Abstract
Colon cancer is one of the most common types of gastrointestinal cancers and the fourth cause of cancer death worldwide. To discover novel diagnostic biomarkers for colon cancer and investigate potential mechanisms of oncogenesis, quantitative proteomic approach using iTRAQ-tagging and 2D-LC-MS/MS was performed to characterize proteins alterations in colon cancer and non-neoplastic colonic mucosa (NNCM) using laser capture microdissection-harvested from the two types of tissues, respectively. As a result, 188 DEPs were identified, and the differential expression of two DEPs (DCN and HSPD1) was further verified by Western blotting and immunohistochemistry. KEGG pathway analysis disclosed that the DEPs were related to signaling pathways associated with cancer; furthermore, DCN and HSPD1 are in the relative central hub position among protein-protein interaction subnetwork of the DEPs. The results not only shed light on the mechanism by the DEPs contributed to colonic carcinogenesis, but also showed that DCN and HSPD1 are novel potential biomarkers for the diagnosis of colon cancer.
Keywords: Proteomics analysis, colon cancer, DCN, HSPD1, iTRAQ, carcinogenesis.
Introduction
Colorectal cancer (CRC) is one of gastrointestinal cancers that arises from the colon or rectum epithelium through the accumulation of genetic and epigenetic changes1. These cancers can also be designated colon cancer or rectal cancer, relying on where initiating cells of them locate. The two types of cancers are often combined together because they have some features common characteristics2. CRC is the third greatest frequently diagnosed cancer and the fourth most common cause of cancer-related death worldwide, with an approximately 1.4 million cases and 693,900 deaths occurring in 20123, 4. Excluding skin cancers, CRC is the third greatest frequently diagnosed cancer and ranks second as a cause of cancer death in the United States, with an estimated 95,270 new cases of colon cancer, 39,220 new cases of rectal cancer, and 49,190 deaths from CRC occurring in 20162, 5. This disease is one of the most common forms of cancer and the top ranked cause of cancer death in China, accounting for 25,159 cases and 12,161 deaths in 20096, 7. Early-stage detection and clinical stages have the most impact on cancer incidence and mortality in CRC because survival rates of 5 years or more are observed for more than 91.4% of patients diagnosed with localized colon, whereas survival rates drop to only 12.4% for colon cancer that has metastasized to distant organs5, 8. With the development of biological technology, a lot of CRC cancer-associated molecules, including 18q/TGF-β pathway, APC loss-induced Wnt signaling pathway, KRAS,TP53, BRAF, and molecular pathways have been reported9. However, much studies remains to be aimed to further explore the potential carcinogenic mechanism of CRC, allowing promoted prognosis, diagnosis, and therapy9.
Proteomic assessment of differentially expressed proteins (DEPs) in CRC disclosed that expression and modified changes of proteins are closely related to clinical outcome and risk evaluation1, 10-12. Our previous studies using proteomics identified the colonic epithelium differential proteins between differential aging and stromal DEPs of colon cancer leading to elucidate potential biomarkers for diagnosis or prognosis of colon cancer13, 14. Although some proteomic studies on CRC have been reported1, 10-12, 15 ,there are more clinically approved biomarkers available for understanding of the ability to present useful clues concerning oncogenesis, prognosis, and even response to targeted therapy for CRC.
Quantitative proteomic analysis of cancer and normal tissues help us discover more diagnostic and prognostic biomarkers. Isobaric tags for relative and absolute quantitation (iTRAQ) coupled with two-dimensional liquid chromatography tandem mass spectrometry (2D LC-MS/MS) technique is a powerful quantitative proteomics method to identify novel cancer biomarkers and understand signal networks16-22. With clinical samples from cancer tissues, undoubtedly proteomic approaches may lead the discovery of novel valuable diagnostic or therapeutic biomarkers. However, a major challenge in analyzing cancer specimens is tissue heterogeneity. Laser capture microdissection (LCM) is an powerful new tool to harvest target cells, solving the problem of tissue heterogeneity and contamination of non-cancer cells23.
In this study, we investigated the samples of LCM-harvested colon tissues from the patients of colon cancer by iTRAQ isobaric tags with 2D LC-MS/MS to discover potential biomarkers which may play important roles in colonic carcinogenesis. A total of 188 DEPs were identified. The underexpression of DCN and overexpression of HSPD1 in colon cancer were evaluated by Western blotting and immunohistochemistry. For the first time, our study indicates that DCN and HSPD1 are new promising biomarkers to detect colon cancer and demonstrate that some signaling pathways participate in human colonic carcinogenesis.
Materials and Methods
Tissue Collection, Laser Capture Microdissection, and Protein Extraction
A total of eight different tissues from each of patients undergoing curative surgery and receiving neither chemotherapy nor radiotherapy for colon cancer were acquired from 2011 to 2012 at the Department of General Surgery, Xiangya Hospital, Central South University, China, and used for proteomics and Western analysis. A total of eight colon cancer and eight NNCM tissues were processed for iTRAQ-based quantitative proteomics analysis. After surgery, pairs of colon cancer and NNCM tissues were removed from the resected colon cancer, then stored at -80°C, and diagnosed by pathologic evaluation of hematoxylin and eosin-stained frozen tissue sections. NNCM tissues were dissected from a distance of at least 10cm away the cancer. every patient signed a written informed consent form for the present study, with the approval of the Medical Research Ethics Committee of Xiangya Hospital (Changsha, China).
LCM was performed with a Leica AS LMD Microsystem (Mannhein, Germany) to harvest the cell populations of interest directly from tissues, as described previously24.After pathological evaluation, frozen sections (8 μm thick) from each colon cancer and NNCM were cut on a cryostat, CM 1900 microtome (Leica, Heidelberger, Germany), at -25°C. The sections were thaw mounted on membrane-coated glass slides (Leica), and then fixed at 20°C in 75% alcohol for 30 seconds. The sections were immersed in 0.5% violet-free methyl green (Sigma-Aldrich) and air dried. The stained sections were isolated using LCM, then stored at -80°C until use. Each captured cell population was to be over 95% homogeneous as determined by direct microscopic visualization of the collected cells.
