferization were established individually for each PDX model (Supplemental Table 1). In general, ascites from established PDX models were implanted intraperitoneally in NOD-SCID IL2Rγnull mice (NSG, Jackson Laboratory) after a comparative DF14-Luc tumor growth rate study demonstrated that latency and growth rates were superior in NSG mice, as compared to SCID or irradiated nude mice (data not shown). Fresh ascites-derived tumor cells from these PDX tumor-bearing NSG mice were then plated ex vivo. They were transduced with FmC Lentiviral vector at a multiplicity of infection of ~10 in medium containing polybrene at 8 µg/ml and selected in puromycin-containing media for 5 to 7 days. The selected cells, once confirmed to be expressing RFP by fluorescent microscopy (Leica) were directly injected into NSG mice intraperitoneally and further expanded (Supplemental Figure 1, Schema). Luciferized PDX models were then further expanded (to a maximum of six passages), banked, characterized, and utilized for drug efficacy and biomarker evaluation studies.
Histological evaluation of tumor xenografts
Major organ tissues collected from mice were fixed overnight in 10% buffered formalin (Fisher) and processed in the Rodent Core Facility at Harvard Medical School (HMS), Boston. 5 µm sections were deparaffinized and rehydrated and then pressure cooked (Biocare Medical) for 30 minutes in Citrate Buffer (DAKO Target Retrieval Solution, S1699) at 120°C. Primary Abs purchased from Abcam (WT-1), Epitomic (P53) and DAKO North America (pan cytokeratin) were incubated 40 minutes at room temperature (RT). Secondary Ab (DAKO Envision+ Rabbit (K401) was applied for 30 minutes at RT. Chromogenic protein detection was determined in the presence of DAB (3,3’-diaminobenzidine) and visualized by Leica Microscopy.
FDG-PET studies
FDG-PET analysis was performed at the Lurie Family Imaging Center of the Center for Biomedical Imaging in Oncology (CBIO/LFIC) DFCI as previously reported (McCall et al. 2015). Four DF86-Luc tumor bearing mice were imaged at 36 days post-implantation by intraperitoneal injection by [18F]-FDG-PET/CT. [18F]-FDG was manufactured by a commercial radiopharmaceutical manufacturer (PETNET Solutions Inc) and supplied in ethanol-stabilized sodium chloride solution. All images were acquired using an Inveon Multi-Modality scanner (Siemens Medical Solutions USA, Inc), a small-animal PET/CT system.
Copy number variation (CNV) and analysis
DNA from patient material and matched established PDX models was isolated using Gentra Puregene Tissue Kit (Qiagen). Array CGH was performed using a whole genome Affymetrix CytoscanTM HD microarray platform with 1 µg of total genomic DNA from each sample. PDX DNAs were tested on the Affymetrix Cytoscan™ HD microarray platform. Data was visualized and analyzed using a Chromosome Analysis Suite (ChAS) software package (Affymetrix, USA) with a minimal cutoff of 20 consecutive markers for CNV calling. All CNVs reported were based on NCBI human genome build 37 (9).
For copy number variation analysis, copy number values were reported as ratios of the PDX sample (original patient sample and luciferized PDX sample) to a reference normal value and were log-2 transformed for further analysis. For evaluating the fidelity of PDX models, we calculated the mean log2 intensity value for each gene and Pearson correlation coefficients were calculated for each PDX sample to its matched patient tumor. We also compared the similarity of the PDX models to tumor samples in the TCGA dataset by calculating Pearson correlation coefficients for each PDX sample to the median ovarian patient CNV score in The Cancer Genome Atlas (TCGA) (10). Analysis was performed in R 3.2.2.
Targeted sequencing analyses
Targeted genomic analyses were performed on a research basis at the UW Department of Laboratory Medicine Genetics and Solid Tumors Laboratory, as previously described (11, 12). DNA samples from 11 PDXs and their corresponding patient’s ascites or pleural fluid-derived ovarian cancer cells were characterized by a BROCA panel including 48 genes (AKT1, APC, ATM, ATR, BAP1, BARD1, BMPR1A, BRCA1, BRCA2, BRIP1, CDH1, CDK4, CDKN2A, CHEK1, CHEK2, CTNNA1, FAM175A (Abraxas), GALNT12, GEN1, GREM1, HOXB13, MEN1, MLH1, MRE11A, MSH2 (+EPCAM), MSH6, MUTYH, NBN, PALB2, PIK3CA, PMS2, POLD1, POLE, PPM1D, PRSS1, PTEN, RAD51B, RAD51C, RAD51D, RET, SDHB, SDHC, SDHD, SMAD4, STK11, TP53, VHL, and XRCC2).
Whole-exome next generation sequencing analyses
Sample library construction, exonic capture, next generation sequencing, and bioinformatic analyses of samples were performed as previously described (13, 14). In brief, fragmented genomic DNA from the patient’s initial ascites-derived ovarian cancer cells, early-passage non-luciferized PDX tumors and matched normal blood samples were used for analysis of exonic regions using custom Agilent SureSelect probes according to the manufacturer’s instructions (Agilent, Santa Clara, CA). Captured DNA libraries were sequenced with the Illumina HiSeq System (Illumina, San Diego, CA). Sequence reads were analyzed and aligned to the human genome sequence (hg18) with the Eland v.2 algorithm in CASAVA 1.7 software (Illumina, San Diego, CA). Potential somatic mutations and copy number alterations excluding mouse-specific variants were identified using VariantDx custom software as described previously (13, 14).
Proteomic analysis
Proteomic analysis using Reverse Phase Protein Microarrays (RPPA) was performed at the RPPA core facility (MD Anderson Cancer Center) using standard operating procedures (15). Each sample was assayed in triplicate and data were normalized using a log2-followed by double z-score- transformation. PI3 kinase pathway proteins and phosphoproteins were analyzed by correlation matrices generated using Spearman correlation and agglomerative clustering by Pearson similarity.
