Abstract
Castration of male pigs is a common practice used to reduce boar taint in commercial pork production, but it also significantly results in fat accumulation in carcass. Our previous study revealed a miRNA gene, ssc-miR-7134-3p that was implicated in adipogenesis. However, the relationship between ssc-miR-7134-3p and fat deposition due to castration is unknown. In the present study, we observed that ssc-miR-7134-3p targets the coding sequence (CDS) region of MARK4 based on bioinformatics analysis and dual-luciferase assays. Experiments using silent mutations and sub-cloning showed that ssc-miR-7134-3p binds independently to two adjacent sites in the MARK4 CDS. Subsequently, ssc-miR-7134-3p inhibits MARK4 protein expression in pig fibroblast cells, being consistent with the targeting demonstrated in vitro. We found higher MARK4 protein levels in the back fat of castrated pigs than in intact pigs, providing further evidence that MARK4 is involved in regulation of fat deposition. In addition, one SNP (g.2581A>G) in MARK4 was significantly associated with the back fat trait in Chinese and European pig populations. Taken together, we would conclude that ssc-miR-7134-3p targets the MARK4 gene for fat accumulation in the castrated male pigs.
Keywords: castration, pig, fat deposition, ssc-miR-7134-3p, MARK4 gene
Introduction
Low testosterone is known to result in male obesity in both humans and animals. Multiple population-based studies demonstrate that low testosterone is associated with obesity in older men affected by diabetes 1. Castration has been widely reported to induce obesity in male pigs, because it totally eliminates secretion of testosterone 2, 3. Although the linkage between testosterone and obesity is well supported, the molecular basis underlying this association remains unclear.
MicroRNAs (miRNAs) are a family of small (21-25 nucleotide), non-coding RNAs that regulate gene expression by targeting mRNAs for cleavage or translational repression 4, 5. Many miRNAs (such as miR-21, miR-27, and miR-143) play crucial roles in adipose deposition or adipogenesis in humans and animals 6-9. In pigs, miR-33b, miR-103 and miR-125a participate in the differentiation of preadipocytes 10-12. Moreover, evidence from high-throughput experiments shows that miRNAs are differentially expressed in adipose tissue during adipogenesis and among different breeds 13-15. For example, 215 unique miRNAs in the back fat of large white and Meishan pigs were identified using Illumina Solexa sequencing technology 13. In our previous study, 177 differentially expressed miRNAs, including ssc-miR-7134-3p, were identified by Solexa sequencing in a comparison between back fat tissues from castrated and intact full-sib pair male pigs 15. ssc-miR-7134-3p was also detected in other areas of pig fat and lung tissues 16, 17 and is expressed at significantly higher levels in back fat tissue from intact males than from castrated males 15. The marked difference in ssc-miR-7134-3p levels between castrated and intact male pigs suggests that this miRNA is a potential regulator of the process of fat deposition caused by castration.
MARK4, together with MARK1, MARK2 and MARK3, constitute a family of microtubule affinity regulatory kinase proteins (MARKs) 18, 19. The function of the MARK family has been related to obesity and hypermetabolism 20, 21. MARK4-deficient male mice exhibit hyperactivity and hypermetabolism, conferring protection from obesity caused by hyperphagia 22. The insulin resistance associated with obesity is attenuated in MARK4-deficient male mice by an increase in insulin-stimulated AKT phosphorylation in white adipose tissue, skeletal muscle and liver. Moreover, ablation of MARK4 also significantly improved glucose homeostasis by up-regulating the activity and expression of AMPK kinase in white adipose tissue 22. These phenotypes are observed only in male mice, not females. MARK4 is known to promote adipogenesis by inhibiting p38MAPK phosphorylation in 3T3-L1 cells 23. P38MAPK, one of the members of the MAP kinase family, is activated by dual kinases known as MAP kinase kinases (MKKs) 24. Inhibition of p38 increases adipogenesis in mice from the embryonic through adult stages 25.
Because our earlier work implicated ssc-miR-7134-3p as a potential regulator in fat deposition, we hypothesized that this miRNA functions by targeting the MARK4 gene. In the present study, dual luciferase and western blot analyses were performed to evaluate MARK4 as a target of ssc-miR-7134-3p and identify the specific target sites. MARK4 protein and p38MAPK phosphorylation levels were also compared in castrated and intact male pigs, and single nucleotide polymorphisms in the MARK4 gene and their association with fat deposition were investigated in different pig populations.
Materials and methods
Animals and sample collection
Eleven pairs of male full sibs from Pietrain sire × Landrace dam crossed pigs were used in this study. Castrated and intact male pigs were processed as described previously 15. Subcutaneous fat tissue and abdominal fat samples were collected for fat-related trait analysis, and subcutaneous fat samples was rapidly frozen in liquid nitrogen, and stored at -80 ℃.
Cell culture and histological analysis
HEK293T cells were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Science, P. R. China, and primary fibroblast cells were isolated from the ear tissue of a piglet. All cells were cultured in DMEM plus 10% FBS and penicillin/streptomycin. Cells were maintained at 37 ℃ with 5% CO2. All reagents used for cell culture were purchased from Invitrogen/Gibco. Adipose samples from back fat tissue were embedded in O.C.T. tissue freezing medium (Leica, Germany) and serially sectioned (6 μm thickness). Sections were stained with hematoxylin and eosin (H&E) (Zhongshan Golden Bridge Biotechnology Co, Beijing, China).