The collected cell populations were completely lysed in protein lysis buffer containing 7 M urea, 2 M thiourea, 0.1 mM phenylmethylsulfonyl fluoride and 65 mM dithiothreitol for 1 hour on ice. After centrifugation(30 minutes, 4°C, 12,000 rpm), the supernatant was harvested, and the total protein concentration of each sample was accurately estimated using a 2D Quant Kit (GE Healthcare, Buckinghamshire, UK). To better understand sample biological variation on proteomic results, the same amount of protein from the collected cells of eight different samples were mixed to produce single sample for each kind of tissues (colon cancer and NNCM), sequentially obtaining the two mixed protein samples used for protein labeling by iTRAQ.
After pathological diagnosis, two types of formalin-fixed and paraffin-embedded archival samples, composed of 20 cases of NNCM and 30 cases of colon cancer were obtained from surgical intervention and stored at Department of Pathology of Xiangya Hospital, Central South University, China, and processed for immunohistochemical analysis. None of the patients in present study received chemotherapy or radiotherapy.
Protein Digestion and Labeling with iTRAQ Reagents
Trypsin digestion of two mixed protein samples and iTRAQ labeling of peptides were performed in accordance with the manufacturer's protocols (Applied Biosystems, Foster City, CA, USA). In brief, 100 μg proteins of each pooled sample were precipitated with cold acetone and reduced with tris-(2-carboxyethyl) phosphine(TCEP,5mM) at 60°C for 1 hour. After protein alkylation with iodoacetamide(84 mM) in the dark at room temperature for 30 minutes and overnight digestion on a shaker at 37°C with trypsin (MS grade; Promega, Madison, WI,USA) , the peptides were labeled with iTRAQ reagents (Applied Biosystems, USA) as follows: colon cancer, iTRAQ reagent 114, 116; and NNCM, iTRAQ reagents 115, 117. Four labeled samples were mixed into one tube and dried by evaporation in a centrifugal vacuum concentrator.
Off-line 2D LC-MS/MS
Firstly, the iTRAQ labeling mixed peptides were separated by strong cation exchange column into ten fractions, as we previously described19. Prior to MS analysis, the peptides mixture was desalted with solid phase extraction cartridge (Sep-Pak C18 Cartridge, vac 1cc, Waters, Milford, MA,USA), added to the loading buffer (25% acetonitrile,10 mM KH2PO4, pH 2.8), and loaded onto a polysulfoethyl column (2.1 × 100 mm, 5 μm, 300 Å; The Nest Group Inc., Southborough, USA).Separation was performed at a flow rate of 200 μl/min for 60 minutes with a linear binary gradient of 0-80% buffer B(25% acetonitrile,10 mM KH2PO4) in buffer A(25% acetonitrile,10 mM KH2PO4, 350 mM KCl).
Each strong cation exchange fraction was dried down by evaporation in the rotary vacuum concentrator, then resuspended in buffer C (0.1% formic acid, 5% acetonitrile,), and loaded on Qstar XL (Applied Biosystems, Warrington, UK), as we previously described19. In brief, the resulting peptides were separated on an Eksigent 1D PLUS system (AB SCIEX, Framingham, MA,USA) equipped with a reverse-phase column (ZORBAX 300SB-C18 column, 5 μm, 300Å, 0.1×15 mm; Waters Micromass, Manchester, UK) by using a linear gradient extending from 5 to 35% buffer D (0.1% formic acid, 95% acetonitrile,) in buffer C at an analytical flow velocity of 200nl/min for 70 minutes. Survey scans were obtained in ranges m/z 400-1800,with up to 4 precursors selected for MS/MS in range m/z 100-2000 with dynamic exclusion.
Data Analysis
Data was acquired using Analyst QS Software 1.1 (version, Applied Biosystems) for the QSTAR XL System. Protein identification, characterization and quantification of the iTRAQ data were performed using Paragon and Pro Group algorithm in ProteinPilot Software (version 4.2, Applied Biosystems) with a precursor tolerance of 150ppm, and a fragment tolerance of 0.2 Da. The data search parameters were described as follows: iTRAQ (peptide labeled) as sample type, fixed modification of methyl methanethiosulfonate tagged cysteine, trypsin as digestive enzyme, Triple TOF 5600 instrument system, Homo sapiens, biological modifications, automatical bias correction, International Protein Index(IPI) human database (version: 3.45; 143,958 entries) as database searching of MS/MS data, thorough identification search, max missed cleavages up to 2, FDR(false discovery rate) analysis, background correction, and no modified parameters. The identified proteins from the ProGroup algorithm were automatically grouped by sharing the same sequences to minimize redundancy. Only peptides unique for a give protein or proteins within the group were used for calculate protein rations, determining true protein isoforms from subsets. The ProteinPilot cutoff score was set as 1.3 (unused ProtScore) with at least one peptide from 95% confidence. A reverse-concatenated database search strategy was investigated using the false discovery rate(FDR) for protein identification. Protein quantified with the relative protein abundances of more than 100%( the iTRAQ ratio ≥2 and ≤0.5) that were analyzed by the signature ion ratio (m/z, 114/115; 116/117), were identified as differentially expressed proteins.
Western Blotting
Western blotting was used to detect changes in the expression of DCN and HSPD1 between NNCM and colon cancer tissues. Briefly, 30-50 μg of cell lysate samples was eletrophoresed on 12% SDS-PAGE and transferred to polyvinylidene difluoride membranes by eletroblotting. Membranes were blocked in 5% milk for 2 hours and then incubated with different primary antibody (anti-DCN antibody, 1:100; anti-HSPD1 antibody, 1:2000, Abcam, Cambridge, MA, USA) for 12 hours at 4°C, followed by incubation with a horseradish peroxidase-coupled mouse, rabbit, and goat secondary antibody for 2 hours at room temperature. After extensive washing, the membranes were placed in contact with ECL Western Blotting Detection Reagent (GE Healthcare, Buckinghamshire, UK), and the images were quantified by densitometry using Imagequant Image Analysis System (Storm Optical Scanner, Molecular Dynamics, Sunnyvale, CA, USA). β-actin was used normalize loading variations and was assessed using a mouse anti-β-actin antibody (1:4,000; Sigma-Aldrich).
