Abstract
The field of RNA nanotechnology has advanced rapidly during the past decade. A variety of programmable RNA nanoparticles with defined shape, size, and stoichiometry have been developed for diverse applications in nanobiotechnology. The rising popularity of RNA nanoparticles is due to a number of factors: (1) removing the concern of RNA degradation in vitro and in vivo by introducing chemical modification into nucleotides without significant alteration of the RNA property in folding and self-assembly; (2) confirming the concept that RNA displays very high thermodynamic stability and is suitable for in vivo trafficking and other applications; (3) obtaining the knowledge to tune the immunogenic properties of synthetic RNA constructs for in vivo applications; (4) increased understanding of the 4D structure and intermolecular interaction of RNA molecules; (5) developing methods to control shape, size, and stoichiometry of RNA nanoparticles; (6) increasing knowledge of regulation and processing functions of RNA in cells; (7) decreasing cost of RNA production by biological and chemical synthesis; and (8) proving the concept that RNA is a safe and specific therapeutic modality for cancer and other diseases with little or no accumulation in vital organs. Other applications of RNA nanotechnology, such as adapting them to construct 2D, 3D, and 4D structures for use in tissue engineering, biosensing, resistive biomemory, and potential computer logic gate modules, have stimulated the interest of the scientific community. This review aims to outline the current state of the art of RNA nanoparticles as programmable smart complexes and offers perspectives on the promising avenues of research in this fast-growing field.
Keywords: RNA nanoparticles, pRNA 3WJ motif, nanotechnology, nanobiotechnology, siRNA
The field of RNA nanotechnology1 has advanced rapidly over the past decade.2−6 RNA’s versatility in structure and function, propensity for bottom-up self-assembly, defined size and structure, favorable in vivo attributes, and large potential as a therapeutic modality make it an attractive candidate as a biomaterial for nanoparticle drug delivery. RNA’s ability to adopt complex quaternary structures,2,7−11 to base stack,12,13 to form canonical Watson–Crick (A–T, G–C) and noncanonical (G–U wobble, sheared G–A pair, G–A imino pair, A–U reverse Hoogsteen) base pairing14,15 leads to a variety of natural structural motifs.16,17 RNA nanotechnology uses such properties to construct nanoparticles for use in nanomedicine and bionanotechnology applications.
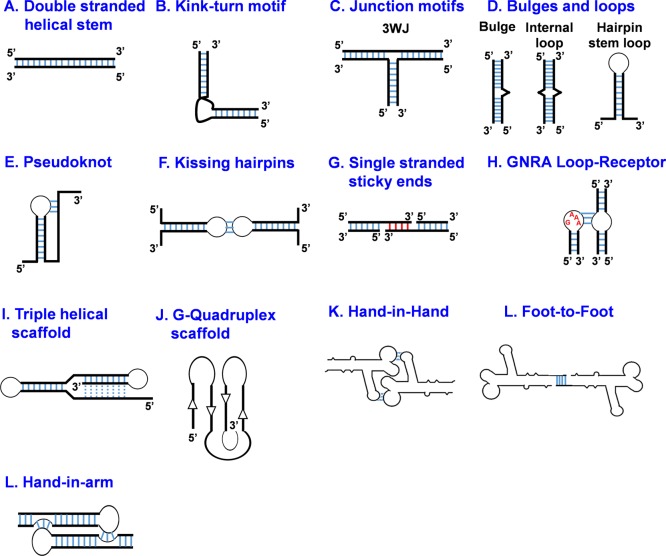
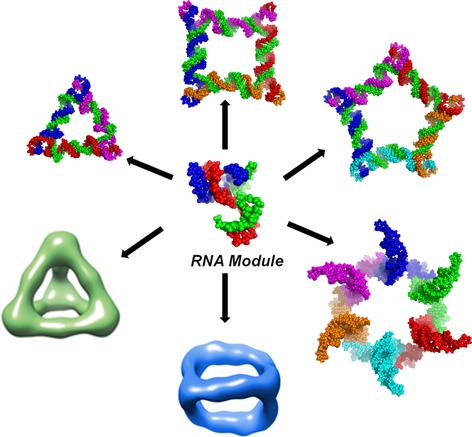
The RNA designer can easily scan through varieties of RNA secondary and tertiary structural motifs (Figure 1) (e.g., bulges, stems, hairpin, loops, and junctions). Such motifs are used as building blocks to design nanoparticles with diverse size and shape by engineering parameters such as motif angle and sequence length.18−22 Other parameters such as nucleotide type can alter nanoparticle properties, including enhancing chemical stability using base or backbone modifications or incorporating fluorescent dyes for nanoparticle tracking. The physiochemical properties of RNA nanoparticles can easily be fine-tuned to tailor-make the nanoparticle for desired applications in vitro and in vivo.
Figure 1.
Motifs for constructing RNA nanoparticles. A multitude of RNA motifs are available for the construction of RNA nanoparticles. RNA motifs are extracted from biological RNAs and, after in-depth structural analysis, can be used to generate higher-order structures. The resulting RNA nanoparticles can be functionalized with targeting, imaging, and therapeutic modules for diverse applications in nanobiotechnology.
The rapid growth of the RNA nanotechnology field necessitates the preparation of an updated review. This article will outline recent advancements in RNA nanotechnology including recent technologies implemented to construct a stable RNA nanoparticle with diverse structure and function. Finally, we will discuss the potential of RNA in medicine and nanotechnology.
Definition of RNA Nanotechnology
RNA nanotechnology is the study of nanometer-scale RNA architectures with their major frame being composed of RNA. The core scaffold, targeting ligand, regulatory moieties, and therapeutic modules can be all composed exclusively of RNA. RNA nanoparticles display the simplistic characteristic of DNA canonical base pairing, while containing the structural flexibility and functional diversity characteristics of proteins. Noncanonical base pairing, base stacking, and elaborate networks of tertiary contacts increase RNA structure versatility while also increasing thermodynamic stability.
Unlike traditional RNA biology research, RNA nanotechnology focuses on utilizing the properties of RNA to build architectures with nanomedicine applications. Classical studies on RNA structure and function focuses on intra-RNA interactions and 2D/3D structure–function relationships, whereas RNA nanotechnology focuses on inter-RNA interactions and quaternary interactions of RNA motifs. However, the fields of RNA research are not exclusive in their information. Much of the pre-existing RNA biology research is utilized extensively in RNA nanotechnology. For example, many RNA nanoparticles use functional RNAs such as ribozymes,23 riboswitches,24 and miRNAs25 discovered previously by traditional RNA biology. Additionally, RNA nanoparticles take advantage of RNA motifs discovered by RNA structural biologists.
Emergence of the RNA Nanotechnology Field
The emergence and advancement of the RNA nanotechnology field is not a simple incidence of the work by one single person but rather a collective effort of many insightful individuals. In 1998, Peixuan submitted a manuscript2 to Cell, reporting his finding of the assembly of pRNA (packaging RNA) dimers, trimers, and hexamers using re-engineered RNA fragments; the Cell associate editor, Vivian Siegel, and the founding editor of Cell, Benjamin Lewin, immediately were intrigued. They recognized that this important finding would promote the visibility of their newly initiated journal Molecular Cell. Thus, this significant discovery was published in Molecular Cell(2) with a mini-review in Cell to feature the work. The editors at Cell asked Guo to recommend an authority in the field to review this finding, and Roger Hendrix was chosen.26 This Molecular Cell paper revealed the ability to engineer RNA into precise constructs to build concise RNA architecture such as dimers, trimers, and hexamers via bottom-up self-assembly, thus showing the concept of RNA nanotechnology.
In the early 2000s, a group led by Eric Westhof predicted that the RNA kissing loop would promote the formation of special RNA structures.27,28 The original concept of “TectoRNA” or RNA “Tetonics” has led to empirical results in RNA nanotechnology.29−31 In 2004, when another empirical paper on RNA nanotechnology was published in Nano Letters, the editor and reporters of MSNBC published a groundbreaking news story entitled “Scientists build tiny structures out of RNA” to promote this concept of RNA nanotechnology. Subsequently, the editors of Science perceived the importance of RNA nanotechnology and published Luc Jaeger’s paper on tectoRNA32 with commentary by Hao Yan.33 More importantly, when three papers exemplifying the use of RNA nanotechnology to treat cancer were published,34−36 the NCI Alliance in Cancer Nanotechnology, led by Piotr Grodzinski, recognized the potential of RNA nanotechnology in cancer treatment (http://nano.cancer.gov/action/news/featurestories/monthly_feature_2006_august.pdf). As part of the NCI’s effort to promote the RNA nanotechnology field, a Workshop on RNA and Disease was organized by the pioneer of computational RNA nanotechnology, Bruce Shapiro. A strong boost to the RNA nanotechnology field can be credited to an invited review by Nature Nanotechnology(1) and a subsequent publication detailing the finding of a stable phi29 pRNA three-way junction implemented as an in vivo delivery system.35 The promoters of the RNA nanotechnology field also include editors of ACS Nano and Nano Today.
Abundance of RNA in Cells Reveals Third Milestone in Pharmaceutical Development
To date, there have been two milestones in pharmaceutical sciences: (1) Chemical drugs and (2) protein drugs, such as antibodies, enzymes, hormones, or chemicals targeting proteins.37 However, human genome sequencing revealed that only a small fraction of the human genome, about 2%, codes for protein, with the remaining 98% thought of as “junk DNA”. Subsequent studies revealed that a portion of the “junk DNA” actually codes for small and long noncoding RNAs.38 Noncoding RNAs are now well-recognized as playing prominent roles in cellular processes such as translation regulation, RNA splicing, DNA replication, and so forth.39 As a result, RNA is predicted to be the third milestone in drug development, including RNA nanoparticles, small therapeutic RNAs, and chemicals targeting RNAs.
Properties of RNA Structure and Chemistry Favorable for Nanoparticle Construction
Nanotechnology that revolves around the manipulation of biological materials is referred to as nanobiotechnology, which includes the field of nucleic acid nanotechnology. First envisioned over 30 years ago,40 DNA self-assembly was exploited to form nanoparticles using the base-pairing mechanism (A–T, G–C) (reviewed in refs (41) and (42)). DNA nanostructures including nanocapsules and other nanocarriers for drug delivery applications have since been constructed.43−45 DNA origami was developed as a powerful tool to build large 2D46 and 3D architectures including tetrahedrons,47 nanorobots,48 helix bundles,49 and tensegrity-based shapes.50 Although DNA nanostructures have exemplified the power of the base-pairing mechanism for structure design, DNA cannot match the thermostability and structural and functional diversity of its nucleic acid counterpart, RNA.
Similar to DNA, RNA is a chain-like biopolymer composed of nucleotide subunits joined by phosphodiester bonds. Each nucleotide is composed of a ribose sugar, phosphate group, and nitrogenous base. The most common bases are cytosine (C), guanine (G), adenine (A), and uracil (U). The basis of RNA secondary structure is hydrogen bonding occurring between complementary nucleotides, G–C and A–U, analogous to that of canonical base pairing found in DNA. Besides base paring being important for RNA secondary structure, base stacking is equally important for nucleic acid stability. Furthermore, the presence of the 2′-OH in RNA has dramatic effects on its properties. The C3′-endo sugar conformation in RNA leads to A-type helical formation (11 bp/turn), offering improved thermostability over DNA B-type helix.
RNA was first characterized as the link between genomic DNA and protein, transferring the genomic code for proteins to the cell’s translation machinery. Investigation of RNA structure revealed the catalytic nature of RNA with the discovery of ribozymes.51 The revolutionary finding of catalytic RNA, an attribute thought only to belong to proteins, led to a shift in the thought of the function of RNA. In addition to functions generated by elaborately structured RNA molecules, specific base-pairing interactions mediated by single-stranded regions of unstructured RNA have further expanded RNA’s functional repertoire.52 Years of research found that RNA performs diverse functions in biology, juggling tasks from catalysis to protein synthesis to gene regulation.
Negative Charge of RNA Nanoparticles Disallows Nonspecific Cell Entry and Minimizes Toxicity
Many nanoparticle systems rely solely on passive targeting, such as the enhanced permeability and retention (EPR) effects.53,54 Targeting the negatively charged cell membrane, positively charged nanoparticles enter cells via fusion and charge interactions. However, many positively charged nanoparticles cause toxicity and lead to unnecessary off-target effects.54,55 RNA’s negatively charged phosphate backbone renders RNA nanoparticles highly anionic, disallowing nonspecific targeting. Furthermore, this polyanionic charge density leads to extensive hydration and minimizes formation of a protein corona that can affect targeted delivery. RNA nanoparticles are functionalized with targeting molecules, such as chemical ligands or RNA aptamers, and therefore utilize receptor-mediated endocytosis to enter cells.
RNA Nanoparticles Can Harbor Multiple Functionalities While Retaining Their Authentic Folding
Functional modules composed of RNA, such as RNA aptamers, ribozymes, riboswitches, siRNA, and a series of noncoding RNAs, are available and can be seamlessly integrated into RNA nanoparticles. These motifs are simply fused to the core sequences of the RNA scaffold. Since RNA nanoparticles are inherently modular, each of the components self-assemble into the multifunctional architectures. Additionally, each helical branch of the RNA motif can be decorated with different subunits. One example is the pRNA-3WJ scaffold,35 which can harbor three functional modules. The driving force from the thermodynamically stable 3WJ scaffold ensures correct folding of RNA functional modules, retaining their functionality.56 Additionally, chemical ligands, such as fluorescent dyes, chemotherapeutic drugs, or biotin, can be easily incorporated to one RNA strand by well-established chemical conjugation strategies.57 Thus, multiple functional units for targeting, therapy, and tracking can be combined into one nanoparticle. Alternatively, multiple units of the same function, such as identical siRNAs or different siRNAs targeting different genes, can be combined on the same nanoparticle for enhanced or synergistic therapeutic effects.
In medical applications, it is paramount to properly characterize the payload. RNA nanoparticles are constructed bottom-up; each step along the way is precisely controlled by the designer. From size, shape, and oligomer selection to functional units and tracking molecules, each property of the nanoparticle is tailored to a specific application. The properties of RNA nanoparticles have the potential to improve upon current delivery systems, while also generating new routes of therapy, due to the distinct mechanism of RNA therapeutics.
RNA Nanoparticles Display Favorable Pharmacological Profiles
Pharmacokinetic and pharmacodynamic (PK/PD) profiles of nanoparticles are perhaps their most important characteristics besides therapeutic efficacy. PK/PD profiles depend on a number of factors, including size, shape, and surface charge. Controlled RNA nanoparticle synthesis results in consistent assembly with narrow size and shape distributions. Therefore, a reproducible set of PK/PD factors is achieved, allowing systematic study of size and shape effect on PK/PD and some predictability of PK/PD profiles. The homogeneous RNA nanoparticle assembly will help to expedite FDA approval as they reach the clinic.
One major factor for in vivo applications is the thermodynamic stability of RNA nanoparticles. Typically, RNA nanoparticles assemble through intermolecular interactions, requiring metal ions in the tens of millimolar. Recently, this limitation was circumvented by the pRNA-3WJ that can assemble in the absence of metal ions, is resistant to denaturation by urea, and will remain intact at ultralow concentrations.35
RNA nanoparticles also display an advantageous size for in vivo applications. They are larger than 10 nm cutoff for rapid renal excretion yet small enough to enter cells via receptor-mediated endocytosis while avoiding entrapment by liver Kupffer cells and lung/liver/spleen macrophages. If macrophages engulf RNA nanoparticles, autoimmunity would be a problem. Additionally, negatively charged RNAs further minimize interactions with negatively charged macrophage membranes. Depending on the proportion of chemically modified nucleotides in the sequence, such as 2′-F, RNA nanoparticles typically have an extended in vivo half-life of around 5–12 h,34 compared to 0.25–0.75 h for naked/unformulated siRNA.58 More importantly, after systemic injection in an orthotopic xenograft, subcutaneous xenograft, or metastatic tumor-bearing mice, pRNA-3WJ nanoparticles are able to target cancer cells specifically with little or no accumulation in healthy vital organs and tissues.34−36,59−63
Many RNA nanoparticles are designed to be ratchet-shaped to favor migration of RNA nanoparticles toward tumors.35,36,59 Additionally, RNA nanoparticles display rubber-like elastic property, akin to amoebae, exhibiting strong elasticity.64 This elastic property allows RNA nanoparticles to “squeeze” through cancer vasculatures by blood pressure while retaining thermodynamic stability at the tumor microenvironment. This enhances the EPR effect, while the ratchet shape prevents RNA nanoparticles from returning to circulation.
RNA Nanoparticle Size and Shape Tunes Immunogenic Potency for Use in Cancer Immunotherapy and Enhanced Drug Efficacy
In general, immune reactions are a defense mechanism of the body. Cancer cells mutate extremely fast and can develop resistance to protein therapies through repeated administration. Traditional chemotherapy targets only one route of tumor proliferation, resulting in chemo-resistant tumors. Cocktail therapies and repeated administration can overcome this challenge. Despite increased circulation time, RNA nanoparticles avoid antibody induction, such as those caused by protein therapies. Therefore, repeated treatment of chronic disease is possible using RNA nanoparticles.
