Abstract
The cell interior is a crowded chemical space, which limits the diffusion of molecules and organelles within the cytoplasm, affecting the rates of chemical reactions. We provide insight into the relationship between non-specific intracellular diffusion and cytoskeletal integrity. Quantum dots entered the cell through microinjection and their spatial coordinates were captured by tracking their fluorescence signature as they diffused within the cell cytoplasm. Particle tracking revealed significant enhancement in the mobility of biocompatible quantum dots within fibrosarcoma cells versus their healthy counterparts, fibroblasts, as well as in actin destabilized fibroblasts versus untreated fibroblasts. Analyzing the displacement distributions provided insight into how the heterogeneity of the cell cytoskeleton influences intracellular particle diffusion. We demonstrate that intracellular diffusion of non-specific nanoparticles is enhanced by disrupting the actin network, which has implications for drug delivery efficacy and trafficking.
Introduction
Actin and tubulin form highly versatile, dynamic polymers that are fundamental to creating the spatial organization of eukaryotic cells through cytoskeletal structures. These cytoskeletal systems are dynamic and adaptable, capable of organizing intracellular compartments, and are critical to a number of cell processes including cell migration and division. Inside these compartments is a crowded chemical space in which the volume fraction of macromolecules can reach up to 40%.1 This large volume fraction limits diffusion of molecules and organelles within the cytoplasm and therefore affects the rates of chemical reactions in the cell. A contributor to this confinement is the cytoskeleton itself, which features a mesh-like network with open spaces that direct the diffusion of macromolecules. The structural role of the actin and microtubule network within cell mechanics has been previously investigated2–4 and together with the whole cytoskeleton network, cytoplasmic organelles, and molecular crowding, serves as obstacles to non-directed intracellular motion. Intracellular particle diffusion has become increasingly important within nanomedicine, where nano-scale photodynamic therapies5–7 and gene or drug delivery systems8–11 have made significant advances in disease treatment options. The effect of the cytoskeleton on the diffusion of nanoparticles brings into question the relative size scales between the mesh and the translocating objects. The actin network has been reported to have a heterogeneous mesh size ranging from 30–100 nm (Luby-Phelps12) to 300–600 nm (Kusumi et al.13) across mammalian cells. More recently, Kronlage et al.14 measured the open pore size within the actin cytoskeleton of endothelial cells under control conditions of approximately 10,000 nm2 (about 100 nm square) and under cytochalasin D treatment, the open area increases to above 30,000 nm2 (173 nm square). When particle size is similar to or greater than the cytoskeletal mesh, the resulting confinement allows for the measuring of the microrheological properties of the cell interior.15, 16 Alternatively, particles smaller than the mesh are subjected to a different microenvironment than larger particles. The different local environment will affect the diffusivity of small particles, critical in nanoparticle-based therapies or diagnostics. For example, in endosomal release of theranostic nanomaterials (e.g., nanoparticle mediated siRNA delivery8, 9), particles must translocate to an alternative intracellular site (e.g., the nucleus), where they accumulate for efficacy of the therapy. To determine the potential for accumulation and therefore the efficacy of these nanoparticle therapies, we must understand the dynamics involved in intracellular nanoparticle diffusion. Here we discuss the influence of cytoskeletal integrity, namely filamentous actin and microtubules, on intracellular nanoparticle diffusion manifested in different cell conditions. Motivated by prior elasticity measurements,2 we identify differences between two cell types, one cancer (fibrosarcoma cells HT 1080) and one non-cancer (human dermal fibroblasts), as well as drug-altered cytoskeletons within those two lines by cytochalasin D treatment (disrupts filamentous actin) and nocodazole treatment (disrupts microtubules). Within the cytoplasm of these cells, the nanoparticles act as internal probes that reflect the influence of the cytoskeleton and its caged network on transport. Analysis of the nanoparticle paths further provides quantitative values of diffusion coefficients that drug carrier and similarly sized biomaterials have inside healthy and diseased cells.
Candidates for assessing intracellular trafficking include tracking small fluorescent molecules using FRAP17 or FCS18, or tracking micron and sub micron particles using single particle tracking (SPT)19 or extensions thereof.15 Particle mobility has also been studied as a probe of local viscosity in the field of microrheology.20 Labeling a few molecules by fluorescent dyes can achieve high temporal resolution but lack the lateral/spatial resolution that SPT can achieve. SPT can be limited by photobleaching of fluorophores if used, and the inability to track particles that travel perpendicular to the imaging plane. However, SPT has been shown to be well suited to the study of living cell interiors.19, 21–25 Quantum dots (QDs) present a unique opportunity for stable particle tracking as they are not subject to photobleaching and can be synthesized to respond to a variety of chosen wavelengths as well as include a chemically grafted outer shell either to confer, or to prevent, specific biological interactions. SPT of QDs within the cellular environment has been used to investigate specific cellular processes such as insulin receptor activity,26 virus infection,27, 28 calcium flux detection,29 and kinesin motor transport.30 Polymer coated nanoparticles have also been used in treatment regimes such as targeted killing of tumor cells5, 6 and alleviating liver damage.31 Once inside the cell, SPT has revealed dynamic properties of delivered particles. Freely diffusive particles (Brownian motion) have a linear relationship between time and mean square displacement (MSD), however, intracellular transport typically results in subdiffusive or confined behavior, i.e., movement of particles with less than t1 dependence.4 The distinction between Brownian motion and subdiffusive motion in the cellular environment is extremely important: if particles are confined, the time and even ability to reach a desired intracellular destination will be affected. Identifying which cellular components contribute to this confinement is critical to understanding intracellular transport.
