Abstract
Purpose
To propose and validate Structural Correlation based Outlier REjection (SCORE), a novel algorithm for removal of artifacts arising from outlier control-label pairs in 2D Arterial Spin Labeling (ASL) data.
Materials and Methods
The proposed method was assessed with respect to other state-of-the-art ASL signal processing approaches using 2D pulsed ASL data obtained with 3T Siemens scanner from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. Longitudinal data from control participants acquired 3 months apart were used to assess within subject coefficient of variation (wsCV) based on the assumption that the optimal signal processing strategy will minimize control subject retest variability. SCORE was further evaluated by determining its sensitivity for distinguishing patients with Alzheimer’s disease (AD) from Controls based on hypoperfusion in predefined ROIs that are known to be sensitive to AD related changes.
Results
SCORE coupled with a preprocessing step to discard few extreme outliers (combined algorithm referred to as SCORE+) reduced wsCV up to 21% in grey matter and 39% in smaller ROIs compared to the reference algorithms. It also provided an average increase in effect size for patient-control differences of 50% compared to other algorithms in a priori ROIs sensitive to AD related changes. This increase was statistically significant (p<0.05) for the majority of the ROIs and methods as evaluated by permutation tests.
Conclusion
CBF maps generated with SCORE or SCORE+ provide improved retest reliability in control subjects while simultaneously increasing sensitivity to pathological CBF effects between controls and patients.
Keywords: Cerebral Blood Flow, Arterial Spin Labeling, Outlier rejection, Alzheimer’s disease, ADNI
INTRODUCTION
Cerebral Blood Flow (CBF)1, 2 is defined as the volume of blood flowing through a specific region of brain tissue per unit time. It is a key physiological quantity used as a biomarker for brain function.3, 4 Arterial Spin Labeled (ASL) perfusion MRI5, 6 provides a non-invasive approach for quantifying regional CBF without the use of exogenous tracers or radioactivity. Instead, ASL uses arterial blood water as an endogenous tracer to measure CBF by magnetically labeling the inflowing arterial blood with radiofrequency pulses. The perfusion signal is acquired as the difference between images with and without arterial spin labeling (referred as tagged or labeled and control images respectively), which is subsequently converted into quantitative CBF based on established models of blood flow.7, 8 Absolute quantification can also be obtained by calibrating the measurements using CBF measured from independently acquired method like phase contrast MRI.9 The difference signal between the tagged and control images usually accounts for around 1% of the background signal,10 resulting in a low signal to noise ratio (SNR). To compensate for the low SNR, multiple tag-control pairs are typically acquired and averaged.
Arterial spin labeling can be achieved using various approaches, the most popular and widely used of which are pulsed ASL (PASL)11 and pseudo continuous ASL (PCASL).12 Because of its higher SNR, PCASL is presently the recommended labeling strategy.13 The effects of ASL can be sampled using a variety of imaging strategies. Most ASL data has been acquired with echo-planar imaging (EPI) owing to its speed and sensitivity, however 3D ASL using stack-of-spiral fast spin echo (FSE)12 or gradient and spin echo (GRASE)14 imaging are increasingly used and preferred. Background suppression (BS) of static brain signals can dramatically increase the temporal signal to noise ratio (TSNR) of ASL MRI15–17 and is optimally combined with a 3D imaging scheme.
CBF quantification using ASL is susceptible to artifacts originating from head motion or other contaminating sources. Typical ASL scans last for several minutes, resulting in several individual label and control image pairs, and the mean CBF map is obtained by averaging these pairs. Within this limited number of perfusion difference images, outliers with large positive or negative values can dominate the signal averaging process and the resulting mean CBF map.18, 19 Various signal processing strategies have been proposed to reduce artifacts in ASL CBF maps.18–28 Miranda et al.28 proposed to discard outliers originating from motion by estimating motion in the label-control time series and rejecting images with more than 2 mm translation or 1.5° rotation between successive images. Head motion (both the absolute motions and motion difference between each control and its corresponding label image) and global signal deviations were also used to identify outlier time points in Wang et al.20 Tan et al.18 proposed to detect outliers based on mean and standard deviation within individual CBF volumes and reject the volume if the quantities are outside some predefined range from the mean of those quantities across time. Maumet et al.24 used a robust statistical approach to compute the representative map. An adaptive outlier cleaning approach (AOC) proposed by Wang et al.19 used the mean CBF as the reference and iteratively removed time-points based on the degree to which they vary from the reference. Outliers identified based on head motion estimations were removed before calculating the initial mean in the AOC.
Several conditions can limit the efficacy of the above methods for rejecting outliers. First, outliers may not all be associated with significant head motion. In addition, motion correction algorithms already at least partially reduce errors in the raw time series data introduced by motion. Hence, it is the residual uncorrected motion that should be considered for outlier detection and not necessarily the motion estimated by the motion correction algorithm, which have already been compensated. Second, when severe artifactual contamination is present in individual maps in the time series, they can dominate the mean CBF map. Consequently, application of methods based on similarity with mean CBF map might result in the undesirable effect of preserving the contaminating tag-control pair, or even worse removing “good” tag-control pairs.
Artifacts originating from outliers is most common when sub-optimal labeling techniques, such as the PASL, are applied and when background suppression15 is not available. While a number of groups have begun to use 3D ASL acquisitions with BS and better labeling properties,29 less optimal strategies are still being used because of their wide availability and experience in implementation. In addition, there is a considerable amount of data acquired with these approaches, e.g. the extensive Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu), for which improved signal processing schemes are needed to derive optimal physiological information.
The goal of this work is to present a novel strategy for detecting and discarding individual CBF volumes in ASL time series that contaminate mean CBF maps. The algorithm is dubbed as structural correlation based outlier rejection (SCORE) based on the principle from which it was derived. The performance of the proposed method was compared with several state-of-the-art reference algorithms using the premise that the best algorithm should provide maximum repeatability in control subjects, while simultaneously maximizing the differences between control and patient cohort expected to be differing in CBF values.
MATERIALS AND METHODS
Cohort
The data acquired in this study were acquired in ADNI-2, which is the third phase of ADNI (after ADNI and ADNI-GO). More information can be obtained at http://adni-info.org. ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). Baseline scans for 60 “amyloid-β negative” controls (age: 71.4±6.5 years, range: 56–85 years, Gender: 53% female) and 49 “amyloid-β positive” AD patients (age: 74.4±8.7 years, range: 56–89 years, gender: 47% female) were used for assessing group differences. 51 of these amyloid-β negative controls had a second MRI 3 months after their baseline scan, and these subjects were considered for assessing repeatability. All participants had signed written, informed consent for the brain MRI and for all other procedures, as reviewed and approved by the institutional review boards at each clinical site.
MRI Data Acquisition
ADNI ASL data were acquired using the Siemens product PICORE PASL sequence with the Q2TIPs technique for defining the spin bolus. The acquisition parameters are TR/TE=3400/12 ms, TI1/TI=700/1900 ms, FOV=256 mm, 24 sequential 4 mm thick slices with a 25% gap between the adjacent slices, partial Fourier factor = 6/8, bandwidth=2368 Hz/px, and imaging matrix = 64 × 64. The first volume of the 105 ASL acquisitions was used as the M0 image with the remaining 104 volumes used as 52 control-label pairs. Structural images in ADNI were acquired using a 3D MPRAGE T1-weighted sequence with TR/TE=2300/2.98 ms, 176 sagittal slices, within plane FOV=256×240mm2, voxel size=1.1×1.1×1.2 mm3, flip angle=90, bandwidth=240 Hz/px.