Dual luciferase assay
HEK293T cells were used to validate the miRNA target. Cells were seeded into 24-cell plates (Corning Incorporated, NY) and transfected 24 hours later with Lipofectamine 2000 (Invitrogen). The ssc-miR-7134-3p was co-transfected with 100 ng of psicheck2-MARK4-fragment and 40 pmol of ssc-miR-7134-3p mimics, or negative control mimics (Genepharma Company, Suzhou, China). Thirty-six hours after transfection, firefly and renlilla luciferase activities were measured using the dual-luciferase reporter kit (Promega). Assays were repeated three times.
RNA extraction and quantitative PCR
Total RNA was extracted from back fat tissue using TRIzol reagent according to the manufacturer's instructions (Life Technologies). cDNA synthesis was performed with 1 ug of total RNA, following the protocol accompanying the FastQuant RT Kit (Tiangen). Quantitative PCR (qPCR) amplification was conducted as described previously 15. To detect the relative expression of MARK4 mRNA, real-time PCR was performed with the primers shown in Table S1, using HPRT as the reference gene. Primers for MARK4 and HPRT were synthesized by Sangon (Shanghai, China).
Protein isolation and western blot analysis
Total protein was extracted from the back fat tissue of pigs with ice-cold lysis buffer containing PMSF (phenylmethylsulfonyl fluoride). Samples were centrifuged at 4 ℃ for 30 min at 12000 × g. Total protein concentration was measured using a Braford protein assay kit (Tiangen- PA102). 30 ug total protein was separated by SDS-PAGE and transferred to PVDF membranes (Millipore). The membranes were probed with the specific primary antibody (anti-MARK4, anti-p38 (phosphor T180 + Y 182), and anti-tubulin) and appropriate secondary antibody (anti-rabbit-HRP or anti-mouse-HRP). The MARK4, p38 and tubulin primary antibodies were diluted to 1:3000, 1:3000 and 1:1000 before use, respectively. The secondary antibodies were diluted to 1:5000.
Gene cloning and vector construction
The pig MARK4 coding region was cloned using primer CLONE-1 (Table S1) and sequenced by BGI (Beijing, China). RACE was performed to amplify the 5'UTR and 3'UTR of MARK4 with the SMARTer® RACE 5'/3'Kit (Clontech-634858) and the gene-specific primers GSP-5 and GSP-3, respectively (Table S1). The MARK4 fragment containing the predicted target site was cloned into the psiCHECK2 vector. We denote these constructs as X/Y-Z, in which X and Y represent the start and stop positions, respectively, in base pairs from the 5' end of the MARK4 mRNA transcription initiation site. Z describes the targeted site as either mutant (Mut) or wild-type (Wt). Fragments spanning coordinates 2235 to 3007 (isolated using AsiSI and PmeI) and from 871 to 1467 (isolated using XhoI and NotI) were cloned into plasmid psiCHECK2 using the primers 2235/3007-p and 871/1467-p. The fragment 2235/3007-Mut was synthesized by Genepharma (Suzhou, China). psiCHECK2-2235/2508-Mut and psiCHECK2-2235/2508-Wt were cloned using primers 2235/2508-2482-Mut-p and 2235/2508-2482-Wt-p (Table S1). The plasmids psiCHECK2-2235/2502-Mut (hereafter 2235/2502-Mut; for convenience psiCHECK2 in all plasmid names have been omitted below) and 2235/2502-Wt, were constructed using the primer 2235/2502-p with the plasmids 2235/3007-Mut and 2235/3007-Wt. Four plasmids, 2434/2533-2482Wt-2502Wt, 2434/2533-2482Wt-2502Mut, 2434/2533-2482Mut-2502Wt and 2434/2533-2482Mut-2502Mut, were constructed using the primers 2434/2533-2502Mut-p and 2434/2533-2502Wt-p with plasmids 2235/3007-Mut and 2235/3007-Wt (Table S1). 2434/2533-R1 and 2434/2533-R2 was constructed using primers 2434/2533-R1-p and 2434/2533-R2-p (Table S1). All fragments were inserted into psiCHECK2 with AsiSI and PmeI (New England BioLabs).
SNP screening and genotyping
PCR amplification for SNP identification was performed using pooled DNA from 10 pigs of each breed, with the primers shown in Table S2. The DNA templates were prepared from the Jinhua, Neijiang, Erhualian, Wuzhishan, Bamei, Tibet, Duroc and Yorkshire breeds. SNP g.2581A>G was detected in seven pig breeds by sequencing MARK4 PCR products with the primers P1-P13 (Table S2). SNP g.2581A>G was genotyped in 6 Chinese indigenous breeds and 2 foreign breeds by restriction fragment length polymorphism (RFLP) using mismatched primers P14 and restriction enzyme PstI (New England BioLabs) (Table S3). Association analyses between genotype and three phenotypes were performed, with back fat thickness, feed conversion ratio (FCR) and average daily gain (ADG). Three traits were adjusted to 100 kg or 115 kg, respectively, and collected from 288 male Duroc pigs, born at the end of 2011 and raised in identical conditions.
Bioinformatics and statistical analyses
TargetScan, along with the ssc-miR-7134-3p seed sequence, was used to predict the putative target gene for ssc-miR-7134-3p (http://www.TargetScan.org/vert_50/seedmatch.html). Data were analyzed using SPASS (IBM Corporation) software with Student's t-test to determine statistical significance.