The immune response elicited by RNA nanoparticles is highly dependent on RNA sequence, chemical modifications, size, and shape. This is exemplified by pRNA nanoparticles, which by themselves are completely nonimmunogenic.21,34,61 Neither unmodified nor 2′-F-modified pRNA nanoparticles induced detectable interferon or cytokine induction. RNA nanoparticles can also be designed as strongly immunogenic21 through the incorporation of CpG DNA, an FDA-approved immunological adjuvant.65−67 TNF-α and IL-6 induction depended upon the shape of the RNA polygons and number of CpG per polygon. The results suggest that CpG coupled to RNA polygons with different shapes have notable immunostimulatory effects and can be used for more effective cancer immunotherapy.68
Generally speaking, RNA nanoparticles are primarily recognized by Toll-like receptors (TLR) expressed on cell surfaces (TLR3), endosomes (TLR3/7/8), and cytoplasmic immunoreceptors, such as protein kinase R (PKR) and helicases (RIG-1 and MDA5). The degree of immune activation via different immunoreceptors is not completely understood but is known to depend on nanoparticle design, method of delivery, and type of cells, such as immune cells.69−71 Different chemical modifications can significantly reduce the immunostimulatory properties of RNA from different receptors. For instance, siRNA constructs with 2′-F and/or 2′-O-Me caused minimal induction of interferon or cytokines compared to unmodified counterparts.72 Strikingly, RNA constructs with 2′-F- or 2′-O-Me-modified U nucleotides were sufficient to eliminate immune off-target effects including TLR-dependent and TLR-independent pathways.73,74 Modification of the siRNA terminal ends with LNA can also effectively block interferon-α immunostimulatory activity while retaining potent silencing activity.75 Similarly, base modifications with pseudouracil or 2-thiouracil can abolish RIG-1-mediated immune stimulation due to presence of 5′-triphosphate.73 Chemical modifications thus provide a powerful tool for not only eliciting gene knockdown without stimulating the immune system but also recruiting immunoresponse for the treatment of cancer and chronic viral infections.
RNA Chemical Modifications: Implications for Serum Stability and Beyond
RNA’s inherent instability once hindered RNA’s use as a construction material. Although RNA nanoparticle size and structure provides some degree of nuclease resistance, it is not sufficient for use in vivo. Modifications to RNA’s natural structure can overcome the susceptibility of RNA therapeutics to serum exo- and endonucleases. Some of the most popular strategies include chemical modifications to the ribose sugar of the bases, base modification, and modification of the link between bases (backbone modification). Chemical modifications can impart higher chemical and thermal stability to RNA structure, thereby allowing in vivo use of RNA therapeutics and nanoparticles.
Sugar Modifications
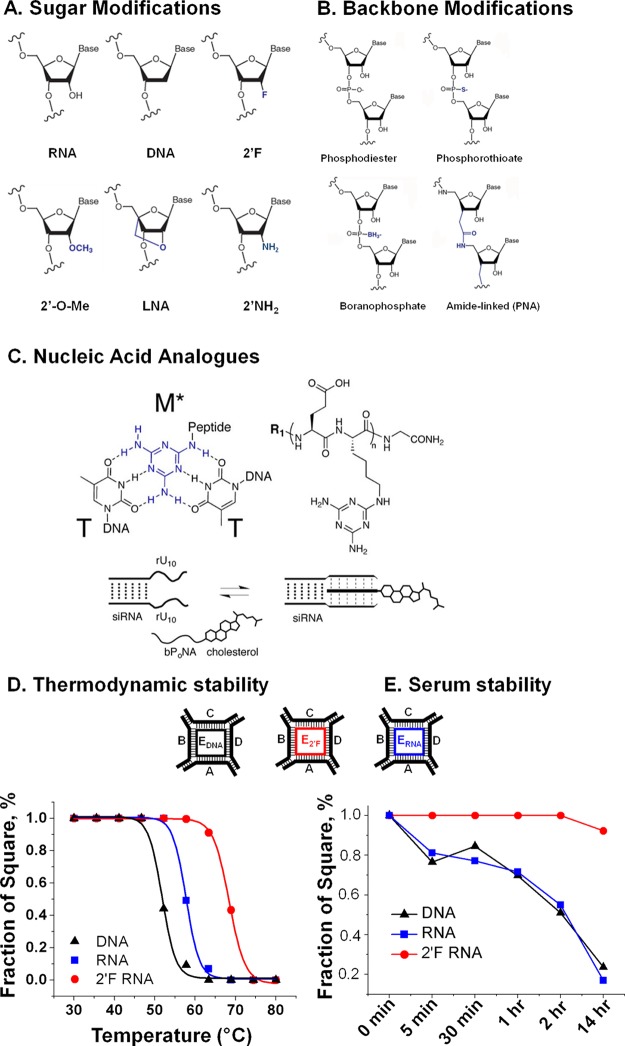
The most widely used modifications of RNA are on the sugar moiety of the nucleotides, as most modifications impart higher thermal76 and enzymatic stability while not affecting folding into the A-form RNA helix (Figure 2A). While the A-form helix is imperative to RNAi silencing efficacy, the 2′-OH is not essential. Thus, when siRNA is modified, it retains its silencing activity.77 2′-Fluorine- (2′-F), 2′-O-methyl- (2′-O-Me), and 2′-amine-modified (2′-NH2) RNA are small in size, compared to that of the 2′-OH native RNA, allowing for modified duplexes to retain their folding.78,79 Bulkier substitutions, such as 2′-O-methoxyethyl (2′-O-MOE) have seen more use as termini modifiers, as internal modifications affect RNA folding. LNA is a sugar modification in that the ribose sugar is structurally constrained (locked) into the A-form helix (3′-endo) by a methylene bridge between the 2′-oxygen and the 4′-carbon.80 While a high percentage of LNA modification to the RNA duplex will affect folding, only a small number of modifications is typically needed.
Figure 2.
RNA modifications to impact the stability of RNA nanoparticles. (A) Modification to RNA’s sugar moiety. Reprinted with permission from ref (78). Copyright 2008 Elsevier. (B) Backbone modifications. Reprinted with permission from ref (78). Copyright 2008 Elsevier. (C) Synthetic RNA triplex to replace native duplex without compromising original function. Reprinted from ref (85). Copyright 2015 American Chemical Society. (D) Tm changes of RNA nanoparticles resulted from 2′ alteration. Reprinted from ref (22). Copyright 2014 American Chemical Society. (E) The 2′ alteration resulted in different levels of serum stability. Reprinted from ref (22). Copyright 2014 American Chemical Society.
Backbone Modifications
Modification of the linkage between bases also increases RNA’s resistance to nucleases (Figure 2B). Phosphorothioate modification, substitution of a sulfur on the phosphodiester backbone for a nonbridging oxygen, is commonly used.81 Phosphorothioate modification increases thermostability and siRNA efficiency when modified at the correct positions on the siRNA sequence. Phosphorothioate modification has also been used to generate a library of DNA aptamers, called X-aptamers, used for cell-specific targeting.82 Boranophosphate modification, substituting borane for oxygen on the phosphate backbone, shows increased siRNA efficiency and provides significant increase in serum stability over native RNA.83 A more significant modification uses amide-linked bases, peptide nucleic acids (PNA).84 Because the negatively charged phosphate is replaced with the neutrally charged amide linkage, affinity between PNA and its complementary RNA or DNA is increased. In addition, PNA’s foreign structure evades detection by both nucleases and proteases, increasing PNA’s resistance to degradation.
Many nucleic acid analogues are being actively explored, and some of the more promising analogues recently developed are synthetic nucleic acid mimics that feature an α peptide backbone and a triaminotriazine base displayed at alternate residues (Figure 2C). The triazine (melamine) base can engage with two Watson–Crick faces of thymine or uracil, thus forming an obligate triplex structure with two T/U-rich strands of DNA/RNA; for this reason, they term this class of macromolecules “bifacial peptide nucleic acid” (bPNA). These bPNA hybrid stems can structurally replace native duplexes, allowing allosteric regulation of aptamer and ribozyme function.85−87 The triplex hybridization is functional with different backbones: peptides,88,89 peptoids,90 and polyacrylates.85 Bifacial polyacrylate nucleic acid (bPoNA), produced by functionalizing polyacrylate backbones with melamine bases, hybridize with DNA/RNA to load polymer nanoparticles and display targeting modalities on siRNA duplexes. These constructs are effectively delivered into HEK293 and MCF7 cells through passive nanoparticle uptake or ligand-driven entry. Using this method, a bPoNA and cholesterol-modified siRNA duplex targeting firefly luciferase was delivered into HeLa-Luc cells (expressing both firefly and renilla luciferase) to yield up to 40% luciferase silencing.85−87
Base Modifications
Modified bases have been explored and are relatively well-tolerated in the RNA duplex. Examples include 5-bromouracil and 5-iodouracil instead of uracil and diaminopurine instead of adenine.78 Other common therapeutics include 2-thiouracil, 4-thiouracil, C-linked base pseudouracil, 5-methylation of pyrimidines, and nonaromatic base, dihydrouracil. In most cases, the chemical stability is significantly enhanced; however, the biochemical function can be affected depending on the location of the modified bases within the sequence. For an in-depth review of RNA modifications, readers are encouraged to read the following review.78
Structural and Functional Implications
The primary goal of the aforementioned chemical modifications of RNA was to overcome the susceptibility of RNA therapeutics to nucleases and in the process extend in vivo circulation time for therapeutic efficacy. Over the past decade, more and more useful structural and functional implications of these chemical modifications have come to light, some of which are outlined below.
Modulation of Thermal Stability
The ability to control in vivo circulation time is one advantage of RNA nanoparticles as a delivery system. For example, substitution of DNA for an RNA strand in RNA nanoparticle construction decreases thermal stability, while substitution of 2′-F for unmodified RNA increases thermal stability (Figure 2D).76 In many therapeutic scenarios, it will be of great benefit to tune circulation time of the delivery vector, a possibility when using nucleic acid nanoparticles. Additionally, for other applications, it may be beneficial for an RNA nanoparticle to display exceptional thermal stability. Extensive investigations revealed that thermodynamic stability follows the trend: LNA/LNA > LNA/2′-F-RNA > 2′-F-RNA/2′-F-RNA > 2′-F-RNA/RNA > RNA/RNA > RNA/DNA > DNA/DNA.1,76,91−93 When diverse thermodynamic parameters are utilized within the same RNA nanoparticle, delivery and drug release mechanisms can be designed into the system. This is exemplified by a recent paper showing incorporation of 8 nt LNA-labeled RNA fragment, which is complementary to the “seed region” of miRNA,60,94 into RNA nanoparticles. Upon delivery in the cytosol, the 8 nt LNA thermodynamically competed and bound to the 8 nt seed in the miRNA and inhibited its function.60
Modulation of Serum Stability
Low stability of RNA in blood serum is one of the most challenging aspects of utilizing RNA as a nanoparticle scaffold. When modifications to the RNA chemical structure are utilized, enhancing stability of RNA in serum is possible. For example, 2′-F modification to the pRNA monomer structure increased serum stability from less than 10 min to more than 36 h.79 Additionally, modifying the backbone connectivity of RNA enhances its stability. Replacing the traditional phosphodiester bond with phosphorothioate, boranophosphate, or amide-linked (PNA) connectivity drastically increase circulation time in a degrading serum environment.78 Therefore, stability in serum of nanoparticles can be tuned by selecting predetermined ratios of oligonucleotide. This was shown by the substitution of DNA and 2′-F oligonucleotide during RNA nanosquare construction, which destabilized and stabilized, respectively, the nanosquares in serum (Figure 2E).22
Increase in Potency
The potency of RNA therapeutics primarily depends on cell type, target, and sequence. Chemical modifications can further increase the potency by boosting target-binding affinity, conformational organization, and flexibility. This is exemplified by a study showing that a siRNA construct built of 2′-O-Me and 2′-F nucleotides was 500× more potent than unmodified constructs using the same sequence.95 Another study showed that concurrent presence of a low-affinity dihydrouracil base at the 3′-end of the sense strand and a high-affinity 2′-thiouracil base at the 3′-end of the antisense strand can generate potent siRNAs.96
RNA Nanoparticle Construction Methods
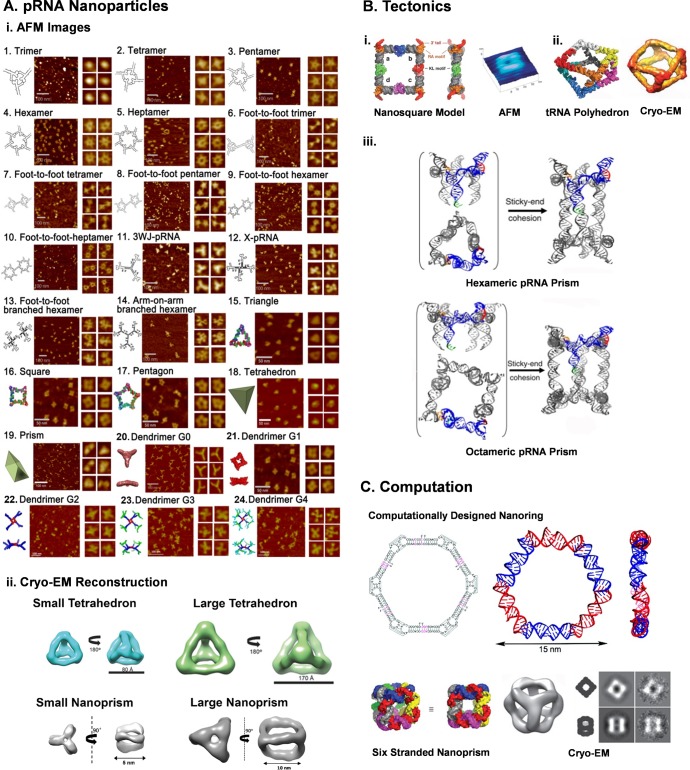
This review aims to describe the latest methods of RNA nanoparticle construction and RNA nanotechnology’s fastest growing topics. We suggest readers refer to the following reviews for detailed information on the following construction methods: (1) Construction of RNA Nanoparticle via Hand–Hand Interactions. The design principles are exemplified by the structural features of the packaging RNA (pRNA) derived from bacteriophage phi2997 to produce dimers, trimers, tetramers, pentamers, hexamers, and heptamers using “hand-in-hand” interactions via interlocking loops (Figure 3A-i:1–5).2,59,98,99 (2) Construction of RNA Nanoparticle via Foot-to-Foot Interactions. pRNA molecules are bridged using single-stranded palindrome sequences to promote self-assembly via “foot-to-foot” interactions (Figure 3A-i:6–10).59,98,99 (3) Rational Design Utilizing Stable Natural RNA Motifs. RNA nanoparticles can be constructed using motifs such as kissing loops, dovetails, pseudoknots, kink turns, and multiway junctions.16,17,28,31,100 (4) Construction of RNA Nanoparticle via Extension of Robust pRNA-3WJ Motif. The robust 3WJ drives the folding of pRNA modules attached to each vertex of the 3WJ (Figure 3A-i:11–14).35 (5) Construction of RNA Nanoparticle via Tectonics. Increasing knowledge of RNA folding and availability of databases such as the nucleic acid database (NAD)101 and the RNAjunction database10 have increased the power of tectonics, as the number of motifs that can be used as potential tectoRNAs is quite large. Examples include the tectoRNA squares and cubes designed from tRNA (Figure 3B-i) and nanoprisms designed from pRNA (Figure 3B-ii).29,31,32,98,102−110 (6) Construction of RNA Nanoparticles via Computational Design. Several software programs have been developed over the years, such as NanoTiler,111 Assemble2,112 RNA2D3D,113 INFO-RNA,114 and NUPACK,115 which are useful to design de novo RNA nanoparticles. Examples are a six-membered RNA nanoring116 and an RNA nanocube generated using Nanotiler (Figure 3C).117 Long sequences as well as individual monomer units that contain internal structures can be computationally designed and experimentally assembled in a one-pot manner including co-transcriptional assembly.
Figure 3.
Methods for constructing RNA nanoparticles. (A-i) RNA nanoparticles constructed based on pRNA-3WJ from bacteriophage phi29. Adapted from ref (59). Reprinted with permission from ref (21) (copyright 2014 Oxford University Press) and ref (126) (copyright 2015 Elsevier). (A-ii) Cryo-EM reconstruction of RNA tetrahedron nanoparticles and RNA nanoprisms based on the pRNA-3WJ motif. Reprinted with permission from refs (18) and (125). Copyright 2016 Wiley Publishing. (B) Tectonics method to construct RNA nanosquares, polyhedron from tRNA, and nanoprisms from phi29 pRNA. Reprinted with permission from ref (103) (copyright 2009 American Chemical Society), ref (109) (copyright 2010 Nature Publishing Group), and ref (110) (copyright 2015 Nature Publishing Group). (C) Computational approaches to expedite RNA nanoparticle manufacture and optimization. Adapted from ref (116) and printed with permission from ref (117). Copyright 2010 Nature Publishing Group.
Most Recent Development and Advances in RNA Nanoparticle Construction Approaches
As described in the previous section, information on RNA nanoparticle construction has been reviewed extensively.1,105,118−123 Readers are encouraged to refer to these reviews if they are interested. Here, we only review the latest developments, advantages, and progression of RNA nanoparticle technologies that have not been well-covered in previous reviews.
Construction of Precise Size RNA Nanoparticles with Using phi29 pRNA-3WJ
Precise size control is of paramount importance for clinical scenarios and one of the hardest aspects to control during nanoparticle formulation. It is a well-known fact that changing the size or molecular weight of nanoparticles can have drastic effects on PK/PD in an in vivo system. Random, and therefore often irreproducible, assembly of nanoparticles is commonly seen.124 The assembly process of RNA nanoparticles can be controlled precisely by the designer. Every nucleotide in the nanoparticle is specifically chosen to fit together in an explicit design scheme, similar to a puzzle piece fitting in one particular location in a puzzle. Precise assembly processes ensure a high yield and consistent production of RNA nanoparticles. Any shape and size of RNA nanoparticles can be broken down to component bases (“puzzle pieces”).