Our results indicate that the diffusion of small, polymer-grafted QDs is sensitive to cytoskeletal modifications, particularly actin network connectivity. Unlike preceding QD intracellular tracking experiments32 these QDs have no specific affinity for binding within the cell. The PEG coating on our QDs has been shown repeatedly to be the most highly non-interacting polymer coating for cellular environments.21, 33 We demonstrate that heterogeneity in the actin network either manifested by cell type or by disruption of cytoarchitecture within healthy cell types results in a change in nanoparticle dynamics thus signifying that the cytoskeleton organization can act as a barrier for non-motor driven intracellular trafficking.
Results and Discussion
In our work, CdSe quantum dots (4 nm) were grafted with a biocompatible PEG34 brush (Mw=5 kDa) that neither interacts with intracellular components nor is toxic to the cell. For example, prior work shows that Ag nanoparticles are toxic to THP-1 cells resulting in a change in cell viscoelasticity with a two fold increase in complex modulus.35 The PEG grafted QDs have a hydrodynamic diameter of 10 nm, determined by diffusion in glycerol:water with known viscosity (supplementary materials). QDs entered each cell by microinjection using an Eppendorf Injection system (supplementary materials), which provides a fast introduction (0.5 s) of QDs by pressure-driven injection of small liquid volumes. The micropipettes had an outer diameter of 1 μm. The delivery and diffusion of QDs were monitored throughout the cell cytoplasm. We manipulated the needle by using a common piezoelectric micromanipulator integrated with a Nikon inverted epifluorescence microscope with 100× oil-immersion objective. The needle was manipulated to approach and penetrate the cell at an angle of 30 degrees from the surface plane. During QD delivery, we could precisely locate the needle in the cellular environment in brightfield and fluorescence imaging mode. The photo stability of QDs allowed continuous observation of QDs for the duration of video capture, which was between 5 and 10 minutes.
To quantify the QD dynamics, we carried out SPT using free software to connect particle centers of mass across images, obtaining particle trajectories.36 Figure 1a–c includes a subset of 50 particle trajectories from QDs in the cytoplasm of fibroblasts, fibrosarcoma cells, and cytochalasin D treated fibroblasts as well as an overlay of all 699, 634, and 353 trajectories, respectively.
Figure 1.
Individual QD particle trajectories (50) within (a) fibroblasts, (b) fibrosarcoma cells, and (c) fibroblasts with cytochalasin D treatment with overlay insets where trajectories are initialized at (0,0) and include all 699, 634, and 353 trajectories recorded, respectively. Mean squared displacements (MSD) of QDs within (d) fibroblasts, (e) fibrosarcoma cells and (f) fibroblasts with cytochalasin D treatment. Color scale used to indicate higher MSD is carried through for individual particle tracks in (a–c).
The QDs within the fibrosarcoma cell cytoplasm had greater mobility than within the fibroblast possibly because the cytoskeletal network is more disorganized. For example, in model mouse ovarian cancer cells, in addition to changes in gene expression levels, the cytoskeleton structure became increasingly disorganized during neoplastic progression.37 Other reports confirm the deformation capacity of cancer cells (and metastatic potential) is due to differences in the cytoskeleton and nuclear protein organization.38–42
To test if the particular organization of microfilaments within the fibrosarcoma cell line contributed to a change in the mobility of the QDs, cytochalasin D was used to disrupt the filamentous actin network in fibroblasts to achieve less organization. This treatment resulted in a longer mean free path of the QD consistent with the comparison between fibroblasts and fibrosarcoma cells (see supplementary materials for actin staining). We also investigated how QD diffusion was influenced by the breakdown of the microtubule network induced by nocodazole treatment. We found that the mobility of QDs within fibroblasts was less affected by nocodazole treatment than cytochalasin D treatement, which is consistent with prior studies showing that the mechanical properties of fibroblasts do not change significantly when exposed to nocodazole.2 From the particle tracks, MSD was determined using MATLAB43. MSDs for QDs within fibroblasts, fibrosarcoma cells, and cytochalasin D treated fibroblasts are shown in Figure 1d–f respectively. To compare ensemble differences, we analyzed the particle trajectories using (1) the center of the MSD distributions at a range of delay times to compare overall mobility, (2) a power law fit to the data to extract diffusion parameters, and (3) step sizes at particular temporal windows to probe local dynamic heterogeneity.