Immunohistochemical Assays
Immunohistochemical staining for DCN and HSPD1 was performed on formalin-fixed, paraffin-embedded (FFPE) tissue sections (4 μm thickness) according to standard protocols. Briefly, each slide was dewaxed, rehydrated, and microwaved in sodium citrate buffer (10 mmol/l, pH 6.0) for antigen retrieval. The sections were incubated with primary antibody (anti-DCN antibody, 1:100,Bioss; anti-HSPD1 antibody,1:250; Abcam) antibody for 12 hours at 4°C, and then were incubated with a biotinylated secondary antibody (1:1,000 dilution) followed by addition of avidin-biotin peroxidase complex (DAKO) in accordance with the manufacturer's protocols. Lastly, FFPE sections were covered with 3′, 3′-diaminobenzidine (Sigma-Aldrich) until a brown color was visble, and subsequently the sections were counterstained with Harris' modified hematoxylin dehydrated and mounted. In negative controls, anti-DCN antibody or anti-HSPD1 was omitted. For the evaluation of DCN and HSPD1 expression, the sections were analyzed independently by two surgical pathologists making great effort to come to a consensus on staining expressions by light microscopy. A semi-quantitative score of histopathological characteristic was used to assess staining intensity and staining area for each case to detect the expression levels of the proteins, as we previously described25. At least 10 high-power fields were selected randomly, and >1,000 cells were counted for each immunohistochemical section. The numerical value for positive intensity of staining was based on a 4 point system: 0, 1+, 2+, 3+ (for none, pale yellow, tan-yellow stained, or brown staining). Similarly, the numerical value for negative intensity of staining was given a value from 0 to 4: 0(<5% of tissue stained negative), 1+(5-25%), 2+( 25%-50%), 3+(50-75%), 4+(>75%). A total staining score (positive stained + negative stained) recorded as negative staining (-, ≤2), weak staining (+,2-3), moderate staining (++,3-6), and strong staining (+++,6-7).
Gene Ontology, KEGG and Protein-Protein Interaction Network Analysis
The DEPs were firstly annotated by gene ontology (GO) from annotated using the PANTHER database (http://www.pantherdb.org/)26.The GO terms were considered statistically significant and enriched when the corrected p-value was <0.05. The GO terms with computed p-values <0.05 were considered as significantly enriched. The DEPs were subjected to gene-pathway annotation by Kyoto Encyclopedia of Genes and Genomes (KEGG) 27. KEGG pathway analysis was performed with the protein-protein interaction network of the DEPs using Cytoscape (V2.4.1)28.For investigation the possibly PPI (protein-protein interaction) of those proteins, the PPI prediction process has been executed by String(V9.05) 29.
Statistical Analysis
Statistical analyses were performed on a computer using SPSS18.0. Student t-test and χ2 test were both used to evaluate significant differences between different groups. A two-sided p <0.05 was considered significant.
Results
Identification of Differentially Expressed Proteins in NNCM and Colon Cancer Using iTRAQ Tagging with 2D LC-MS/MS
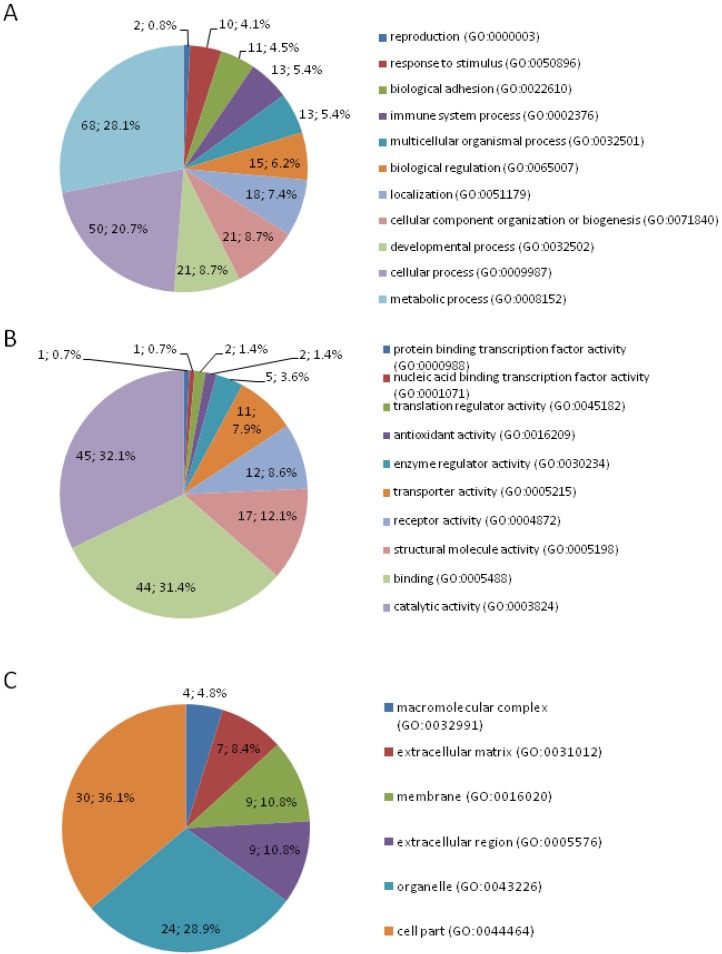
A total of 188 DEPs were identified by quantitative proteomic approach of iTRAQ-labeling coupled 2D LC-MS/MS, among which 77 upregulated and 111 downregulated proteins in colon cancer (Table S1). DEPs were classified according to GO term at the biologic process, molecular function, and cellular compartment level, respectively using the PANTHER GO classification system (Figure 1). In the aspect of the biologic process, the majority of the proteins were involved in metabolic process (28.10%), cellular process (20.70%), developmental process (8.70%), cellular component organization or biogenesis (8.70%), localization (7.4%), and biological regulation (6.2%). In respect to molecular function, the most common differential expression proteins were associated with catalytic activity, followed by binding, structural molecule activity, receptor activity, and transporter activity. According to the subcellular distribution, DEPs were cell part proteins (36.10%), organelle proteins (28.90%), extracellular region proteins (10.80%), membrane proteins (10.80%), extracellular matrix proteins (8.40%), and macromolecular complex proteins (4.80%). The DEPs were correlated with a wide range of biological processes and molecular functions.
Figure 1.
Classification of differentially expressed proteins by gene ontology biological process (A), molecular function (B), and subcellular component (C).