Results
Back fat and abdominal fat increased after castration in male pigs
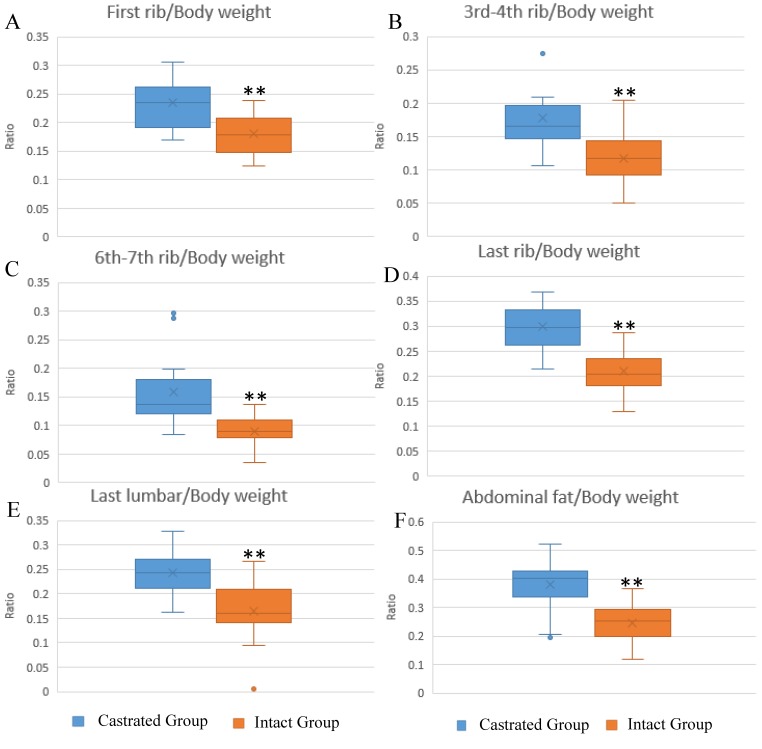
The influence of castration on back fat and abdominal fat accumulation is shown in Figure 1. Subcutaneous and abdominal fat increased significantly in response to castration. The ratios of back fat thickness to body weight at slaughter, measured at the 1st rib, 3rd-4th rib, 6th-7th rib, last rib and the last lumbar, were augmented significantly in castrated animals compared to their intact full-sibs (Figure 1A-1E). The left leaf fat ratio (the ratio of the weight of left leaf fat to body weight at slaughter) was also significantly higher in castrated males (Figure 1F). To help understand the changes underlying the phenotypic differences, fat tissues were subjected to histological analysis. Larger adipocytes were observed in castrated males, consistent with the results described above (Figure S1A). The average adipocyte number per visual field was 33.75±8.32 in castrated males, significantly lower than the number observed in intact animals (63.4±5.99) (Figure S1B).
Figure 1.
Differences in fat deposition between castrated and intact male pigs. (A-E) The ratio at first-rib, 3rd-4th rib, 6th-7th rib, last rib and the last lumbar, respectively. The ratio is the thickness of back fat relative to body weight at slaughter. (F) The ratio of left leaf fat weight relative to body weight at slaughter. **, p< 0.01.
Identification of the ssc-miR-7134-3p target
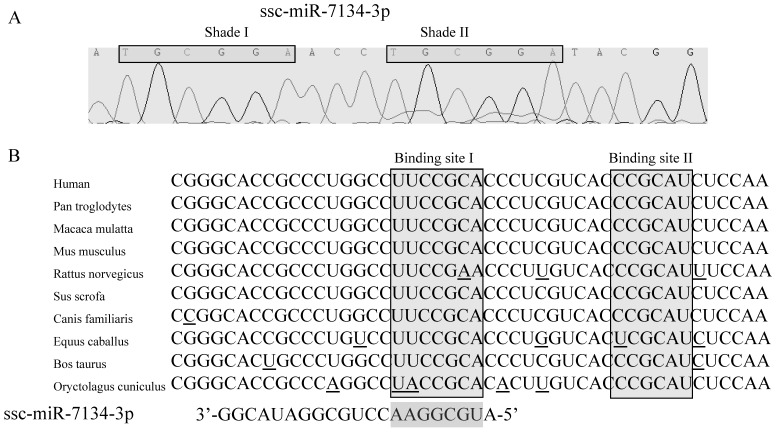
In our previous study, results from SOLiD sequencing and miRNA stem-loop specific qPCR revealed that the level of ssc-miR-7134-3p decreases significantly in castrated male pigs compared to intact males 15. To identify the target for this miRNA, we first confirmed the sequence of ssc-miR-7134-3p using stem-loop qPCR (Figure 2A). Interestingly, a subsequence (TGCGGA) within this seed region is repeated within Shade II (Figure 2A). Next, the TargetScan analysis website was used to predict putative target genes with the ssc-miR-7134-3p seed region sequence. A potential binding site was identified at the end of the MARK4 CDS at coordinate 2482 from the 5' end (Binding site I in Figures 2B).
Figure 2.
The ssc-miR-7134-3p and its homology to the MARK4 target site as predicted by TargetScan. (A) Sequence of ssc-miR-7134-3p miRNA isolated from the back fat tissue of male pigs. (B) MARK4 mRNA sequences from 10 species are aligned 5' to 3'. Potential base pairing between ssc-miR-7134-3p (3' to 5') and MARK4 is shown in the Binding site I region. The Binding site I target was predicted by TargetScan; Binding site II was considered as another possible target site in our analysis.