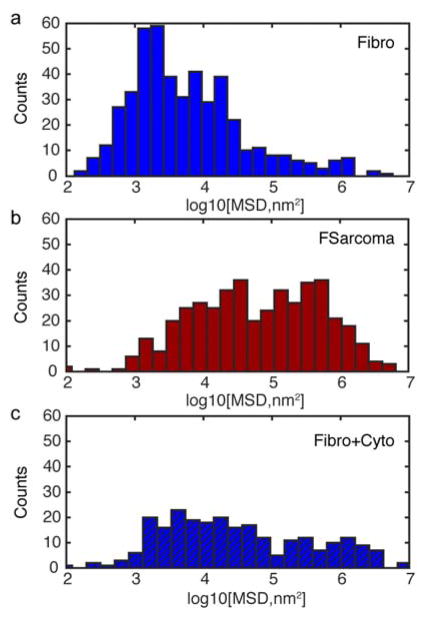
At a delay time of 1 s, Figure 2 shows the MSD for QDs in fibroblasts, fibrosarcoma cells, and fibroblasts treated with cytochalasin D. Although the MSD has a broad distribution in all cases, QDs within the fibrosarcoma cell cytoplasm travel the furthest with a MSD geometric average near 6 × 104 nm2 compared to the fibroblast case centered near 6 × 103 nm2. After exposing fibroblasts to cytochalasin D, the MSD values broadened and the geometric center value increased to 3 × 104 nm2 an order of magnitude increase over the fibroblast case and only 50% less than the fibrosarcoma cell value. These relationships persist at shorter and longer delay times, namely t = 0.5 s and 3 s. For fibroblasts, the breadth and number of tracks with MSDs greater than 104 nm2 significantly increased upon exposure to cytochalasin D suggesting an increase in mobility and less confinement of QDs during diffusion. Interestingly, MSD values for fibroblasts and cytochalasin D treatment correspond to the open spaces within the actin network measured by Kronalge14 within endothelial cells.
Figure 2.
Histogram of log of MSDs at a delay time of 1 s for QDs within (a) fibroblasts, (b) fibrosarcoma cells, and (c) fibroblasts with cytochalasin D treatment.
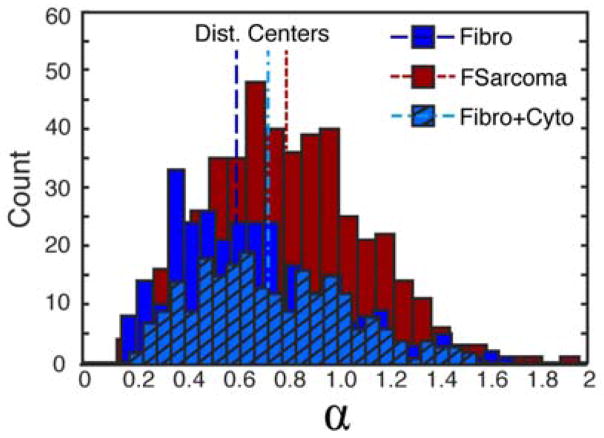
As demonstrated by previous particle dynamics researchers, particles within cells tended toward subdiffusive rather than Brownian motion with a few exceptions.4 The MSD for diffusion can be given by MSD = 2Dtα where D is the diffusion coefficient, t is time, and α is the power law exponent. A power law was fit to each condition and sorted for those trajectories with r2 values greater than 0.7 (43% of trajectories within fibroblasts and 72% of trajectories within fibrosarcoma cells fit this criteria). For QDs diffusing in fibrosarcoma cells, the power law exponents centered around 0.75 whereas for fibroblasts, alpha was lower, centered around 0.60 as shown in Figure 3. Thus, QDs within the cytoplasm of fibrosarcoma cells experienced less confinement. When the actin network of fibroblasts was disrupted using cytochalasin D, decreasing network confinement, the power law exponent increased to 0.70 representing a shift toward Brownian motion but still exhibiting sub-diffusive behaviour. Alternatively, when the actin network of fibrosarcoma cells was disrupted, power law exponents for QDs within fibrosarcoma cells were minimally affected by cytochalasin D treatment (supplementary materials). Diffusion coefficients were determined from tracks with alphas between 0.75 and 1.25. The percentage of tracks that fall into this range are 12% and 33% for fibroblasts and fibrosarcoma cells, respectively. For QDs within fibroblasts, the diffusion coefficient was 4 × 104 nm2/s, whereas QD diffusion was twice as high in fibrosarcoma cells, 9 × 104 nm2/s.
Figure 3.
Histogram of power law diffusion exponent, α, for QDs within control fibroblasts (Fibro, blue solid), fibrosarcoma cells (Fsarcoma, red), and fibroblasts with cytochalasin D treatment (Fibro+Cyto, blue hash). Distribution centers are noted with vertical lines for each distribution.
Destabilizing the actin of the fibroblasts increased the diffusion coefficient to 13 × 104 nm2/s, which is three times higher than in the control cells and larger than fibrosarcoma cells. These diffusion coefficients are smaller than expected for QD diffusion in water, (the main solvent of the cytosol) which is attributed to obstacles such as organelles and cytoskeletal network in the cytoplasm12. A significant finding of this study is that small particles move faster and further upon disrupting the actin cytoskeletal network. While diffusion coefficients are extracted from particles that followed Brownian motion, these parameters represent a smaller subset of the particles. Most particles experienced confined motion and a more detailed analysis of particle step size provides further insight into the overall response of QDs to the cell cytoplasm.