Western blots were also performed from the same snap-frozen ascites-derived tumor cells used for RPPA analysis. Tumor cell lysates were generated after lysing cells in RIPA buffer and protease inhibitor cocktail (Roche). Protein samples were probed with antibodies (Cell Signaling) to phospho-ERK, total ERK, phospho-AKT, total AKT, phospho-S6, and total S6.
qRT-PCR was performed by extracting total RNA from snap frozen ascites-derived tumor cells using human-specific primers and probes for PIK3CA and 18sRNA (Applied Biosystems) on an ABI-PRISM 7900 thermal cycle (Applied Biosystems). Data analysis was performed by the comparative threshold cycle method (16).
PIK3CA copy number was assessed by fluorescence in situ hybridization per standard protocols on ascites-derived tumor cells. BAC clone RP11-386L21 (CHORI; http://bacpac.chori.org) containing PIK3CA was labeled with SpectrumGreen dUTP using nick translation to generate the PIK3CA probe. CEP3 reference probe labeled with SpectrumRed was purchased from Abbott Molecular (Des Plaines, IL). FISH signal evaluation and acquisition were performed manually using filter sets and software developed by Applied Spectral Imaging (Carlsbad, CA). Several fields with at least 25 cells total were captured and ratio of PIK3CA to CEP3 signal numbers was calculated. A PIK3CA:CEP3 signal ratio of 2 or greater was defined as PIK3CA amplification. Samples having a PIK3CA:CEP3 ratio between 1.5 and 2 were defined as having relative PIK3CA gain.
SOC efficacy studies
NSG mice were implanted intraperitoneally with ~5 × 106 ascites-derived luciferized PDX cells, and tumor burden was assessed by bioluminescence imaging (BLI) as previously described (17). Animals were imaged a week after injection, and mice with established tumor burden as documented by BLI were randomized and grouped into cohorts that were treated once weekly for 3 weeks with either saline, carboplatin (80 mg/kg intraperitoneally), paclitaxel (20 mg/kg intravenously), or combination of carboplatin and paclitaxel. Serial imaging was used to assess disease burden, and data plotted as the mean ± SEM for each group. One way ANOVA analysis with Tukey post-test was used to determine the significance of all pairwise comparisons. For the evaluation of platinum-sensitivity across models, tumor bearing mice were treated weekly with carboplatin (80 mg/kg intraperitoneally) for 3 weeks and subsequent tumor regrowth was monitored by serial BLI imaging at regular intervals for up to 70 weeks.
Evaluation of CA125 and LINE1 plasma assays in PDX models
Blood (20–150 µL) was collected either at terminal ascites endpoint from individual PDXs or serially over a defined duration,via retro-orbital bleeding under an institutionally-approved animal protocol. Whole blood was centrifuged for 10 minutes at 1200g, and supernatant plasma was further cleared by centrifugation for 10 minutes at 3000g. Cell free DNA for LINE-1 assay was isolated using the QIAmp Circulating Nucleic Acid Kit (Qiagen) according to the manufacturer’s protocol. DNA was eluted in AVE buffer (20 µL) and stored at −80°C until use.
Mouse plasma CA125 levels were measured via a custom assay using BioScale’s Acoustic Membrane Micro Particle (AMMP) technology (18). Universal Detection Kit, diluent, regeneration solution, and magnetic beads were purchased from BioScale, and complementary detection and capture antibodies were purchased from CalBioreagents (Cat# M184 and M185). The detection antibody was labeled with fluorescein via a standard NHS fluorescein labeling protocol. Capture antibodies were conjugated to Bioscale’s Type II magnetic microparticles. The final optimized antibody concentrations for this assay were determined to be 1.5 × 105 beads and 0.2 ng/ml of fluorescein antibody per reaction well. Serially diluted plasma and capture and detection antibodies were incubated under constant, gentle agitation for 4 hours at room temperature after which the bead-fluorescein-analyte complex was captured by the anti-fluorescein-coated acoustic membrane, and read on the ViBE after stringent washing. Concentrations were determined against a concomitantly assayed standard from recombinant CA125 (R&D Systems).
Human LINE-1 was quantified by qRT-PCR using a modified version developed by Rago et al.(19), using the forward primer FWD 5′-TCACTCAAAGCCGCTCAACTAC-3′ (Operon) and reverse primer REV 5′-TCTGCCTTCATTTCGTTATGTACC-3′ (Operon). The reaction was monitored on a StepOnePlus (Applied Biosystems), and the threshold cycle number was determined using Applied Biosystems’ analysis software. Standard curves were generated using DNA from A549 cells starting at 1000 pg/µL and serially diluted down tenfold for five data points with an additional sixth point of 0 pg/µL (water). All samples and standard curves were assayed in triplicates.
Results
Establishment of a panel of primary ovarian cancer PDXs
Between August 2005 and December 2012, a total of 94 separate clinical samples were collected and implanted intraperitoneally in mice. A total of 29 PDX models that successfully grew through at least 3 serial passages were established for a take rate of 31%. The latency time to development of clinically apparent disease from the time of initial implantation varied from 2 to 12 months. 14 models with growth kinetics suitable for robust in vivo experiments were selected for further luciferization and characterization. Clinical annotation and PDX characterization for these 14 models is shown in Table 1.
Table 1.