ASL Pre-Processing
ASL data were pre-processed using custom MATLAB scripts (The Mathworks Inc. Natick, Massachusetts, USA), SPM8 (Wellcome Department of Imaging Neuroscience, London, UK, http://www.fil.ion.ucl.ac.uk/spm/software/spm8/) and ASL toolbox.20 Each raw EPI time series was motion corrected using the method by Wang21. The method consists of estimating a 6-parameter rigid body motion spatial transformation, and subsequently regressing out the spurious motion component caused by the systematic label/control alternation from the motion parameters before applying the transformation on the images. The mean EPI images were automatically coregistered to the high-resolution T1 images using SPM8. In parallel, the structural images were segmented into grey matter (GM), white matter (WM) and cerebrospinal fluid (CSF) tissue probability maps (TPMs) using the segmentation tool in SPM8. Binary masks comprising of GM, WM and ventricular CSF were created to restrict the computation of CBF within these masks. Image masks and the TPMs were subsequently resliced to the native ASL space.
The M0 image of each subject was co-registered to the mean EPI image and smoothed using an isotropic Gaussian kernel with full-width-half-max (FWHM)=5mm to avoid noise propagation13. Pairwise subtraction of the resulting images was performed and the difference images were converted to absolute CBF maps using the formula13
where ΔM is control-label difference, λ is the blood:brain partition coefficient, ω is the post labeling delay, T1,blood is the T1 of blood, α is the tagging efficiency, M0 is the equilibrium magnetization of the brain (separately acquired, coregistered to mean EPI and smoothed) and τ is the labeling duration. In the present work, λ=0.9ml/g, ω = 1.9s, T(1,blood)=1650 ms, α=0.98, and τ=700ms. Voxels outside the brain mask were set to zero. The CBF time series was subsequently used for the proposed outlier rejection scheme, as well as the other reference methods.
A local template for all subjects was generated using Diffeomorphic Anatomical Registration Through Exponentiated Lie Algebra (DARTEL)30 based on their segmented grey and white matter probability maps. The local template was subsequently registered to the MNI space using a linear affine transformation. These two transformations were used to map the CBF maps and grey matter TPM to the MNI space. The images in the MNI space were smoothed with a FWHM=8mm isotropic Gaussian kernel to avoid potential errors in transformation and to enhance matching of voxels. The MNI-normalized GM TPM was binarized with a threshold of 0.4 to create a grey matter mask. Region of interest (ROI) analyses for each subject were performed only within this mask.
SCORE Algorithm
The SCORE algorithm iteratively discards outlier CBF volumes and is detailed below.
Identification Of A Potential Outlier
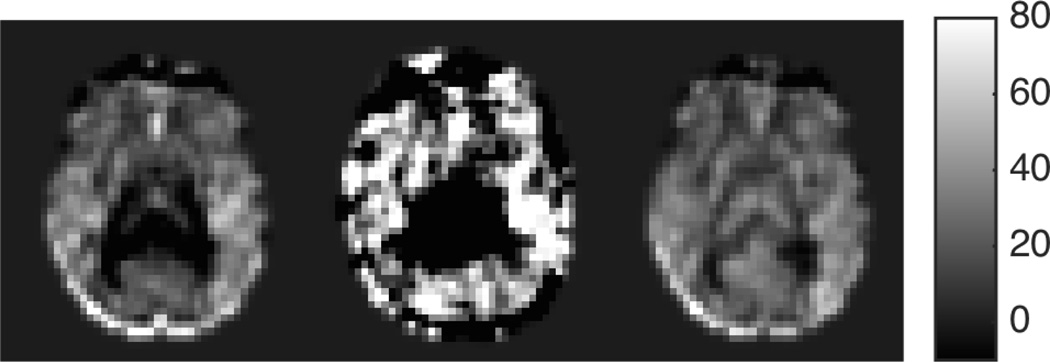
Outlier identification in SCORE is based on our hypothesis that for a mean CBF map with significant spatially constrained artifacts caused by a few corrupted volumes, the individual volume in the time series contributing most to the artifact of the mean CBF is the one that shows the highest spatial correlation with the mean. Non-outliers on the other hand represent a mixture of true mean CBF map, random noise and physiological CBF fluctuations, and show a smaller correlation because of the dominance of the random noise in individual volumes. While temporal averaging reduces the random noise in the resulting mean, the artifacts present in the outliers would remain in the mean CBF map. So the artifact-containing mean CBF would show a higher similarity to outliers compared to non-outliers. Hence the CBF volume having highest spatial correlation with the intermediate mean CBF map is iteratively removed, followed by an update of the intermediate mean using the remaining CBF volumes until no more outliers are detected. An example of mean CBF map, most correlated CBF volume and the output after removing 18 most correlated CBF images for a sample dataset is shown in the left, middle and right subplots of Fig. 1 respectively.
Fig. 1.
(LEFT) Mean CBF in a representative slice from all label/control pairs for a particular subject; (MIDDLE) the most structurally correlated pair to the mean CBF; and (RIGHT) Output of SCORE after discarding 18 CBF volumes based on structural correlation.
The above iterative procedure of discarding the most correlated volume with the mean CBF requires a stopping criterion. Hence, categorization of the most correlated volume as outlier or non-outlier is an important step of this process and is considered next.
Iteration Stopping Criterion
The stopping criterion used in SCORE assumes that the variability within each tissue type has three components: within tissue heterogeneity, random noise and artifact. Under ideal conditions, individual CBF maps in the time series are contaminated by random noise only and not by structured artifacts of large magnitude. Temporal averaging of the CBF maps reduces the random noise and in turn reduces the variance within each tissue type, viz. GM, WM, and CSF. On the other hand, addition of an outlier volume containing spatially constrained artifact to this mean CBF will increase the variance within the tissue where the artifact is present. Hence, the spatial variance within each tissue type is an indicator of the presence of artifact. An increase in the variance from the preceding iteration after removing the most correlated pair implies an effect of removal of random noise rather than structured artifact. In contrast, a decrease in variance from the previous iteration would imply removal of an outlier volume containing structured noise. Therefore, as a criterion of stopping the iteration of removing the most correlated pair, the variance within each tissue type can be monitored. In this work, we considered the pooled variance, defined as
where Vk is the variance and Nk is the number of voxels in the kth tissue (k=GM,WM,CSF), and the iteration is stopped when there is an increase in the pooled variance from the previous iteration.
