Abstract
The discovery of long non-coding RNAs that control the liaisons between a transcription factor with a key role in prostate cancer and its target genes sheds light on how RNAs dictate information flow in the cell nucleus.
The human genome generates more than 10,000 long non-coding RNA (lncRNA) molecules, yet the functions of only several dozen of these transcripts are known. On page 598 of this issue, Yang et al.1 describe two lncRNAs that bind to, and govern the function of, the androgen receptor — a transcription factor that activates the expression of thousands of genes in response to the hormone androgen*. The authors find that inhibition of these two lncRNAs can block the growth of prostate-cancer cells that are resistant to hormone therapy owing to a mutation in the androgen receptor.
Androgen signalling has been recognized as the principle driver of prostate-cancer growth since the 1940s, when castration was first shown to slow the progression of prostate cancer2. Consequently, androgen-deprivation therapy, which can involve castration, has been the mainstay of treatment for advanced prostate cancer and achieves excellent response rates. However, in most men the cancer becomes resistant within two years of starting this therapy. It is now clear that such resistance arises by various mechanisms that reconstitute androgen-receptor signalling2.
A large proportion of lncRNAs associate with regulators of chromatin (DNA–protein complexes)3, and several are known to ‘tag’ specific chromosomal regions with particular chromatin marks that modulate the availability of the associated genes for expression4. Several prostate-cancer-specific lncRNAs have been identified, and some are associated with distinct subtypes of the disease5. In 2012, the US Food and Drug Administration approved the use of the lncRNA PCA3 for the detection of prostate cancer6. But, despite the discovery of multiple cancer-related lncRNAs, functions for most of these remain unknown.
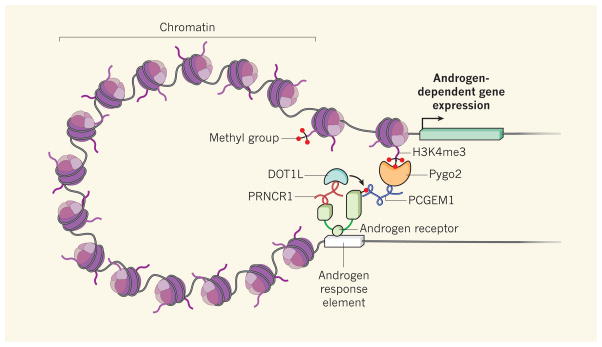
Yang et al. report that PRNCR1 and PCGEM1, two lncRNAs seen at high levels in many aggressive prostate cancers, enhance androgen-receptor-associated transcriptional programs to promote this cancer’s growth. The authors’ ChIRP analysis (a recently developed method of RNA localization7) reveals that these lncRNAs localize to distal androgen-response elements on chromatin, co-localizing with the androgen receptor (Fig. 1). In a surprisingly intricate series of events, PRNCR1 interacts with the acetylated carboxy terminus of the androgen receptor and recruits the enzyme DOT1L to methylate the amino terminus of this receptor; this step is necessary for the subsequent association of the androgen receptor with PCGEM1. PCGEM1 then interacts with the protein Pygo2, which can bind to H3K4me3 — a methyl mark on the chromatin-associated histone H3 protein that is prevalent at gene-promoter sequences.
Figure 1. Long non-coding RNAs and prostate-cancer growth.
Yang et al.1 report that two long non-coding RNAs (lncRNAs) — PRNCR1 and PCGEM1 — activate the androgen receptor. Interaction of PRNCR1 with this receptor at androgen-response genomic elements allows recruitment of DOT1L, an enzyme that methylates and so activates the receptor. PCGEM1 can now bind to the active androgen receptor and recruit the enzyme Pygo2, which allows communication between this receptor and its target genes by binding to H3K4me3 chromatin marks in the genes’ promoter sequences. Many androgen-receptor target genes have been implicated in prostate-cancer growth10. (Only one molecule of the androgen-receptor dimer is shown.)
Yang and colleagues further show that PCGEM1 and Pygo2 facilitate ‘looping’ between enhancer and promoter sequences — a recently recognized role for lncRNAs8 — and thus activation of androgen-receptor target genes. Moreover, even a truncated androgen receptor that is always active regardless of the presence of androgen depends on these lncRNAs. And lncRNA inhibition blocks the growth of both androgen-dependent and androgen-independent cancer cells. If these results can be replicated in subsequent studies, they offer a notable advance in the fight against advanced prostate cancer, by highlighting lncRNAs as a class of drug targets.
In addition to their relevance to disease, the current results illuminate several fundamental molecular mechanisms. PRNCR1 and PCGEM1 underscore a new role of RNA interaction with sequence-specific DNA-binding proteins — modification of transcription-factor activity. The liaisons between lncRNAs and transcription factors can program stepwise chemical modifications on transcription factors, gating the successive flow of information from enhancer engagement to target-gene activation.
The insights that these findings provide into how lncRNAs can mediate enhancer–promoter looping are also intriguing. The RNA-mediated recruitment of a protein with intrinsic avidity for a promoter-associated histone mark to distantly located enhancer elements could stabilize DNA looping and promote communication over three-dimensional space. This would mean that, rather than being simple scaffolds, lncRNAs are more akin to a complex computer circuit board, linking together various disparate molecular components and dictating the logical operation of the system.
Yang and co-workers’ study also raises a plethora of questions. For example, the researchers report a series of apparently specific RNA–protein interactions, the molecular details of which should be investigated. It is not clear whether all of the components involved in these interactions have been identified, and whether similar mechanisms operate for other steroid-hormone receptors or transcription factors.
Moreover, how PRNCR1 and PCGEM1 levels are controlled remains unclear. The authors’ data suggest that defined ranges of PRNCR1 and PCGEM1 levels are crucial for proper androgen signalling, but mechanisms by which lncRNA biogenesis and increases in the levels of these RNAs occur in prostate cancer are not known. With the recent recognition that many lncRNAs are dysregulated in human cancers9, understanding how these transcripts are generated and regulated is likely to take a central place in this field of research. The intricacies of the RNA circuits motivate the development of precise RNA-engineering technologies to reprogram nuclear signalling and for cancer diagnostic and prevention purposes, rather than simply modulating lncRNA levels.
Footnotes
This article and the paper under discussion1 were published online on 14 August 2013.
Contributor Information
ADAM M. SCHMITT, Howard Hughes Medical Institute and the Program in Epithelial Biology, Stanford University School of Medicine, Stanford, California 94305, USA. Department of Radiation Oncology, Stanford University School of Medicine
HOWARD Y. CHANG, Howard Hughes Medical Institute and the Program in Epithelial Biology, Stanford University School of Medicine, Stanford, California 94305, USA
References
- 1.Yang L, et al. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan CJ, Tindall DJ. J Clin Oncol. 2011;29:3651–3658. doi: 10.1200/JCO.2011.35.2005. [DOI] [PubMed] [Google Scholar]
- 3.Guttman M, et al. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batista PJ, Chang HY. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prensner JR, et al. Nature Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/DeviceApprovalsandClearances/Recently-ApprovedDevices/ucm294907.htm
- 7.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai F, et al. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunner AL, et al. Genome Biol. 2012;13:R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma NL, et al. Cancer Cell. 2013;23:35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]