Clinical annotation and PDX characteristics
| PDX model |
Clinical Annotation | PDX characterization | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Histologic subtype |
Source | # of prior lines of chemotherapy |
Prior platinum |
Germline BRCA status |
PAX8 (IHC) |
WT1 (IHC) |
Pan CK (IHC) |
BROCA mutations |
PDX model plasma CA125 |
|
| DF09 | HGSOC | Ascites | 0 | No | Unknown | Positive | Positive | Positive | Not performed | Positive |
| DF14 | HGSOC | Ascites | 5 | Yes | Unknown | Positive | Positive | Positive | Not performed | Positive |
| DF20 | HGSOC | Ascites | 0 | No | Unknown | Positive | Positive | Positive | TP53, PTEN, PPM1D |
Positive |
| DF59 | HGSOC | Ascites | 7 | Yes | BRCA1 5385insC |
Positive | Positive | Positive | Not performed | Positive |
| DF68 | HGSOC | Ascites | 5 | Yes | BRCA1 Q563X |
Positive | Positive | Positive | TP53, BRCA1, PTEN (copy loss) |
Positive |
| DF83 | HGSOC | Ascites | 4 | Yes | Unknown | Positive | Negative | Positive | TP53, CDKN2A (copy loss) |
Positive |
| DF86 | HGSOC | Ascites | 6 | Yes | BRCA1 del exons 21–24 |
Positive | Positive | Positive | TP53, BRCA1, APC |
Positive |
| DF101 | HGSOC | Pleural fluid |
2 | Yes | BRCA1 187delAG |
Positive | Positive | Positive | TP53, BRCA1, NBN, PTEN (copy loss) |
Positive |
| DF106 | HGSOC | Ascites | 1 | Yes | Unknown | Positive | Positive | Positive | TP53, CDKN2A (copy loss) |
Positive |
| DF118 | HGSOC | Ascites | 1 | Yes | Unknown | Positive | Positive | Positive | TP53 | Positive |
| DF149 | HGSOC | Ascites | 0 (previously treated for breast cancer) |
No | Wild type | Positive | Patchy | Positive | TP53 | Positive |
| DF172 | Mixed serous and endometrioid |
Ascites | 2 | Yes | Unknown | Positive | Patchy | Positive | TP53, RET, RAD51C |
Positive |
| DF181 | HGSOC | Ascites | 7 | Yes | Wild type | Negative | Negative | Positive | TP53, BRIP1 | Negative |
| DF216 | Adenoca | Ascites | 2 | Yes | Wild type | Positive | Positive | Positive | TP53 | Positive |
Ascites-derived ovarian PDX models reflect clinical ovarian cancer
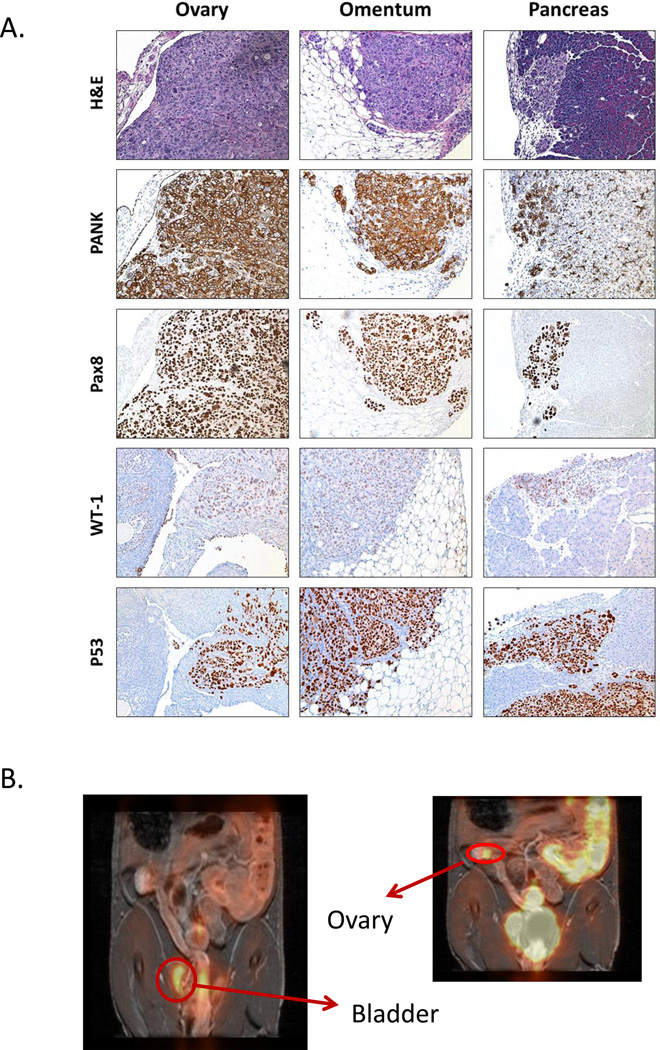
We performed necropsy on each of the luciferized, orthotopic PDX tumor-bearing mice upon reaching an ascites endpoint (distended abdomen or ~40% body weight gain) and major organs were analyzed for histopathology. All the PDX tumors exhibited diffusely disseminated peritoneal disease with tumor cell infiltration of the omentum, ovaries, pancreas, bowel, mesentery, spleen, pancreas, liver and diaphragm along with ascites and abdominal distention, consistent with clinical ovarian cancer. Representative images of luciferized DF216 (DF216-Luc) PDX tumor infiltration to the pancreas, ovary, and omental tissues are shown in Figure 1A. Disease dissemination was also assessed by FDG-PET in a model of DF86-Luc and demonstrated presence of disease in the ovary and near the bladder (Figure 1B).
Figure 1.
PDX models of ovarian cancer demonstrate same pattern of metastasis as clinical ovarian cancer, as seen on histology (A) or imaging by FDG-PET (B). PDX models demonstrate IHC marker expression patterns consistent with HGSOC (A).