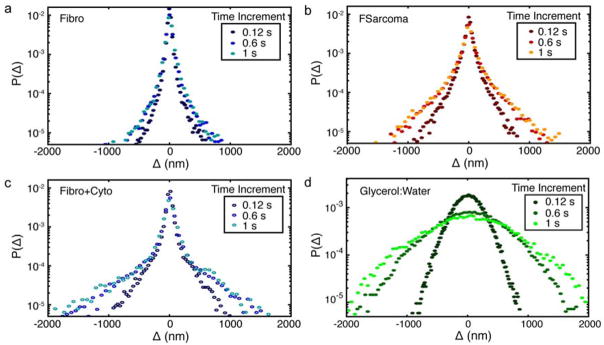
The distribution of QD displacements provides further insight into the nanoscale environment and dynamic heterogeneity experienced by the QDs. The van Hove correlation function, Δ, describes the probability of a particle moving a distance along a single axis within a specific time interval, τ.44 The self-portion, the average motion of the particle initially at the origin, is given by Δ=x(t+τ)−x(t) and y(t+τ) −y(t) for steps in the × and y directions, respectively.45 The van Hove distributions were calculated using the above equation for Δ and normalizing the area under the curves to one. The van Hove distributions for fibroblast, fibroblast cells exposed to cytochalasin D, fibrosarcoma cells, and glycerol:water are displayed in Figure 4. The van Hove distribution is Gaussian for spatially homogenous environments46 and was observed for QDs within homogeneous glycerol:water solutions (Figure 4d). While all cell distributions (Figure 4a–c) clearly deviate from Gaussian and exhibit central peaks at all time intervals, the larger displacement tails are qualitatively and quantitatively different. As seen by the area under the displacement distribution in Figure 4a, the majority of QDs within fibroblast cells were confined to displacements of 200 nm, while larger displacements had a low likelihood of occurring. Additionally, displacements larger than 1000 nm were not realized in fibroblast cells at a time interval of 1 s. The limited time dependence, evidenced by the nearly overlapping values at all time intervals, indicates QDs in fibroblast cells were exploring a confined spatial region. Displacement distributions of QDs within fibroblast cells exposed to cytochalasin D display a greater likelihood of large displacements, as seen by the increased area under the curve in Figure 4c. The distributions display an increasing range of motion with increasing time from 0.12 to 0.6 s, indicating that the QDs explored a greater spatial area. A smaller change is seen from 0.6 to 1 s, commensurate with the MSD and alpha distributions, which display greater, but still confined, QD mobility. Similar behavior is seen for QDs within the fibrosarcoma cells, Figure 4b. QD displacements were larger, even at 0.12 s, than QD displacements within fibroblast cells. While the central Gaussian peaks correspond to displacements bounded by approximately ±200 nm for all conditions, the tails of the distribution exhibit an exponential dependence that can be quantified by characteristic lengths, Table 1. Studies on particle dynamics in F-actin solutions47 have established that the exponential tails reflect the interaction between particle and environment. For our study, these length scales indicate the degree of confinement of QDs by their local environment. The characteristic lengths of the QD displacement distribution tails within fibroblasts changed negligibly with increasing time, from 96 nm to 115 nm at time intervals of 0.12 s and 1 s, respectively. For fibroblast cells exposed to cytochalasin D, however, the characteristic length increased from 193 to 328 nm from 0.12 to 1 s. Fibrosarcoma cells show an increase in characteristic length from 130 to 224 nm over the same time interval. The characteristic length of the fibrosarcoma cells exposed to nocodazole decreased to 98 nm and 140 nm from control values of 130 nm and 224 nm, respectively at time intervals of 0.12 and 1 s corresponding to a decrease in mobility (Supplementary Material). The displacement distributions and characteristic lengths of QDs within fibroblast cells exposed to nocodazole and QDs within fibrosarcoma cells exposed cytochalasin D, display behaviors similar to their control condition (Supplementary Material).
Figure 4.
van Hove distributions for QDs within (a) fibroblasts, (b) fibrosarcoma cells, (c) fibroblasts with cytochalasin D treatment, and (d) 90% glycerol:water solution at three time increments of 0.12 s, 0.6 s, and 1 s. Step size, Δ, combines steps in both × and y into a single van Hove distribution.
Table 1.