Preprocessing to Remove Extreme Outliers
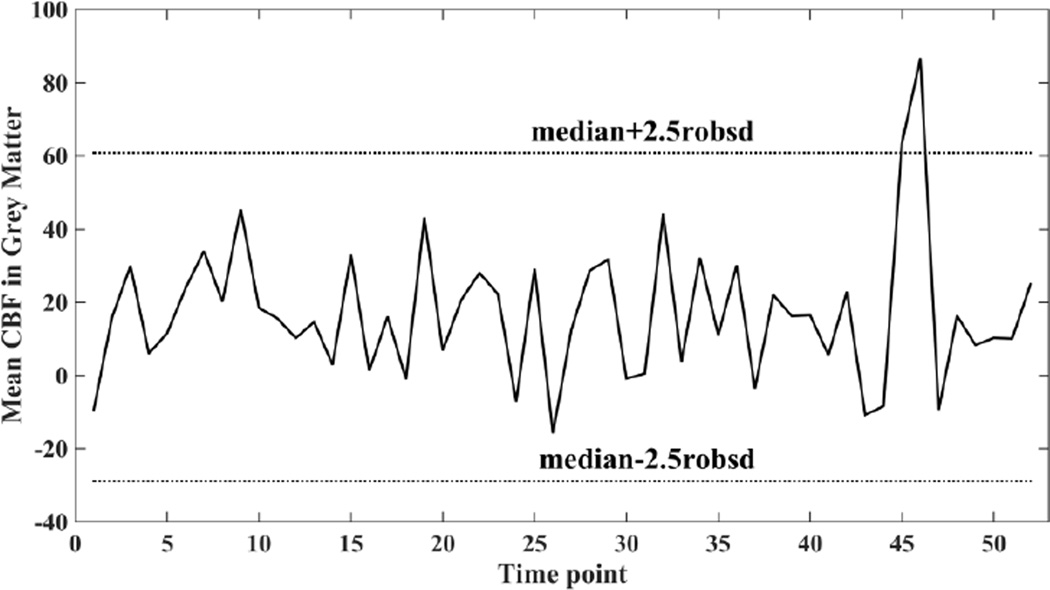
Using a preprocessing step that discards a small number of extreme outliers before running SCORE can further enhance its performance. Such outliers are identified as CBF volumes with mean CBF abnormally different compared to overall distribution of their values in the time series. We computed mean CBF in GM (referred to as mean GM-CBF below) of the CBF volumes in the time series and discarded volumes with mean GM-CBF outside 2.5 standard deviations of their means. Since the mean and standard deviation can be impacted by large outliers, instead median and 1.4826 × median absolute deviation31 were used as robust measures of distribution center and spread respectively. Fig. 2 shows a plot of mean GM-CBF of the CBF volumes in the time series for a sample subject with the exclusion limits indicated in dotted lines. Two outlier volumes having noticeably larger mean GM-CBF values compared to the other volumes were identified as outliers and discarded. Note that this procedure can potentially retain volumes having non-physiological negative mean GM-CBF. However, discarding all volumes having negative mean GM-CBF can systematically bias the mean CBF to a higher value, since some negative CBF values are expected at the level of individual control-label volumes due to noise.
Fig. 2.
Plot of mean CBF in grey matter in different volumes of the CBF time series of a representative subject along with median ± 2.5 robsd thresholds. Robsd stands for robust standard deviation and is computed as 1.4826 × median absolute deviation.
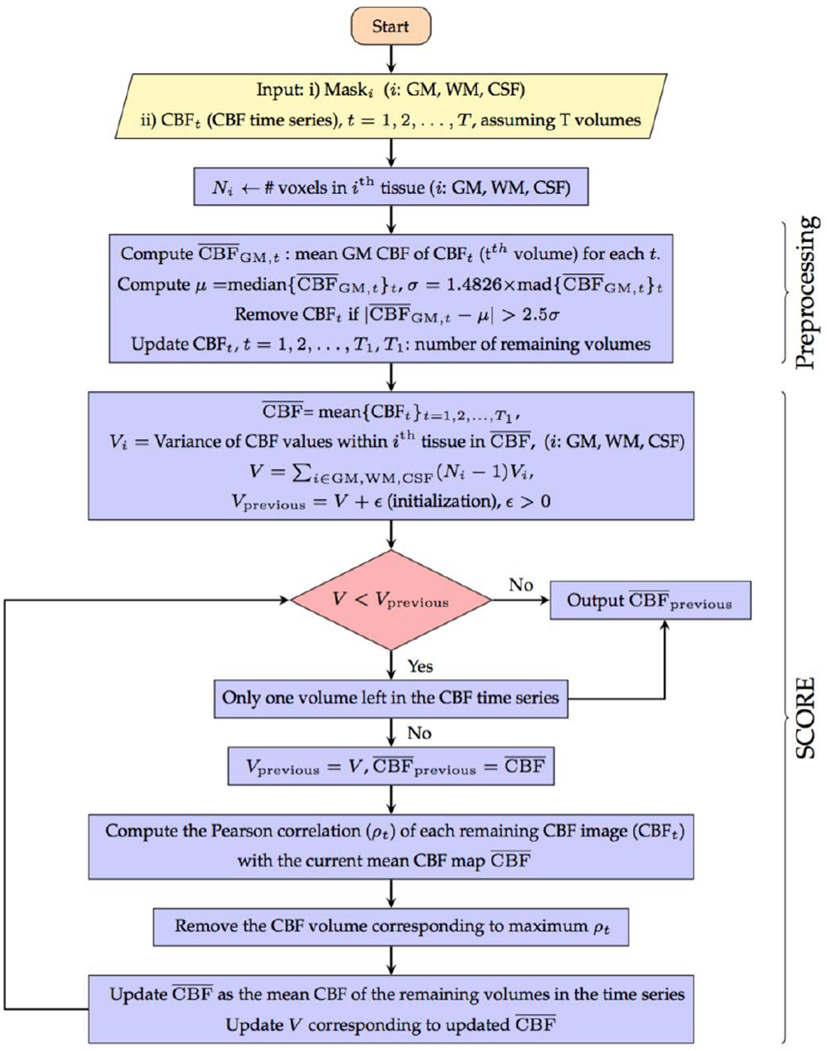
SCORE along with the preprocessing step has been referred to as SCORE+ in the remainder of this paper. The complete flow chart of SCORE+ is shown in Fig. 3.
Fig. 3.
Flow chart of SCORE+
Comparison Methods
The performance of SCORE and SCORE+ was compared with several alternative algorithms that represent the state-of-the-art in published signal processing strategies. As the first reference method, the basic simple average (labeled as SA) of all the CBF volumes in the time series was considered for comparison. Second, the mean and standard deviation based filter18 was implemented (labeled as MSD). The method computes the mean and standard deviation of the intensities within the brain for each CBF volume (as explained above in ASL pre-processing section) in the time series and removes those volumes as outliers, if the z-scores of the means and the standard deviations are outside some predefined range. Specifically, denoting mi and si as the global mean and standard deviations of each CBF volume in the time series, a volume is detected as outlier if it satisfies one of the following criteria18
The remaining volumes are averaged to compute the final CBF map. Third, the method proposed by Maumet et al.24 was used. This method estimates the intensity at each voxel of the final CBF map by using Huber’s M-estimation (labeled as HME) of the CBF values along the temporal direction corresponding to that voxel. Fourth, a pipeline proposed by Fazlollahi et al.27 was used that adapts a GLM analysis for confound (nuisance parameter) removal from the raw EPI time series, similar to the work by Wang and others19, 21. Confounds consist of estimated motion parameters, global signal and also signal from CSF in which there is no neural activity.23 The method is labeled as nuisance cleaning (NC). The final comparison method considered in this paper was the adaptive outlier cleaning (labeled as AOC) approach19, which can be considered as a further refinement of NC. After application of nuisance cleaning, it removes CBF volumes based on head motion, global signal, and finally employs an iterative approach to remove the CBF volumes that are least correlated with the mean CBF map. Conceptually the last step of AOC is opposite to SCORE. The abbreviations of the reference algorithms are summarized in Table 1.
Table 1.
Abbreviations of the reference algorithms.