Immunohistochemistry (IHC) of all luciferized PDX models revealed pan-cytokeratin staining, confirming epithelial origin. In addition, tumor tissue in most models demonstrated PAX8 and WT1 expression, consistent with epithelial ovarian cancer (Table 1). Comparison of IHC from multiple passages of a representative PDX, DF68, to the original patient tumor demonstrated preservation of histologic features, including positive staining for PAX8, p53, CK7 and Ki67, implying that histological fidelity of the model to the original patient sample is conserved across multiple serial passages (Supplemental Figure 2).
PDX models maintain molecular fidelity to primary ovarian tumors
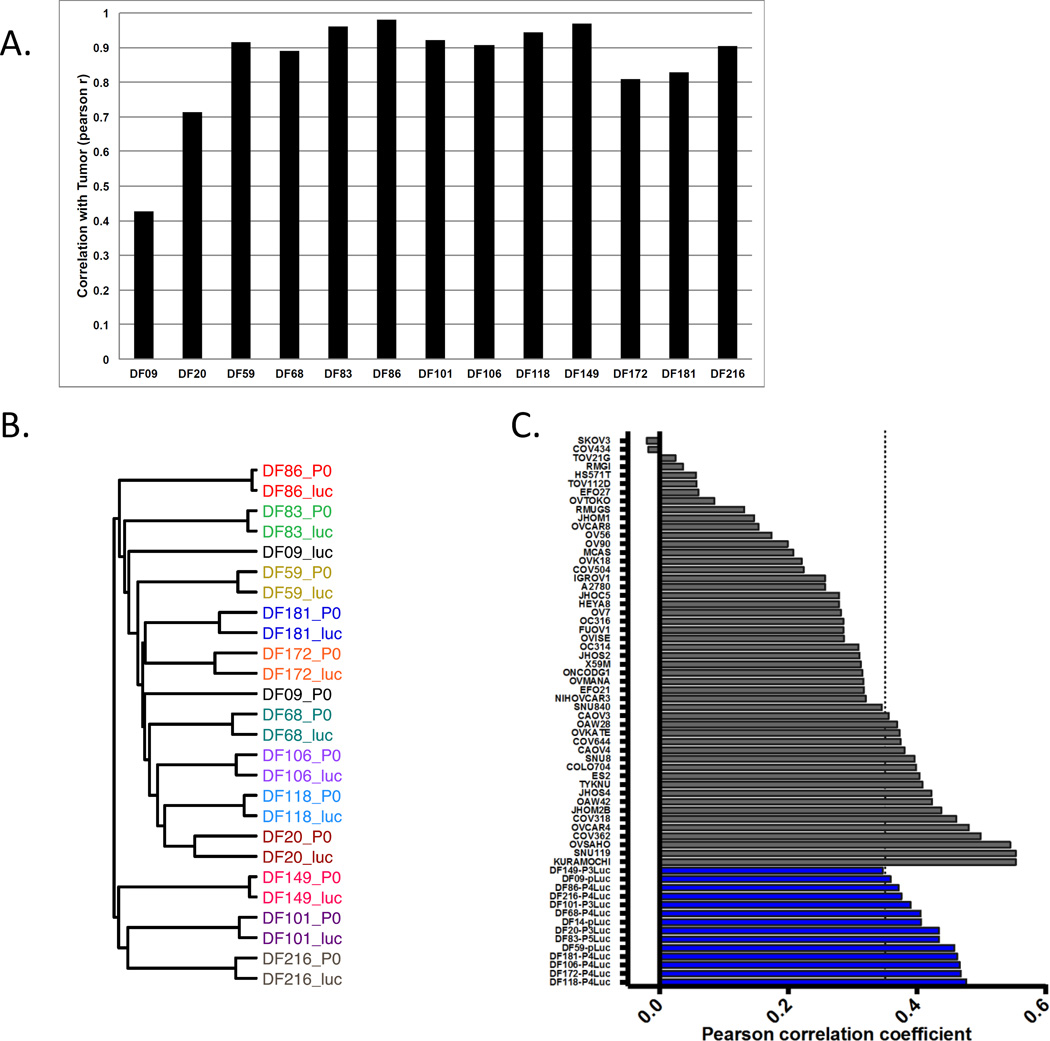
Genomic copy number variations (CNVs) can change during establishment of luciferized PDX lines. To compare the tumor and PDX genomes, patient tumor and luciferized PDX DNA from 13 sample pairs were subjected to array CGH. We found that the CNV profiles of 11 luciferized PDX lines are highly correlated (Pearsons r = >0.8) with their matched tumor sample (Figure 2A, Supplemental Table 2). A representative karyotype view of CNV profiles of DF86-Luc and its matched pre-luciferized PDX and initial patient tumor is shown in Supplemental Figure 3. Two PDX lines, DF09 and DF20, have moderate correlation with coefficients of 0.4 and 0.7, respectively. These results indicate that most of the PDX lines maintain the CNV profile of the original tumor with high fidelity. Unsupervised hierarchical clustering of the CNV data (Figure 2B) demonstrates that, with the exception of DF09, all of the samples from the same patient cluster more closely than unrelated samples. Clustering also reveals that the CNV profiles display inter-tumor heterogeneity between patients. This heterogeneity is maintained in the PDX lines, suggesting that this panel of PDX lines reflects the diversity in CNV profiles of the HGSOC patients. To assess the suitability of these lines to model HGSOC, we used the method described in Domcke et al. (2). In this analysis, the luciferized PDX CNV profiles were compared to the mean CNV profile of all HGSOC samples in The Cancer Genome Atlas (TCGA). We found that the PDX lines display high copy number Pearson correlation coefficients indicating that they are suitable HGSOC models (Figure 2C, blue bars). In contrast, most publicly available ovarian lines (Figure 2C, gray bars) display lower coefficients, suggesting that the luciferized PDX lines are more suitable models of HGSOC than most established ovarian cell lines.
Figure 2.