Characteristic lengths extracted from van Hove distributions for fibroblasts (Fibro), fibrosarcoma cells (FSarcoma), and fibroblasts treated with cytochalasin D (Fibro+Cyto).
| Cell Type | Time (s) | Characteristic Length (nm) |
|---|---|---|
| Fibro | 0.12 | 96.0 |
| 0.6 | 101.2 | |
| 1.0 | 114.9 | |
|
| ||
| FSarcoma | 0.12 | 129.4 |
| 0.6 | 187.0 | |
| 1.0 | 224.4 | |
|
| ||
| Fibro+Cyto | 0.12 | 192.9 |
| 0.6 | 297.6 | |
| 1.0 | 328.4 | |
The differences observed in the displacement distributions can be explained by changes in the nanoscale environment the QDs experience. In fibroblasts, QDs were confined by the actin network, which is heterogeneous, yet has a mesh dimension on the order of 400 nm consistent with the central peak of the distribution, ±200 nm, that confines the majority of the QD displacements. Studies on nanoparticle mobility within gels48 have used the central Gaussian portion of the van Hove to characterize caging lengths. In the case of QDs within the cell cytoplasm, we assert that the relevant caging length scale is the average mesh size. In fibroblasts, escaping a primary mesh and traveling to a neighbouring mesh is a rare event, resulting in a drastically different environment experienced by the QD. In comparison to published research on the mesh size of the actin network, values of 100–200 nm12,14 are more prevalent than values of 300–600 nm13 for fibroblast cell lines. The actin mesh size is cell type dependent as well as locally heterogeneous as the network undergoes densification toward the cell periphery. Our experiments avoid the cell periphery and thus would expect a slightly larger mesh size as a result. In fibroblasts exposed to cytochalasin D, the actin network was disrupted (supplementary materials). While a primary mesh of 400 nm exists in some areas demonstrated by the van Hove distribution width, the connectivity of the network has decreased, homogenizing the distribution of mesh sizes. This loss of connectivity increases the probability of QDs moving between primary meshes, allows QDs to explore more area with increasing time, and decreases the confinement experienced by QDs within the cytoplasm, ultimately leading to increased QD mobility. In fibrosarcoma cells, the disorganized actin cytoskeleton (supplementary materials) allows QDs to move between primary meshes, to explore more area with increasing time, and to experience less confinement from the actin cytoskeleton. These disparate dynamic behaviors of nanoparticles in different cell lines and conditions indicate differences in the local microenvironment.
The use of nanoparticles as probes to understand complex environments spans many fields, from industrially relevant polymer gels to extracellular polymer networks.49, 50 Additionally, non-Gaussian dynamics, specifically displacement distributions with exponentially decaying tails, have been observed in a wide variety of nano and micron scale systems, such as colloidal suspensions,51 solutions of f-actin,47 and recently, live muscle cell membranes.11 Our study shows nanoparticles within the cell cytoplasm also move in a non-Gaussian manner, indicating the ubiquitous nature of non-Gaussian dynamics and calling for enhanced models to determine diffusion properties when Gaussian relationships do not hold. Also, the dynamic behavior of QDs provided spatial information about the altered cytoskeletal networks, including average mesh size and changes in the mesh hindrance.
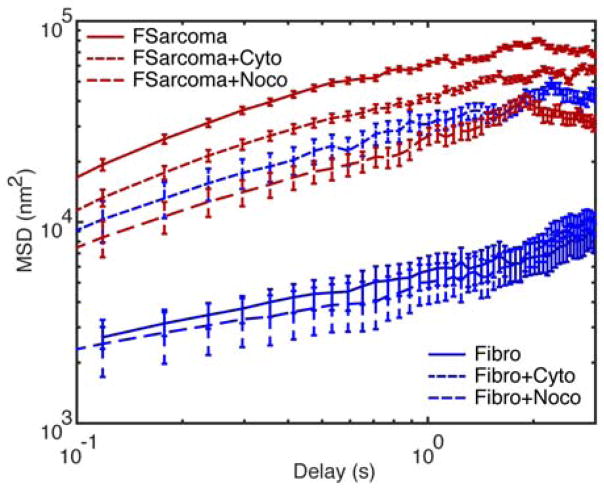
Geometric mean MSDs are shown in Figure 5 for all QD particle tracks within this study, which summarizes QD diffusion within each cell line and drug condition. The geometric mean was chosen to represent the central tendency because the distribution of values did not follow a standard normal distribution. QDs within fibroblasts and fibroblasts treated with nocodazole (microtubule disruption) exhibited the lowest MSDs. Disrupting the actin network with cytochalasin D treatment resulted in a significant increase (order of magnitude) in MSD whose behavior becomes similar to the fibrosarcoma results. Disruption of the cytoskeletal elements within fibrosarcoma cells with either nocodazole or cytochalasin D resulted in a decrease in MSD. QD mobility increased by an order of magnitude within fibrosarcoma cell cytoplasm compared to within the fibroblast cytoplasm.
Figure 5.
Geometric mean MSD for each of six conditions. Nomenclature is as follows: fibrosarcoma cells (FSarcoma), fibroblasts (Fibro), cytochalasin D treatment (Cyto), nocodazole treatment (Noco). Error bars are standard error.