One recent method for the precise control of RNA nanoparticle size was developed using phi29 pRNA-3WJ nanoparticles. This 3WJ can serve as a robust scaffold for the design and construction of RNA polygons using a multistrand assembly process (Figure 3A-i:15–17).20−22 The native 3WJ inner angle of 60° was used to construct equilateral triangles. By simply adding one additional external strand and increasing the length of the inner strand, the inner angle of the 3WJ can be increased to 90° and induce square geometry.22 This same procedure can be followed to induce pentagon geometry.21 The angle stretching of the 3WJ can occur without disturbing the high thermal stability of the RNA polygons. Following the same design process, RNA squares were designed and constructed at sizes of 5, 10, and 20 nm along each edge. Their sizes were characterized by atomic force microscopy and dynamic light scattering and showed narrow size distributions. Changing the size of the RNA squares was easily done by modifying the number of base pairs connecting each 3WJ at the corners of the squares.22
Construction of 3D RNA Architectures and Containers Using phi29 pRNA 3WJ
More recently, the 2D architectures have been extended to 3D RNA nanoparticles with tetrahedral geometry (four triangular faces and six edges) (Figure 3A-i:18 and Figure 4A-ii top panel).125 As RNA tetrahedrons display high mechanical rigidity and structural stability, they are envisioned to have utility in a broad range of applications in nanomedicine and nanomaterials. Furthermore, the RNA tetrahedrons showed effective targeting of triple-negative breast cancer using the EFGR aptamer fused to the tetrahedron structure.
Figure 4.
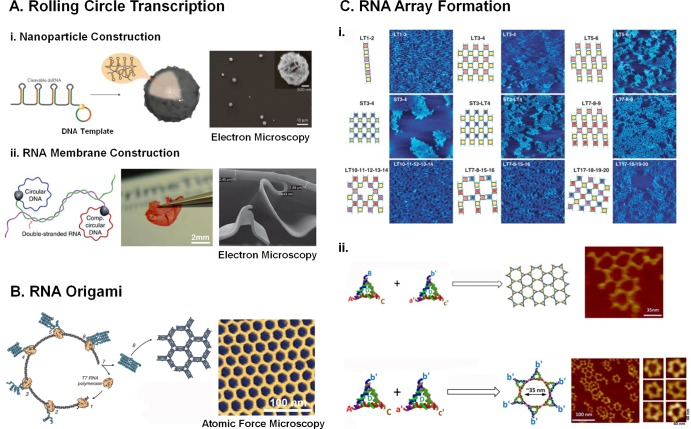
Methods for constructing RNA nanoparticles. (A) Rolling circle transcription to construct RNA architectures and membrane. Reprinted with permission from ref (5) (copyright 2012 Nature Publishing Group) and ref (132) (copyright 2014 Nature Publishing Group). (B) RNA origami utilizing co-transcriptional folding to produce large RNA nanostructures, such as hexameric arrays. Reprinted with permission from ref (127). Copyright 2014 AAAS. (C) RNA arrays. Reprinted with permission from (C-i) ref (32) (copyright 2004 AAAS) and (C-ii) ref (20) (copyright 2014 American Chemical Society).
Additional applications of 3D nanostructures were shown by the encapsulation of a model small molecule drug inside a nanoprism constructed from two pRNA triangles (Figure 3A-i:19 and Figure 4A-ii bottom panel). Two different size nanoprisms were constructed, with the smaller prism protecting a fragile RNA aptamer holding a fluorogenic malachite green molecule. Upon binding of the malachite green molecule to its aptamer, fluorescence drastically increases. The small nanoprism blocks the entry of degrading proteins by steric hindrance, almost doubling the fluorescent half-life of malachite green dye.18 This encapsulation mechanism will serve to advance the RNA nanotechnology field by protecting small molecule drugs as they circulate to cancerous cells, as well as preventing the leaking of cancer drugs that can sometimes be toxic to healthy cells.
Construction of RNA Dendrimer Structures Using phi29 pRNA-3WJ
The 3WJ-driven design method was also used to construct highly branched RNA dendrimer architectures (Figure 3A-i:20–24).126 The pRNA nanosquare was used as a symmetrical core for generating RNA dendrimers. The square shape reduces the possibility of steric hindrance that may arise while building higher-ordered structures. A stepwise iterative assembly approach was then applied via intramolecular interactions between pRNA-3WJ motifs to construct highly branched generation-4 (G4) RNA dendrimers. Resulting particles displayed precise control of size (∼65 nm), shape (3D globular), and stoichiometry (32 terminal units). RNA dendrimers have high loading capacity and can be easily functionalized with a large copy number of RNA aptamers, chemical ligands, RNAi modules, and chemotherapeutics. In addition, dense loading of imaging agents will allow high-resolution visualization of targeted cells/tumors in vivo by MRI or SPECT/PET imaging.
Construction of RNA Nanoparticles Co-transcriptionally and Intracellularly Using phi29 pRNA-3WJ
One ability of RNA nanoparticles is their ability to form co-transcriptionally, folding of the nanoparticle as it is being transcribed in vitro.127−129 This was exemplified using the phi29 pRNA-3WJ fused with RNA functionalities including malachite green fluorogenic aptamer, spinach fluorogenic aptamer, HBV ribozyme, streptavidin aptamer, and survivin siRNA.56 In different multifunctional nanoparticles, each functional modality retained its authentic function while assembling co-transcriptionally and intracellularly.
Intracellular fabrication will allow for large-scale production of RNA nanoparticles, as plasmids encoding for nanoparticle components can easily be cloned and expressed within bacterial cells. In the future, large-scale production of RNA nanoparticles will be possible through fermentation of bacteria encoding for RNA nanoparticles.
In summary, rational design requires prior knowledge of the 3D folding of individual motifs that will be used as RNA nanoparticle scaffolds as well as the folding of the functional modules that will be incorporated onto the scaffold. Care must be taken while altering any of the nucleic acid sequences as this could affect the global folding of RNA molecules. Several RNA folding programs are available to facilitate prediction of RNA structure or folding for nanoparticle assembly. Since this approach relies on modular building blocks, each component strand can be chemically synthesized with high yield and then self-assembled in a stepwise manner or in one pot. Given the overall simplicity, the method may be generally applied for a wide range of functional molecules.
Construction of RNA Nanoparticles by Rolling Circle Transcription
Getting high doses of therapeutic RNAi to the cell is one problem that could hinder the use of RNA nanoparticles. Much like rolling circle amplification (RCA) for isothermal DNA amplification,130 rolling circle transcription (RCT)131 produces large amounts of concatemeric RNA sequences from a circular DNA template using T7 in vitro transcription. A ssDNA oligomer antisense to the target sequence is circularized through ligation, then a short splint ssDNA containing the T7 RNA polymerase promoter sequence is annealed to the circular DNA, allowing RNA polymerase initiation. After ligation, in vitro transcription occurs in a continuous fashion.
One example is the construction of a circular DNA harboring the siRNA gene that is driven by a T7 promoter without any terminators (Figure 4A-i).5 During in vitro transcription, T7 RNA polymerase progressively works along the circular DNA thousands of times to generate many repeats of siRNA. As the RNA strand grew in length, it adopted a fiber-like configuration, followed by lamellar sheet-like formation, and finally into spherical sponge-like RNA nanoparticles. The features of the microsponges protected the unmodified RNA from degradation in serum. The multimeric transcript was successfully processed by Dicer, yielding a large amount of siRNAs, resulting in successful gene knockdown.
RCT was also used to construct membranes composed solely of RNA (Figure 4A-ii).132 Multiple circular templates were constructed containing complementary sequences to one another. During the RCT reaction, long ssRNA oligomers assemble into large complexes, and after evaporation-induced self-assembly, uniform RNA sheets form. The final dimensions measured by electron microscopy were about 2.5 μm thick and a few millimeters in width. It is foreseeable that RNA sheets could be used to load large amounts of therapeutic RNAs or carry a large payload of intercalating small molecules, such as anticancer agent doxorubicin.
While less precise than tectonics or computer-aided designs, many of the properties of RNA nanoparticles generated from RCT can be controlled. RCT-based RNA nanoparticles are morphologically round, due to aggregative formation with metal salts produced during transcription. The size of the RNA nanoparticles is tuned by increasing or decreasing the amount of polymerase during the RCT reaction. The size of the RNA nanoparticles can be further condensed by using synthetic polycations, such as polyethylenimine to facilitate entry into cells.5 The obvious advantage is that multiple siRNA targeting the same gene locus or different genes can be incorporated into one RNA nanoparticle. However, introduction of too much siRNA into cells can have toxic effects;133 therefore, the amount of RNA being delivered to cells must be taken under careful consideration.
Construction of RNA Nanoparticle via Origami
The popularity of DNA origami46 has led RNA researchers to attempt to employ the same methods for RNA nanoparticle construction. RNA origami has been achieved despite increased difficulty due to a more complicated folding pathway in RNA.127 Single-stranded RNA tiles were prepared by co-transcriptional folding of a single RNA strand into predesigned tiles (Figure 4B). The tiles then formed into complex shapes through kissing loop interactions. The shape of the origami was tuned by modulating the angle of the kissing loop used for tile association. This approach overcomes costly chemical synthesis and lengthy annealing steps associated with DNA origami. Additionally, the real advantage lies in their great potential to be cloned and expressed in large quantities in vivo for potential applications in synthetic biology.
Construction of RNA Nanoparticle Arrays
In a cellular environment, enzymatic pathways are spatially arranged onto scaffolds or subdivided into specially designed compartments, such as organelles. Spatial organization expedites the enzymatic processes by arranging proteins and their substrates in close proximity, while limiting potential cross-talk between enzymes. DNA nanotechnology has focused on spatial arrangement of biomolecules; however, it is limited to in vitro applications due to stability issues. RNA nanoparticles’ in vivo stability allows for cellular control at a more basal level. One prominent example of this is the isothermal assembly of RNA molecules in vivo into predefined discrete 1D and 2D structures with distinct protein-docking sites used as scaffolds for the arrangement of a hydrogen-producing enzymatic pathway.4 In contrast to protein-based approaches, RNA nanotechnology enables the formation of complex multidimensional architectures with nanometer precision to engineer biological pathways through spatial constraints.
Examples of RNA array formation include the arrangement of tectosquares into a pattern resembling a checkerboard.32,134 The corners of the tectosquares are functionalized with sticky end overhangs in different patterns to control the assembly of the arrays (Figure 4C-i). Patterns include linear one-dimensional arrays, symmetrical arrays from two and four different tectosquares, as well as arrays with specifically designed holes, all based on sticky end geometry.
Triangles constructed from the pRNA-3WJ were also used to construct supramolecular patterned arrays (Figure 4C-ii top panel).20 Two triangles were constructed containing three distinct sticky ends at each vertex: A, B, C on one triangle and a′, b′, and c′ on the other. A is complementary to a′, B is complementary to b′, and C is complementary to c′. This resulted in the formation of honeycomb-patterned arrays. Blocking formation of one of the sticky ends by changing b′ to B resulted in the formation of hexamer-shaped particles constructed from triangles (Figure 4C-ii bottom panel).
Applications in Nanomedicine and Nanobiotechnology
Due to advances in nanotechnology, many nanoparticles platforms have been developed over the years including liposomes,135−137 polymers,138,139 viral nanoparticles,140,141 dendrimers,142−145 and inorganic nanoparticles.143,146 These sub-micron-sized platforms show potential for improving the performance of therapeutic modalities.55,147 Nonetheless, nonspecific accumulation of therapeutic nanoparticles in healthy vital organs (i.e., liver, lungs, kidneys, and spleen) remains as a major challenge. Low specificity reduces the fraction of nanoparticles that reach diseased cells while increasing the toxicity and side effects. These adverse effects are often related to particle heterogeneity, aggregation, dissociation, unfavorable PK/PD profiles, and difficulty in penetrating biological barriers surrounding diseased cells, such as the tumor microenvironment.124,148,149 In addition, high production cost, unstable thermodynamic and chemical properties, and lack of controlled-release mechanisms impede clinical translation.150 Complex compositions of nanocarriers with diverse functional modules (inorganic/organic nanoscaffolds, RNAi/protein targeting antibodies, chemical drugs/antibodies) may cause regulatory approval issues of these technologies.124,151 Recent studies have shown that RNA nanoparticles can address several of these issues to overcome critical hurdles in nanomedicine for cancer therapy,34−36,59−63,152 viral infections,8,153,154 and eye disease.155
Application of RNA Biochemical Properties in RNA Nanotechnology
Applying RNA Aptamers for Targeted Therapeutic Delivery or as Potent Inhibitors
Targeted delivery of nanoparticles greatly reduces off-target toxicity and nanoparticle accumulation in healthy organs generally associated with passive mechanisms relying solely on EPR effects. An emerging class of targeted therapeutic molecules has been developed based on RNA aptamers:156 single-stranded RNA sequences that fold into specific 3D configurations and bind with high selectivity and affinity to extracellular domains of cell surface receptors. A combination of electrostatic interactions, hydrogen bonding, van der Waals forces, and base stacking mediates binding. Aptamers possess many advantages over protein antibodies as targeting reagents including low cost, faster selection and optimization process, convenient synthesis and modification with high batch fidelity, low immunogenicity, rapid tissue penetration, and long-term stability.1,157 The development of the systematic evolution of ligands by an exponential enrichment (SELEX)158,159 process has made aptamer development possible for many proteins and peptides. The broad applicability of SELEX means that, theoretically, an aptamer for any target can be selected. Currently, tens of RNA aptamers are available for targeting specific cell surface receptors followed by subsequent internalization of RNA nanoparticles: glioblastoma (e.g., EGFRvIII),160 breast cancer (e.g., EGFR, HER2, HER3) (Figure 5A-ii),161−163 prostate cancer (e.g., PSMA) (Figure 5A-iv),164 ovarian cancer (e.g., E-selectin),165 colon cancer (e.g., EpCAM),166 and lymphoma (e.g., CD19)167 cells as well as for viral infected cells, such as HIV (e.g., CD4).168 By sequence fusion, cell-internalizing RNA aptamers are incorporated into RNA nanoparticles for targeted delivery of therapeutic agents such as siRNA, miRNA, and chemotherapeutics into the cytosol.
Figure 5.
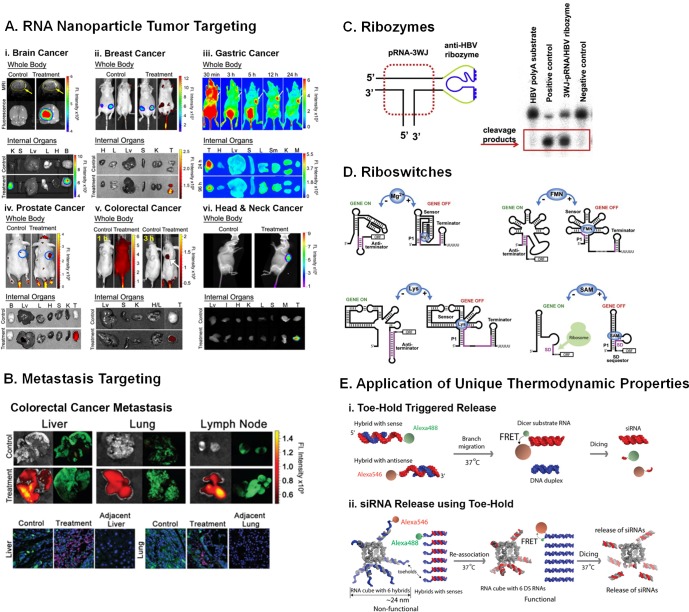
RNA nanoparticles for therapy. (A) RNA aptamers and chemical ligands are used to specifically target tumors in vivo without accumulation in healthy organs, targeting glioblastoma,63 breast cancer,60 gastric cancer,61 prostate cancer,152 colorectal cancer,62 and head and neck cancer.59 Adapted with permission from indicated references. (B) Targeting colorectal cancer metastasis utilizing RNA nanoparticles. Adapted from ref (62). Copyright 2015 American Chemical Society. (C) Ribozymes for cleaving specific RNA substrates similar to RNAi. Adapted with permission from ref (35). Copyright 2011 Nature Publishing Group. (D) Riboswitches to regulate gene expression. Reprinted with permission from ref (175). Copyright 2009 Elsevier. (E) Thermodynamic properties of nucleic acids used for activation and reagent release. Reprinted from (E-i) ref (181) (copyright 2013 Nature Publishing Group) and (E-ii) ref (180) (copyright 2014 American Chemical Society).
Additional strategies for cell-specific targeting include the conjugation of small molecule ligands to the RNA nanoparticles, for example, folic acid, which bind to receptors on the surfaces of cells.169 Many epithelial cancer cells overexpress the folate receptor on their cell surface, allowing for folic acid conjugated nanoparticles to target these cells at a rate higher than that of normal cells.169 Cancer types such as glioblastoma (Figure 5A-i),63 gastric cancer (Figure 5A-iii),61 colorectal cancer (Figure 5A-v),62 and head and neck cancer (Figure 5A-vi)59 have been targeted using pRNA-3WJ-folate nanoparticles.
Targeting cancer metastasis is another area in which RNA nanoparticles have shown considerable promise. Many metastatic cancers are hard to target due to the spread of cancerous cells to distant organs and the lymph nodes.170 Utilizing the folic acid as a targeting agent, RNA nanoparticles were able to simultaneously target colon cancer cells in the major sites of metastasis including the liver, lymph nodes, and lungs (Figure 5B).62
Aptamer-based therapeutics rely on direct binding of an aptamer to a therapeutic target to modulate downstream signaling. Leading examples include those that target receptor tyrosine kinases (e.g., anti-EGFR inhibitors), cell adherence factors (e.g., E-selectin), modulators of the immune system (e.g., anti-CTLA-4 inhibitors), and cell growth (e.g., α-fetoprotein) for cancer therapy.156,171,172 Tens of aptamers are in preclinical and clinical testing.172 Aptamers have also been developed for the treatment of neurological disorders including multiple sclerosis (e.g., IL-17), Alzheimer’s disease (e.g., β-scretase1), Parkinson’s disease (e.g., AMPA), and stroke (e.g., Factor IX-a).156,171,172 RNA nanotechnology offers an avenue to enhance the thermodynamic stability and functionality of therapeutic aptamers by inclusion of robust scaffolds during the in vitro selection process.