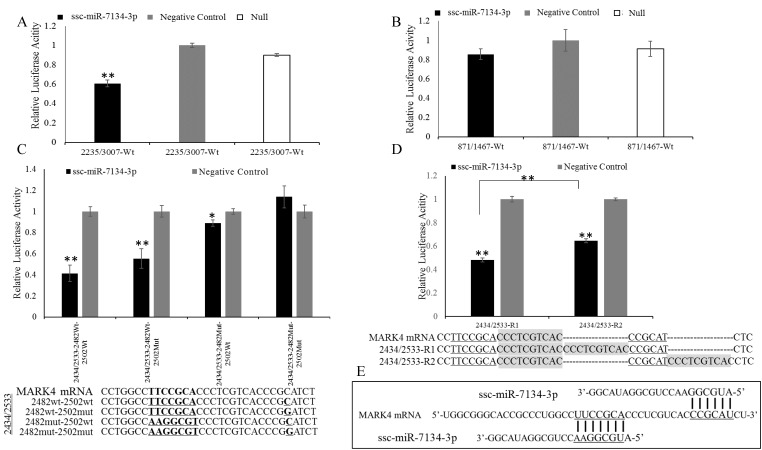
To confirm the ssc-miR-7134-3p target on MARK4, a plasmid (2235/3007) containing the site adjacent to coordinate 2482 was transfected into HEK293T cells together with ssc-miR-7134-3p mimics or negative control mimics. The 871/1467 plasmid was used as a negative control. ssc-miR-7134-3p significantly decreased the luciferase activity of 2235/3007, but did not affect the activity of 871/1467 (Figures 3A-3B). The result indicates that ssc-miR-7134-3p binds to the MARK4 region of plasmid 2235/3007.
Figure 3.
Inhibitory effect of ssc-miR-7134-3p on MARK4 fragments using the dual-luciferase system. (A-B) The assay was conducted with fragments spanning coordinates 2235 to 3007 and 871 to 1467 relative to the 5' end of the MARK4 transcription initiation site. (C) The assay was conducted with a fragment spanning coordinates 2434 to 2533 in which mutations had been introduced as follows: 2482wt-2502wt, 2482wt-2502mut, 2482mut-2502wt, and 2482mut-2502mut. (D) A 10 bp duplication was inserted between the two binding sites (2373/2533-R1), or downstream of binding site 2502 (2373/2533-R2). (E) The schematic diagram of ssc-miR-7134-3p binding at the two sites in MARK4. All relative luciferase activity values were normalized to the negative control mimics. **, p<0.01; *, p<0.05.
Identification of target sites
To localize the ssc-miR-7134-3p target site within plasmid 2235/3007, silent mutations were designed in the MARK4 region to disrupt putative base pairing between ssc-miR-7134-3p and MARK4 mRNA. The constructs 2235/3007-2482Wt and 2235/3007-2482Mut were co-transfected with ssc-miR-7134-3p or negative control mimics. The luciferase activities of 2235/3007-2482Wt and 2235/3007-2482Mut were both suppressed by ssc-miR-7134-3p, but the suppression was weaker in 2235/3007-2482Mut (Figure S2A). To confirm the predicted target site and reduce noise from other possible binding sites, the MARK4 insert size was reduced. First, we decreased the MARK4 region within plasmid 2235/3007 to 2235/2508 and repeated the co-transfection experiment, but obtained similar results (Figure S2B). The 2235/2508 insert was then shortened to 2235/2502. In this experiment, reduced luciferase activity was observed from 2235/2502-2482Wt but not 2235/2502-2482Mut (Figure S2C), confirming that ssc-miR-7134-3p targets site 2482 in MARK4.
Because luciferase emission from 2235/3007-2482Mut was detected in the presence of ssc-miR-7134-3p, we re-examined the MARK4 sequence and identified site 2502 as a second potential target (Binding site II in Figure 2B). To confirm its ability to bind the miRNA, four versions of plasmid 2434/2533 were constructed with different mutations (Figure 3C) and then analyzed by co-transfection. The results showed that the luciferase activity of 2434/2533-2482Wt-2502Wt is the most sensitive to ssc-miR-7134-3p, followed by 2434/2533-2482Wt-2502Mut. The luciferase activity of 2434/2533-2482Mut-2502Mut was not suppressed by ssc-miR-7134-3p (Figure 3C). This result supports the classification of site 2502 as a target for ssc-miR-7134-3p on MARK4.
To determine whether the 2482 and 2502 sites are targeted by independent miRNAs, two plasmids were constructed (2434/2533-R1 and 2434/2533-R2) in which the sites were positioned at different distances from one another. 2434/2533-R1 generated less luciferase activity than 2434/2533-R2 (Figure 3D). The result suggests that sites 2482 and 2502 are each binding an ssc-miR-7134-3p molecule (Figure 3E).