Details of Statistical Analyses
We report the average number of CBF volumes discarded by SCORE, preprocessing step of SCORE+, MSD, and AOC. The other algorithms do not discard volumes explicitly. For the SCORE+ preprocessing step, we also report the average absolute difference between median of the time series and the mean GM-CBF of the discarded volume expressed in terms of robust standard deviation of the series. Furthermore, for each method, we compared the number of volumes discarded in controls and AD patients based on two sample T tests. For all statistical hypothesis testing, p<0.05 was considered significant.
Motion is usually considered as the primary source of artifact present in ASL CBF volumes. We investigated how relative motion between controls and their corresponding label images are associated with outliers in the time series based on the estimated motion parameters used to realign the raw EPI time series. The rigid-body motion estimation algorithm estimates 6 parameters, 3 translational (x, y, z) and 3 rotational (pitch, roll, yaw), corresponding to each EPI volume. We computed relative motion as the absolute difference between SPM generated motion parameters of the corresponding control and label images. These relative motion parameters in retained and SCORE+ discarded volumes were compared using two sample T tests.
SCORE and SCORE+ were first compared with the reference algorithms based on visual inspection. Artifacts generally manifested as extensive negative or extremely high CBF values in resulting CBF maps. In addition, quantitative comparisons were performed based on the retest variability and patient-control differences described below.
Retest Variability
Retest variability using repeated scans at an interval of 3 months from 51 ADNI control subjects was used as a metric for evaluating the effectiveness of the signal-processing algorithms. The assumption was that an optimal strategy will result in minimum variation between CBF values within the same ROI in two sets of scans of controls obtained only a few months apart. Denoting CBFt,k and CBFr,k as the mean CBF within a specific ROI for the test and retest sessions of the kth subject, the coefficient of variation for each subject (CVk for the kth subject) and the within-subject CV (wsCV) for that specific ROI were computed as
where std denotes the standard deviation operation and N is the number of subjects. The denominator in the expression of CV used the mean value across subjects and sessions to make it less susceptible to outliers. We report the results of wsCV for each method-ROI combination. In addition, to analyze the distribution of CV across methods and ROIs, we compared the boxplots of CV for each method-ROI combination.
Sensitivity to Patient-Control Group Difference
Although improved test-retest reliability is a desirable property, it cannot be considered as the sole criterion to validate a signal processing strategy. For example, a homogenous map of fixed value for all the subjects will result in maximum agreement, but is meaningless physiologically. Hence, in addition to test-retest reliability, we assessed the efficacy of the algorithms by examining their sensitivities in evaluating control-patient differences. In particular, we looked at the differences in CBF values between controls and AD patients within hippocampus, precuneus and posterior cingulate cortex, regions that are known to be sensitive to AD-related CBF changes32–34. The motor cortex was also included in the comparison as a control region, as mean CBF in this region is relatively unchanged in AD relative to controls. We computed effect sizes between controls and patients within each ROI as the difference between mean CBFs of the two groups divided by the pooled standard deviation of the two groups. Subsequently, the effect sizes obtained from the different algorithms were compared based on permutation testing with 10,000 permutations. We also report the mean CBFs for each group-ROI combination and p values corresponding to group differences based on two-sample t tests.
RESULTS
We analyzed a total of 160 data sets (60 controls, 51 of which repeated, and 49 AD patients), each having 52 CBF volumes (or equivalently control-label pairs). Out of these 52 volumes, an average of 9.7±5.8 (min: 0, max: 31) volumes were discarded based on SCORE. In addition, 2.1±1.7 (min: 0, max: 7) volumes were discarded in the preprocessing step of SCORE+. The mean GM-CBF in these volumes deviated from their median in the time series by 3.7±1.4 robust standard deviations. There were no statistically significant differences between volumes rejected in Controls and AD patients based on mean GM-CBF (p=0.45) or structural correlation (p=0.83) criteria. The number of volumes discarded by SCORE+ was significantly higher (p<0.0001) than in MSD (3.9±1.5) or AOC (7.9±3.2). Only AOC discarded higher number of volumes in patients than in controls (p=0.01).
Both translational and rotational relative motion components associated with volumes discarded by SCORE+ were found to be significantly higher (p<0.001) than in retained volumes. Translation in y direction and rotational pitch showed the largest differences between the two groups. The small number of volumes discarded by the preprocessing step were associated with the largest motion parameters.
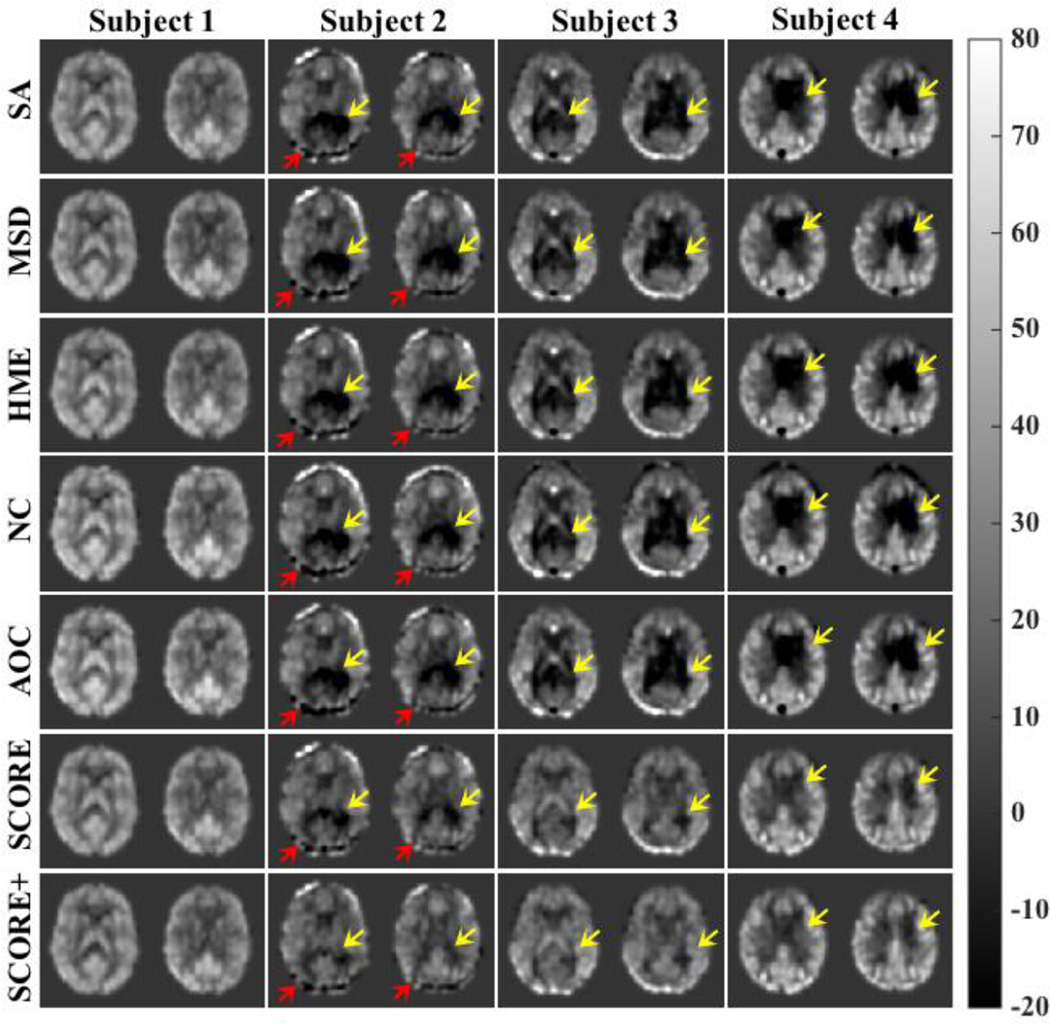
Fig. 4 shows representative slices from CBF maps of selected subjects obtained using different algorithms. The first column shows example of a CBF map with no visible artifacts and all the algorithms performed comparably. CBF maps of subjects 2–4 obtained using SA had noticable artifacts manifested as extensive negative CBF values. The yellow arrows point to locations of these artifacts. While the artifacts are largely present in the output of the other comparing algorithms, SCORE and SCORE+ showed reduced artifacts in the resulting maps. For example, the subcortical grey matter structures are not visible in the CBF maps of subject 3 obtained from any of the reference algorithms, whereas SCORE and SCORE+ provided a much better visualization of these structures. Red arrows in the CBF map of subject 2 provides an example of artifact that was not removed by any of the algorithms, including SCORE and SCORE+.