Applying Ribozymes for Targeted Therapy
Ribozyme-based therapeutic strategies have emerged as powerful tools for the treatment of cancer and viral infections. Ribozymes are RNA motifs that possess catalytic properties much like those of proteins. Ribozymes targeting mRNA show promise for gene therapy of breast cancer and hepatocellular carcinoma in animal models as they display similar effectiveness as RNAi, while exhibiting less off-target effects.173 Ribozyme-based therapies are considered as viable alternatives to anti-retroviral therapies, which are often plagued with toxicity and the emergence of resistant phenotypes.174 However, the challenge lies in developing nonviral vectors for in vivo delivery of ribozymes to specific cells. Ribozyme sequences are fused with RNA scaffolds creating a targeting platform to specifically deliver ribozymes. This is exemplified by pRNA nanoparticles harboring hepatitis B virus (HBV) hammerhead ribozyme that retains its enzymatic activity to cleave the poly(A) signal on HBV mRNA and in the process inhibited HBV replication (Figure 5C).8,35
Applying Riboswitches for Modulating Gene Expression and as Antimicrobial Targets
Riboswitches offer therapeutic avenues for addressing a wide range of diseases by regulating gene expression. They are widespread in numerous evolutionarily distant bacteria, with counterparts in archaea, plants, fungi, and algae. Riboswitches are composed of two domains, a sensor portion (a natural aptamer) for ligand recognition and an expression platform that can adopt two mutually exclusive conformations based on ligand binding for controlling gene expression (without the need for protein cofactors) (Figure 5D).175 Metabolite-binding riboswitches can potentially address multiple drug resistance as they can be considered as antimicrobial targets.176 Examples include the TPP riboswitch, which is involved in the biosynthesis and transport of thiamine in bacteria; the lysine riboswitch, which is involved in the control of biosynthesis and transport of lysine; the FMN riboswitch, which is involved in the control of riboflavin biosynthesis; and the guanine riboswitch, which controls the expression of genes involved in purine biosynthesis and transport.177,178 In eukaryotes, TPP riboswitches regulate genes via alternative splicing to generate mRNAs containing internal stop codons that cause either translation of aberrant peptides or premature translation. Since riboswitches control vital metabolic and virulence genes in pathogenic species, they represent an attractive strategy for therapeutic intervention without inflicting side effects on the eukaryotic host. Riboswitches can also be used to engineer artificial genetic circuits controlled by introducing synthetic ligands in human cells. Based on simplistic design principles, RNA nanotechnology can be applied not only to deliver the RNA switches in vivo but also to rationally engineer synthetic riboswitches to repress or activate gene expression in a ligand-dependent fashion.
Applying Thermodynamic Properties for Controlled Release of Functional Agents
The thermodynamic property displayed by different types of nucleic acids can be programmed into RNA nanoparticles to trigger the release or activation of functional modules. As discussed before, nucleotide selection provides tunable nanoparticle stability and thus controllable release. This is evident in pRNA-based nanosquares that can be tuned by substituting their “core” strands for DNA and 2′-F RNA22 (Figure 2D,E). 2′-F RNA as the core strand increases both melting temperature and its resistance to serum degradation, whereas DNA substitution does the exact opposite. Strategies utilizing differences in nucleic acid thermodynamic stability can be used to tune the release of drugs from the particle, whether the drugs are intercalated or conjugated to the vertices of the particles.
RNA/DNA hybrid duplex and strand displacement have also been used to design triggers for the conditional release of dicer substrate RNA.179−182 An RNA nanoparticle consisting of a partial RNA/DNA duplex region, which is complementary to a target mRNA, can conditionally release the Dicer substrate upon binding to its target mRNA, thereby avoiding off-target toxicity associated with RNAi. Additionally, functionalities can be split among different helices, and upon toe-hold interaction, their functions are triggered. To monitor the release, each piece of the split-functionality is labeled with a FRET pair molecule; therefore, upon association of the functionalities, FRET signal drastically increases (Figure 5E).
Applying RNA Interference Therapy: siRNA, miRNA, and LNA
The discovery of the RNA interference (RNAi) pathway in 1998183 created an avenue of treatments for many diseases, including cancer. RNAi is the process by which the expression of a gene is modulated by short double-stranded RNA sequences, typically 21–23 nucleotides in length. There are two central molecules to RNAi, miRNA and siRNA.184 While miRNAs are endogenous to cells and target multiple genes, siRNAs are mostly artificial sequences (some endogenous siRNAs do exist in cells185) that are designed to target a specific mRNA. Theoretically, any known mRNA sequence can be targeted using siRNAs and miRNAs, making diseases previously thought to be undruggable susceptible to RNAi therapy. RNA nanoparticles are designed such that endonuclease Dicer can access the dsRNA sequences encoding for the siRNAs and miRNAs and cleave them to generate functional miRNA and siRNA, which are then loaded into the RNA-induced silencing complex (RISC) and thereby regulate gene expression by selectively targeting mRNAs.
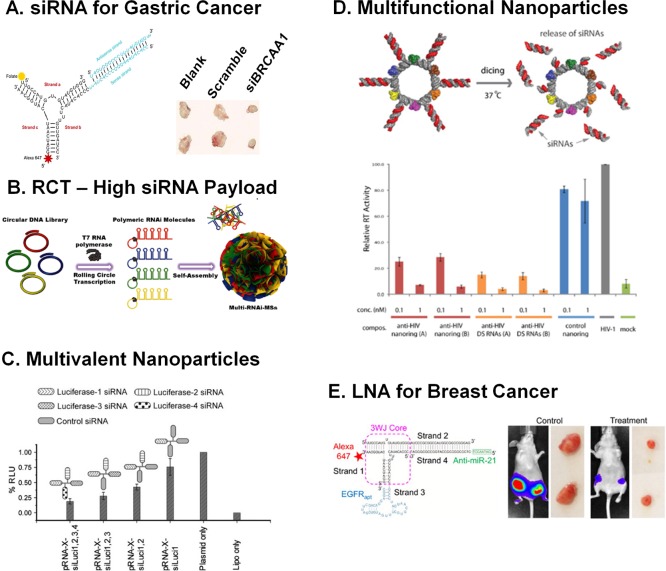
The delivery of siRNA in vivo is exemplified by pRNA-3WJ nanoparticles.60,63 In one study, pRNA-3WJ harboring folate as a targeting ligand and BRCAA1 siRNA as therapeutic module demonstrated potentials for gastric cancer therapy (Figure 6A).61 Upon systemic injection in subcutaneous gastric tumor xenograft-bearing mice, the RNA nanoparticles were internalized into tumors via folate-receptor-mediated endocytosis and the siRNA silenced the expression of BRCAA1. BRCAA1 knockdown down-regulated the expression of antiapoptotic Bcl-2 gene and up-regulated the expression of pro-apoptotic Rb and Bax genes, thereby regressing the growth of the tumors. Another study demonstrated that pRNA-3WJ nanoparticles harboring folate and luciferase siRNA can efficiently silence the expression of the luciferase gene after systemic injection in intracranial glioblastoma xenograft-bearing mice.63 In an alternative strategy, RNAi microsponges, composed of tandem repeats of luciferase siRNA synthesized by RCT, were efficiently cleaved by Dicer, converting stable hairpin RNA to siRNA only after cellular uptake (Figure 6B). Luciferase expression was successfully silenced in subcutaneous ovarian cancer xenograft-bearing mice.5 This system was further developed to contain multiple siRNAs for targeting of multiple genes.186 Another study showed the potentials for viral therapy by demonstrating that intravaginal application of RNA nanoparticles, composed of CD4 aptamer and siRNAs targeting HIV coreceptor CCR5, gag, and vif, protected humanized mice from sexual transmission of HIV.187
Figure 6.
RNA nanoparticles for RNAi. (A) pRNA-3WJ to deliver BRCAA1 siRNA in vivo and inhibit the growth of gastric tumors. Adapted with permission from ref (61). Copyright 2015 Nature Publishing Group. (B) RCT used to generate kilobase concatemeric RNA oligomers for a high payload delivery of multiple siRNA. Adapted with permission from ref (186). Copyright 2016 Wiley Publishing Group. (C) Multivalency of siRNA showing a synergistic effect on gene knockdown. Increasing the copy number of luciferase siRNA shows drastic decrease in luminescence units. Reprinted with permission from ref (36). Copyright 2012 Elsevier. (D) RNA nanorings to carry six siRNAs for different targets in HIV for gene knockdown. Adapted from ref (188). Copyright 2014 American Chemical Society. (E) pRNA-3WJ to deliver anti-miRNA, LNA, to slow the growth of tumors in triple negative breast cancer. Adapted from ref (60). Copyright 2015 American Chemical Society.
Effective gene knockdown using siRNA is sometimes limited, casting doubt on the future of siRNA therapy. To circumvent limited efficacy, multivalent siRNA delivery can be implemented. Increasing the copy number of luciferase siRNA on the pRNA X-way motif was shown to cause a synergistic effect on gene knockdown (Figure 6C).36 In an alternative approach, RNA nanorings were functionalized with six different siRNAs, each targeting a different region of HIV-1.188 The nanoring drastically reduced rates of virus production in HeLa cells, as tested by reverse transcriptase measurements (Figure 6D).
RNA nanoparticles can effectively deliver anti-miRNAs to down-regulate oncogenic miRNAs or miRNAs to increase endogenous downstream tumor suppressor miRNAs. pRNA-3WJ nanoparticles harboring EGFR-targeting RNA aptamers and anti-miR-21 efficiently inhibited the growth of triple-negative breast cancer (TNBC) (Figure 6E). Upon systemic injection in orthotopic TNBC tumor-bearing mice, EGFR aptamer-harboring RNA nanoparticles effectively targeted and internalized into tumor cells via receptor-mediated endocytosis.60 After entering cells, an 8-mer anti-miR-21 LNA agent bound to the miR-21 seed region,94 thereby inhibiting miR-21’s oncogenic properties. Similarly, pRNA-3WJ nanoparticles harboring PSMA aptamer and anti-miR-21 targeted subcutaneous prostate xenografts and delivered anti-miR-21 to inhibit tumor growth in animal models.152 The pRNA-3WJ was also shown to be an effective carrier of miRNA to silence viral genes by targeting the 3′-untranslated region (UTR) of the coxsackievirus genome.189,190
Using RNA Nanoparticles for Immunotherapy
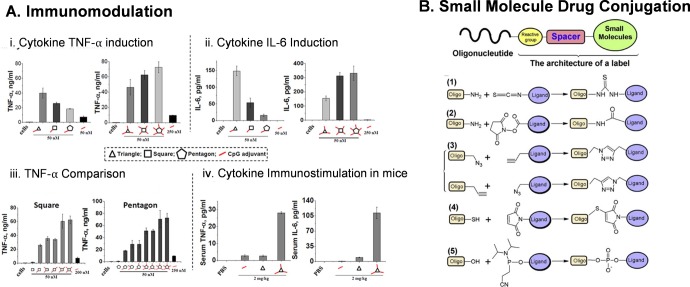
The versatility of RNA nanoparticles allows sequence integration of immune system stimulating motifs, such as CpG,191 acting as immuno-adjuvants. This is exemplified by RNA polygons functionalized with CpG DNA that induce a potent immune response in mice, displaying 100-fold increase in cytokine TNF-α and IL-6 induction (Figure 7A).21 Recent studies showed that sequence modification coupled with structural modification can generate potent immunostimulatory RNA sequences. For example, introduction of a miRNA-like nonpairing uridine bulge in the passenger strand of siRNA that can induce immunostimulatory activity on human immune cells.192 These immune-cell-activating sequences have potential applications in antiviral therapies and cancer immunotherapy.
Figure 7.
RNA nanoparticles for immunotherapy and chemotherapeutic delivery. (A) RNA nanoparticles display little to no immune response under normal conditions, but addition of immunostimulatory CpG sequences results in huge increases in the immune response for cytokine production. Reprinted with permission from ref (21). Copyright 2014 Oxford University Press. (B) Methods for conjugation of chemicals and drugs to RNA nanoparticles. Reprinted with permission from ref (119). Copyright 2014 Elsevier.
Recently, aptamers were developed to target proteins involved in cancer immunotherapy preventing tumor cells from escaping the host immune system.193 Aptamer-based immunotherapy can potentially replace traditional monoclonal antibody therapy primarily because they can overcome dose-limiting autoimmune responses. Aptamers are available for secretory targets such as cytokines and chemokines (e.g., IFN-γ, TGF-β), co-stimulatory molecules (e.g., CTLA-4, CD28), adhesion molecules (e.g., VCAM-1, Selectin-P/L), tumor immunosuppressive induction molecules, activation and inhibitory receptors (for agonist and antagonist targeting) on T cells, NK cells, macrophages, and dendritic cells. Bispecific RNA aptamers, mimicking bispecific antibodies, and siRNA have have been used to induce HIV resistance.194 These are re-engineered RNA fragments with two different variable regions binding to two different molecular targets. Dual aptamers can form a bridge to enable simultaneous binding to two cell surface markers or recruit T-cells to the proximity of tumor cells, leading to destruction of diseased cells. One example of a bispecific aptamer is one that simultaneously binds to CD16α on natural killer (NK) cells and c-Met receptor of receptor tyrosine kinases present on tumors.195 This results in recruitment of NK cells to c-Met-positive tumor cells, thereby inducing antibody-dependent cellular cytotoxicity (ADCC) mediated antitumor response. RNA nanotechnology can help overcome challenges associated with developing bispecific aptamers such as refolding, instability, loss of binding specificity after fusing two different aptamers into one RNA sequence, and size modulation to enable binding to both targets simultaneously.35
Chemotherapeutic Drug Delivery
Small molecule chemotherapy drugs are routinely used for the treatment of cancers. However, chemotherapeutic drug delivery traditionally relies on passive targeting via EPR effects. Small molecule drugs pass kidneys and blood vessels quickly due to their low molecular weight. After systemic administration, however, distribution is typically observed throughout the whole body with poor accumulation at tumor sites, thus producing suboptimal antitumor potency while increasing off-target effects.55,124 Due to tumor heterogeneities, not all cancers exhibit EPR effects, thus losing drug efficacy in some tumors. RNA nanotechnology is an attractive strategy to improve all aspects of traditional chemotherapy, including enhanced circulation time and blood plasma concentration, reduction in the administered dose, lowering of off-target effects by delivering payloads specifically to diseased cells and tissues, and enabling release mechanisms to decrease systemic toxicity. Many well-established conjugation methods are available for linking chemical drugs to chemically synthesized end-labeled RNA nanoparticles in high yield including thiourea linkage, click chemistry, amide linkage through NHS-ester chemistry, and thioether linkage, among others (Figure 7B).57 Conjugation of hydrophobic chemical drugs to the highly negatively charged and water-soluble RNA oligomers further solubilizes the drugs, helping to avoid potentially toxic organic delivery solutes.
Controlled release of drug conjugates is one approach to minimize off-target toxicity. Many factors present in cancer cells, but not healthy cells, can be exploited for controlled release mechanisms. For example, high esterase activity in some cancer cell types has been exploited by synthesizing a drug linker sensitive to esterase degradation.196 Additionally, increased acidity in tumor microenvironment and more importantly in endosomes, a location where many nanoparticles are trapped, has been extensively utilized. Incorporation of an acid-labile linker, where low pH causes cleavage, releases the drugs so that they can diffuse out of the endosomes.196 Intercalating drugs have the potential for a much higher ratio of drug to RNA nanoparticle. Doxorubicin, for example, can intercalate into sequence-specific regions in the RNA and DNA backbone, 5′-GC-3′ specifically.197 Drug release profiles have been tuned based on intercalator density and oligonucleotide properties. Intercalation serves as a viable alternative to drug conjugation, as long as minute amounts of drugs are released in the circulation prior to reaching the tumor microenvironment. Encapsulation of drugs in 3D RNA architectures is another feasible alternative without changing intrinsic drug properties.
Other Promising and Upcoming Applications
Molecular Beacons
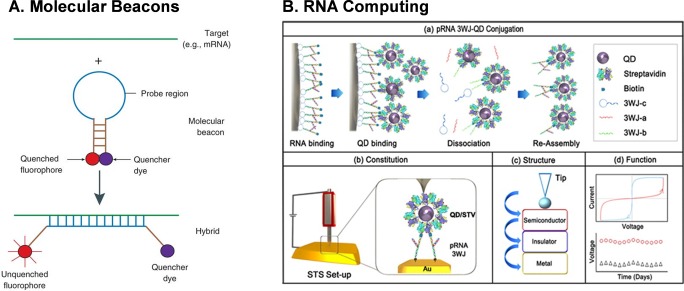
There is a significant interest in developing image-guided vectors for detecting RNA trafficking, endogenous gene expression, and silencing in vivo. Molecular beacons to image intracellular RNA expression in vitro by competitive hybridization methods is a popular detection method.198−206 These are stem-loop (hairpin) structures of oligonucleotide dual-labeled with a fluorophore on one end and quencher on the other. Initially, the fluorophore and quencher are in close proximity, resulting in little to no fluorescence (Figure 8A). Upon hybridization with a complementary RNA target, the “reporter” fluorophore and quencher separate and fluorescence is restored.207 Several studies have shown the feasibility of detecting mRNAs and monitoring the transportation of RNAs in cells,208−210 but robust in vivo delivery vectors are needed to apply this promising technology into broad research areas or into a routine clinical procedure. RNA nanotechnology can significantly advance the development of molecular-beacon-based RNA nanoparticles, especially in regards to improving their thermodynamic and chemical stability for efficient competitive hybridization to the mRNA targets. For more detailed information on the design and application of molecular beacons, readers are encouraged to refer to articles referenced in this review.211,212
Figure 8.
Application of RNA nanotechnology in beacons (A) and resistive biomemory (B). Panel A reprinted with permission from ref (207). Copyright 2006 Nature Publishing Group. Panel B reprinted from ref (220). Copyright 2015 American Chemical Society.