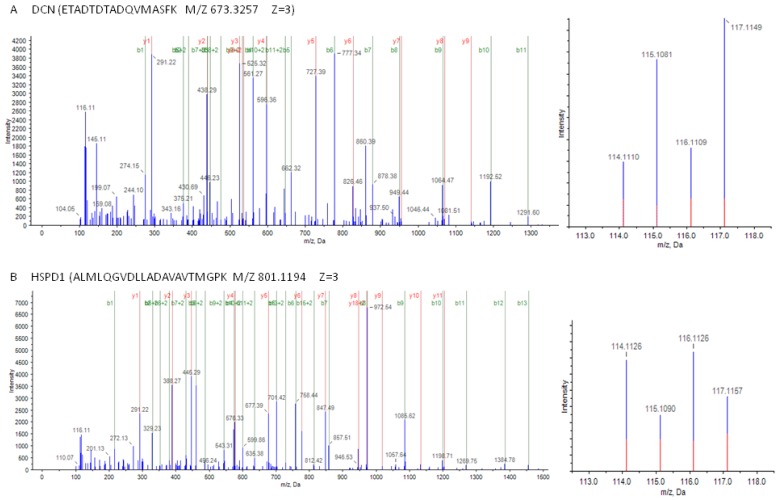
As shown in Figure 2, MS/MS spectra consistent with peptides that are part of DCN and HSPD1.
Figure 2.
Mass spectrometry (MS) spectra of precursor ions in consistent with peptides are part of DCN and HSPD1. (A) (left) The sequence ETADTDTADQVMASFK allows the identification of DCN; (right) the released iTRAQ reporter ions provide the relative quantitation of DCN from the two tissues evaluated. (B) (left) The sequence ALMLQGVDLLADAVAVTMGPK allows the identification of HSPD1; (right) the released iTRAQ reporter ions provide the relative quantitation of HSPD1 from the two tissues evaluated. Non-neoplastic colonic mucosa, labeled with iTRAQ reagent 115,117; colon cancer, labeled with iTRAQ reagents 114,116.
Validation of Differentially Expressed Proteins Identified by Proteomics
Two proteins (DCN and HSPD1) identified by mass spectrometry analysis (MS) were selected for verification. Western blotting was performed to assess the expressional levels of the two proteins in 10 cases of LCM-purified NNCM and colon cancer. As illustrated in Figure 3, DCN was downregulated, whereas HSPD1 was upregulated in the colon cancer versus NNCM (p <0.01), which correspond to the findings in MS analysis.
Figure 3.
Expressional levels of DCN and HSPD1 in non-neoplastic colonic mucosa (NNCM) (N) and colon cancer (C). (left) Representative results of Western blotting analysis of DCN and HSPD1 in NNCM (N) and colon cancer (C). (right) Histogram shows the expression levels of the two proteins in these tissues as determined by densitometric analysis. β-actin was used as a loading control. Columns, mean from 10 cases of tissues; bars, S.D.
Detection of the Expression of DCN and HSPD1 by Immunohistochemistry
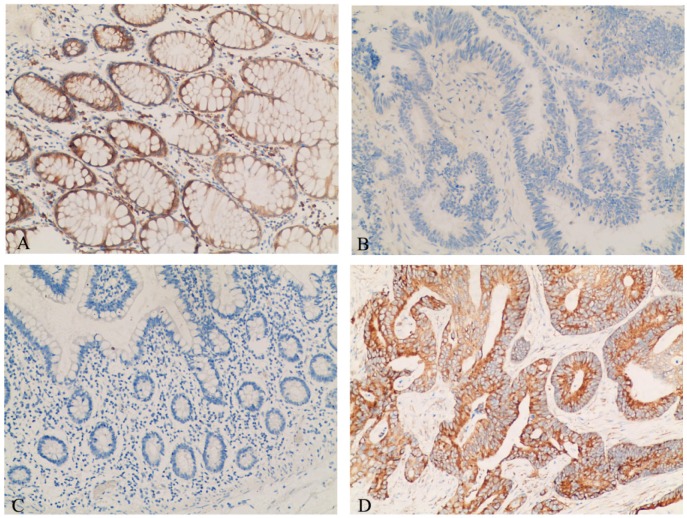
The expressional levels of DCN and HSPD1 in 30 NNCM and 20 colon cancer were evaluated by immunohistochemisry. As observed in Figure 4A,B and Table 1, the expression of DCN in colon cancer was significantly reduced than that in NNCM (p <0.025). As shown in Figure 4C,D and Table 1, the expression of HSPD1 was upregulated in colon cancer tissues compared with NNCM tissues (P <0.05).
Figure 4.
Representative results of immunohistochemistry of DCN and HSPD1 in tissue specimens. (A) DCN-non-neoplastic colonic mucosa. (B) DCN-colon cancer. (C) HSPD1-non-neoplastic colonic mucosa (D) HSPD1-colon cancer. Main image, ×100.
Table 1.
DCN and HSPD1 expression in various tissues of in non-neoplastic colonic mucosa and colon cancer by immunohistochemistry.
| Classification | Number | Score | P-value | ||
|---|---|---|---|---|---|
| - | + | ++-+++ | |||
| DCN | |||||
| NNCM | 30 | 22 | 6 | 2 | <0.025 |
| Colon cancer | 20 | 8 | 11 | 1 | |
| HSPD1 | |||||
| NNCM | 30 | 2 | 4 | 24 | <0.05 |
| Colon cancer | 20 | 12 | 5 | 3 | |
NNCM, non-neoplastic colonic mucosa.