MARK4 expression and p38MAPK phosphorylation in intact and castrated male pigs
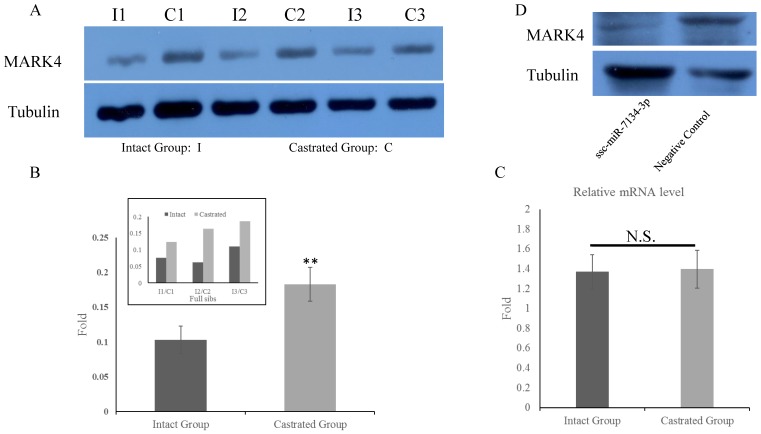
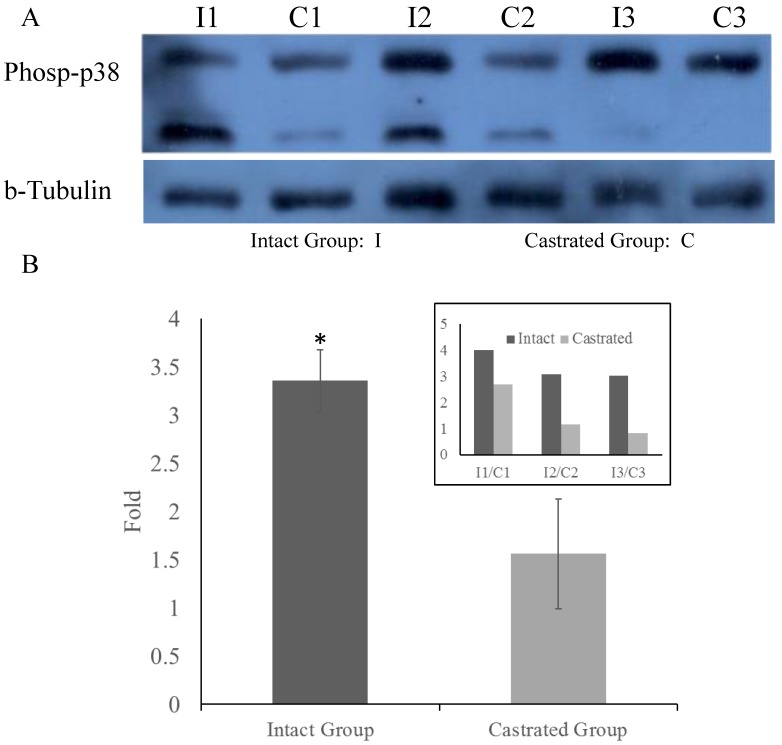
To determine whether MARK4 levels in back fat tissue differ between intact and castrated pigs, both mRNA and protein levels were compared. Based on band intensities, MARK4 protein is markedly less abundant in intact males than in the corresponding castrated full-sib male (Figure 4A). The difference is statistically significant when protein levels are normalized against tubulin (Figure 4B). In contrast, there is no significant difference in MARK4 mRNA levels between intact and castrated male pigs (Figure 4C). These results are consistent with a model in which ssc-miR-7134-3p downregulates MARK4 expression at the translational level in male pigs. To test this possibility, ssc-miR-7134-3p was transfected into pig primary fibroblast cells. As predicted, MARK4 protein levels are suppressed by ssc-miR-7134-3p in comparison with the negative control miRNA mimics (Figure 4D). In addition, it is well known that the p38MAPK pathway is regulated by MARK4. In this study, the phosphorylation level of p38MAPK was significantly lower in castrated males relative to intact males (Figure 5A and 5B).
Figure 4.
Expression of MARK4 protein and mRNA in back fat tissue and primary fibroblast cells. MARK4 expression in back fat tissue by western blot (A) and qPCR (C). (B) Relative expression in intact and castrated pigs, based on the data in (A). (D) Expression of MARK4 detected by western blot in primary fibroblast cells transfected with ssc-miR-7134-3p and negative control. N.S., not significant. **, p<0.01.
Figure 5.
Phosphorylation of p38MAPK in back fat tissue. Comparison of p38MAPK phosphorylation levels in back fat tissue from castrated and intact full-sib males (A). Comparison of relative levels in intact and castrated animals (B). The inset on the upper right shows the values measured for each individual. *, p<0.05.
MARK4 mRNA sequence and SNP association analysis
Using RACE, we cloned the full-length pig MARK4 mRNA, which is 3527 bp in length and includes the CDS region (2259 bp), the 5'UTR (267 bp), and the 3'UTR (1001 bp). The MARK4 CDS encodes 752 amino acids and shares high homology (over 97.89%) to human, mouse and rat MARK4 sequences (Figure S3A and S3B). Human and pig MARK4 amino acid sequences share 99.08% identity. Phylogenetic analysis classifies the amino acid sequences from the four species into two subfamilies (Figure S3C).
MARK4 exon sequences were obtained from a variety of pig breeds and examined for SNPs. The analysis covered all 16 exons in the mRNA, but omitted the first 51 bp at the 5' end because this region is not included in the current version of the pig genome. One SNP (g.2581A>G) was identified. The GG genotype frequency occurs at frequencies ≥ 0.92 in Chinese indigenous breeds, except in Bamei pigs (0.72). The frequency of the AA genotype is dominant in Yorkshire pigs, occurring at a frequency of 0.80, but the AA genotype was absent in the Chinese indigenous breeds Jinhua, Neijiang, Erhualian, Wuzhishan, Bamei and Tibet. The frequency of the AG genotype is 0.52 in the Duroc population. Furthermore, the frequency of the G allele exceeds 0.86 among the 6 Chinese breeds, whereas the frequency of the A allele is 0.57 in Duroc pigs, and 0.90 in Yorkshire pigs (Table S3).