Copy number variation (CNV) analysis to evaluate fidelity of luciferized PDX models and relevance to TCGA tumors. (A): The graph depicts the correlation coefficients comparing CNV profiles of the original patient tumors and luciferized PDX (black bars) models over all genes. Most models have very high correlation coefficients. (B): PDX models maintain the heterogeneity of CNV profiles of the original patient samples. The dendrogram was derived from unsupervised hierarchical clustering of CNV data from the patient tumors (p0) and luciferized lines (luc) using Pearson’s distance and average linkage. Nearly all of the samples from the same patient cluster more closely than unrelated samples. (C): Luciferized PDX models are suitable models for HGSOC using the method developed by Domcke et al (2). Luciferized PDX models have a high copy number Pearson correlation coefficient with the mean CNV profile derived from all TCGA HGSOC samples. Dotted line represents the threshold for suitability of established cell lines as models for HGSOC (2). Established ovarian cancer cell lines are shown in gray; PDX-Luc models are shown in blue.
Matched patient tumor and luciferized PDX DNA samples from 11 PDX sample sets were subjected to BROCA panel targeted DNA sequencing analysis (Table 1 and Supplemental Table 3). Although this targeted panel does not definitively distinguish between tumor-specific (somatic) and germline alterations, candidate somatic mutations in a tumor can be compared to the PDX derived from the same patient. Luciferized PDX models demonstrated close fidelity to the primary patient tumors in terms of gene alterations detected by BROCA panel analysis. Where new mutations were detected in the PDX, these mutations generally represented a small fraction of the tumor cells based on variant allele fraction. Of note, all 11 samples demonstrated the presence of TP53 mutation in both the primary patient tumor and in the luciferized PDX models, consistent with HGSOC phenotype. Where available, BROCA data were compared to clinical annotation. As germline BRCA mutation testing was not standard of care when many of the specimens were collected, BRCA status was available in only 7 models. In the three models with a known germline BRCA deletion where BROCA testing was performed, the presence of this mutation was detected in both the patient tumor and the luciferized PDX sample.
We also performed whole-exome sequencing analyses to compare genetic alterations between two non-luciferized PDXs and the patient tumor cells from which the PDXs were derived, as well as from matched normal blood (Supplemental Tables 4–5). We observed 82 somatic mutations in the DF101 patient tumor and 86 somatic alterations in the matched early passage PDX. All 82 of the mutations from the patient tumor were present in the PDX, while the four additional somatic mutations not observed in the patient tumor were present at a low mutant allele frequency (<20%) in the PDX. Somatic alteration of TP53 and homozygous deletion of PTEN was detected in both the DF101 PDX and the corresponding patient tumor. These data highlight that the DF101 patient tumor and matched PDX showed high concordance among the sequence alterations identified, and essentially perfect concordance for sequence alterations with moderate to high mutant allele frequencies (>20%). We observed similar results for the DF149 PDX and its patient tumor.