KEGG Pathways and protein-protein interaction Analysis of Differentially Expressed Proteins
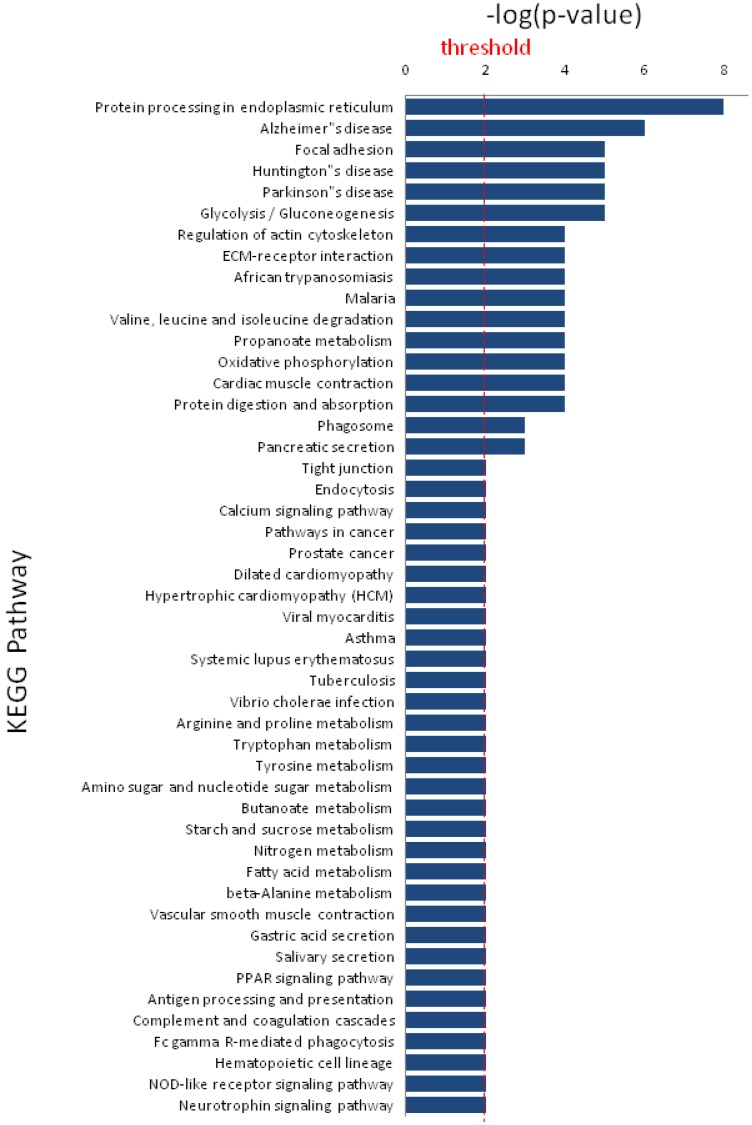
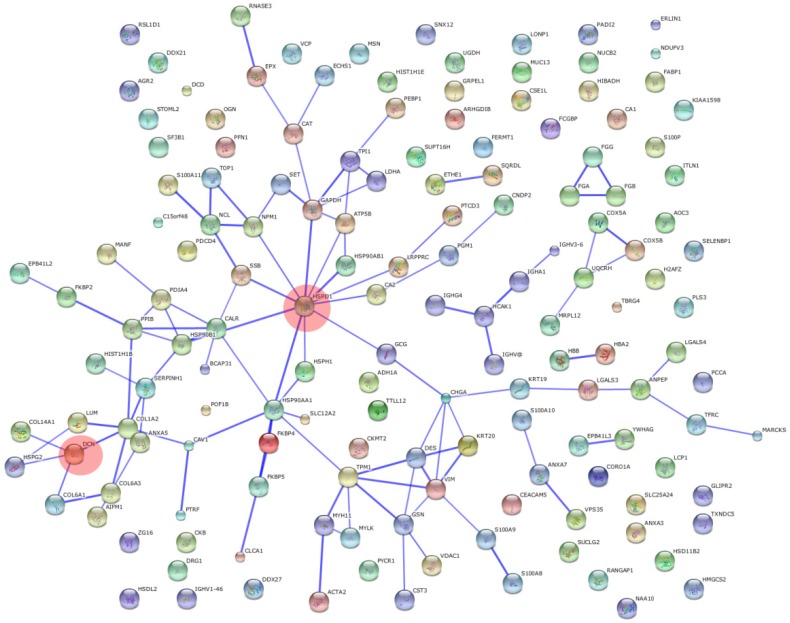
KEGG pathway analysis revealed that DEPs are implicated in cancer-related signaling pathways such as protein processing in endoplasmic reticulum, focal adhesion, glycolysis/gluconeogenesis, regulation of actin cytoskeleton, and extracellular matrix-receptor interaction (Figure 5). The DEPs may play a great role in colonic epithelial carcinogenesis by these signaling pathways. DCN and HSPD1 play an important role in the protein-protein interaction networks of the DEPs (Figure 6). DCN is associated with COL14A1, COL6A1, COL1A2, and HSPG2, which are involved in TGF-β signaling pathway and ECM-receptor interaction. HSPD1 is connected with HSP90AA1, HSPH1, CALR, HSPAB1, HSP90B1, etc., which are involved in chaperone activity and folding, sorting and degradation.
Figure 5.
Significant KEGG pathways that are involved in protein-protein interaction network of differentially expressed proteins.
Figure 6.
Protein-protein interaction networks of differentially expressed proteins. Opaque red circles, HSPD1 and DCN of nodes.
Discussion
The development of iTRAQ-based quantitative proteomics has offered the option to identified differentially expressed proteins, which may be correlated with colonic carcinogenesis and be used for biomarkers. In this report, iTRAQ-coupled 2D LC-MS/MS was used to identify DEPs in NNCM and colon cancer. As a result, 188 DEPs were identified, and two differential proteins (DCN and HSPD1) showing altered expression between NNCM and colon cancer were selectively validated. These results indicated that the two proteins were potentials biomarkers for the detection of colon cancer.
DCN (also called decorin), which belongs to a member of the small leucine-rich proteoglycans family, exists and plays multifaceted roles in stromal and epithelial cells30. Some studies have shown that DCN is involved in multifunctional biological and physiological processes, such as extracellular matrix organization, cell signal transduction, cell proliferation, wound healing, and cell differentiation31-33, whereas accumulating evidence shows that DCN is dysregulated expression and targets a number of crucial signaling molecules in a wide variety of human tumors, for example, oral, pancreatic, lung, prostate, and breast cancers30, 34-36. DCN expression was reduced in cancerous tissues of renal cell carcinoma and an ectopic expression of DCN significantly upregulated P21 and E-cadherin expression that revealed DCN could be used as a potential therapeutic target in renal cell carcinoma37. It was found that DCN over expression correlated with decreased adhesion and migration of U87MG glioma cells by downregulation of TGF-β signaling38. Additionally, it was indicated that DCN inhibited the growth arrest and metastasis of non-small-cell lung cancer with E-cadherin and vascular endothelial growth factor30. Genetic deletion of DCN can facilitate intestinal tumor formation in mice by attenuating of p21, p27(kip1), and E-cadherin, and enhancing of β-catenin signaling39. Further studies have found that DCN-induced attenuated CRC growth and migration through the interaction of E-cadherin using a decorin knockout mouse model40. Our study indicated a significant downregulated of DCN in colon cancer compared to NNCM, could offer a clue that DCN might be related to colonic carcinogenesis.