Altering the connectivity of the actin network within fibroblasts resulted in QD mobility similar to that of QDs within the untreated fibrosarcoma cell cytoplasm. The intrinsic differences between the two cell lines result in vastly different intracellular particle mobility. What this research demonstrates is the sensitivity of intracellular QD diffusion to actin cytoskeletal modifications. Not all cytoskeletal modifications had equal effects, as nocodazole treatment, which attacks the microtubule network, had little effect on the MSD of QDs within the fibroblast cytoplasm. Interestingly, actin and microtubule disruption of fibrosarcoma cells resulted in slightly decreased mobility of QDs within the cytoplasm. One possibility is that the local microviscosity of the fibrosarcoma cells experience a greater change in response to the toxins than the fibroblasts. Measures of elasticity of the same cell lines indicated a stiffening behavior in the fibrosarcoma line in response to microtubule disruption.2
Experimental Methods
Cell culture
Human dermal fibroblasts and HT 1080 (fibrosarcoma cells). Fibroblasts were cultured in Fibrolife (Lifeline Cell Technologies) were cultured for this study. All cells were incubated at 37 °C in a humidified atmosphere with 5% CO2. Dishes and coverslips were coated for 30–40 min with 5 μg/mL fibronectin (BD Biosciences) dissolved in PBS prior to plating. Cells were initially plated at 75–100k density on glass coverslips (22 × 40 mm) for all studies, and were incubated for 48 hrs prior to experiments. Three conditions were studied: control (recording buffer only), 2.5 μM Cytochalasin D (Sigma Aldrich) in recording buffer for 30 min, and 10 μM Nocodazole (Sigma Aldrich) in recording buffer for 30 min.
Cell injection
A Nikon Eclipse Ti inverted fluorescence microscope was paired with an Eppendorf InjectMan4 micromanipulator and Eppendorf FemtoJet4i pressure injector. QDs were dispersed in DI water at 0.2 nM concentration and loaded into Eppendorf Femtotips. Typical input pressure values used were 200–400 hPa. Injections lasted 0.5 to 1 s. An x–y stage mounted to the microscope platform allowed the movement of the cell-coated coverslip into different viewing windows.
Particle tracking
Videos were recorded in brightfield before injection, then fluorescence just after injection, and finally brightfield again to examine the cell. Particle tracking software, FIESTA, was used to extract particle tracks from the fluorescence signature of QDs within the recorded videos. A frame rate of 16.9 fps was used and the calibration of the objective was 110 nm per pixel. The FIESTA program applies a Gaussian fit to the fluorescence to determine the center of the fluorescence with nm precision. A selection box was used to identify approximate cell boundaries and to limit the run time of the program by excluding areas outside of the cell from analysis. Drift was evaluated using immobilized QDs and found to be on the order of error in positioning. Particle tracks with an average positioning error greater than 25 nm were excluded from analysis.
Conclusions
Although our method could be employed to observe a variety of dynamic biological events, we have placed an emphasis on evaluating the effect of the cytoskeletal network on non-directed diffusion as opposed to active motor-driven transport. We have compared fibroblasts and fibrosarcoma cells with two cytoskeletal destabilizers acting on distinct elements. The results indicate that non-specific transport of nanoscale particles is inhibited by confinement due to the actin network. This is an intriguing phenomena brought on by small size scales; the nanoscale particles are affected by a mesoscale network, which shifted nanoparticle mobility toward subdiffusive, non-Gaussian dynamics more than would be expected simply due to the viscous material present between the mesh. In our work, the influence of intermediate filaments has not been explored. According to Guo et al.,52 intermediate filaments may play a role in intracellular dispersion of larger organelles within cells. Specifically, the tracking of endogenous vesicles within mouse embryonic fibroblasts revealed increased motility in cells that lack vimentin intermediate filaments. We anticipate further studies to elucidate the effects of intermediate filaments on small particle transport. Further extensions of our work include probing the effects of bridging proteins (e.g., plectin, septins, myosin filaments), DNA linking, motor driven transport, and implications for understanding nanoparticle trafficking and therapeutics in diseased cells. Specifically, many anti-cancer drugs target the destabilization of filamentous actin and microtubule spindle dynamics.10 From our study, anti-cancer drugs that disrupt the cell architecture would not greatly affect the dynamics of nanoparticle drug carriers within cancerous cells, but would increase transport of the nanoparticles within the surrounding healthy tissues should the drug carriers also enter these cells. In conclusion, we have demonstrated that the relationship between nanoparticle dynamics and cytoskeletal integrity is of critical importance to particles smaller than the mesh size, which leads to changes in confinement, diffusion parameters, and even the ability of a nanoparticle to reach a desired intracellular destination.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge our funding sources: ONR Grant N000141410538 (DME), NIH U01 EB016027 (DME), the Provost’s Postdoctoral Fellowship for Academic Diversity (MEG), and URF 4-000002-4820 (DME), which made this work possible. Support was also provided by the NSF PIRE OISE-1545884 (RJC, DME), ACS/PRF 54028-ND7 (RJC, EP), NSF/MWN DMR-1210379 (RJC, EP) and NSF/DMR 1507713 (RJC). The work was performed at and supported by the Nano Bio Interface Center (NBIC) at the University of Pennsylvania through NSF NSEC DMR08-32802, NSF MRI DBI-0721913, and NSF NSEC DMR-0425780. The authors thank Nadia Krook for TEM and FTIR assistance and Ben Lindsay for MATLAB coding assistance. We also thank Dr. Matt Brukman for NBIC instrument support. We thank Dr. Bruce Malkowicz for the generous donation of HT-1080 cells.