Overall, data from these analyses indicated that the PDXs maintain high fidelity with regards to the genetic alterations and copy number variation profiles of the patient tumors.
Molecular diversity in HGSOC PDX Tumors
It is increasingly recognized that significant molecular diversity exists, even within more narrowly defined subtypes such as HGSOC. Targeted sequencing analyses of the PDX models demonstrated multiple alterations within the BROCA panel, including three BRCA mutations, one BRIP1 mutation, two PTEN copy losses, and two CDKN2A losses (Table 1).
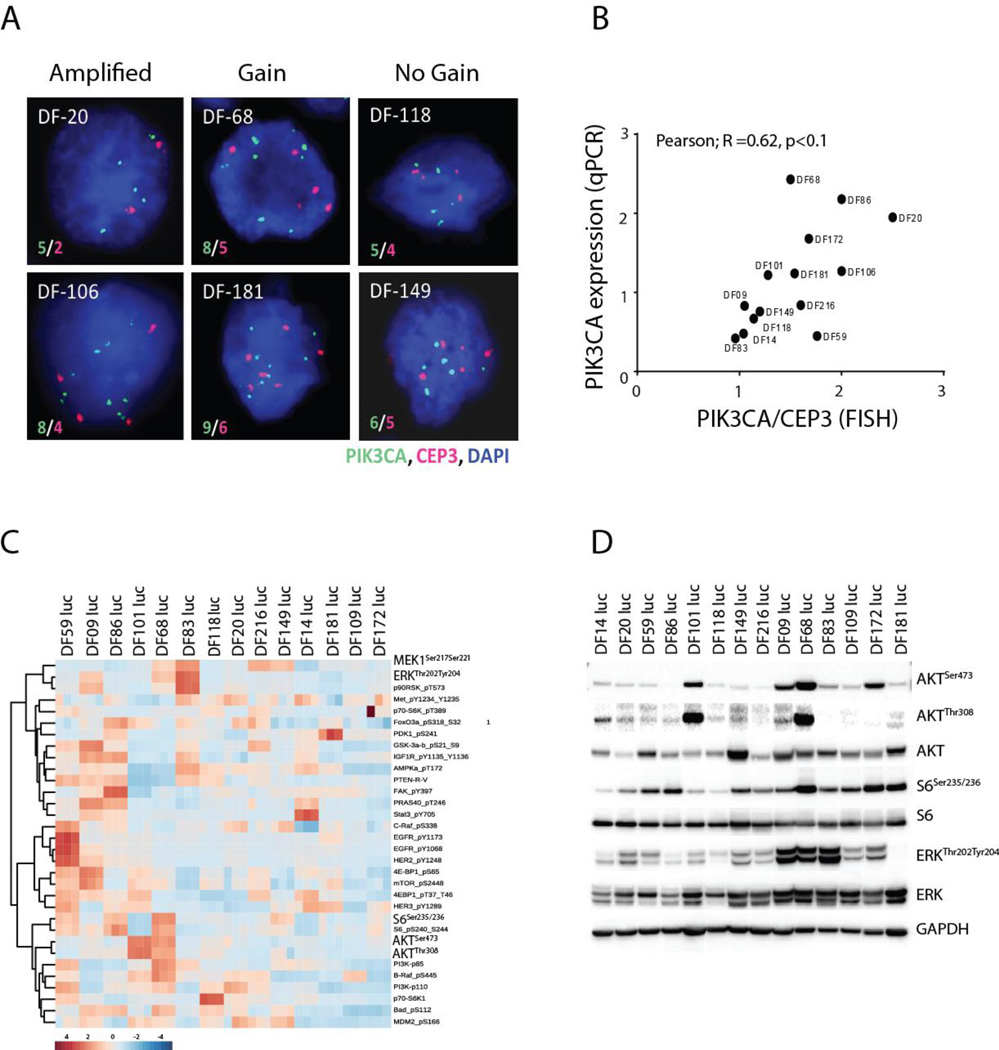
To further assess diversity within the PDX models with regard to potential druggable targets, and as PI3K pathway signaling is frequently altered in HGSOC (10), we assessed the activation of the PI3K and other canonical signaling pathways (Figure 3). FISH analysis demonstrated that certain models demonstrated amplification, gain, or no gain of PIK3CA (Figure 3A, Supplemental Table 6), and that FISH PIK3CA score correlated with expression of PIK3CA as assessed by qRT-PCR (Figure 3B). Across the PDX models, there was wide variability in extent of PI3K pathway activation as well as other signaling pathways, as assessed by RPPA (Figure 3C). The phosphorylation levels of pERK, pS6K, and pAKT were assessed by Western blot on the same protein lysates and correlated with levels reported by RPPA (Figure 3D). Clustering of activated proteins within known canonical pathways was observed, with major clusters showing co-phosphorylation between AKT and its downstream targets, the ERK pathway, and the EGFR pathway (Supplemental Figure 4). In addition, phospho-proteins representing adjacent nodes in a given signaling pathway (e.g. phospho-MEK and phospho-ERK) were also highly correlated (Spearman r=0.61, p <0.0001)) (Supplemental Figure 4), supporting the internal validity of the RPPA data. These results demonstrate that the PDX models demonstrate diversity on a genetic and signal transduction pathway level.
Figure 3.
PIK3CA amplification by FISH (representative images in (A)) correlates with expression by qRT-PCR (B) and varies across PDX models. Activation of PI3K pathway proteins, as assessed by RPPA, also varies extensively across models. Each sample was assayed in triplicate, log-2 transformed followed by z-score transformation across samples and phosphoproteins (C) and correlates with assessment by Western blot (D).
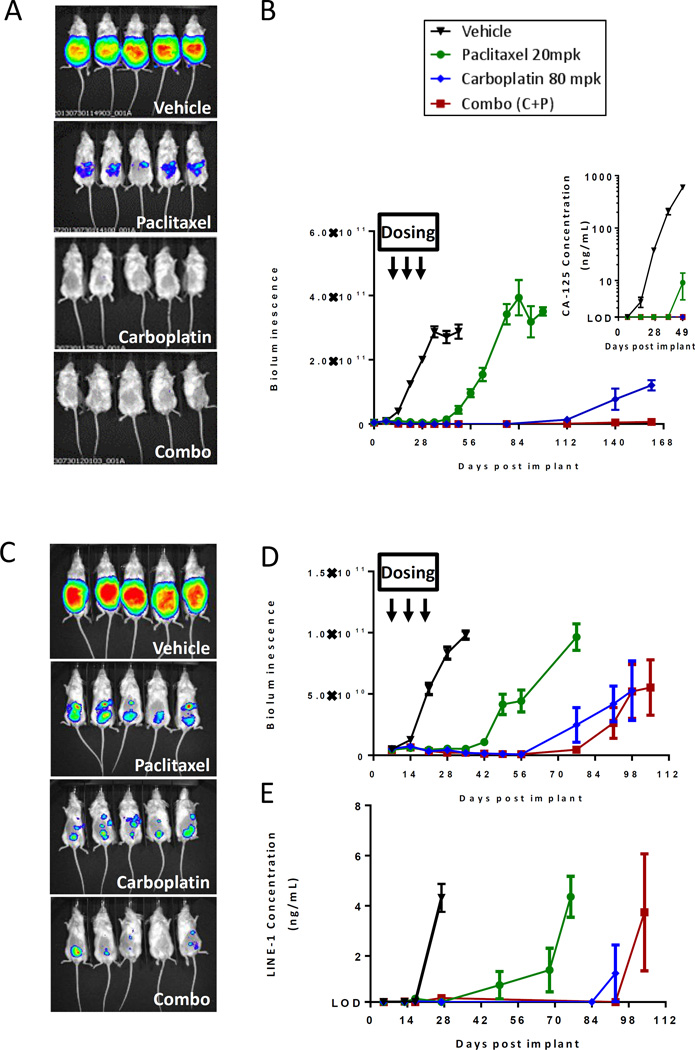
Luciferized PDX models can reproducibly model response to cytotoxic chemotherapy agents
A key goal of this study was to generate models where tumor growth or response could be modeled in a reproducible manner by bioluminescent imaging (BLI), thereby avoiding the need to employ potentially more time-intensive or less reproducible methods of animal imaging, such as MRI or ultrasound. Cohorts of 10 NSG mice bearing luciferized PDX tumors were therefore treated with vehicle, carboplatin, or paclitaxel, either as monotherapy or in combination, and followed by weekly BLI measurements. As illustrated in Figure 4, BLI reproducibly demonstrated the effectiveness of therapy in the DF14-Luc and DF181-Luc PDX models. Serial plasma CA125 levels or LINE-1 biomarkers served as surrogate biomarkers and changes within these values correlated with BLI signal (Figures 4B inset, 4E, Supplemental Figure 5), demonstrating the consistency of response evaluation across different assay platforms. Of note, detectable plasma CA125 levels were present in 13 of the 14 luciferized PDX models at the terminal ascites endpoint (Table 1).