Heat shock proteins(HSPs) were originally discovered as a member of stress-induced proteins characterized by chaperoning functions. It is thought indicated that HSPs play an vital role in the control of immune responses by various cell stresses41. HSPD1(heat shock protein family D [Hsp60[ member 1) is essential for the folding and assembly of imported proteins in the mitochondria as a signaling molecule in the innate and adaptive immune system42. HSPD1 is involved in tumorigenesis and its levels are dysregulated in various cancers43-47. Furthermore, it has been shown that HSPD1 can utilize tumor cell apoptosis, relying on the mechanisms of its accumulation in the cytosol48. HSPD1 has been considered as a promising prognostic biomarker because its overexpression was correlated with the development and invasion in gastric cancer47. Moreover, it has been reported that downregulation of HSP60-induced apoptosis in gastric cancer cells was negatively correlated with the MEK/ERK signaling in vitro49. It has been reported that downregulation of HSP60 suppressed the proliferation of glioblastoma cells through the ROS/AMPK/mTOR pathway50. Our results are in accordance with the reports that HSPD1 is overexpressed in colon cancer compared with NNCM. Although many potential functions have been presented for HSPD1 in colon cancer, its biological role still remains to be defined.
The diversity of signaling pathways play an essential role in cancer initiation and progression associated with cell cycle checkpoints, transcriptional regulation, environmental cues, angiogenic signaling, metabolic coercion, epigenetic regulation, cell survival, immune suppression, and more51, 52. Stress induced by accumulation of misfolded proteins in the endoplasmic reticulum is found in many physiologic and pathologic conditions53. The unfolded protein response, one of the proteins processing in endoplasmic reticulum, is the cellular response to endoplasmic reticulum stress and plays a pivotal role in oncogenesis54. Focal adhesion kinase(FAK), a cytoplasmic tyrosine kinase that participated in tumor initiation, progression, and metastasis by triggering multiple downstream signaling pathways to regulate some cellular functions55, 56. The last decade has proved a renaissance of the Warburg effect, that mitochondrial malfunction and subsequent stimulation of cellular glucose utilization including glycolysis and gluconeogenesis lead to the development of cancer57, 58. Some studies suggest that actin remodeling is an upstream regulator of epithelial mesenchymal transition associated with the development and spread of cancer using extracellular matrix-receptor interaction in metastatic cancer cells59-61. In the present study, we found that KEGG pathway analysis showed that the DEPs are implicated in cancer-associated signaling pathways. We also showed that the proteins interacted with DCN or HSPD1 have many different and varied biological functions and the alternation of expression level of the two proteins have influence on many biological processes. These indicate that DCN and HSPD1 play central roles of in the PPI subnetwork of the DEPs was true. The studies may offer valuable clues for further in-depth study of molecular mechanisms that lead to the normal-to-cancer conversion of human colonic epithelium.
In brief, 188 DEPs were identified in NNCM and colon cancer by iTRAQ-labeling coupled with 2D LC-MS/MS, and two differential proteins (DCN and HSPD1) with expressional changes were selectively verified. The DEPs are correlated with cancer-associated signaling pathways, such as unfolded protein response, glucose utilization, and epithelial mesenchymal transition.
Clinical Perspectives
The current study provides comprehensive insights into the novel biomarkers and the molecular signaling pathways that are involved in colonic oncogenesis, which may help to identify new strategies to improve colon cancer survival.
Supplementary Material
Table S1.
Acknowledgments
Grants were provided by the National Natural Science Foundation of China (No. 81272971, No. 81202129, and No. 81372516), the National Key Basic Research Program of China (973 Program) (No. 2011CB910704 and No. 2014CBA02004), the Project of Scientific Research Fund of Hunan Provincial Education Department (No. 16C1409), and the Natural Science Foundation of Hunan Province of China (No. 2016JJ5013).
Abbreviations
- 2D LC-MS/MS
two-dimensional liquid chromatography tandem mass spectrometry
- CRC
Colorectal cancer
- DEPs
Differentially expressed proteins
- FFPE
Formalin-fixed, paraffin-embedded
- GO
Gene ontology
- iTRAQ
Isobaric tags for relative and absolute quantitation
- KEGG
Kyoto encyclopedia of genes and genomes
- LCM
Laser capture microdissection
- MS
mass spectrometry, NNCM: Non-neoplastic colonic mucosa.
Biographies
Dr. Zhuchu Chen is a professor and director of Key Laboratory of Cancer Proteomics of Chinese Ministry of Health, Xiangya Hospital, Central South University, China. He has coauthored over 200 publications including J Pathol, Cell Mol Life Sci, J Proteome Res, BMC Bioinformatics, Oncotarget. The current research interests in Professor Chen's group include: (1) carcinogenic mechanism; (2)cancer proteomics and structural biology.
Guoqing Li obtained his Master's degree from School of Life Science, South China Normal University in 2005. He is currently a Ph. D. student under the supervision of Prof. Zhuchu Chen. At the same time, he is a lecturer in Hunan Province Cooperative Innovation Center for Molecular Target New Drug Study, School of Pharmacy and Life Science, University of South China. His research is centered on cancer proteomics and cell signal transduction.