Footnotes
Electronic Supplementary Information (ESI) available: [Supporting information is available along with this manuscript]. See DOI: 10.1039/x0xx00000x
References
- 1.Długosz M, Trylska J. BMC Biophysics. 2011;4:3–3. doi: 10.1186/2046-1682-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grady ME, Composto RJ, Eckmann DM. J Mech Behav Biomed Mater. 2016;61:197–207. doi: 10.1016/j.jmbbm.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Brangwynne CP, Koenderink GH, MacKintosh FC, Weitz DA. J Cell Biol. 2008;183:583–587. doi: 10.1083/jcb.200806149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofling F, Franosch T. Rep Prog Phys. 2013;76:046602. doi: 10.1088/0034-4885/76/4/046602. [DOI] [PubMed] [Google Scholar]
- 5.Feng L, Zhu J, Wang Z. Acs Appl Mater Inter. 2016;8:19364–19370. doi: 10.1021/acsami.6b06642. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy LC, Bickford LR, Lewinski NA, Coughlin AJ, Hu Y, Day ES, West JL, Drezek RA. Small. 2011;7:169–183. doi: 10.1002/smll.201000134. [DOI] [PubMed] [Google Scholar]
- 7.Lucky SS, Soo KC, Zhang Y. Chem Rev. 2015;115:1990–2042. doi: 10.1021/cr5004198. [DOI] [PubMed] [Google Scholar]
- 8.Ma D. Nanoscale. 2014;6:6415–6425. doi: 10.1039/c4nr00018h. [DOI] [PubMed] [Google Scholar]
- 9.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M. Nat Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 10.Veeranarayanan S, Poulose AC, Mohamed MS, Varghese SH, Nagaoka Y, Yoshida Y, Maekawa T, Kumar DS. Small. 2012;8:3476–3489. doi: 10.1002/smll.201200874. [DOI] [PubMed] [Google Scholar]
- 11.Yao VJ, D’Angelo S, Butler KS, Theron C, Smith TL, Marchio S, Gelovani JG, Sidman RL, Dobroff AS, Brinker CJ, Bradbury AR, Arap W, Pasqualini R. J Controlled Release. 2016;240:267–286. doi: 10.1016/j.jconrel.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luby-Phelps K. Int Rev Cytol. 1999;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 13.Kusumi A, Sako Y, Yamamoto M. Biophys J. 1993;65:2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kronlage C, Schafer-Herte M, Boning D, Oberleithner H, Fels J. Biophys J. 2015;109:687–698. doi: 10.1016/j.bpj.2015.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crocker JC, Hoffman BD. Methods Cell Biol. 2007;83:141–178. doi: 10.1016/S0091-679X(07)83007-X. [DOI] [PubMed] [Google Scholar]
- 16.Baker EL, Bonnecaze RT, Zaman MH. Biophys J. 2009;97:1013–1021. doi: 10.1016/j.bpj.2009.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reits EAJ, Neefjes JJ. Nat Cell Biol. 2001;3:E145–E147. doi: 10.1038/35078615. [DOI] [PubMed] [Google Scholar]
- 18.Hess ST, Huang SH, Heikal AA, Webb WW. Biochemistry (Mosc) 2002;41:697–705. doi: 10.1021/bi0118512. [DOI] [PubMed] [Google Scholar]
- 19.Saxton MJ, Jacobson K. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 20.Wirtz D. Annu Rev Biophys. 2009;38:301–326. doi: 10.1146/annurev.biophys.050708.133724. [DOI] [PubMed] [Google Scholar]
- 21.Schuster BS, Ensign LM, Allan DB, Suk JS, Hanes J. Adv Drug Deliver Rev. 2015;91:70–91. doi: 10.1016/j.addr.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cognet L, Leduc C, Lounis B. Curr Opin Chem Biol. 2014;20:78–85. doi: 10.1016/j.cbpa.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 23.Pack CG, Song MR, Lee Tae E, Hiroshima M, Byun KH, Kim JS, Sako Y. J Controlled Release. 2012;163:315–321. doi: 10.1016/j.jconrel.2012.07.036. [DOI] [PubMed] [Google Scholar]
- 24.Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruthardt N, Lamb DC, Brauchle C. Mol Ther. 2011;19:1199–1211. doi: 10.1038/mt.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng XT, Than A, Ananthanaraya A, Kim DH, Chen P. Acs Nano. 2013;7:6278–6286. doi: 10.1021/nn4023137. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Ke X, Zheng Z, Zhang C, Zhang Z, Zhang F, Hu Q, He Z, Wang H. Acs Nano. 2013;7:3896–3904. doi: 10.1021/nn305189n. [DOI] [PubMed] [Google Scholar]
- 28.Wang ZG, Liu SL, Zhang ZL, Tian ZQ, Tang HW, Pang DW. Small. 2014;10:2712–2720. doi: 10.1002/smll.201303532. [DOI] [PubMed] [Google Scholar]