Fig. 4.
CBF maps from 4 subjects obtained with the reference algorithms, SCORE, and SCORE+. The CBF map of Subject 1 shows no visible artifact and all the algorithms performed comparably. The CBF maps of subjects 2,3 and 4 obtained from each of the reference algorithms have visible artifacts manifested by negative CBF values (indicated by yellow arrows). SCORE and SCORE+ showed reduced artifact in the resulting maps. However, artifacts indicated by red arrows in the CBF maps of subject 2 are not removed by any of the algorithms.
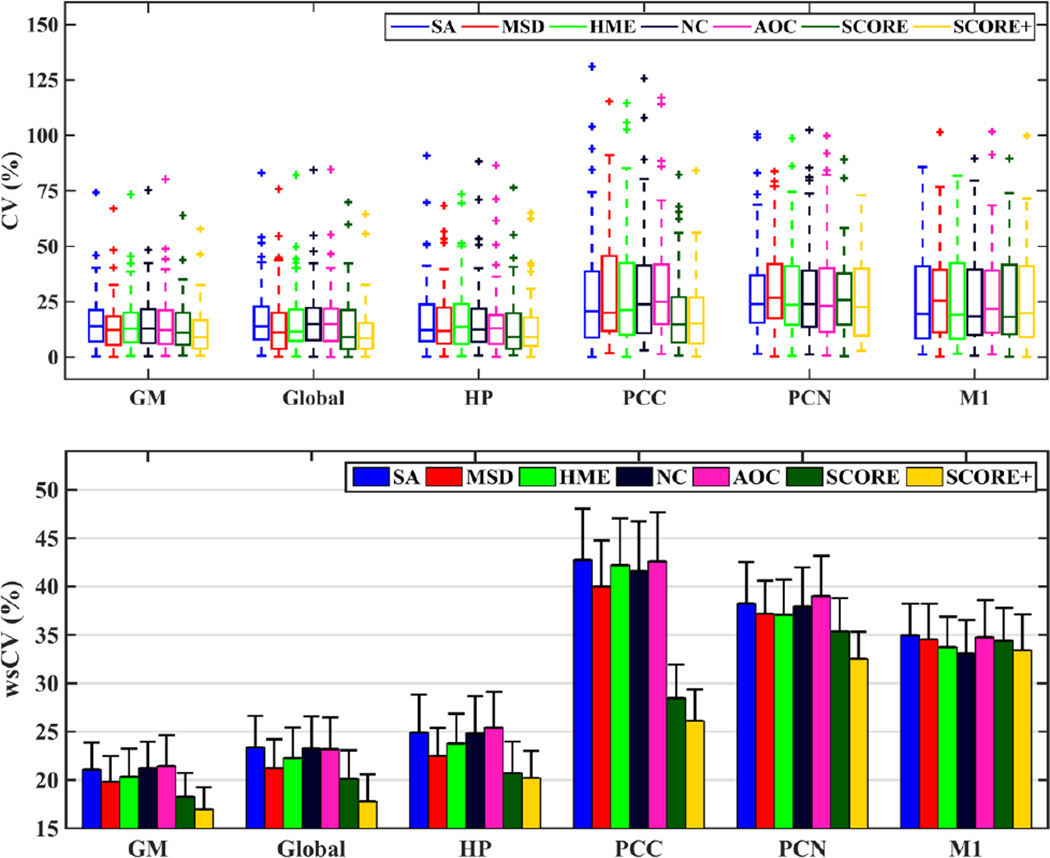
Fig. 5 shows the performance of each algorithm in the repeatability study. The top subplot shows the coefficients of variation of mean CBF values obtained from control subjects scanned twice, 3 months apart. The box plots are grouped based on the ROIs while the performance of each algorithm for that ROI is demonstrated within each group. SCORE (dark green) provided lowest maximum CV among the different methods while producing lowest number of outliers in majority of the cases. SCORE+ (yellow) provided further improvement on top of SCORE. The bottom subplot shows wsCVs grouped similarly. Except in motor cortex, both SCORE and SCORE+ provided noticeable reductions in wsCVs with SCORE+ being consistently lowest. SCORE provided an improvement of 5–15% in larger ROIs such as GM and whole brain while improvement was 5–33% in the smaller ROIs. Improvement with SCORE+ ranged between 14% and 24% in larger ROIs and 10–39% in smaller ROIs. In the motor cortex, wsCV with SCORE increased by 2% compared to HME and 4% compared to NC and decreased by about 1% compared to the other algorithms. SCORE+ had 1% increase in wsCV compared to NC, but 1–4% decrease compared to the other algorithms. The other algorithms produced mixed results with MSD providing marginally improved performance compared to the other algorithms in most of the cases. AOC provided marginally weaker agreement compared to NC, indicating that potentially incorrect volumes were removed in the outlier rejection process.
Fig. 5.
(TOP) Box plots of coefficients of variation (CV) for test-retest of CBF values in grey matter (GM), whole brain (global), hippocampus (HP), posterior cingulate cortex (PCC), precuneus (PCN), and primary motor cortex (M1) obtained using different algorithms. (BOTTOM) wsCV for different ROIs and for each method. The error bars show the standard errors in each case.