In Vitro Diagnostics
Early detection of diseases is beneficial as it helps doctors react quickly to patient’s needs. Importantly, survival prognosis increases with early detection. To achieve early detection, a system needs to detect as low amounts as possible of diseases markers, such as a small molecule or protein indicators. Aptamer-based biosensors (aptasensors) have emerged as powerful alternatives to traditional antibodies and enzymes, especially with the advent of nanotechnology.213 To increase target sensitivity, after aptasensor binding, amplification steps such as real-time PCR and rolling circle amplification have been used, allowing picomolar sensitivity of the analyte. These platforms can be integrated with microarrays to quantify thousands of targets.214 The folding conformation of aptamers can be modulated to facilitate either adsorption or desorption of a target by competitive interaction among aptamer, target, and complementary sequence. These properties of aptamers coupled with methods in signal generation (e.g., labeling with dyes or functional group for oriented immobilization on solid supports) enable the development of sophisticated biosensors for the detection of small molecule targets and protein/peptide biomarkers by SPR or quartz crystal microbalance. RNA nanotechnology could facilitate further development of aptasensors given that the tunable size and shape, optical properties, surface properties, switchable modules, ease of labeling, and catalytic activities of RNA nanoparticles can be useful not only for signal generation but also for signal amplification. For more detailed information on RNA based in vitro diagnostics, readers are encouraged to refer to articles referenced in this review.215−217
RNA Computing and Biomemory
As computers become faster and transistors become smaller, more methods for computing are needed to keep pace with Moore’s law prediction of double the capacity of transistors in the same unit area every 2 years. Many researchers are looking to use computation to analyze nucleic acids. Because of the atomic size of nucleic acids, it is possible that DNA or RNA computing can keep pace with Moore’s law prediction. The first DNA logic gate was designed in 1989,218 followed by the beginning of molecular scale computing in 1994 when a DNA-based computational design was used to solve a seven-city Hamiltonian math equation.219 Nucleic-acid-based molecular computing works by hybridization of an input DNA to a chip-immobilized DNA. The stability of DNA and on/off controllability of DNA hybridization makes it an ideal material for molecular-scale computation. Since RNA displays many different structural motifs compared to DNA, it could be advantageous to employ the structural diversity and complexity of RNA in nucleic acid computation. In addition, RNA can act as an insulator for biomemory applications. The pRNA-3WJ motif was used as both insulator and mediator in an RNA–quantum dot conjugate for biomemory applications (Figure 8B).220 Because the size of RNA nanoparticles is tunable, the distance between the conducting surfaces of a circuit is precisely controlled, leading to consistent current across a chip’s surface.
RNA Nanotechnology and CRISPR
The gene editing system “clustered regularly interspaced short palindromic repeats” (CRISPR)-associated nuclease Cas9 has attracted much attention recently as it may have tremendous potential to specifically edit, add, or remove genes from a genome.221 An appropriate guide RNA can specifically edit genes, rendering them benign, or can deliver genes imparting genetic effects.222,223 One example is the suppression of hepatitis B virus in chronically infected cells.224,225 However, similar to delivering naked siRNA in vivo, there are potential stability issues with delivering naked, unmodified guide RNA into cells.226 In the future, RNA nanotechnology may aid in the design of guide RNAs for the CRISPR/Cas system. Similar to how RNA nanoparticles and chemically modified RNAs confer stability into native RNA strands, RNA nanotechnology techniques may contribute to increasing the stability and effectiveness of CRISPR guide RNAs.
Challenges and Outlook
RNA nanotechnology for drug delivery has garnered much attention recently as it has tremendous potential to help treat many diseases, especially cancer. Certainly, improvements are necessary to push this innovative platform toward clinical trials and bring the product to the market. Some of the challenges and possible solutions are discussed below.
Limited Small Molecule Payload
Currently, RNA nanoparticles have limited small molecule drug loading capacity. Single drugs can be labeled onto the terminal ends of RNA strands with exquisite drug release mechanisms. Whole chain labeling methods can increase chemical drug payload but can cause misfolding of RNA nanoparticles due to steric hindrance and also compromise the release of the drugs. Computational approaches can help to identify locations where drugs can be introduced without disrupting the folding of the nanoparticle. Ensuring the drugs are orientated out of plane from the RNA nanostructure can also minimize any structural defects in the scaffold.
Intercalation is another viable approach to increase drug loading capacity, but drug release profiles need to be closely evaluated as premature release of the drug leads to nonspecific side effects. For instance, if the pRNA-3WJ nanoparticle is used as a delivery vector for targeting solid tumors, the projected half-life of the intercalated drug release should be more than 4 h, as extensive biodistribution studies revealed that the vast majority of the systemically administered pRNA-3WJ nanoparticles localize in the tumor site within 1–4 h.
Recently, RNA dendrimers have been constructed up to generation-4.126 Dendrimers can potentially overcome the loading challenges due to their branched architecture with repetitive concentric layers and hollow cavities suitable for loading drugs as well as peripheral/terminal units for harboring multiple drugs and other targeting modules. More recently, 3D RNA polygons have been developed that can potentially serve as RNA cages or containers to encapsulate hundreds of small molecule drugs.18,125 Plus, the RNA cages can assemble in organic or aqueous solvent without changing intrinsic RNA properties and are biocompatible and biodegradable compared to synthetic nanocarriers.
RNAi is considered the next frontier of cancer therapy and is thought by many to have potential to “drug the undruggable”. However, like chemical drug conjugation, RNA nanoparticles are limited in the number of RNAi molecules that can be delivered to cells. However, this limitation may not be an issue as studies report that there are only about 103–105 Dicer molecules per cell, and oversaturation of therapeutic RNAs can lead to nonspecific binding and off-target gene knockdown.133 Too much shRNA delivered to cells can cause cytotoxic effects.227 Therefore, it is possible that too much of a good thing is actually bad, and RNA nanoparticles’ lower payload may eventually be an advantage, evidenced in animal studies showing that RNA nanoparticles have minimal toxicity. RNA microsponges have the potential for delivering high payloads of siRNAs,5 and their utility in broader preclinical studies will be improved if the particle size can be reduced to avoid organ accumulation.
Endosome Escape To Fulfill the Promise of RNAi
Similar to most nanoparticle platforms, RNA nanoparticles enter cells through receptor-mediated endocytosis. Thus, intracellular RNA nanoparticle trafficking becomes the next challenge. Early endosomal vesicles are the first destination of RNA nanoparticles. Once sorted, RNA nanoparticles are transferred to late endosomes and lysosomes, where they are trapped without reaching their intended target. Fortunately, endosomal escape using small 8 nt anti-miRNA LNA fragments in RNA nanoparticle delivery was successful and cancer regression efficient,60 but the efficacy of endosomal escape of siRNA in RNA nanoparticles is still unknown as cancer regression after siRNA delivery via receptor-mediated endocytosis is relatively low.3,61 To date, there is very limited knowledge on different cellular endocytosis or internalization pathways that govern subsequent intracellular processing and endosomal escape of RNA nanoparticles. Nevertheless, there are well-characterized tools available to enhance endosome escape such as chemical functional groups including acid-cleavable linkers such as acetal, hydrazone, and maleic amides, or other acid protonating groups such as amino esters and sulfonamide.228 The pH-sensitive materials undergo high amounts of protonation, thereby inducing an influx of ions into the endosome, resulting in osmosis and endosome rupture—referred to as “proton-sponge” effect. As discussed earlier, a wide range of chemical conjugation strategies are available for site-specific incorporation of the endosome disrupting chemicals to RNA nanoparticles.
Large-Scale Production and Purification of RNA Nanoparticles
One of the major bottlenecks for future clinical applications is the large-scale production, large-scale purification, and cost of RNA nanoparticle production. Typically, RNA nanoparticles are designed to be modular composed of multiple short strands that are well within the limits of chemical synthesis (maximum of 80 nt). Over the years, the cost of RNA oligosynthesis has progressively decreased due to major improvements in chemical synthesis efficiency based on 2′-protecting groups, such as ether, acetal, orthoester, ester, O-acetalester, and pivaloyloxymethyl (PivOM).229,230 Commercial vendors have developed production facilities capable of producing tens of grams of RNA per synthesis cycle in GMP grade production capabilities. However, large-scale purification remains a challenge. HPLC and gel electrophoresis have limited capabilities with somewhat low yields. Due to the special nature of RNA nanotechnology, the size of the assembled nanoparticle is significantly different from its building blocks, thus, preparative ultracentrifugation has recently been employed231 and looks to be a promising approach for purification of fully assembled RNA nanoparticles with high yield.
Rational Incorporation of Chemically Modified Nucleotides
While chemical modifications offer many advantages concerning serum stability, thermodynamic stability, and immunogenicity, in some cases they do affect the folding of RNA motifs. For example, the malachite green (MG) fluorogenic RNA aptamer232 is nonfunctional after 2′-F modification due to misfolding. Rational modification is a case by case issue and is RNA-sequence- and structure-dependent. Specific patterns of chemical modifications can work well with one system but may not for another. The challenge is to find universal chemical modification strategies that can work for any given sequence. General consideration of improvement in RNA nanoparticle designs related to chemical modification is still necessary for clinical translation of a wide range of RNA-based drugs.
Computational Guide to RNA Nanoparticle Assembly
Understanding the correct global folding of RNA constructs is of paramount importance to ensure authentic function of multifunctional RNA nanoparticles. RNA molecules are dynamic and can alternate between different 3D conformations that display small differences in ΔG. Larger assemblies pose more constraints on the thermodynamic and kinetic aspects of RNA folding. Although several user-friendly online resources have been developed including Mfold,233 RNA designer,234 Sfold,235 NUPACK,115 Nanofolder,236 and Hyperfold,182 accurate prediction of RNA structure and folding for nanoparticle assembly remains a great challenge. RNA nanotechnology necessitates the development of prediction software capable of predicting inter-RNA interaction and analyzing 3D and 4D structures for nanoparticle designs. Some progress has been made in RNA 3D computation from the traditional intramolecular interactions to intermolecular interactions,111,116,117 but the field is still in its infancy.
Conclusions
RNA nanotechnology has come a long way since its inception in 1998.2 Some of the most exciting advancements happened only within the last 5 years. The design of elegant nanostructures with precise arrangement of functional modules in 3D space and self-assembly in a programmable 4D structure with controlled manner have greatly advanced the field. Progress in the understanding of the principle for structure-based design will enhance the capacity to make more intricate RNA nanoparticles with diverse function that 1 day can mimic naturally occurring RNA nanomachines like the ribosome. It is clear that RNA nanoparticles display many advantages over other nanoparticle systems; however, the development of RNA nanoparticles as drugs lags behind that of liposome and polymer systems, which have been in clinical trials for years. To reach clinical applications, the development of RNA nanoparticles should broaden beyond that of just their construction and focus more on applying the advantages of RNA nanotechnology. RNA nanotechnology has certainly sparked interest from investors evidenced by the establishment of several start-up companies whose goal is to rapidly advance the technology from the bench to the clinic.
Acknowledgments
The research in P.G.’s lab was supported by NIH Grants R01EB019036, U01CA151648, and U01CA207946.
Glossary
Vocabulary
- RNA building block
a central core RNA structure allowing for the construction of RNA nanoparticles, resulting in a more complex shape and particles than the individual building block
- RNA motif
a single-stranded RNA sequence that folds into a complex structure, resulting in a specific and defined functionality much like that of a protein; examples are RNA aptamers, riboswitches, and ribozymes
- quaternary interactions (4D)
interactions between at least two RNA monomer motifs building a multiunit molecule such as the complexing of the Phi29 pRNA into a hexameric ring via hand-in-hand interactions
- immuno-adjuvant
a substance that creates an enhanced immune response which may or may not be directed to a specific antigen; in RNA nanotechnology, specific nucleic acids can result in the stimulation of the immune system
- thermostability
a governing principle in RNA nanoparticle design, in which the base-pairing thermodynamics results in the construction of a stable structure for in vivo applications
- RNAi
(or RNA interference) the result of short nucleic acid strands blocking a specific gene expression through the complexation with RNA-induced silencing complex (RISC) and degradation of messenger RNA (mRNA)
Author Contributions
† D.J. and F.H. contributed equally to this work.
The authors declare the following competing financial interest(s): P.G.'s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery is funded by the CM Chen Foundation. P.G.'s inventions at the University of Kentucky have been licensed to Matt Holding LTD. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. P.G. is the consultant of Oxford Nanopore Technologies and Nanobio Delivery Pharmaceutical Co. Ltd, as well as the cofounder of Shenzhen P&Z Bio-medical Co. Ltd and its subsidiary US P&Z Biological Technology LLC.