References
- 1.Luo Y, Wang L, Wang J. Developing proteomics-based biomarkers for colorectal neoplasms for clinical practice: opportunities and challenges. Proteomics. Clinical applications. 2013;7(1-2):30–41. doi: 10.1002/prca.201200071. [DOI] [PubMed] [Google Scholar]
- 2.Colorectal Cancer 2015. Society AC; http://www.cancer.org/acs/groups/cid/documents/webcontent/003096-pdf.pdf. [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Ferlay J, Soerjomataram I, Dikshit R. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. Journal international du cancer. 2015;136(5):E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 5.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(2):104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 6.CHEN Wan-qing ZR-s, ZENG Hong-mei, ZOU Xiao-nong, ZHANG Si-wei, HAO Jie. Report of Cancer Incidence and Mortality in China,2011. China Cancer. 2015;24(1):1–10. [Google Scholar]
- 7.WANG Ning ST-t, ZHENG Rong-shou, ZHANG Si-wei, CHEN Wang-qing. An Analysis of Incidence and Mortality of Colorectal Cancer in China,2009. China Cancer. 2013;22(7):516–520. [Google Scholar]
- 8.van Rossum LG, van Rijn AF, Laheij RJ. et al. Random comparison of guaiac and immunochemical fecal occult blood tests for colorectal cancer in a screening population. Gastroenterology. 2008;135(1):82–90. doi: 10.1053/j.gastro.2008.03.040. [DOI] [PubMed] [Google Scholar]
- 9.Herzig DO, Tsikitis VL. Molecular markers for colon diagnosis, prognosis and targeted therapy. Journal of surgical oncology. 2015;111(1):96–102. doi: 10.1002/jso.23806. [DOI] [PubMed] [Google Scholar]
- 10.Mazzanti R, Solazzo M, Fantappie O. et al. Differential expression proteomics of human colon cancer. American journal of physiology. Gastrointestinal and liver physiology. 2006;290(6):G1329–1338. doi: 10.1152/ajpgi.00563.2005. [DOI] [PubMed] [Google Scholar]
- 11.Ma Y, Peng J, Liu W. et al. Proteomics identification of desmin as a potential oncofetal diagnostic and prognostic biomarker in colorectal cancer. Molecular & cellular proteomics: MCP. 2009;8(8):1878–1890. doi: 10.1074/mcp.M800541-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Song MQ, Zhu JS. et al. Identification of differentially-expressed proteins between early submucosal non-invasive and invasive colorectal cancer using 2D-DIGE and mass spectrometry. International journal of immunopathology and pharmacology. 2011;24(4):849–859. doi: 10.1177/039463201102400404. [DOI] [PubMed] [Google Scholar]
- 13.Zhu G, Xiao ZQ, Chen ZC. et al. [Identification of aging related proteins in human normal colonic epithelium]. Zhong nan da xue xue bao. Yi xue ban = Journal of Central South University. Medical sciences. 2005;30(6):625–630. [PubMed] [Google Scholar]
- 14.Mu Y, Chen Y, Zhang G. et al. Identification of stromal differentially expressed proteins in the colon carcinoma by quantitative proteomics. Electrophoresis. 2013;34(11):1679–1692. doi: 10.1002/elps.201200596. [DOI] [PubMed] [Google Scholar]
- 15.Chen D, Chen F, Lu X. et al. Identification of prohibitin as a potential biomarker for colorectal carcinoma based on proteomics technology. International journal of oncology. 2010;37(2):355–365. doi: 10.3892/ijo_00000684. [DOI] [PubMed] [Google Scholar]
- 16.Zieske LR. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. Journal of experimental botany. 2006;57(7):1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 17.Ralhan R, Desouza LV, Matta A. et al. Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Molecular & cellular proteomics: MCP. 2008;7(6):1162–1173. doi: 10.1074/mcp.M700500-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeSouza LV, Grigull J, Ghanny S. et al. Endometrial carcinoma biomarker discovery and verification using differentially tagged clinical samples with multidimensional liquid chromatography and tandem mass spectrometry. Molecular & cellular proteomics: MCP. 2007;6(7):1170–1182. doi: 10.1074/mcp.M600378-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Xiao Z, Li G, Chen Y. et al. Quantitative proteomic analysis of formalin-fixed and paraffin-embedded nasopharyngeal carcinoma using iTRAQ labeling, two-dimensional liquid chromatography, and tandem mass spectrometry. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 2010;58(6):517–527. doi: 10.1369/jhc.2010.955526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Y, Yang M, She S. et al. iTRAQ-based quantitative proteomic analysis of cervical cancer. International journal of oncology. 2015;46(4):1748–1758. doi: 10.3892/ijo.2015.2859. [DOI] [PubMed] [Google Scholar]
- 21.Zhang PF, Zeng GQ, Yi LZ. et al. Identification of integrin beta1 as a prognostic biomarker for human lung adenocarcinoma using 2D-LC-MS/MS combined with iTRAQ technology. Oncology reports. 2013;30(1):341–349. doi: 10.3892/or.2013.2477. [DOI] [PubMed] [Google Scholar]
- 22.Zhu G, Cai G, Liu Y. et al. Quantitative iTRAQ LC-MS/MS Proteomics Reveals Transcription Factor Crosstalk and Regulatory Networks in Hypopharyngeal Squamous Cell Carcinoma. Journal of Cancer. 2014;5(7):525–536. doi: 10.7150/jca.9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu YF, Xiao ZQ, Li MX. et al. Quantitative proteome analysis reveals annexin A3 as a novel biomarker in lung adenocarcinoma. The Journal of pathology. 2009;217(1):54–64. doi: 10.1002/path.2429. [DOI] [PubMed] [Google Scholar]
- 24.Cheng AL, Huang WG, Chen ZC. et al. Identificating cathepsin D as a biomarker for differentiation and prognosis of nasopharyngeal carcinoma by laser capture microdissection and proteomic analysis. Journal of proteome research. 2008;7(6):2415–2426. doi: 10.1021/pr7008548. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Cao LQ, Jin LY. et al. Quantitative proteomic study of human lung squamous carcinoma and normal bronchial epithelial acquired by laser capture microdissection. Journal of biomedicine & biotechnology. 2012;2012:510418. doi: 10.1155/2012/510418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi H, Lazareva-Ulitsky B, Loo R. et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic acids research. 2005;33(Database issue):D284–288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S, Kawashima S. et al. The KEGG resource for deciphering the genome. Nucleic acids research. 2004;32(Database issue):D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopes CT, Franz M, Kazi F. et al. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26(18):2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szklarczyk D, Franceschini A, Kuhn M. et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic acids research. 2011;39(Database issue):D561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi X, Liang W, Yang W. et al. Decorin is responsible for progression of non-small-cell lung cancer by promoting cell proliferation and metastasis. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(5):3345–3354. doi: 10.1007/s13277-014-2968-8. [DOI] [PubMed] [Google Scholar]