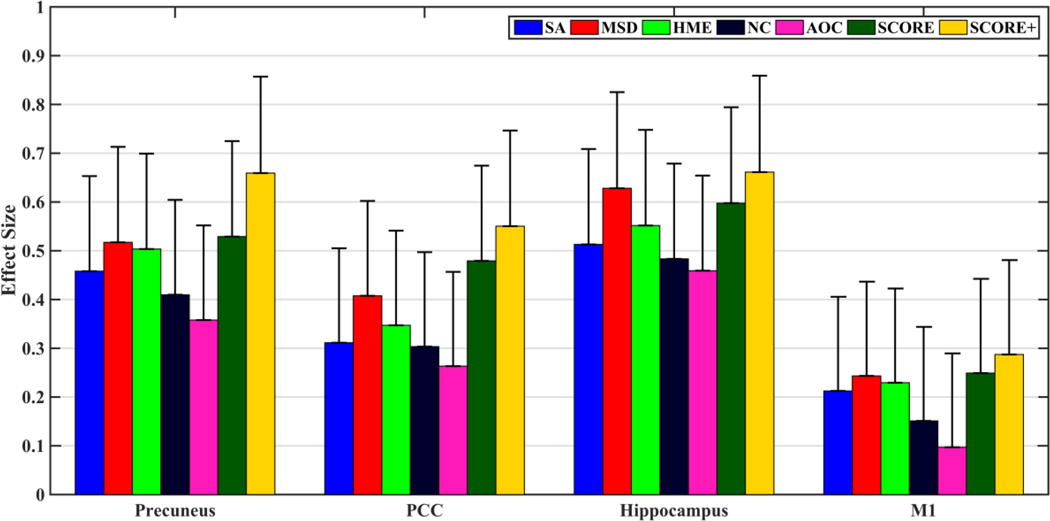
Fig. 6 shows effect sizes for discriminating 49 AD patients from 60 controls based on mean CBF in precuneus, posterior cingulate cortex, hippocampus and motor cortex ROIs for each of the data cleaning methods, while Table 2 lists the relevant statistics. An increased effect size using SCORE+ is readily observable for all ROIs. Based on permutation tests, SCORE+ performed either significantly better (p<0.05) or trended towards significance (P<0.12) for all methods and regions except hippocampus for MSD, where the performance was comparable (p=0.36). SCORE also showed improvements in precuneus and PCC while performance was worse than MSD in hippocampus. Effect sizes in M1 are much smaller compared to the other ROIs implying relative stability of this region in the AD population. The first two columns of Table 2 provide the means and standard deviation values for the controls and the patients. In addition, the table also shows the effect sizes (same as that shown in Fig. 6) and the p values corresponding to two sample T tests for each method corresponding to different ROIs. The standard deviations were lowest in the case of SCORE and SCORE+ in almost all the cases. In the case of hippocampus, all the algorithms provided statistically significant group differences. Similar results were obtained for precuneus as well, except for AOC. For PCC, only MSD, SCORE and SCORE+ produced group differences that were statistically significant. As expected, none of the algorithms show statistically significant group differences in the motor cortex. As with the test-retest example, AOC demonstrated weaker performance compared to NC.
Figure 6.
Effect sizes in Control-AD discrimination based on CBF in precuneus, posterior cingulate cortex (PCC), hippocampus and motor cortex (M1). The error bars show the standard errors in each case. SCORE produced largest effect sizes compared to the reference algorithms.
Table 2.
ROI CBF values and effect sizes for patient-control differences for the evaluated signal processing strategies (Mean±standard deviation and other statistics as specified)
| Mean CBF in Control |
Mean CBF in AD |
Effect Size |
p value in two sample T test |
|
|---|---|---|---|---|
| Precuneus | ||||
| SA | 21.22±8.83 | 16.80±10.61 | 0.46 | 0.019 |
| MSD | 21.52±9.38 | 16.34±10.73 | 0.52 | 0.008 |
| HME | 21.75±8.85 | 16.81±10.86 | 0.50 | 0.010 |
| NC | 22.43±8.86 | 18.19±11.93 | 0.41 | 0.036 |
| AOC | 21.91±8.88 | 18.28±11.52 | 0.36 | 0.066 |
| SCORE | 21.52±9.28 | 16.31±10.50 | 0.53 | 0.007 |
| SCORE+ | 22.01±9.15 | 15.71±10.03 | 0.66 | 0.001 |
| PCC | ||||
| SA | 30.57±12.10 | 26.38±15.00 | 0.31 | 0.109 |
| MSD | 30.92±11.83 | 25.62±14.30 | 0.41 | 0.037 |
| HME | 30.92±12.25 | 26.12±15.53 | 0.35 | 0.074 |
| NC | 32.34±12.46 | 28.08±15.80 | 0.30 | 0.118 |
| AOC | 31.75±12.65 | 28.00±15.95 | 0.26 | 0.174 |
| SCORE | 31.55±10.79 | 26.09±12.08 | 0.48 | 0.014 |
| SCORE+ | 31.93±10.66 | 25.77±11.83 | 0.55 | 0.005 |
| Hippocampus | ||||
| SA | 26.43±8.65 | 22.17±7.88 | 0.51 | 0.009 |
| MSD | 26.87±8.10 | 22.02±7.24 | 0.63 | 0.002 |
| HME | 26.54±8.32 | 22.11±7.64 | 0.55 | 0.005 |
| NC | 27.81±9.08 | 23.58±8.32 | 0.48 | 0.014 |
| AOC | 27.63±8.91 | 23.64±8.40 | 0.46 | 0.019 |
| SCORE | 26.24±7.91 | 21.70±7.21 | 0.60 | 0.002 |
| SCORE+ | 26.43±7.83 | 21.63±6.46 | 0.66 | 0.001 |
| Motor Cortex | ||||
| SA | 21.89±11.71 | 19.36±12.11 | 0.21 | 0.272 |
| MSD | 22.15±11.81 | 19.32±11.38 | 0.24 | 0.209 |
| HME | 22.25±11.75 | 19.58±11.56 | 0.23 | 0.236 |
| NC | 22.98±11.77 | 21.09±13.29 | 0.15 | 0.434 |
| AOC | 22.26±11.73 | 21.08±12.86 | 0.10 | 0.616 |
| SCORE | 21.86±11.37 | 19.05±11.15 | 0.25 | 0.199 |
| SCORE+ | 22.43±11.42 | 19.20±10.95 | 0.29 | 0.139 |
Discussion
Using 2D PASL data obtained from the ADNI2 database, the results of this study demonstrate that both repeatability in controls and patient-control discrimination can be improved through data cleaning, with the SCORE and SCORE+ algorithm outperforming comparison methods from the literature18, 19, 24, 27. SCORE exploits the notion that artifacts may dominate the mean CBF map derived from ASL time series data and hence individual volumes containing artifact show a high spatial correlation with the mean map. Although counterintuitive, SCORE considers the most similar volume with the mean CBF as a potential outlier. All the other algorithms evaluated instead rely on the mean as the reference. For example, MSD is dependent on the overall mean of the individual means and standard deviations for each volume in the time series. Since similar artifacts might be present in multiple tag-control pairs, mean values may be dominated by artifact leading to retention of contaminating volumes. The HME method uses a voxel-wise outlier removal and is not constrained by the spatial structure of the outliers. The NC approach improves the temporal standard deviation by removing variability across time series, but this does not necessarily improve the mean CBF map. AOC did not improve on NC suggesting an inability to identify the outliers correctly and potentially deleting non-outlier volumes for very noisy data because of its dependence of the corrupted mean CBF map as the reference. The number of volumes discarded in MSD and AOC were significantly lower than SCORE+, indicating possible retention of artifacts in the data.
Although SCORE is based on the idea of removing the volume most similar to the mean, SCORE+ additionally incorporates a preprocessing step that relies on the dissimilarity with the median of the distribution of the mean GM-CBF values. The use of median and robust standard deviation makes this dissimilarity approach less susceptible to outliers than approaches based on mean differences.35 Although the preprocessing step only removes a small number of volumes (2 on an average), its use seems to improve the performance of SCORE. A possible reason for this is that while SCORE is designed to remove spatially constrained artifacts, the correlation criterion is agnostic to any large global bias present in individual CBF volumes. For example, SCORE might not discard a CBF volume with notably different mean GM-CBF compared to the other volumes if the extreme values are not caused at least in part by spatially constrained artifacts. On the other hand, inclusion of such a volume will result in a different value of mean CBF. It should be noted that MSD, which performed comparably to SCORE in distinguishing controls and patients, also relies on discarding volumes with extreme global CBF values, although MSD uses mean and standard deviations and not their robust versions to identify outliers. In summary, the two steps of SCORE+ seem to be complementary and reduce errors in mean CBF maps differently.