References
- Guo P. The Emerging Field of RNA Nanotechnology. Nat. Nanotechnol. 2010, 5, 833–842. 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P.; Zhang C.; Chen C.; Trottier M.; Garver K. Inter-RNA Interaction of Phage Phi29 PRNA to Form a Hexameric Complex for Viral DNA Transportation. Mol. Cell 1998, 2, 149–155. 10.1016/S1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- Guo S.; Tschammer N.; Mohammed S.; Guo P. Specific Delivery of Therapeutic RNAs to Cancer Cells Via the Dimerization Mechanism of Phi29 Motor PRNA. Hum. Gene Ther. 2005, 16, 1097–1109. 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delebecque C. J.; Lindner A. B.; Silver P. A.; Aldaye F. A. Organization of Intracellular Reactions With Rationally Designed RNA Assemblies. Science 2011, 333, 470–474. 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- Lee J. B.; Hong J.; Bonner D. K.; Poon Z.; Hammond P. T. Self-Assembled RNA Interference Microsponges for Efficient SiRNA Delivery. Nat. Mater. 2012, 11, 316–322. 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo H.; Wu S.; Li M.; Li Y.; Jiang W.; Mao C. A Case Study of the Likes and Dislikes of DNA and RNA in Self-Assembly. Angew. Chem., Int. Ed. 2015, 54, 15118–15121. 10.1002/anie.201507375. [DOI] [PubMed] [Google Scholar]
- Laurenti E.; Barde I.; Verp S.; Offner S.; Wilson A.; Quenneville S.; Wiznerowicz M.; MacDonald H. R.; Trono D.; Trumpp A. Inducible Gene and ShRNA Expression in Resident Hematopoietic Stem Cells in Vivo. Stem Cells 2010, 28, 1390–1398. 10.1002/stem.460. [DOI] [PubMed] [Google Scholar]
- Hoeprich S.; ZHou Q.; Guo S.; Qi G.; Wang Y.; Guo P. Bacterial Virus Phi29 PRNA As a Hammerhead Ribozyme Escort to Destroy Hepatitis B Virus. Gene Ther. 2003, 10, 1258–1267. 10.1038/sj.gt.3302002. [DOI] [PubMed] [Google Scholar]
- Chang K. Y.; Tinoco I. Jr. Characterization of a ″Kissing″ Hairpin Complex Derived From the Human Immunodeficiency Virus Genome. Proc. Natl. Acad. Sci. U. S. A. 1994, 91 (18), 8705–8709. 10.1073/pnas.91.18.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindewald E.; Hayes R.; Yingling Y. G.; Kasprzak W.; Shapiro B. A. RNAJunction: a Database of RNA Junctions and Kissing Loops for Three-Dimensional Structural Analysis and Nanodesign. Nucleic Acids Res. 2008, 36, D392–D397. 10.1093/nar/gkm842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C.; Ehresmann C.; Ehresmann B.; Brunel C. Mechanism of Dimerization of Bicoid MRNA: Initiation and Stabilization. J. Biol. Chem. 2004, 279, 4560–4569. 10.1074/jbc.M306511200. [DOI] [PubMed] [Google Scholar]
- Sugimoto N.; Nakano S.; Katoh M.; Matsumura A.; Nakamuta H.; Ohmichi T.; Yoneyama M.; Sasaki M. Thermodynamic Parameters to Predict Stability of RNA/DNA Hybrid Duplexes. Biochemistry 1995, 34, 11211–11216. 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- Searle M. S.; Williams D. H. On the Stability of Nucleic Acid Structures in Solution: Enthalpy-Entropy Compensations, Internal Rotations and Reversibility. Nucleic Acids Res. 1993, 21, 2051–2056. 10.1093/nar/21.9.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux S.; Major F. RNA Canonical and Non-Canonical Base Pairing Types: a Recognition Method and Complete Repertoire. Nucleic Acids Res. 2002, 30, 4250–4263. 10.1093/nar/gkf540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N. B.; Westhof E. Geometric Nomenclature and Classification of RNA Base Pairs. RNA 2001, 7, 499–512. 10.1017/S1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N. B.; Lescoute A.; Westhof E. The Building Blocks and Motifs of RNA Architecture. Curr. Opin. Struct. Biol. 2006, 16, 279–287. 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N. B.; Westhof E. Analysis of RNA Motifs. Curr. Opin. Struct. Biol. 2003, 13, 300–308. 10.1016/S0959-440X(03)00076-9. [DOI] [PubMed] [Google Scholar]
- Khisamutdinov E. F.; Jasinski D. L.; Li H.; Zhang K.; Chiu W.; Guo P. Fabrication of RNA 3D Nanoprism for Loading and Protection of Small RNAs and Model Drugs. Adv. Mater. 2016, 28, 10079–10087. 10.1002/adma.201603180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khisamutdinov E. F.; Bui M. N.; Jasinski D.; Zhao Z.; Cui Z.; Guo P. Simple Method for Constructing RNA Triangle, Square, Pentagon by Tuning Interior RNA 3WJ Angle From 60 Degrees to 90 Degrees or 108 Degrees. Methods Mol. Biol. 2015, 1316, 181–193. 10.1007/978-1-4939-2730-2_15. [DOI] [PubMed] [Google Scholar]
- Khisamutdinov E. F.; Jasinski D. L.; Guo P. RNA As a Boiling-Resistant Anionic Polymer Material to Build Robust Structures With Defined Shape and Stoichiometry. ACS Nano 2014, 8, 4771–4781. 10.1021/nn5006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khisamutdinov E.; Li H.; Jasinski D.; Chen J.; Fu J.; Guo P. Enhancing Immunomodulation on Innate Immunity by Shape Transition Among RNA Triangle, Square, and Pentagon Nanovehicles. Nucleic Acids Res. 2014, 42, 9996–10004. 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski D.; Khisamutdinov E. F.; Lyubchenko Y. L.; Guo P. Physicochemically Tunable Poly-Functionalized RNA Square Architecture With Fluorogenic and Ribozymatic Properties. ACS Nano 2014, 8, 7620–7629. 10.1021/nn502160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott W. G. Ribozymes. Curr. Opin. Struct. Biol. 2007, 17, 280–286. 10.1016/j.sbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Breaker R. R. Riboswitches and the RNA World. Cold Spring Harbor Perspect. Biol. 2012, 4, a003566. 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M.; Kim V. N. Regulation of MicroRNA Biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Hendrix R. W. Bacteriophage DNA Packaging: RNA Gears in a DNA Transport Machine (Minireview). Cell 1998, 94, 147–150. 10.1016/S0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- Leontis N. B.; Stombaugh J.; Westhof E. The Non-Watson-Crick Base Pairs and Their Associated Isostericity Matrices. Nucleic Acids Res. 2002, 30, 3497–3531. 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L.; Westhof E.; Leontis N. B. TectoRNA: Modular Assembly Units for the Construction of RNA Nano-Objects. Nucleic Acids Res. 2001, 29, 455–463. 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa J.; Furuta H.; Ikawa Y. RNA Tectonics (TectoRNA) for RNA Nanostructure Design and Its Application in Synthetic Biology. Wiley. Interdiscip. Rev. RNA 2013, 4, 651–664. 10.1002/wrna.1185. [DOI] [PubMed] [Google Scholar]
- Jaeger L.; Chworos A. The Architectonics of Programmable RNA and DNA Nanostructures. Curr. Opin. Struct. Biol. 2006, 16, 531–543. 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Westhof E.; Masquida B.; Jaeger L. RNA Tectonics: Towards RNA Design. Folding Des. 1996, 1, R78–R88. 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- Chworos A.; Severcan I.; Koyfman A. Y.; Weinkam P.; Oroudjev E.; Hansma H. G.; Jaeger L. Building Programmable Jigsaw Puzzles With RNA. Science 2004, 306, 2068–2072. 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- Yan H. Nucleic Acid Nanotechnology. Science 2004, 306, 2048–2049. 10.1126/science.1106754. [DOI] [PubMed] [Google Scholar]
- Abdelmawla S.; Guo S.; Zhang L.; Pulukuri S.; Patankar P.; Conley P.; Trebley J.; Guo P.; Li Q. X. Pharmacological Characterization of Chemically Synthesized Monomeric PRNA Nanoparticles for Systemic Delivery. Mol. Ther. 2011, 19, 1312–1322. 10.1038/mt.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D.; Shu Y.; Haque F.; Abdelmawla S.; Guo P. Thermodynamically Stable RNA Three-Way Junctions for Constructing Multifuntional Nanoparticles for Delivery of Therapeutics. Nat. Nanotechnol. 2011, 6, 658–667. 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F.; Shu D.; Shu Y.; Shlyakhtenko L.; Rychahou P.; Evers M.; Guo P. Ultrastable Synergistic Tetravalent RNA Nanoparticles for Targeting to Cancers. Nano Today 2012, 7, 245–257. 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S.; Linton L. M.; Birren B.; Nusbaum C.; Zody M. C.; Baldwin J.; Devon K.; Dewar K.; Doyle M.; FitzHugh W.; Funke R.; Gage D.; Harris K.; Heaford A.; Howland J.; Kann L.; Lehoczky J.; Levine R.; McEwan P.; McKernan K.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Claverie J. M. Fewer Genes, More Noncoding RNA. Science 2005, 309, 1529–1530. 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- Lieberman J.; Slack F.; Pandolfi P. P.; Chinnaiyan A.; Agami R.; Mendell J. T. Noncoding RNAs and Cancer. Cell 2013, 153, 9–10. 10.1016/j.cell.2013.03.019. [DOI] [PubMed] [Google Scholar]
- Seeman N. C. Nucleic Acid Junctions and Lattices. J. Theor. Biol. 1982, 99, 237–247. 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- Jones M. R.; Seeman N. C.; Mirkin C. A. Nanomaterials. Programmable Materials and the Nature of the DNA Bond. Science 2015, 347, 1260901. 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- Seeman N. C. Nanomaterials Based on DNA. Annu. Rev. Biochem. 2010, 79, 65–87. 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E. S.; Dong M.; Nielsen M. M.; Jahn K.; Subramani R.; Mamdouh W.; Golas M. M.; Sander B.; Stark H.; Oliveira C. L.; Pedersen J. S.; Birkedal V.; Besenbacher F.; Gothelf K. V.; Kjems J. Self-Assembly of a Nanoscale DNA Box With a Controllable Lid. Nature 2009, 459, 73–76. 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- Goodman R. P.; Heilemann M.; Doose S.; Erben C. M.; Kapanidis A. N.; Turberfield A. J. Reconfigurable, Braced, Three-Dimensional DNA Nanostructures. Nat. Nanotechnol. 2008, 3, 93–96. 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- Yang Y. R.; Liu Y.; Yan H. DNA Nanostructures As Programmable Biomolecular Scaffolds. Bioconjugate Chem. 2015, 26, 1381. 10.1021/acs.bioconjchem.5b00194. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W. K. Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Ke Y.; Sharma J.; Liu M.; Jahn K.; Liu Y.; Yan H. Scaffolded DNA Origami of a DNA Tetrahedron Molecular Container. Nano Lett. 2009, 9, 2445–2447. 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Bachelet I.; Church G. M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- Dietz H.; Douglas S. M.; Shih W. M. Folding DNA into Twisted and Curved Nanoscale Shapes. Science 2009, 325, 725–730. 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedl T.; Hogberg B.; Tytell J.; Ingber D. E.; Shih W. M. Self-Assembly of Three-Dimensional Prestressed Tensegrity Structures From DNA. Nat. Nanotechnol. 2010, 5, 520–524. 10.1038/nnano.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A. J.; Grabowski P. J.; Cech T. R. Autocatalytic Cyclization of an Excised Intervening Sequence RNA Is a Cleavage-Ligation Reaction. Nature 1983, 301, 578–583. 10.1038/301578a0. [DOI] [PubMed] [Google Scholar]
- Jady B. E.; Kiss T. A Small Nucleolar Guide RNA Functions Both in 2’-O-Ribose Methylation and Pseudouridylation of the U5 Spliceosomal RNA. EMBO J. 2001, 20, 541–551. 10.1093/emboj/20.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y.; Maeda H. A New Concept for Macromolecular Therapeutics in Cancer Chemotherapy: Mechanism of Tumoritropic Accumulation of Proteins and the Antitumor Agent Smancs. Cancer Res. 1986, 46, 6387–6392. [PubMed] [Google Scholar]
- Prabhakar U.; Maeda H.; Jain R. K.; Sevick-Muraca E. M.; Zamboni W.; Farokhzad O. C.; Barry S. T.; Gabizon A.; Grodzinski P.; Blakey D. C. Challenges and Key Considerations of the Enhanced Permeability and Retention Effect for Nanomedicine Drug Delivery in Oncology. Cancer Res. 2013, 73, 2412–2417. 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers T.; Kiessling F.; Hennink W. E.; Storm G. Drug Targeting to Tumors: Principles, Pitfalls and (Pre-) Clinical Progress. J. Controlled Release 2012, 161, 175–187. 10.1016/j.jconrel.2011.09.063. [DOI] [PubMed] [Google Scholar]
- Shu D.; Khisamutdinov E.; Zhang L.; Guo P. Programmable Folding of Fusion RNA Complex Driven by the 3WJ Motif of Phi29 Motor PRNA. Nucleic Acids Res. 2014, 42, e10. 10.1093/nar/gkt885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes E.; Evans M.; Das S. R. RNA Labeling, Conjugation and Ligation. Methods 2011, 54, 251–259. 10.1016/j.ymeth.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Behlke M. A. Progress Towards in Vivo Use of SiRNAs. Mol. Ther. 2006, 13, 644–670. 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y.; Haque F.; Shu D.; Li W.; Zhu Z.; Kotb M.; Lyubchenko Y.; Guo P. Fabrication of 14 Different RNA Nanoparticles for Specific Tumor Targeting Without Accumulation in Normal Organs. RNA 2013, 19, 767–777. 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D.; Li H.; Shu Y.; Xiong G.; Carson W. E.; Haque F.; Xu R.; Guo P. Systemic Delivery of Anti-MiRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology. ACS Nano 2015, 9, 9731–9740. 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui D.; Zhang C.; Liu B.; Shu Y.; Du T.; Shu D.; Wang K.; Dai F.; Liu Y.; Li C.; Pan F.; Yang Y.; Ni J.; Li H.; Brand-Saberi B.; Guo P. Regression of Gastric Cancer by Systemic Injection of RNA Nanoparticles Carrying Both Ligand and SiRNA. Sci. Rep. 2015, 5, 10726. 10.1038/srep10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychahou P.; Haque F.; Shu Y.; Zaytseva Y.; Weiss H. L.; Lee E. Y.; Mustain W.; Valentino J.; Guo P.; Evers B. M. Delivery of RNA Nanoparticles into Colorectal Cancer Metastases Following Systemic Administration. ACS Nano 2015, 9, 1108–1116. 10.1021/acsnano.5b00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. J.; Haque F.; Shu D.; Yoo J. Y.; Li H.; Yokel R. A.; Horbinski C.; Kim T. H.; Kim S.-H.; Nakano I.; Kaur B.; Croce C. M.; Guo P. RNA Nanoparticles As a Vector for Targeted SiRNA Delivery into Glioblastoma Mouse Model. Oncotarget 2015, 6, 14766–14776. 10.18632/oncotarget.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H. C.; Koh K.; Evich M.; Lesiak A.; Germann M. W.; Bongiorno A.; Riedo E.; Storici F. RNA Intrusions Change DNA Elastic Properties and Structure. Nanoscale 2014, 6, 10009–10017. 10.1039/C4NR01794C. [DOI] [PubMed] [Google Scholar]
- Krieg A. M. CpG Motifs in Bacterial DNA and Their Immune Effects. Annu. Rev. Immunol. 2002, 20, 709–760. 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- Krieg A. M. Therapeutic Potential of Toll-Like Receptor 9 Activation. Nat. Rev. Drug Discovery 2006, 5, 471–484. 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- Vollmer J.; Krieg A. M. Immunotherapeutic Applications of CpG Oligodeoxynucleotide TLR9 Agonists. Adv. Drug Delivery Rev. 2009, 61, 195–204. 10.1016/j.addr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Vanneman M.; Dranoff G. Combining Immunotherapy and Targeted Therapies in Cancer Treatment. Nat. Rev. Cancer 2012, 12, 237–251. 10.1038/nrc3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead K. A.; Dahlman J. E.; Langer R. S.; Anderson D. G. Silencing or Stimulation? SiRNA Delivery and the Immune System. Annu. Rev. Chem. Biomol. Eng. 2011, 2, 77–96. 10.1146/annurev-chembioeng-061010-114133. [DOI] [PubMed] [Google Scholar]
- Judge A. D.; Sood V.; Shaw J. R.; Fang D.; McClintock K.; MacLachlan I. Sequence-Dependent Stimulation of the Mammalian Innate Immune Response by Synthetic SiRNA. Nat. Biotechnol. 2005, 23, 457–462. 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Judge A. D.; Bola G.; Lee A. C. H.; Maclachlan I. Design of Noninflammatory Synthetic SiRNA Mediating Potent Gene Silencing in Vivo. Mol. Ther. 2006, 13, 494–505. 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Morrissey D. V.; Lockridge J. A.; Shaw L.; Blanchard K.; Jensen K.; Breen W.; Hartsough K.; Machemer L.; Radka S.; Jadhav V.; Vaish N.; Zinnen S.; Vargeese C.; Bowman K.; Shaffer C. S.; Jeffs L. B.; Judge A.; Maclachlan I.; Polisky B. Potent and Persistent in Vivo Anti-HBV Activity of Chemically Modified SiRNAs. Nat. Biotechnol. 2005, 23, 1002–1007. 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Hornung V.; Ellegast J.; Kim S.; Brzozka K.; Jung A.; Kato H.; Poeck H.; Akira S.; Conzelmann K. K.; Schlee M.; Endres S.; Hartmann G. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science 2006, 314, 994–997. 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Cekaite L.; Furset G.; Hovig E.; Sioud M. Gene Expression Analysis in Blood Cells in Response to Unmodified and 2’-Modified SiRNAs Reveals TLR-Dependent and Independent Effects. J. Mol. Biol. 2007, 365, 90–108. 10.1016/j.jmb.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Hornung V.; Guenthner-Biller M.; Bourquin C.; Ablasser A.; Schlee M.; Uematsu S.; Noronha A.; Manoharan M.; Akira S.; de Fougerolles A.; Endres S.; Hartmann G. Sequence-Specific Potent Induction of IFN-[Alpha] by Short Interfering RNA in Plasmacytoid Dendritic Cells Through TLR7. Nat. Med. 2005, 11, 263–270. 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Binzel D. W.; Khisamutdinov E. F.; Guo P. Entropy-Driven One-Step Formation of Phi29 PRNA 3WJ From Three RNA Fragments. Biochemistry 2014, 53, 2221–2231. 10.1021/bi4017022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlke M. A. Chemical Modification of SiRNAs for in Vivo Use. Oligonucleotides 2008, 18, 305–319. 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- Watts J. K.; Deleavey G. F.; Damha M. J. Chemically Modified SiRNA: Tools and Applications. Drug Discovery Today 2008, 13, 842–855. 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Liu J.; Guo S.; Cinier M.; Shlyakhtenko L. S.; Shu Y.; Chen C.; Shen G.; Guo P. Fabrication of Stable and RNase-Resistant RNA Nanoparticles Active in Gearing the Nanomotors for Viral DNA Packaging. ACS Nano 2011, 5, 237–246. 10.1021/nn1024658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J.; Thonberg H. Ã.; Ljungberg K.; Frieden M.; Westergaard M.; Xu Y.; Wahren B.; Liang Z.; Ã̃rum H.; Koch T. Locked Nucleic Acid (LNA) Mediated Improvements in SiRNA Stability and Functionality. Nucleic Acids Res. 2005, 33, 439–447. 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein F. Phosphorothioates, Essential Components of Therapeutic Oligonucleotides. Nucleic Acid Ther. 2014, 24, 374–387. 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- Somasunderam A.; Thiviyanathan V.; Tanaka T.; Li X.; Neerathilingam M.; Lokesh G. L.; Mann A.; Peng Y.; Ferrari M.; Klostergaard J.; Gorenstein D. G. Combinatorial Selection of DNA Thioaptamers Targeted to the HA Binding Domain of Human CD44. Biochemistry 2010, 49, 9106–9112. 10.1021/bi1009503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. H.; Wan J.; Shaughnessy E. E.; Ramsay S. B.; Alexander K. A. RNA Interference Using Boranophosphate SiRNAs: Structure-Activity Relationships. Nucleic Acids Res. 2004, 32, 5991–6000. 10.1093/nar/gkh936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcheddu A.; Giacomelli G. Peptide Nucleic Acids (PNAs), a Chemical Overview. Curr. Med. Chem. 2005, 12, 2561–2599. 10.2174/092986705774370664. [DOI] [PubMed] [Google Scholar]