Figure 4.
Luciferized PDX models DF14-Luc (A-B) and DF181-Luc (C-E) can reproducibly model response to standard of care chemotherapy agents and can be serially followed by BLI imaging (A, C), by serum CA125 (B insert), or by serum LINE-1 assay (E). All measurements are represented as Mean + SEM.
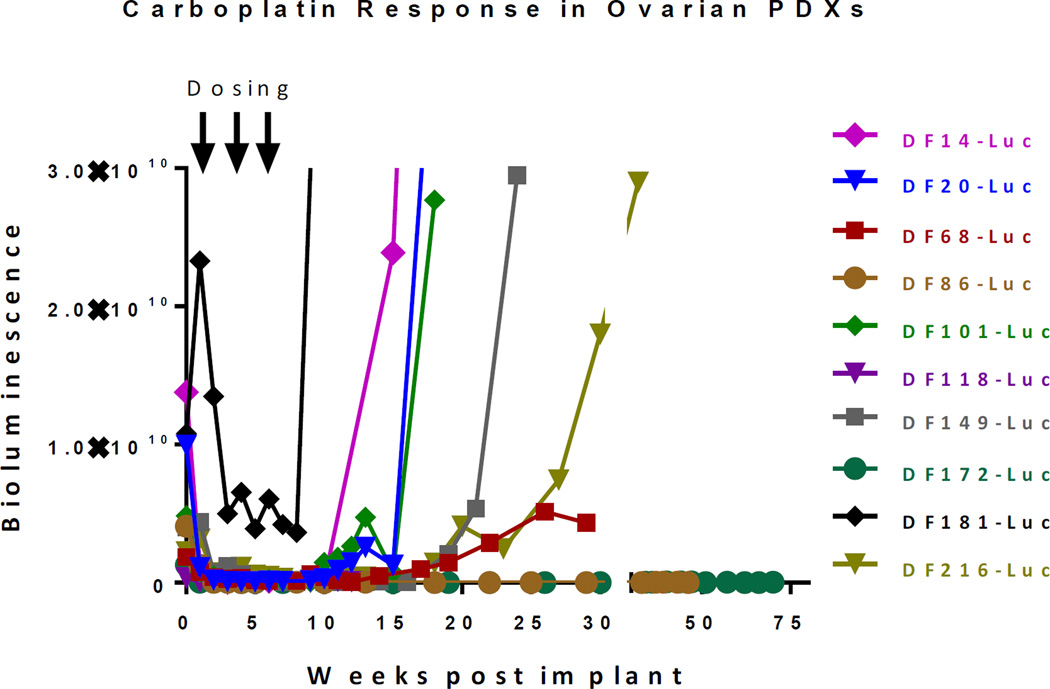
Although all models except for DF20 were obtained from patients who had clinically platinum-resistant disease at the time of tumor sample collection (defined as growth on platinum or within 6 months of the last platinum regimen), differential sensitivity to carboplatin was still observed in the panel of PDX models. To model the degree of platinum sensitivity, PDX models were treated with three doses of weekly carboplatin (80 mg/kg) and the degree of response and time to recurrence following treatment were assessed. As seen in Figure 5, variability was seen within the models in terms of sensitivity to carboplatin, with some models demonstrating early recurrences and higher degree of platinum-resistance (DF181-Luc), while others demonstrated sustained remission following treatment (DF86-Luc, DF172-Luc). Of note, two of the three models which were derived from patients with platinum-refractory disease (DF14-Luc, DF181-Luc, DF216-Luc) demonstrated the most resistance to carboplatin.
Figure 5.
Sensitivity to carboplatin varies across PDX models. 3–4 mice were implanted with each of the luciferized PDX models. Mice with established BLI signals were treated with 3 weekly treatments of carboplatin (arrows) and followed for subsequent tumor regrowth.
Discussion
In this paper, we describe the establishment of a panel of 14 molecularly characterized and clinically annotated luciferized PDX models in which tumor growth and kinetics can be reproducibly followed by BLI as well as plasma biomarker assays. We have found that histologic and molecular features are preserved through multiple passages of PDX models as well as post-luciferization, suggesting that these PDX models, despite their high degree of genomic instability, continue to faithfully reflect the genomic characteristics and pathophysiology of HGSOC through serial passages. Consistently, these PDX models respond to standard of care chemotherapy in a manner reflective of the clinical behavior of ovarian cancer. The PDX models span a range of platinum sensitivity, and their responses can be followed either by BLI or plasma CA125 and LINE-1 biomarker assays.
Our findings are consistent with those seen in other PDX models of ovarian cancer (7, 8) in terms of histologic and molecular fidelity; however, they differ in other aspects. While engraftment rates of 74% (8) and 83% (7) have been described in other collections of ovarian cancer PDXs, our engraftment rate was notably lower (31%). This may be because our protocol utilized tumor cells isolated from ascites to establish PDX models, while both Weroha et al. and Topp et al. utilized tumor fragments obtained during surgery. Additionally, we implanted tumor cells in irradiated nude mice, while other PDX model collections were generated with either SCID or NSG mice, which may also alter engraftment rates. Despite the lower engraftment rate, our evidence that it is feasible to generate robust clinically-relevant orthotopic ovarian PDX models from ovarian cancer cells isolated from human ascites is important. As surgery is frequently not clinically indicated in advanced recurrent disease, tumor fragments may be difficult to obtain in this setting. In contrast, the accumulation of ascites is a common event in recurrent disease and is frequently removed for palliation. Thus, this methodology allows for the generation of PDX models that may better reflect the biology of recurrent treatment-resistant disease.