Because most Chinese indigenous pig breeds have higher fat deposition than European commercial breeds, we hypothesized that the G allele is associated with an increase in back fat thickness. Association analysis in a group of 268 male Duroc pigs confirmed that back fat thinness is significantly higher in GG genotype than in AA genotype pigs, after back fat thickness is adjusted to 100 kg and 115 kg (p<0.05) (Table 1). The analysis also showed that a significantly higher adjusted FCR (feed conversion ratio) was observed in GG and AG pigs, compared to AA pigs (FCR adjusted from 30 to 100 kg and 115 kg; p<0.05) (Table 1). No significant difference in adjusted ADG (average daily gain) was observed between AA genotype and GG genotype animals (Table 1). Together, these results demonstrate that the MARK4 SNP g.2581A>G is associated with back fat thickness and FCR in male Duroc pigs.
Table 1.
Association analysis between the MARK4 g.2581A>G polymorphism and back fat thinness, FCR and ADG in male Duroc pigs.
| No. | Genotype | Backfat-100 kg (mm) | Backfat-115 kg (mm) | FCR 1 | FCR 2 | ADG 1 | ADG 2 |
|---|---|---|---|---|---|---|---|
| 83 | AA | 10.49±0.21a | 11.94±0.24a | 2.04±0.02a | 2.19±0.02a | 952.86±15.04 | 959.65±13.90 |
| 140 | AG | 11.17±0.19ab | 12.70±0.22ab | 2.09±0.01b | 2.24±0.01b | 973.08±9.76 | 982.01±9.08 |
| 45 | GG | 11.33±0.30b | 12.89±0.34b | 2.15±0.04b | 2.30±0.04b | 990.29±20.52 | 988.90±18.17 |
Notes: Values are expressed as mean ± standard error. Different lowercase characters (a and b) indicate significant differences between genotypes (p<0.05). Superscript 1 denotes 30-100 kg body weight. Superscript 2 denotes 30-115 kg body weight.
Discussion
Castration in the male pig is a standard procedure in commercial pork production for reducing boar taint, but it increases the deposit of adipose tissue 26, 27. We also observed this effect in our study (Figure 1 and Figure S1). However, the molecular mechanism underlying the increase in adipose tissue in castrated pigs is unclear. In recent years, miRNAs have recently been reported as crucial regulatory factors involved in important phenotypes or traits, including adipogenesis and lipid metabolism in mammals 28.
Our previous study identified 177 differentially expressed miRNAs in back fat tissue in a comparison between castrated male pigs and intact full-sib pair male pigs, and we hypothesized that miRNAs may be important regulators in fat deposition after castration. The most differentially expressed miRNA, ssc-miR-7134-3p, was selected in this study to examine its potential contribution to obesity caused by castration. However, because ssc-miR-7134-3p orthologs have not been identified in human or other animals, miRNA target predication was based entirely on the seed region sequence (nucleotides 2-8 from the 5'end of the miRNA) and in silico analysis using TargetScan. The results suggested that ssc-miR-7134-3p targets the MARK4 coding region. miRNA target sites can be located in CDS as well as in 3'UTRs, and multiple target sites for one miRNA can be located within one CDS 29-31. To test the target site prediction, silent mutations at the putative target site were introduced, and subregions containing the target were examined. Two adjacent target sites within the MARK4 CDS were subsequently identified (Figure 3E). CDS-localized miRNA targets do not reduce mRNA levels effectively, but are highly efficient inhibitors of translation 32. In our study, lower MARK4 protein levels were observed in back fat tissue in intact male pigs (Figure 4A-4B). These tissues also exhibited higher ssc-miR-7134-3p levels in our previous study, relative to castrated full-sib males. In contrast, no differences in MARK4 mRNA levels were detected between castrated and intact male pigs (Figure 4C). These results suggest that the ssc-miR-7134-3p target sites in the MARK4 CDS function by inhibiting translation, although we cannot eliminate the possibility that other miRNAs are also involved in suppressing MARK4 protein levels. Because miRNA target sites located in coding regions show significantly low evolutionary rates in mammals, the relationship between ssc-miR-7134-3p and MARK4 may be evolutionarily stable in the pig 32.
Interestingly, the two target sites in the MARK4 CDS for ssc-miR-7134-3p are in close proximity (15 bases). We originally speculated that duplication of the seed region in ssc-miR-7134-3p might contribute to stabilizing the miRNA-mRNA complex (Figure 2A). However, a 10 base insertion between the two targets increased the inhibitory effect, consistent with independent binding of an ssc-miR-7134-3p molecule at each target site (Figure 3C-3E). This is in agreement with results reported by Tay et al., who found that miR-470 targets the Oct4 CDS at two sites that require a spacing of at least 9 bases between them 31.
MARK4-deficient male mice exhibit hyperphagia, hyperactivity, hypermetabolism and resistance to obesity 22. In male mice, inactivation of MARK4 results in lower weight gain, entirely as a result of decreased fat mass, and insulin sensitivity is enhanced compared to wild-type mice 22. These phenotypes are only observed in male mice, not in females 22. In our study, the GG genotype of SNP g.2581A>G is significantly associated with higher back fat thickness and FCR in Duroc male pigs (Table 1). The GG genotype predominates in 6 indigenous Chinese breeds characterized as fat-type, and is rarely observed in Duroc and Yorkshire populations characterized as lean-type (Table S3). Because MARK4 sequences are highly similar in pig, human, rat and mouse, it is likely that MARK4 gene function has been conserved among these species (Figure S3). In particular, both pig and mouse MARK4 may promote fat accumulation. Because MARK4 participates in adipogenesis in 3T3-L1 cells by reducing p38MAPK phosphorylation 23, we compared p38MAPK phosphorylation levels the back fat tissue from castrated and intact males by western blot (Figure 5A). p38MAPK phosphorylation was significantly lower in castrated males (Figure 5B), which is consistent with higher MARK4 expression in these animals. Together, our results suggest that the decrease in ssc-miR-7134-3p levels induced by castration causes an increase in MARK4 protein levels. In turn, increased MARK4 levels inhibit p38MAPK phosphorylation, thereby enhancing fat accumulation.