- 29.Zamaleeva AI, Collot M, Bahembera E, Tisseyre C, Rostaing P, Yakovlev AV, Oheim M, de Waard M, Mallet JM, Feltz A. Nano Lett. 2014;14:2994–3001. doi: 10.1021/nl500733g. [DOI] [PubMed] [Google Scholar]
- 30.Courty S, Luccardini C, Bellaiche Y, Cappello G, Dahan M. Nano Lett. 2006;6:1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- 31.Volarevic V, Paunovic V, Markovic Z, Simovic Markovic B, Misirkic-Marjanovic M, Todorovic-Markovic B, Bojic S, Vucicevic L, Jovanovic S, Arsenijevic N, Holclajtner-Antunovic I, Milosavljevic M, Dramicanin M, Kravic-Stevovic T, Ciric D, Lukic ML, Trajkovic V. Acs Nano. 2014;8:12098–12109. doi: 10.1021/nn502466z. [DOI] [PubMed] [Google Scholar]
- 32.Yum K, Na S, Xiang Y, Wang N, Yu MF. Nano Lett. 2009;9:2193–2198. doi: 10.1021/nl901047u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Z, He C, Tian H, Ding J, Hsiao BS, Chu B, Chen X. Prog Polym Sci. 2016;60:86–128. [Google Scholar]
- 34.Discher DE, Ahmed F. Annu Rev Biomed Eng. 2006;8:323–341. doi: 10.1146/annurev.bioeng.8.061505.095838. [DOI] [PubMed] [Google Scholar]
- 35.Caporizzo MA, Roco CM, Ferrer MCC, Grady ME, Parrish E, Eckmann DM, Composto RJ. Nanobiomedicine. 2015:2. doi: 10.5772/61328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruhnow F, Zwicker D, Diez S. Biophys J. 2011;100:2820–2828. doi: 10.1016/j.bpj.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts PC, Mottillo EP, Baxa AC, Heng HH, Doyon-Reale N, Gregoire L, Lancaster WD, Rabah R, Schmelz EM. Neoplasia. 2005;7:944–956. doi: 10.1593/neo.05358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Badique F, Stamov DR, Davidson PM, Veuillet M, Reiter G, Freund JN, Franz CM, Anselme K. Biomaterials. 2013;34:2991–3001. doi: 10.1016/j.biomaterials.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 39.Rolli CG, Seufferlein T, Kemkemer R, Spatz JP. Plos One. 2010;5:e8726. doi: 10.1371/journal.pone.0008726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi H, Condeelis J. Biochim Biophys Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kallergi G, Agelaki S, Markomanolaki H, Georgoulias V, Stournaras C. Cell Physiol Biochem. 2007;20:977–986. doi: 10.1159/000110458. [DOI] [PubMed] [Google Scholar]
- 42.Salker MS, Schierbaum N, Alowayed N, Singh Y, Mack AF, Stournaras C, Schaffer TE, Lang F. Sci Rep-Uk. 2016;6:29370. doi: 10.1038/srep29370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarantino N, Tinevez JY, Crowell EF, Boisson B, Henriques R, Mhlanga M, Agou F, Israël A, Laplantine E. J Cell Biol. 2014;204:231–245. doi: 10.1083/jcb.201307172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hopkins P, Fortini A, Archer AJ, Schmidt M. J Chem Phys. 2010;133:224505. doi: 10.1063/1.3511719. [DOI] [PubMed] [Google Scholar]
- 45.Aufderhorst-Roberts A, Frith WJ, Donald AM. Soft Matter. 2012;8:5940–5946. [Google Scholar]
- 46.Kegel WK, van Blaaderen A. Science. 2000;287:290–293. doi: 10.1126/science.287.5451.290. [DOI] [PubMed] [Google Scholar]
- 47.Wang B, Anthony SM, Bae SC, Granick S. Proc Natl Acad Sci U S A. 2009;106:15160–15164. doi: 10.1073/pnas.0903554106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee CH, Crosby AJ, Emrick T, Hayward RC. Macromolecules. 2014;47:741–749. [Google Scholar]
- 49.Jee AY, Curtis-Fisk JL, Granick S. Macromolecules. 2014;47:5793–5797. [Google Scholar]
- 50.Chhetri RK, Blackmon RL, Wu WC, Hill DB, Button B, Casbas-Hernandez P, Troester MA, Tracy JB, Oldenburg AL. Proc Natl Acad Sci U S A. 2014;111:E4289–4297. doi: 10.1073/pnas.1409321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guan J, Wang B, Granick S. Acs Nano. 2014;8:3331–3336. doi: 10.1021/nn405476t. [DOI] [PubMed] [Google Scholar]
- 52.Guo M, Ehrlicher Allen J, Mahammad S, Fabich H, Jensen Mikkel H, Moore Jeffrey R, Fredberg Jeffrey J, Goldman Robert D, Weitz David A. Biophys J. 2013;105:1562–1568. doi: 10.1016/j.bpj.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.