- Zhou Z.; Xia X.; Bong D. Synthetic Polymer Hybridization With DNA and RNA Directs Nanoparticle Loading, Silencing Delivery, and Aptamer Function. J. Am. Chem. Soc. 2015, 137, 8920–8923. 10.1021/jacs.5b05481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X.; Piao X.; Bong D. Bifacial Peptide Nucleic Acid As an Allosteric Switch for Aptamer and Ribozyme Function. J. Am. Chem. Soc. 2014, 136, 7265–7268. 10.1021/ja5032584. [DOI] [PubMed] [Google Scholar]
- Piao X.; Xia X.; Mao J.; Bong D. Peptide Ligation and RNA Cleavage Via an Abiotic Template Interface. J. Am. Chem. Soc. 2015, 137, 3751–3754. 10.1021/jacs.5b00236. [DOI] [PubMed] [Google Scholar]
- Zeng Y.; Pratumyot Y.; Piao X.; Bong D. Discrete Assembly of Synthetic Peptide-DNA Triplex Structures From Polyvalent Melamine-Thymine Bifacial Recognition. J. Am. Chem. Soc. 2012, 134, 832–835. 10.1021/ja2099326. [DOI] [PubMed] [Google Scholar]
- Piao X.; Xia X.; Bong D. Bifacial Peptide Nucleic Acid Directs Cooperative Folding and Assembly of Binary, Ternary, and Quaternary DNA Complexes. Biochemistry 2013, 52, 6313–6323. 10.1021/bi4008963. [DOI] [PubMed] [Google Scholar]
- Mao J.; Bong D. Synthesis of DNA-Binding Peptoids. Synlett 2015, 26 (11), 1581–1585. 10.1055/s-0034-1380698. [DOI] [Google Scholar]
- Bondensgaard K.; Petersen M.; Singh S. K.; Rajwanshi V. K.; Kumar R.; Wengel J.; Jacobsen J. P. Structural Studies of LNA:RNA Duplexes by NMR: Conformations and Implications for RNase H Activity. Chem. - Eur. J. 2000, 6, 2687–2695. . [DOI] [PubMed] [Google Scholar]
- Kaur H.; Arora A.; Wengel J.; Maiti S. Thermodynamic, Counterion, and Hydration Effects for the Incorporation of Locked Nucleic Acid Nucleotides into DNA Duplexes. Biochemistry 2006, 45, 7347–7355. 10.1021/bi060307w. [DOI] [PubMed] [Google Scholar]
- Kurreck J.; Wyszko E.; Gillen C.; Erdmann V. A. Design of Antisense Oligonucleotides Stabilized by Locked Nucleic Acids. Nucleic Acids Res. 2002, 30, 1911–1918. 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obad S.; dos Santos C. O.; Petri A.; Heidenblad M.; Broom O.; Ruse C.; Fu C.; Lindow M.; Stenvang J.; Straarup E. M.; Hansen H. F.; Koch T.; Pappin D.; Hannon G. J.; Kauppinen S. Silencing of MicroRNA Families by Seed-Targeting Tiny LNAs. Nat. Genet. 2011, 43, 371–378. 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allerson C. R.; Sioufi N.; Jarres R.; Prakash T. P.; Naik N.; Berdeja A.; Wanders L.; Griffey R. H.; Swayze E. E.; Bhat B. Fully 2’-Modified Oligonucleotide Duplexes With Improved in Vitro Potency and Stability Compared to Unmodified Small Interfering RNA. J. Med. Chem. 2005, 48, 901–904. 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- Sipa K.; Sochacka E.; Kazmierczak-Baranska J.; Maszewska M.; Janicka M.; Nowak G.; Nawrot B. Effect of Base Modifications on Structure, Thermodynamic Stability, and Gene Silencing Activity of Short Interfering RNA. RNA 2007, 13, 1301–1316. 10.1261/rna.538907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P.; Erickson S.; Anderson D. A Small Viral RNA Is Required for in Vitro Packaging of Bacteriophage Phi29 DNA. Science 1987, 236, 690–694. 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- Shu D.; Moll W. D.; Deng Z.; Mao C.; Guo P. Bottom-Up Assembly of RNA Arrays and Superstructures As Potential Parts in Nanotechnology. Nano Lett. 2004, 4, 1717–1723. 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y.; Shu D.; Haque F.; Guo P. Fabrication of PRNA Nanoparticles to Deliver Therapeutic RNAs and Bioactive Compounds into Tumor Cells. Nat. Protoc. 2013, 8, 1635–1659. 10.1038/nprot.2013.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A. I.; Zirbel C. L.; Leontis N. B. Automated Classification of RNA 3D Motifs and the RNA 3D Motif Atlas. RNA 2013, 19, 1327–1340. 10.1261/rna.039438.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham M.; Dror O.; Nussinov R.; Wolfson H. J. Analysis and Classification of RNA Tertiary Structures. RNA 2008, 14, 2274–2289. 10.1261/rna.853208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabow W. W.; Zakrevsky P.; Afonin K. A.; Chworos A.; Shapiro B. A.; Jaeger L. Self-Assembling RNA Nanorings Based on RNAI/II Inverse Kissing Complexes. Nano Lett. 2011, 11, 878–887. 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severcan I.; Geary C.; Verzemnieks E.; Chworos A.; Jaeger L. Square-Shaped RNA Particles From Different RNA Folds. Nano Lett. 2009, 9, 1270–1277. 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibrov S. M.; McLean J.; Parsons J.; Hermann T. Self-Assembling RNA Square. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 6405–6408. 10.1073/pnas.1017999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlea L.; Bindewald E.; Sharan R.; Bartlett N.; Moriarty D.; Oliver J.; Afonin K. A.; Shapiro B. A. Ring Catalog: A Resource for Designing Self-Assembling RNA Nanostructures. Methods 2016, 103, 128–137. 10.1016/j.ymeth.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L.; Leontis N. B. Tecto-RNA: One Dimensional Self-Assembly Through Tertiary Interactions. Angew. Chem., Int. Ed. 2000, 39, 2521–2524. . [DOI] [PubMed] [Google Scholar]
- Nasalean L.; Baudrey S.; Leontis N. B.; Jaeger L. Controlling RNA Self-Assembly to Form Filaments. Nucleic Acids Res. 2006, 34, 1381–1392. 10.1093/nar/gkl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary C.; Chworos A.; Jaeger L. Promoting RNA Helical Stacking Via A-Minor Junctions. Nucleic Acids Res. 2011, 39, 1066–1080. 10.1093/nar/gkq748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severcan I.; Geary C.; Chworos A.; Voss N.; Jacovetty E.; Jaeger L. A Polyhedron Made of TRNAs. Nat. Chem. 2010, 2, 772–779. 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. W.; Liu Z. Y.; Jiang W.; Wang G. S.; Mao C. D. De Novo Design of an RNA Tile That Self-Assembles into a Homo-Octameric Nanoprism. Nat. Commun. 2015, 6, 5724–5729. 10.1038/ncomms6724. [DOI] [PubMed] [Google Scholar]
- Bindewald E.; Grunewald C.; Boyle B.; O’Connor M.; Shapiro B. A. Computational Strategies for the Automated Design of RNA Nanoscale Structures From Building Blocks Using NanoTiler. J. Mol. Graphics Modell. 2008, 27, 299–308. 10.1016/j.jmgm.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossinet F.; Ludwig T. E.; Westhof E. Assemble: an Interactive Graphical Tool to Analyze and Build RNA Architectures at the 2D and 3D Levels. Bioinformatics 2010, 26, 2057–2059. 10.1093/bioinformatics/btq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez H. M.; Maizel J. V.; Shapiro B. A. RNA2D3D: A Program for Generating, Viewing, and Comparing 3-Dimensional Models of RNA. J. Biomol. Struct. Dyn. 2008, 25, 669–683. 10.1080/07391102.2008.10531240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A.; Backofen R. INFO-RNA--a Server for Fast Inverse RNA Folding Satisfying Sequence Constraints. Nucleic Acids Res. 2007, 35, W310–W313. 10.1093/nar/gkm218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadeh J. N.; Steenberg C. D.; Bois J. S.; Wolfe B. R.; Pierce M. B.; Khan A. R.; Dirks R. M.; Pierce N. A. NUPACK: Analysis and Design of Nucleic Acid Systems. J. Comput. Chem. 2011, 32, 170–173. 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- Yingling Y. G.; Shapiro B. A. Computational Design of an RNA Hexagonal Nanoring and an RNA Nanotube. Nano Lett. 2007, 7, 2328–2334. 10.1021/nl070984r. [DOI] [PubMed] [Google Scholar]
- Afonin K. A.; Bindewald E.; Yaghoubian A. J.; Voss N.; Jacovetty E.; Shapiro B. A.; Jaeger L. In Vitro Assembly of Cubic RNA-Based Scaffolds Designed. Nat. Nanotechnol. 2010, 5, 676–682. 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P.; Coban O.; Snead N. M.; Trebley J.; Hoeprich S.; Guo S.; Shu Y. Engineering RNA for Targeted SiRNA Delivery and Medical Application. Adv. Drug Delivery Rev. 2010, 62, 650–666. 10.1016/j.addr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y.; Pi F.; Sharma A.; Rajabi M.; Haque F.; Shu D.; Leggas M.; Evers B. M.; Guo P. Stable RNA Nanoparticles As Potential New Generation Drugs for Cancer Therapy. Adv. Drug Delivery Rev. 2014, 66C, 74–89. 10.1016/j.addr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Lee T.; Dziubla T.; Pi F.; Guo S.; Xu J.; Li C.; Haque F.; Liang X.; Guo P. RNA As a Stable Polymer to Build Controllable and Defined Nanostructures for Material and Biomedical Applications. Nano Today 2015, 10, 631–655. 10.1016/j.nantod.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabow W. W.; Jaeger L. RNA Self-Assembly and RNA Nanotechnology. Acc. Chem. Res. 2014, 47, 1871–1880. 10.1021/ar500076k. [DOI] [PubMed] [Google Scholar]
- Leontis N. B.; Westhof E. Chemistry. Self-Assembled RNA Nanostructures. Science 2014, 345, 732–733. 10.1126/science.1257989. [DOI] [PubMed] [Google Scholar]
- Afonin K. A.; Lindsay B.; Shapiro B. A. Engineered RNA Nanodesigns for Applications in RNA Nanotechnology. DNA and RNA Nanotechnol. 2013, 1, 1–15. 10.2478/rnan-2013-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodzinski P., Torchilin V., Eds. Advanced Drug Delivery Reviews: Cancer Nanotechnology; Elsevier: Amsterdam, 2014; Vol. 66. [Google Scholar]
- Li H.; Zhang K.; Pi F.; Guo S.; Shlyakhtenko L.; Chiu W.; Shu D.; Guo P. Controllable Self-Assembly of RNA Tetrahedrons With Precise Shape and Size for Cancer Targeting. Adv. Mater. 2016, 28, 7501–7507. 10.1002/adma.201601976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A.; Haque F.; Pi F.; Shlyakhtenko L.; Evers B. M.; Guo P. Controllable Self-Assembly of RNA Dendrimers. Nanomedicine 2016, 12, 835–844. 10.1016/j.nano.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary C.; Rothemund P. W.; Andersen E. S. A Single-Stranded Architecture for Co-Transcriptional Folding of RNA Nanostructures. Science 2014, 345, 799–804. 10.1126/science.1253920. [DOI] [PubMed] [Google Scholar]
- Afonin K. A.; Desai R.; Viard M.; Kireeva M. L.; Bindewald E.; Case C. L.; Maciag A. E.; Kasprzak W. K.; Kim T.; Sappe A.; Stepler M.; Kewalramani V. N.; Kashlev M.; Blumenthal R.; Shapiro B. A. Co-Transcriptional Production of RNA-DNA Hybrids for Simultaneous Release of Multiple Split Functionalities. Nucleic Acids Res. 2014, 42, 2085–2097. 10.1093/nar/gkt1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin K. A.; Kireeva M.; Grabow W. W.; Kashlev M.; Jaeger L.; Shapiro B. A. Co-Transcriptional Assembly of Chemically Modified RNA Nanoparticles Functionalized With SiRNAs. Nano Lett. 2012, 12, 5192–5195. 10.1021/nl302302e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean F. B.; Nelson J. R.; Giesler T. L.; Lasken R. S. Rapid Amplification of Plasmid and Phage DNA Using Phi 29 DNA Polymerase and Multiply-Primed Rolling Circle Amplification. Genome Res. 2001, 11, 1095–1099. 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubendiek S.; Ryan K.; Kool E. Rolling-Circle RNA Synthesis: Circular Oligonucleotides As Efficient Substrates for T7 RNA Polymerase. J. Am. Chem. Soc. 1995, 117 (29), 7818–7819. 10.1021/ja00134a032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.; Park Y.; Kim H.; Lee J. B. Self-Assembly of Free-Standing RNA Membranes. Nat. Commun. 2014, 5, 4367. 10.1038/ncomms5367. [DOI] [PubMed] [Google Scholar]
- Wang D.; Zhang Z.; O’Loughlin E.; Lee T.; Houel S.; O’Carroll D.; Tarakhovsky A.; Ahn N. G.; Yi R. Quantitative Functions of Argonaute Proteins in Mammalian Development. Genes Dev. 2012, 26, 693–704. 10.1101/gad.182758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansma H. G.; Oroudjev E.; Baudrey S.; Jaeger L. TectoRNA and ’Kissing-Loop’ RNA: Atomic Force Microscopy of Self-Assembling RNA Structures. J. Microsc. 2003, 212, 273–279. 10.1111/j.1365-2818.2003.01276.x. [DOI] [PubMed] [Google Scholar]
- Yingchoncharoen P.; Kalinowski D. S.; Richardson D. R. Lipid-Based Drug Delivery Systems in Cancer Therapy: What Is Available and What Is Yet to Come. Pharmacol. Rev. 2016, 68, 701–787. 10.1124/pr.115.012070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Z.; Conejos-Sanchez I.; Griffin B. T.; O’Driscoll C. M.; Alonso M. J. Lipid-Based Nanocarriers for Oral Peptide Delivery. Adv. Drug Delivery Rev. 2016, 106, 337–354. 10.1016/j.addr.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Liu D. M.; He C. B.; Wang A. Z.; Lin W. B. Application of Liposomal Technologies for Delivery of Platinum Analogs in Oncology. Int. J. Nanomed. 2013, 8, 3309–3319. 10.2147/IJN.S38354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack D. W.; Hoffman A. S.; Pun S.; Stayton P. S. Design and Development of Polymers for Gene Delivery. Nat. Rev. Drug Discovery 2005, 4, 581–593. 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- Duncan R. Polymer Therapeutics As Nanomedicines: New Perspectives. Curr. Opin. Biotechnol. 2011, 22, 492–501. 10.1016/j.copbio.2011.05.507. [DOI] [PubMed] [Google Scholar]
- Lee K. L.; Twyman R. M.; Fiering S.; Steinmetz N. F. Virus-Based Nanoparticles As Platform Technologies for Modern Vaccines. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2016, 8, 554–578. 10.1002/wnan.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. T.; Hawes A. K.; Bundy B. C. Reengineering Viruses and Virus-Like Particles Through Chemical Functionalization Strategies. Curr. Opin. Biotechnol. 2013, 24, 620–626. 10.1016/j.copbio.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Hong C. A.; Eltoukhy A. A.; Lee H.; Langer R.; Anderson D. G.; Nam Y. S. Dendrimeric SiRNA for Efficient Gene Silencing. Angew. Chem., Int. Ed. 2015, 54, 6740–6744. 10.1002/anie.201412493. [DOI] [PubMed] [Google Scholar]
- Sun W.; Mignani S.; Shen M.; Shi X. Dendrimer-Based Magnetic Iron Oxide Nanoparticles: Their Synthesis and Biomedical Applications. Drug Discovery Today 2016, 21, 1873. 10.1016/j.drudis.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Chaplot S. P.; Rupenthal I. D. Dendrimers for Gene Delivery--a Potential Approach for Ocular Therapy?. J. Pharm. Pharmacol. 2014, 66, 542–556. 10.1111/jphp.12104. [DOI] [PubMed] [Google Scholar]
- Hsu H. J.; Bugno J.; Lee S. R.; Hong S. Dendrimer-Based Nanocarriers: a Versatile Platform for Drug Delivery. Wiley. Interdiscip. Rev.:Nanomed. Nanobiotechnol. 2017, 9, e1409. 10.1002/wnan.1409. [DOI] [PubMed] [Google Scholar]
- Chen S.; Hao X.; Liang X.; Zhang Q.; Zhang C.; Zhou G.; Shen S.; Jia G.; Zhang J. Inorganic Nanomaterials As Carriers for Drug Delivery. J. Biomed. Nanotechnol. 2016, 12, 1–27. 10.1166/jbn.2016.2122. [DOI] [PubMed] [Google Scholar]
- van der Meel R.; Vehmeijer L. J.; Kok R. J.; Storm G.; van Gaal E. V. Ligand-Targeted Particulate Nanomedicines Undergoing Clinical Evaluation: Current Status. Adv. Drug Delivery Rev. 2013, 65, 1284–1298. 10.1016/j.addr.2013.08.012. [DOI] [PubMed] [Google Scholar]