The main difference between castrated and intact male pigs is the absence or presence of testosterone. Low testosterone is reported to be associated with obesity, metabolic syndrome and insulin sensitivity in men 33-36. Similar phenotypes are also observed in androgen receptor knock-out male mice and adult Göttingen minipigs after castration 2, 37, 38. The opposite phenotypes are observed in MARK4-null male mice, including lower fat mass, higher insulin sensitivity, hyperphagia, hyperactivity, hypermetabolism and increased oxygen consumption 22. In our study, the phenotypes observed in castrated male pigs are similar to those exhibited by androgen receptor knock-out male mice and men with low testosterone, and we therefore speculated that MARK4 might play a key role in obesity caused by low testosterone in males.
In summary, our results show that the miRNA ssc-miR-7134-3p targets two sites in the MARK4 CDS in pigs. The decrease in ssc-miR-7134-3p levels, caused by castration, results in an increase in MARK4 protein levels, presumably because ssc-miR-7134-3p functions as a translational inhibitor. MARK4 contributes to fat accumulation, possibly by inhibiting p38MAPK phosphorylation. We conjecture that MARK4 is involved in fat accumulation in testosterone-deficient human males. Further study will be necessary to decipher the molecular mechanisms underlying human MARK4 function to enable drug target identification.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
The authors are grateful to Prof. Changxin Wu, Dr. Keliang Wu, Dr. Chunjiang Zhao, Dr. Yuchang Yao, Dr. Jinming Huang and Dr. Gang Liu for assistance with sample collection. This work was supported by the Natural Science Foundation of China (NSFC, Grant 31372275 & 31471180), National Basic Research Program of China (973 Program, Grant 2014CB138504) and the Program for Changjiang Scholars and Innovation Research Teams in the University (IRT1191).
References
- 1.Grossmann M. Low testosterone in men with type 2 diabetes: significance and treatment. The Journal of clinical endocrinology and metabolism. 2011;96:2341–53. doi: 10.1210/jc.2011-0118. [DOI] [PubMed] [Google Scholar]
- 2.Christoffersen BO, Gade LP, Golozoubova V, Svendsen O, Raun K. Influence of castration-induced testosterone and estradiol deficiency on obesity and glucose metabolism in male Gottingen minipigs. Steroids. 2010;75:676–84. doi: 10.1016/j.steroids.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Li M, Bjorntorp P. Effects of testosterone on triglyceride uptake and mobilization in different adipose tissues in male rats in vivo. Obesity research. 1995;3:113–9. doi: 10.1002/j.1550-8528.1995.tb00128.x. [DOI] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews Genetics. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV. et al. MicroRNA-143 regulates adipocyte differentiation. The Journal of biological chemistry. 2004;279:52361–5. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 7.Kim YJ, Hwang SJ, Bae YC, Jung JS. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem cells. 2009;27:3093–102. doi: 10.1002/stem.235. [DOI] [PubMed] [Google Scholar]
- 8.Hilton C, Neville MJ, Karpe F. MicroRNAs in adipose tissue: their role in adipogenesis and obesity. International journal of obesity. 2013;37:325–32. doi: 10.1038/ijo.2012.59. [DOI] [PubMed] [Google Scholar]
- 9.Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. A role of miR-27 in the regulation of adipogenesis. Febs J. 2009;276:2348–58. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G, Wu Z, Li X, Ning X, Li Y, Yang G. Biological role of microRNA-103 based on expression profile and target genes analysis in pigs. Molecular biology reports. 2011;38:4777–86. doi: 10.1007/s11033-010-0615-z. [DOI] [PubMed] [Google Scholar]
- 11.Taniguchi M, Nakajima I, Chikuni K, Kojima M, Awata T, Mikawa S. MicroRNA-33b downregulates the differentiation and development of porcine preadipocytes. Molecular biology reports. 2014;41:1081–90. doi: 10.1007/s11033-013-2954-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji HL, Song CC, Li YF, He JJ, Li YL, Zheng XL. et al. miR-125a inhibits porcine preadipocytes differentiation by targeting ERRalpha. Molecular and cellular biochemistry. 2014;395:155–65. doi: 10.1007/s11010-014-2121-4. [DOI] [PubMed] [Google Scholar]
- 13.Chen C, Deng B, Qiao M, Zheng R, Chai J, Ding Y. et al. Solexa sequencing identification of conserved and novel microRNAs in backfat of Large White and Chinese Meishan pigs. Plos One. 2012;7:e31426. doi: 10.1371/journal.pone.0031426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li GX, Li YJ, Li XJ, Ning XM, Li MH, Yang GS. MicroRNA Identity and Abundance in Developing Swine Adipose Tissue as Determined by Solexa Sequencing. J Cell Biochem. 2011;112:1318–28. doi: 10.1002/jcb.23045. [DOI] [PubMed] [Google Scholar]