Visual examination of the mean CBF map does not always reveal the superiority of one method over others, so we also relied on alternative quantitative validation strategies. As a potential biomarker of regional brain function, ASL CBF should have the characteristics of i) being reproducible in repeated scans within a homogeneous group, and ii) different between diverse groups. The validation strategies used in this study for the different signal processing algorithms reflect these two scenarios.
Repeatability was assessed in ADNI-2 control subjects scanned after a 3 month interval. Although CBF decreases with age even in healthy controls,36 3 months is a relatively short interval with respect to human aging. While there certainly can be real physiological variations and individual subjects can demonstrate a change in CBF over a period of three months, it is reasonable to assume that CBF remains stable across a group of adult controls within this interval. SCORE+ not only provided the lowest wsCV, which reflects the average deviation between scans across subjects, but it also produced the least number of outliers in majority of the cases. These findings suggest that SCORE+ is an effective and efficient cleaning strategy for 2D ASL data.
Comparison of ADNI-2 cognitively normal adults to patients with Alzheimer’s disease was used to assess for sensitivity to group differences, as differences in CBF between these two groups have been repeatedly demonstrated.32–34 All the algorithms demonstrated similar differences in mean values between controls and AD patients. However, the standard errors in SCORE and SCORE+ were lower than the other methods, which lead to the larger effect sizes for group discrimination, implying greater statistical power and lower sample sized for detecting significant group differences. The reduction in standard error can be attributed to a reduction in artifacts in SCORE and SCORE+.
SCORE+ is based on the idea of removing complete CBF volumes from ASL MRI time series. When a complete volume is discarded, voxels without artifact potentially suffer from loss of SNR. However, the average number of volumes rejected was ~12 out of 52 in the dataset considered in this study. Thus, any voxel lacking artifact would only have a reduction in SNR on average. Accordingly, the effect of loss of SNR can be small compared to the detrimental effect of the larger artifacts in other brain regions. Furthermore, most biomedical applications of ASL MRI primarily focus on group effects in which inter-subject variability (random effects) is the major source of noise. In such applications, sample size is much more important than the SNR of an individual measurement.37
The Control-AD group discrimination provided mostly medium effect sizes and none of the algorithms have been able to achieve a large effect size (>0.8). This is primarily because of the characteristic low SNR of PASL data. Better labeling and acquisition strategies coupled with background suppression may provide a better differentiation of the Control and diseased groups. In addition, although CBF decreases with age and the mean age of the population under study is over 70, the mean absolute CBF values within each ROI shown in Table 2 are all somewhat lower than might be expected. This may reflect lower labeling efficiency than expected and/or incorrect assumption of model parameters used to quantify CBF.
Motion appeared to be a significant source of artifacts as evident from our analysis. Although motion compensation algorithms are employed in ASL pre-processing, potential incorrect estimation and interpolation can result in residual motion effects. Rigid body motion compensation algorithms also cannot effectively remove secondary motion effects resulting from magnetic susceptibility differences. Motion effects between control and corresponding label images were found to contaminate the resulting CBF volumes significantly. Motion in y direction and pitch (both implying nodding motion) seemed to have the highest effect.
Although the results demonstrated in the current study are for PASL, SCORE can be applied to continuous or pseudo-continuous ASL (CASL and PCASL) without any modification. In addition, it can potentially also be applied to background suppressed 3D ASL with possible modification of the stopping criterion, though the expected benefit in the latter will likely be lower due to its higher temporal signal to noise ratio compared to ASL data acquired without BS29.
Since SCORE is based on a comparison between individual control-label volumes as compared to mean CBF across volumes, it does not remove artifacts that cancel out over successive volumes and so do not manifest in the mean CBF map. However, this is not a limitation for studies focusing on mean CBF rather than dynamic CBF changes. In addition, SCORE is an outlier rejection algorithm and hence CBF maps with residual artifacts can still occur when the contamination is not strictly due to artifacts in a small number of volumes. An example is shown in the result section where SCORE or SCORE+ did not remove some artifact in the posterior part of the brain of a subject. When small artifacts are present in almost all the volumes and each of them does not markedly influence the mean CBF, the effect of decrease in artifact due to removal of one time point is less than the increase in random noise due to one less volume available for averaging and these volumes are retained by the algorithm. Further signal processing on individual-voxel basis will be required to counteract this type of scenario.
This study lacks a gold standard for regional CBF quantification, which could have provided an alternate validation strategy.15O H2O PET is currently considered as a gold standard for CBF, but it is difficult to obtain in a large cohort because the method is costly, logistically challenging, and requires exposure to ionizing radiation. Instead, repeatability of CBF values has often been used21, 27 to validate signal-processing algorithms. In this study, we specifically considered the ADNI dataset because of its large cohort size and the availability of both repeated measures and data acquired from well-defined patient and control populations. In addition, ADNI PASL data has low SNR and hence particularly benefits from data cleaning strategies. However, it would be desirable to compare the algorithms using additional data,38 ideally including a gold standard measure of CBF.
In conclusion, the outlier rejection strategy based on structural correlation, ideally coupled with the preprocessing step, was found to provide a superior solution of estimating a mean CBF map from 2D ASL data as compared to previously proposed approaches. The utility of this approach for background-suppressed 3D ASL data remains to be established, and strategies are also needed to remove residual artifacts present throughout ASL time series data rather than in outlier volumes.