One significant feature of the PDX model system described in this manuscript is the ability to follow the burden of intraperitoneal disease in a reproducible and less labor-intensive manner. The luciferization of each of the PDX models allows for the use of BLI, a robust and reproducible in vivo imaging technique, in following the burden of disease in these ovarian PDX models. Additionally, in our PDX models, plasma CA125 could be detected in 13 of 14 models and correlated with tumor response. Of note, the CA125 assay utilized in this study was specifically designed for detection of CA125 from small quantities of blood, allowing for more sensitive detection and the ability to serially monitor CA125 levels over time in mouse models. We also further demonstrate that the use of a separate assay to detect human LINE-1, though not translatable to human studies, may be equally effective in monitoring disease burden in mouse studies and can be followed in models where CA125 levels are below the levels of detection.
Importantly, the models established in this study represent clinically relevant molecular categories of HGSOC, with targeted and whole-exome analyses revealing that the models display a spectrum of alterations in various DNA repair genes, as well as models which do not demonstrate such alterations. Characterization of PI3K and other pathway signaling across these models also supports their diversity and value in modeling multiple subtypes of HGSOC. The accompanying molecular annotation also makes these models a valuable tool to assess the efficacy of targeted agents in specific molecular backgrounds. Their diversity, both with regard to DNA damage repair gene mutations as well as with regard to the PI3K and other signaling pathways, was striking and likely reflects the inter-individual diversity within even a defined histological subtype such as HGSOC. This molecular diversity underscores the necessity to screen a large array of pre-clinical models when planning treatment studies, rather than relying on cell line response data.
In summary, we have now established a collection of 14 luciferized PDX models of ovarian cancer, accompanied by molecular and clinical characterization. These models can now serve as a platform for further therapeutic development and proof-of-concept validation of novel therapeutic strategies in ovarian cancer.
Supplementary Material
Translational Relevance.
We have established a molecularly diverse panel of 14 clinically annotated and luciferized patient-derived xenograft (PDX) models of high-grade serous ovarian cancer which demonstrate immunohistologic and molecular fidelity to the original patient tumor. Unlike previously reported ovarian PDXs that rely on caliper or radiographic measurements of tumor, our PDX models allow for robust orthotopic modeling of ovarian cancer in the intraperitoneal space by bioluminescent imaging as well as by serum biomarkers. Response to standard of care chemotherapies can be reproducibly modeled in these ovarian PDXs. The models have been characterized with regards to DNA repair pathway alterations, copy number variation, and activation of key signaling pathways, such as PI3K. This PDX collection represents a valuable platform for target identification and validation of novel therapies or therapeutic combinations in ovarian cancer.
Acknowledgments
We thank Dr. Lynda Chin for initial array CGH characterization of several non-luciferized PDX models, Dr. George Demetri for support of PDX development, Anh Tran for project management and administrative support, Sanam Dharma, Shruti Shah, John Murgo, Melissa Buttimer and Justin Evangelista for technical assistance, Tanya Tupper for FDG-PET analysis, Barbara Smith and James Folcrum for DFCI Animal Resource Facility support, and Roderick Bronson for rodent pathology.
Funding:
This work was supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (R.D., V.E.V., J.S.B., G.B.M., and I.Z.), US National Institutes of Health grants CA121113 (V.E.V.), CA006973 (V.E.V.), CA083636 (R.D.), CA152990 (R.D.), K12CA08772307 (J.F.L.), Expect Miracles Foundation (Belfer Center for Applied Cancer Science, DFCI), The Commonwealth Foundation (V.E.V.), the Pallotta Investigatorship (J.F.L.), the Ovarian Cancer Research Fund (J.F.L.), and the Honorable Tina Brozman Foundation for Ovarian Cancer Research (R.D.).
Footnotes
Conflicts of Interest: None of the authors report any conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nature communications. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elias KM, Emori MM, Papp E, MacDuffie E, Konecny GE, Velculescu VE, et al. Beyond genomics: critical evaluation of cell line utility for ovarian cancer research. Gynecologic oncology. 2015;139:97–103. doi: 10.1016/j.ygyno.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitra AK, Davis DA, Tomar S, Roy L, Gurler H, Xie J, et al. In vivo tumor growth of high-grade serous ovarian cancer cell lines. Gynecologic oncology. 2015;138:372–377. doi: 10.1016/j.ygyno.2015.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer discovery. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nature reviews Clinical oncology. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topp MD, Hartley L, Cook M, Heong V, Boehm E, McShane L, et al. Molecular correlates of platinum response in human high-grade serous ovarian cancer patient-derived xenografts. Molecular oncology. 2014;8:656–668. doi: 10.1016/j.molonc.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weroha SJ, Becker MA, Enderica-Gonzalez S, Harrington SC, Oberg AL, Maurer MJ, et al. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:1288–1297. doi: 10.1158/1078-0432.CCR-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh T, Lee MK, Casadei S, Thornton AM, Stray SM, Pennil C, et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12629–12633. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Science translational medicine. 2015;7:283ra53. doi: 10.1126/scitranslmed.aaa7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertotti A, Papp E, Jones S, Adleff V, Anagnostou V, Lupo B, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263–267. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular cancer therapeutics. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 16.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer cell. 2003;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 18.Yan ZH, Madison LL, Burkhardt A, Yu J, Tayber O, Li Z, et al. Analysis of two pharmacodynamic biomarkers using acoustic micro magnetic particles on the ViBE bioanalyzer. Analytical biochemistry. 2011;410:13–18. doi: 10.1016/j.ab.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Rago C, Huso DL, Diehl F, Karim B, Liu G, Papadopoulos N, et al. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer research. 2007;67:9364–9370. doi: 10.1158/0008-5472.CAN-07-0605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.