- 15.Bai Y, Huang JM, Liu G, Zhang JB, Wang JY, Liu CK. et al. A comprehensive microRNA expression profile of the backfat tissue from castrated and intact full-sib pair male pigs. BMC genomics. 2014;15:47. doi: 10.1186/1471-2164-15-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mentzel CM, Anthon C, Jacobsen MJ, Karlskov-Mortensen P, Bruun CS, Jorgensen CB. et al. Gender and Obesity Specific MicroRNA Expression in Adipose Tissue from Lean and Obese Pigs. Plos One. 2015;10:e0131650. doi: 10.1371/journal.pone.0131650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Podolska A, Anthon C, Bak M, Tommerup N, Skovgaard K, Heegaard PM. et al. Profiling microRNAs in lung tissue from pigs infected with Actinobacillus pleuropneumoniae. BMC genomics. 2012;13:459. doi: 10.1186/1471-2164-13-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 19.Hurov J, Piwnica-Worms H. The Par-1/MARK family of protein kinases: from polarity to metabolism. Cell cycle. 2007;6:1966–9. doi: 10.4161/cc.6.16.4576. [DOI] [PubMed] [Google Scholar]
- 20.Hurov JB, Huang M, White LS, Lennerz J, Choi CS, Cho YR. et al. Loss of the Par-1b/MARK2 polarity kinase leads to increased metabolic rate, decreased adiposity, and insulin hypersensitivity in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5680–5. doi: 10.1073/pnas.0701179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lennerz JK, Hurov JB, White LS, Lewandowski KT, Prior JL, Planer GJ. et al. Loss of Par-1a/MARK3/C-TAK1 kinase leads to reduced adiposity, resistance to hepatic steatosis, and defective gluconeogenesis. Molecular and cellular biology. 2010;30:5043–56. doi: 10.1128/MCB.01472-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun C, Tian L, Nie J, Zhang H, Han X, Shi Y. Inactivation of MARK4, an AMP-activated protein kinase (AMPK)-related kinase, leads to insulin hypersensitivity and resistance to diet-induced obesity. The Journal of biological chemistry. 2012;287:38305–15. doi: 10.1074/jbc.M112.388934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng M, Tian L, Gan L, Liu Z, Sun C. Mark4 promotes adipogenesis and triggers apoptosis in 3T3-L1 adipocytes by activating JNK1 and inhibiting p38MAPK pathways. Biol Cell. 2014;106:294–307. doi: 10.1111/boc.201400004. [DOI] [PubMed] [Google Scholar]
- 24.Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–8. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 25.Aouadi M, Laurent K, Prot M, Le Marchand-Brustel Y, Binetruy B, Bost F. Inhibition of p38MAPK increases adipogenesis from embryonic to adult stages. Diabetes. 2006;55:281–9. doi: 10.2337/diabetes.55.02.06.db05-0963. [DOI] [PubMed] [Google Scholar]
- 26.Lundstrom K, Matthews KR, Haugen JE. Pig meat quality from entire males. Animal: an international journal of animal bioscience. 2009;3:1497–507. doi: 10.1017/S1751731109990693. [DOI] [PubMed] [Google Scholar]
- 27.Bonneau M. Use of entire males for pig meat in the European Union. Meat Sci. 1998;49:S257–S72. [PubMed] [Google Scholar]
- 28.Peng Y, Yu S, Li H, Xiang H, Peng J, Jiang S. MicroRNAs: emerging roles in adipogenesis and obesity. Cellular signalling. 2014;26:1888–96. doi: 10.1016/j.cellsig.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 32.Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28:771–6. doi: 10.1093/bioinformatics/bts043. [DOI] [PubMed] [Google Scholar]
- 33.Wang C, Jackson G, Jones TH, Matsumoto AM, Nehra A, Perelman MA. et al. Low testosterone associated with obesity and the metabolic syndrome contributes to sexual dysfunction and cardiovascular disease risk in men with type 2 diabetes. Diabetes care. 2011;34:1669–75. doi: 10.2337/dc10-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salam R, Kshetrimayum AS, Keisam R. Testosterone and metabolic syndrome: The link. Indian journal of endocrinology and metabolism. 2012;16(Suppl 1):S12–9. doi: 10.4103/2230-8210.94248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corona G, Monami M, Rastrelli G, Aversa A, Sforza A, Lenzi A. et al. Type 2 diabetes mellitus and testosterone: a meta-analysis study. International journal of andrology. 2011;34:528–40. doi: 10.1111/j.1365-2605.2010.01117.x. [DOI] [PubMed] [Google Scholar]
- 36.Yialamas MA, Dwyer AA, Hanley E, Lee H, Pitteloud N, Hayes FJ. Acute sex steroid withdrawal reduces insulin sensitivity in healthy men with idiopathic hypogonadotropic hypogonadism. The Journal of clinical endocrinology and metabolism. 2007;92:4254–9. doi: 10.1210/jc.2007-0454. [DOI] [PubMed] [Google Scholar]
- 37.Fan W, Yanase T, Nomura M, Okabe T, Goto K, Sato T. et al. Androgen receptor null male mice develop late-onset obesity caused by decreased energy expenditure and lipolytic activity but show normal insulin sensitivity with high adiponectin secretion. Diabetes. 2005;54:1000–8. doi: 10.2337/diabetes.54.4.1000. [DOI] [PubMed] [Google Scholar]
- 38.Lin HY, Xu Q, Yeh S, Wang RS, Sparks JD, Chang C. Insulin and leptin resistance with hyperleptinemia in mice lacking androgen receptor. Diabetes. 2005;54:1717–25. doi: 10.2337/diabetes.54.6.1717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.