- 31.Neill T, Schaefer L, Iozzo RV. Decorin: a guardian from the matrix. The American journal of pathology. 2012;181(2):380–387. doi: 10.1016/j.ajpath.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang JY, Bao HY, Huang SM. et al. [Mechanisms of decorin inhibiting epithelial-to-mesenchymal transition induced by transforming growth factor beta1 in renal tubular epithelial cells]. Zhonghua er ke za zhi. Chinese journal of pediatrics. 2010;48(1):50–54. [PubMed] [Google Scholar]
- 33.Scott RA, Ramaswamy AK, Park K, Decorin mimic promotes endothelial cell health in endothelial monolayers and endothelial-smooth muscle co-cultures. Journal of tissue engineering and regenerative medicine; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasamatsu A, Uzawa K, Minakawa Y. et al. Decorin in human oral cancer: a promising predictive biomarker of S-1 neoadjuvant chemosensitivity. Biochemical and biophysical research communications. 2015;457(1):71–76. doi: 10.1016/j.bbrc.2014.12.093. [DOI] [PubMed] [Google Scholar]
- 35.Na Y, Choi JW, Kasala D. et al. Potent antitumor effect of neurotensin receptor-targeted oncolytic adenovirus co-expressing decorin and Wnt antagonist in an orthotopic pancreatic tumor model. Journal of controlled release: official journal of the Controlled Release Society. 2015;220(Pt B):766–782. doi: 10.1016/j.jconrel.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 36.Xu W, Neill T, Yang Y. et al. The systemic delivery of an oncolytic adenovirus expressing decorin inhibits bone metastasis in a mouse model of human prostate cancer. Gene therapy. 2015;22(3):247–256. doi: 10.1038/gt.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y, Xia Q, Rao Q. et al. DCN deficiency promotes renal cell carcinoma growth and metastasis through downregulation of P21 and E-cadherin. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(4):5171–5183. doi: 10.1007/s13277-015-4160-1. [DOI] [PubMed] [Google Scholar]
- 38.Yao T, Zhang CG, Gong MT. et al. Decorin-mediated inhibition of the migration of U87MG glioma cells involves activation of autophagy and suppression of TGF-beta signaling. FEBS open bio. 2016;6(7):707–719. doi: 10.1002/2211-5463.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bi X, Tong C, Dockendorff A. et al. Genetic deficiency of decorin causes intestinal tumor formation through disruption of intestinal cell maturation. Carcinogenesis. 2008;29(7):1435–1440. doi: 10.1093/carcin/bgn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bi X, Pohl NM, Qian Z. et al. Decorin-mediated inhibition of colorectal cancer growth and migration is associated with E-cadherin in vitro and in mice. Carcinogenesis. 2012;33(2):326–330. doi: 10.1093/carcin/bgr293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin L, Pan S, Zhao J. et al. HSPD1 interacts with IRF3 to facilitate interferon-beta induction. PloS one. 2014;9(12):e114874. doi: 10.1371/journal.pone.0114874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quintana FJ, Cohen IR. The HSP60 immune system network. Trends in immunology. 2011;32(2):89–95. doi: 10.1016/j.it.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 43.Rappa F, Farina F, Zummo G. et al. HSP-molecular chaperones in cancer biogenesis and tumor therapy: an overview. Anticancer research. 2012;32(12):5139–5150. [PubMed] [Google Scholar]
- 44.Jin H, Cheng X, Pei Y. et al. Identification and verification of transgelin-2 as a potential biomarker of tumor-derived lung-cancer endothelial cells by comparative proteomics. Journal of proteomics. 2016;136:77–88. doi: 10.1016/j.jprot.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 45.Davalieva K, Kostovska IM, Kiprijanovska S. et al. Proteomics analysis of malignant and benign prostate tissue by 2D DIGE/MS reveals new insights into proteins involved in prostate cancer. The Prostate. 2015;75(14):1586–1600. doi: 10.1002/pros.23034. [DOI] [PubMed] [Google Scholar]
- 46.Campanella C, Rappa F, Sciume C. et al. Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery. Cancer. 2015;121(18):3230–3239. doi: 10.1002/cncr.29499. [DOI] [PubMed] [Google Scholar]
- 47.Li XS, Xu Q, Fu XY. et al. Heat shock protein 60 overexpression is associated with the progression and prognosis in gastric cancer. PloS one. 2014;9(9):e107507. doi: 10.1371/journal.pone.0107507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandra D, Choy G, Tang DG. Cytosolic accumulation of HSP60 during apoptosis with or without apparent mitochondrial release: evidence that its pro-apoptotic or pro-survival functions involve differential interactions with caspase-3. The Journal of biological chemistry. 2007;282(43):31289–31301. doi: 10.1074/jbc.M702777200. [DOI] [PubMed] [Google Scholar]
- 49.Tong WW, Tong GH, Kong H, The tumor promoting roles of HSP60 and HIF2alpha in gastric cancer cells. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine; 2016. [DOI] [PubMed] [Google Scholar]
- 50.Tang H, Li J, Liu X. et al. Down-regulation of HSP60 Suppresses the Proliferation of Glioblastoma Cells via the ROS/AMPK/mTOR Pathway. Scientific reports. 2016;6:28388. doi: 10.1038/srep28388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 53.Hetz C, Chevet E, Oakes SA. Proteostasis control by the unfolded protein response. Nature cell biology. 2015;17(7):829–838. doi: 10.1038/ncb3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maurel M, McGrath EP, Mnich K. et al. Controlling the unfolded protein response-mediated life and death decisions in cancer. Seminars in cancer biology. 2015;33:57–66. doi: 10.1016/j.semcancer.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 55.Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer metastasis reviews. 2009;28(1-2):35–49. doi: 10.1007/s10555-008-9165-4. [DOI] [PubMed] [Google Scholar]
- 56.Lee BY, Timpson P, Horvath LG. et al. FAK signaling in human cancer as a target for therapeutics. Pharmacology & therapeutics. 2015;146:132–149. doi: 10.1016/j.pharmthera.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Gogvadze V, Zhivotovsky B, Orrenius S. The Warburg effect and mitochondrial stability in cancer cells. Molecular aspects of medicine. 2010;31(1):60–74. doi: 10.1016/j.mam.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 58.Upadhyay M, Samal J, Kandpal M. et al. The Warburg effect: insights from the past decade. Pharmacology & therapeutics. 2013;137(3):318–330. doi: 10.1016/j.pharmthera.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Jiang P, Enomoto A, Takahashi M. Cell biology of the movement of breast cancer cells: intracellular signalling and the actin cytoskeleton. Cancer letters. 2009;284(2):122–130. doi: 10.1016/j.canlet.2009.02.034. [DOI] [PubMed] [Google Scholar]
- 60.Shankar J, Nabi IR. Actin cytoskeleton regulation of epithelial mesenchymal transition in metastatic cancer cells. PloS one. 2015;10(3):e0119954. doi: 10.1371/journal.pone.0119954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benvenuti S, Comoglio PM. The MET receptor tyrosine kinase in invasion and metastasis. Journal of cellular physiology. 2007;213(2):316–325. doi: 10.1002/jcp.21183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.