Acknowledgments
Grant Support:
The study was supported by grants from the National Institutes of Health, R01 MH080729 and P41 EB015893. Dr. Shinohara was partially supported by P30 NS045839. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
References
- 1.Kety SS, Schmidt CF. The determination of cerebral blood flow in man by the use of nitrous oxide in low concentrations. Am J Physiol. 1945;143:53–66. [Google Scholar]
- 2.Herscovitch P, Markham J, Raichle ME. Brain blood flow measured with intravenous H2(15)O. I. Theory and error analysis. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 1983;24:782–789. [PubMed] [Google Scholar]
- 3.Detre JA, Wang J, Wang Z, Rao H. Arterial spin-labeled perfusion MRI in basic and clinical neuroscience. Current opinion in neurology. 2009;22:348–355. doi: 10.1097/WCO.0b013e32832d9505. [DOI] [PubMed] [Google Scholar]
- 4.Mette D, Strunk R, Zuccarello M. Cerebral blood flow measurement in neurosurgery. Translational stroke research. 2011;2:152–158. doi: 10.1007/s12975-010-0064-y. [DOI] [PubMed] [Google Scholar]
- 5.Detre JA, Leigh JS, Williams DS, Koretsky AP. Perfusion imaging. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1992;23:37–45. doi: 10.1002/mrm.1910230106. [DOI] [PubMed] [Google Scholar]
- 6.Williams DS, Detre JA, Leigh JS, Koretsky AP. Magnetic resonance imaging of perfusion using spin inversion of arterial water. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:212–216. doi: 10.1073/pnas.89.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alsop DC, Detre JA. Reduced transit-time sensitivity in noninvasive magnetic resonance imaging of human cerebral blood flow. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1996;16:1236–1249. doi: 10.1097/00004647-199611000-00019. [DOI] [PubMed] [Google Scholar]
- 8.Buxton RB, Frank LR, Wong EC, Siewert B, Warach S, Edelman RR. A general kinetic model for quantitative perfusion imaging with arterial spin labeling. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 1998;40:383–396. doi: 10.1002/mrm.1910400308. [DOI] [PubMed] [Google Scholar]
- 9.Aslan S, Xu F, Wang PL, et al. Estimation of labeling efficiency in pseudocontinuous arterial spin labeling. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2010;63:765–771. doi: 10.1002/mrm.22245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong EC. Potential and pitfalls of arterial spin labeling based perfusion imaging techniques for MRI. In: Bandettini CTWMaPA, editor. Functional MRI. New York. 1999. pp. 63–69. [Google Scholar]
- 11.Wong EC, Buxton RB, Frank LR. Implementation of quantitative perfusion imaging techniques for functional brain mapping using pulsed arterial spin labeling. NMR in biomedicine. 1997;10:237–249. doi: 10.1002/(sici)1099-1492(199706/08)10:4/5<237::aid-nbm475>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 12.Dai W, Garcia D, de Bazelaire C, Alsop DC. Continuous flow-driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2008;60:1488–1497. doi: 10.1002/mrm.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alsop DC, Detre JA, Golay X, et al. Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2015;73:102–116. doi: 10.1002/mrm.25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunther M, Oshio K, Feinberg DA. Single-shot 3D imaging techniques improve arterial spin labeling perfusion measurements. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2005;54:491–498. doi: 10.1002/mrm.20580. [DOI] [PubMed] [Google Scholar]
- 15.Ye FQ, Frank JA, Weinberger DR, McLaughlin AC. Noise reduction in 3D perfusion imaging by attenuating the static signal in arterial spin tagging (ASSIST) Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2000;44:92–100. doi: 10.1002/1522-2594(200007)44:1<92::aid-mrm14>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Seara MA, Wang Z, Wang J, et al. Continuous arterial spin labeling perfusion measurements using single shot 3D GRASE at 3 T. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2005;54:1241–1247. doi: 10.1002/mrm.20674. [DOI] [PubMed] [Google Scholar]
- 17.Maleki N, Dai W, Alsop DC. Optimization of background suppression for arterial spin labeling perfusion imaging. MAGMA. 2012;25:127–133. doi: 10.1007/s10334-011-0286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan H, Maldjian JA, Pollock JM, et al. A fast, effective filtering method for improving clinical pulsed arterial spin labeling MRI. Journal of magnetic resonance imaging : JMRI. 2009;29:1134–1139. doi: 10.1002/jmri.21721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Das SR, Xie SX, et al. Arterial spin labeled MRI in prodromal Alzheimer's disease: A multi-site study. NeuroImage Clinical. 2013;2:630–636. doi: 10.1016/j.nicl.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Aguirre GK, Rao H, et al. Empirical optimization of ASL data analysis using an ASL data processing toolbox: ASLtbx. Magnetic resonance imaging. 2008;26:261–269. doi: 10.1016/j.mri.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z. Improving cerebral blood flow quantification for arterial spin labeled perfusion MRI by removing residual motion artifacts and global signal fluctuations. Magnetic resonance imaging. 2012;30:1409–1415. doi: 10.1016/j.mri.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wells JA, Thomas DL, King MD, Connelly A, Lythgoe MF, Calamante F. Reduction of errors in ASL cerebral perfusion and arterial transit time maps using image de-noising. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2010;64:715–724. doi: 10.1002/mrm.22319. [DOI] [PubMed] [Google Scholar]
- 23.Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage. 2007;37:90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maumet C, Maurel P, Ferre JC, Barillot C. Robust estimation of the cerebral blood flow in arterial spin labelling. Magnetic resonance imaging. 2014;32:497–504. doi: 10.1016/j.mri.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Liang X, Connelly A, Calamante F. Improved partial volume correction for single inversion time arterial spin labeling data. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2013;69:531–537. doi: 10.1002/mrm.24279. [DOI] [PubMed] [Google Scholar]
- 26.Asllani I, Borogovac A, Brown TR. Regression algorithm correcting for partial volume effects in arterial spin labeling MRI. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2008;60:1362–1371. doi: 10.1002/mrm.21670. [DOI] [PubMed] [Google Scholar]
- 27.Fazlollahi A, Bourgeat P, Liang X, et al. Reproducibility of multiphase pseudo-continuous arterial spin labeling and the effect of post-processing analysis methods. NeuroImage. 2015;117:191–201. doi: 10.1016/j.neuroimage.2015.05.048. [DOI] [PubMed] [Google Scholar]
- 28.Miranda MJ, Olofsson K, Sidaros K. Noninvasive measurements of regional cerebral perfusion in preterm and term neonates by magnetic resonance arterial spin labeling. Pediatr Res. 2006;60:359–363. doi: 10.1203/01.pdr.0000232785.00965.b3. [DOI] [PubMed] [Google Scholar]
- 29.Vidorreta M, Wang Z, Rodriguez I, Pastor MA, Detre JA, Fernandez-Seara MA. Comparison of 2D and 3D single-shot ASL perfusion fMRI sequences. NeuroImage. 2013;66C:662–671. doi: 10.1016/j.neuroimage.2012.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Rousseeuw PJ, Croux C. Alternatives to the Median Absolute Deviation. Journal of the American Statistical Association. 1993;88:1273–1283. [Google Scholar]
- 32.Alsop DC, Detre JA, Grossman M. Assessment of cerebral blood flow in Alzheimer's disease by spin-labeled magnetic resonance imaging. Annals of neurology. 2000;47:93–100. [PubMed] [Google Scholar]
- 33.Du AT, Jahng GH, Hayasaka S, et al. Hypoperfusion in frontotemporal dementia and Alzheimer disease by arterial spin labeling MRI. Neurology. 2006;67:1215–1220. doi: 10.1212/01.wnl.0000238163.71349.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Wolk DA, Reddin JS, et al. Voxel-level comparison of arterial spin-labeled perfusion MRI and FDG-PET in Alzheimer disease. Neurology. 2011;77:1977–1985. doi: 10.1212/WNL.0b013e31823a0ef7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leys L, Ley C, Klein O, Bernard P, Licata L. Detecting outliers: Do not use standard deviation around the mean, use absolute deviation around the median. Journal of Experimental Social Psychology. 2013;49:764–766. [Google Scholar]
- 36.Martin AJ, Friston KJ, Colebatch JG, Frackowiak RS. Decreases in regional cerebral blood flow with normal aging. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1991;11:684–689. doi: 10.1038/jcbfm.1991.121. [DOI] [PubMed] [Google Scholar]
- 37.Viviani R. Unbiased ROI selection in neuroimaging studies of individual differences. NeuroImage. 2010;50:184–189. doi: 10.1016/j.neuroimage.2009.10.085. [DOI] [PubMed] [Google Scholar]
- 38.Heijtel DF, Mutsaerts HJ, Bakker E, et al. Accuracy and precision of pseudo-continuous arterial spin labeling perfusion during baseline and hypercapnia: a head-to-head comparison with (1)(5)O H(2)O positron emission tomography. NeuroImage. 2014;92:182–192. doi: 10.1016/j.neuroimage.2014.02.011. [DOI] [PubMed] [Google Scholar]