- Grodzinski P.; Farrell D. Future Opportunities in Cancer Nanotechnology--NCI Strategic Workshop Report. Cancer Res. 2014, 74, 1307–1310. 10.1158/0008-5472.CAN-13-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni W. C.; Torchilin V.; Patri A. K.; Hrkach J.; Stern S.; Lee R.; Nel A.; Panaro N. J.; Grodzinski P. Best Practices in Cancer Nanotechnology: Perspective From NCI Nanotechnology Alliance. Clin. Cancer Res. 2012, 18, 3229–3241. 10.1158/1078-0432.CCR-11-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer D.; Karp J. M.; Hong S.; Farokhzad O. C.; Margalit R.; Langer R. Nanocarriers As an Emerging Platform for Cancer Therapy. Nat. Nanotechnol. 2007, 2, 751–760. 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- Tandon P.; Farahani K. NCI Image-Guided Drug Delivery Summit. Cancer Res. 2011, 71, 314–317. 10.1158/0008-5472.CAN-10-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binzel D.; Shu Y.; Li H.; Sun M.; Zhang Q.; Shu D.; Guo B.; Guo P. Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Mol. Ther. 2016, 24, 1267–1277. 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Shu Y.; Guo P.; Smith D.; Rossi J. Dual Functional RNA Nanoparticles Containing Phi29 Motor PRNA and Anti-Gp120 Aptamer for Cell-Type Specific Delivery and HIV-1 Inhibition. Methods 2011, 54, 284–294. 10.1016/j.ymeth.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.; Wang D.; Wu Y.; Ho R.; Jia L.; Guo P.; Hu L.; Xing G.; Zeng Y.; Liang X. J. AIDS Treatment With Novel Anti-HIV Compounds Improved by Nanotechnology. AAPS J. 2010, 12, 272–278. 10.1208/s12248-010-9187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L.; Li S. K.; Liu H.; Liu C. Y.; LaSance K.; Haque F.; Shu D.; Guo P. Ocular Delivery of PRNA Nanoparticles: Distribution and Clearance After Subconjunctival Injection. Pharm. Res. 2014, 31, 1046–1058. 10.1007/s11095-013-1226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram P.; Kurniawan H.; Byrne M. E.; Wower J. Therapeutic RNA Aptamers in Clinical Trials. Eur. J. Pharm. Sci. 2013, 48, 259–271. 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Guo P.; Haque F.; Hallahan B.; Reif R.; Li H. Uniqueness, Advantages, Challenges, Solutions, and Perspectives in Therapeutics Applying RNA Nanotechnology. Nucleic Acid Ther. 2012, 22, 226–245. 10.1201/b15152-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A. D.; Szostak J. W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Tuerk C.; Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Kuan C. T.; Mi J.; Zhang X.; Clary B. M.; Bigner D. D.; Sullenger B. A. Aptamers Selected Against the Unglycosylated EGFRvIII Ectodomain and Delivered Intracellularly Reduce Membrane-Bound EGFRvIII and Induce Apoptosis. Biol. Chem. 2009, 390, 137–144. 10.1515/BC.2009.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito C. L.; Passaro D.; Longobardo I.; Condorelli G.; Marotta P.; Affuso A.; de Franciscis V.; Cerchia L. A Neutralizing RNA Aptamer Against EGFR Causes Selective Apoptotic Cell Death. PLoS One 2011, 6, e24071. 10.1371/journal.pone.0024071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel K. W.; Hernandez L. I.; Dassie J. P.; Thiel W. H.; Liu X.; Stockdale K. R.; Rothman A. M.; Hernandez F. J.; McNamara J. O.; Giangrande P. H. Delivery of Chemo-Sensitizing SiRNAs to HER2+-Breast Cancer Cells Using RNA Aptamers. Nucleic Acids Res. 2012, 40, 6319–6337. 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H.; Chernis G. A.; Hoang V. Q.; Landgraf R. Inhibition of Heregulin Signaling by an Aptamer That Preferentially Binds to the Oligomeric Form of Human Epidermal Growth Factor Receptor-3. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 9226–9231. 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey W. M.; Hernandez F. J.; Huang S. Y.; Cao S.; Howell C. A.; Thomas G. S.; Liu X. Y.; Lapteva N.; Spencer D. M.; McNamara J. O.; Zou X.; Chen S. J.; Giangrande P. H. Rational Truncation of an RNA Aptamer to Prostate-Specific Membrane Antigen Using Computational Structural Modeling. Nucleic Acid Ther. 2011, 21, 299–314. 10.1089/nat.2011.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann A. P.; Somasunderam A.; Nieves-Alicea R.; Li X.; Hu A.; Sood A. K.; Ferrari M.; Gorenstein D. G.; Tanaka T. Identification of Thioaptamer Ligand Against E-Selectin: Potential Application for Inflamed Vasculature Targeting. PLoS One 2010, 5, e13050. 10.1371/journal.pone.0013050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigdar S.; Lin J.; Yu Y.; Pastuovic M.; Wei M.; Duan W. RNA Aptamer Against a Cancer Stem Cell Marker Epithelial Cell Adhesion Molecule. Cancer Sci. 2011, 102, 991–998. 10.1111/j.1349-7006.2011.01897.x. [DOI] [PubMed] [Google Scholar]
- Mayer G.; Ahmed M. S.; Dolf A.; Endl E.; Knolle P. A.; Famulok M. Fluorescence-Activated Cell Sorting for Aptamer SELEX With Cell Mixtures. Nat. Protoc. 2010, 5, 1993–2004. 10.1038/nprot.2010.163. [DOI] [PubMed] [Google Scholar]
- Davis K. A.; Lin Y.; Abrams B.; Jayasena S. D. Staining of Cell Surface Human CD4 With 2’-F-Pyrimidine-Containing RNA Aptamers for Flow Cytometry. Nucleic Acids Res. 1998, 26, 3915–3924. 10.1093/nar/26.17.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low P. S.; Kularatne S. A. Folate-Targeted Therapeutic and Imaging Agents for Cancer. Curr. Opin. Chem. Biol. 2009, 13, 256–262. 10.1016/j.cbpa.2009.03.022. [DOI] [PubMed] [Google Scholar]
- Schroeder A.; Heller D. A.; Winslow M. M.; Dahlman J. E.; Pratt G. W.; Langer R.; Jacks T.; Anderson D. G. Treating Metastatic Cancer With Nanotechnology. Nat. Rev. Cancer 2012, 12, 39–50. 10.1038/nrc3180. [DOI] [PubMed] [Google Scholar]
- Kang K. N.; Lee Y. S. RNA Aptamers: a Review of Recent Trends and Applications. Adv. Biochem. Eng./Biotechnol. 2012, 131, 153–169. 10.1007/10_2012_136. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Bobbin M. L.; Burnett J. C.; Rossi J. J. Current Progress of RNA Aptamer-Based Therapeutics. Front. Genet. 2012, 3, 234. 10.3389/fgene.2012.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhbacher J.; St-Pierre P.; Lafontaine D. A. Therapeutic Applications of Ribozymes and Riboswitches. Curr. Opin. Pharmacol. 2010, 10, 551–556. 10.1016/j.coph.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Fanning G.; Amado R.; Symonds G. Gene Therapy for HIV/AIDS: the Potential for a New Therapeutic Regimen. J. Gene Med. 2003, 5, 645–653. 10.1002/jgm.436. [DOI] [PubMed] [Google Scholar]
- Serganov A. The Long and the Short of Riboswitches. Curr. Opin. Struct. Biol. 2009, 19, 251–259. 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount K. F.; Breaker R. R. Riboswitches As Antibacterial Drug Targets. Nat. Biotechnol. 2006, 24, 1558–1564. 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Suess B.; Weigand J. E. Engineered Riboswitches - Overview, Problems and Trends. RNA Biol. 2008, 5, 24–29. 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- Berens C.; Groher F.; Suess B. RNA Aptamers As Genetic Control Devices: the Potential of Riboswitches As Synthetic Elements for Regulating Gene Expression. Biotechnol. J. 2015, 10, 246–257. 10.1002/biot.201300498. [DOI] [PubMed] [Google Scholar]
- Afonin K. A.; Bindewald E.; Kireeva M.; Shapiro B. A. Computational and Experimental Studies of Reassociating RNA/DNA Hybrids Containing Split Functionalities. Methods Enzymol. 2015, 553, 313–334. 10.1016/bs.mie.2014.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin K. A.; Viard M.; Kagiampakis I.; Case C. L.; Dobrovolskaia M. A.; Hofmann J.; Vrzak A.; Kireeva M.; Kasprzak W. K.; KewalRamani V. N.; et al. Triggering of RNA Interference With RNA–RNA, RNA–DNA, and DNA–RNA Nanoparticles. ACS Nano 2015, 9, 251–259. 10.1021/nn504508s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin K. A.; Viard M.; Martins A. N.; Lockett S. J.; Maciag A. E.; Freed E. O.; Heldman E.; Jaeger L.; Blumenthal R.; Shapiro B. A. Activation of Different Split Functionalities on Re-Association of RNA-DNA Hybrids. Nat. Nanotechnol. 2013, 8, 296–304. 10.1038/nnano.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindewald E.; Afonin K. A.; Viard M.; Zakrevsky P.; Kim T.; Shapiro B. A. Multistrand Structure Prediction of Nucleic Acid Assemblies and Design of RNA Switches. Nano Lett. 2016, 16, 1726–1735. 10.1021/acs.nanolett.5b04651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A.; Xu S.; Montgomery M. K.; Kostas S. A.; Driver S. E.; Mello C. C. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Aagaard L.; Rossi J. J. RNAi Therapeutics: Principles, Prospects and Challenges. Adv. Drug Delivery Rev. 2007, 59, 75–86. 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Z.; Chen E.; Gu S. G. Complex Coding of Endogenous SiRNA, Transcriptional Silencing and H3K9Methylation on Native Targets of Germline Nuclear RNAi in C. Elegans. BMC Genomics 2014, 15, 1157. 10.1186/1471-2164-15-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh Y. H.; Deng J. Z.; Dreaden E. C.; Park J. H.; Yun D. S.; Shopsowitz K. E.; Hammond P. T. A Multi-RNAi Microsponge Platform for Simultaneous Controlled Delivery of Multiple Small Interfering RNAs. Angew. Chem., Int. Ed. 2016, 55, 3347–3351. 10.1002/anie.201508978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler L. A.; Vrbanac V.; Trifonova R.; Brehm M. A.; Gilboa-Geffen A.; Tanno S.; Greiner D. L.; Luster A. D.; Tager A. M.; Lieberman J. Durable Knockdown and Protection From HIV Transmission in Humanized Mice Treated With Gel-Formulated CD4 Aptamer-SiRNA Chimeras. Mol. Ther. 2013, 21, 1378–1389. 10.1038/mt.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin K. A.; Viard M.; Koyfman A. Y.; Martins A. N.; Kasprzak W. K.; Panigaj M.; Desai R.; Santhanam A.; Grabow W. W.; Jaeger L.; Heldman E.; Reiser J.; Chiu W.; Freed E. O.; Shapiro B. A. Multifunctional RNA Nanoparticles. Nano Lett. 2014, 14, 5662–5671. 10.1021/nl502385k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X.; Hemida M.; Zhang H. M.; Hanson P.; Ye Q.; Yang D. Current Advances in Phi29 PRNA Biology and Its Application in Drug Delivery. Wiley. Interdiscip. Rev. RNA 2012, 3, 469–481. 10.1002/wrna.1111. [DOI] [PubMed] [Google Scholar]
- Ye X.; Liu Z.; Hemida M. G.; Yang D. Targeted Delivery of Mutant Tolerant Anti-Coxsackievirus Artificial MicroRNAs Using Folate Conjugated Bacteriophage Phi29 PRNA. PLoS One 2011, 6, e21215. 10.1371/journal.pone.0021215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirota H.; Klinman D. M. Use of CpG Oligonucleotides for Cancer Immunotherapy and Their Effect on Immunity in the Tumor Microenvironment. Immunotherapy 2013, 5, 787–789. 10.2217/imt.13.70. [DOI] [PubMed] [Google Scholar]
- Gantier M. P.; Tong S.; Behlke M. A.; Irving A. T.; Lappas M.; Nilsson U. W.; Latz E.; McMillan N. A.; Williams B. R. Rational Design of Immunostimulatory SiRNAs. Mol. Ther. 2010, 18, 785–795. 10.1038/mt.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khedri M.; Rafatpanah H.; Abnous K.; Ramezani P.; Ramezani M. Cancer Immunotherapy Via Nucleic Acid Aptamers. Int. Immunopharmacol. 2015, 29, 926–936. 10.1016/j.intimp.2015.10.013. [DOI] [PubMed] [Google Scholar]
- Anderson J.; Akkina R. HIV-1 Resistance Conferred by SiRNA Cosuppression of CXCR4 and CCR5 Coreceptors by a Bispecific Lentiviral Vector. AIDS Res. Ther. 2005, 2, 1–12. 10.1186/1742-6405-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boltz A.; Piater B.; Toleikis L.; Guenther R.; Kolmar H.; Hock B. Bi-Specific Aptamers Mediating Tumor Cell Lysis. J. Biol. Chem. 2011, 286, 21896–21905. 10.1074/jbc.M111.238261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramod P. S.; Deshpande N. U.; Jayakannan M. Real-Time Drug Release Analysis of Enzyme and PH Responsive Polysaccharide Nanovesicles. J. Phys. Chem. B 2015, 119, 10511–10523. 10.1021/acs.jpcb.5b05795. [DOI] [PubMed] [Google Scholar]
- Agudelo D.; Bourassa P.; Berube G.; Tajmir-Riahi H. A. Intercalation of Antitumor Drug Doxorubicin and Its Analogue by DNA Duplex: Structural Features and Biological Implications. Int. J. Biol. Macromol. 2014, 66, 144–150. 10.1016/j.ijbiomac.2014.02.028. [DOI] [PubMed] [Google Scholar]
- Chen A. K.; Behlke M. A.; Tsourkas A. Avoiding False-Positive Signals With Nuclease-Vulnerable Molecular Beacons in Single Living Cells. Nucleic Acids Res. 2007, 35, e105. 10.1093/nar/gkm593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. K.; Behlke M. A.; Tsourkas A. Efficient Cytosolic Delivery of Molecular Beacon Conjugates and Flow Cytometric Analysis of Target RNA. Nucleic Acids Res. 2008, 36, e69. 10.1093/nar/gkn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhlanga M. M.; Vargas D. Y.; Fung C. W.; Kramer F. R.; Tyagi S. TRNA-Linked Molecular Beacons for Imaging MRNAs in the Cytoplasm of Living Cells. Nucleic Acids Res. 2005, 33, 1902–1912. 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee W. J.; Santangelo P. J.; Jo H.; Bao G. Target Accessibility and Signal Specificity in Live-Cell Detection of BMP-4 MRNA Using Molecular Beacons. Nucleic Acids Res. 2008, 36, e30. 10.1093/nar/gkn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. K.; Choi K. J.; Lee M.; Jo M. H.; Kim S. Molecular Imaging of a Cancer-Targeting Theragnostics Probe Using a Nucleolin Aptamer- and MicroRNA-221 Molecular Beacon-Conjugated Nanoparticle. Biomaterials 2012, 33, 207–217. 10.1016/j.biomaterials.2011.09.023. [DOI] [PubMed] [Google Scholar]
- Peng X. H.; Cao Z. H.; Xia J. T.; Carlson G. W.; Lewis M. M.; Wood W. C.; Yang L. Real-Time Detection of Gene Expression in Cancer Cells Using Molecular Beacon Imaging: New Strategies for Cancer Research. Cancer Res. 2005, 65, 1909–1917. 10.1158/0008-5472.CAN-04-3196. [DOI] [PubMed] [Google Scholar]
- Bryson J. M.; Fichter K. M.; Chu W. J.; Lee J. H.; Li J.; Madsen L. A.; McLendon P. M.; Reineke T. M. Polymer Beacons for Luminescence and Magnetic Resonance Imaging of DNA Delivery. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 16913–16918. 10.1073/pnas.0904860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L.; Wu C.; You M.; Han D.; Chen T.; Zhu G.; Jiang J.; Yu R.; Tan W. A Targeted, Self-Delivered, and Photocontrolled Molecular Beacon for MRNA Detection in Living Cells. J. Am. Chem. Soc. 2013, 135, 12952–12955. 10.1021/ja406252w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. W.; Akens M. K.; Chen J.; Wise-Milestone L.; Wilson B. C.; Zheng G. Imaging of Specific Activation of Photodynamic Molecular Beacons in Breast Cancer Vertebral Metastases. Bioconjugate Chem. 2011, 22, 1021–1030. 10.1021/bc200169x. [DOI] [PubMed] [Google Scholar]
- Mhlanga M. M.; Tyagi S. Using TRNA-Linked Molecular Beacons to Image Cytoplasmic MRNAs in Live Cells. Nat. Protoc. 2006, 1, 1392–1398. 10.1038/nprot.2006.242. [DOI] [PubMed] [Google Scholar]
- Amrute-Nayak M.; Bullock S. L. Single-Molecule Assays Reveal That RNA Localization Signals Regulate Dynein-Dynactin Copy Number on Individual Transcript Cargoes. Nat. Cell Biol. 2012, 14, 416–423. 10.1038/ncb2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadakuma H.; Ishihama Y.; Shibuya T.; Tani T.; Funatsu T. Imaging of Single MRNA Molecules Moving Within a Living Cell Nucleus. Biochem. Biophys. Res. Commun. 2006, 344, 772–779. 10.1016/j.bbrc.2006.03.202. [DOI] [PubMed] [Google Scholar]
- Vargas D. Y.; Raj A.; Marras S. A.; Kramer F. R.; Tyagi S. Mechanism of MRNA Transport in the Nucleus. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 17008–17013. 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.; Yang R.; Shi M.; Wu C.; Fang X.; Li Y.; Li J.; Tan W. Rationally Designed Molecular Beacons for Bioanalytical and Biomedical Applications. Chem. Soc. Rev. 2015, 44, 3036–3055. 10.1039/C5CS00020C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. X.; Jia X.; Ma J. L.; Zhu Q. Molecular Beacons: a Novel Optical Diagnostic Tool. Arch. Immunol. Ther. Exp. 2013, 61, 139–148. 10.1007/s00005-012-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. S.; Raston N. H.; Gu M. B. Aptamer-Based Nanobiosensors. Biosens. Bioelectron. 2016, 76, 2–19. 10.1016/j.bios.2015.06.040. [DOI] [PubMed] [Google Scholar]
- Kraemer S.; Vaught J. D.; Bock C.; Gold L.; Katilius E.; Keeney T. R.; Kim N.; Saccomano N. A.; Wilcox S. K.; Zichi D.; Sanders G. M. From SOMAmer-Based Biomarker Discovery to Diagnostic and Clinical Applications: a SOMAmer-Based, Streamlined Multiplex Proteomic Assay. PLoS One 2011, 6, e26332. 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodakov D.; Wang C.; Zhang D. Y. Diagnostics Based on Nucleic Acid Sequence Variant Profiling: PCR, Hybridization, and NGS Approaches. Adv. Drug Delivery Rev. 2016, 105, 3–19. 10.1016/j.addr.2016.04.005. [DOI] [PubMed] [Google Scholar]
- Schulte C.; Zeller T. MicroRNA-Based Diagnostics and Therapy in Cardiovascular Disease-Summing Up the Facts. Cardiovasc. Diagn. Ther. 2015, 5, 17–36. 10.3978/j.issn.2223-3652.2014.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.; Ng T. B. In-Vitro Nanodiagnostic Platform Through Nanoparticles and DNA-RNA Nanotechnology. Appl. Microbiol. Biotechnol. 2015, 99, 3359–3374. 10.1007/s00253-015-6506-4. [DOI] [PubMed] [Google Scholar]
- Okamoto A.; Tanaka K.; Saito I. DNA Logic Gates. J. Am. Chem. Soc. 2004, 126, 9458–9463. 10.1021/ja047628k. [DOI] [PubMed] [Google Scholar]
- Adleman L. M. Molecular Computation of Solutions to Combinatorial Problems. Science 1994, 266, 1021–1024. 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- Lee T.; Yagati A. K.; Pi F.; Sharma A.; Choi J.-W.; Guo P. Construction of RNA-Quantum Dot Chimera for Nanoscale Resistive Biomemory Application. ACS Nano 2015, 9, 6675–6682. 10.1021/acsnano.5b03269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan J. G.; Lee M. E.; Almeida R.; Gilbert L. A.; Whitehead E. H.; La Russa M.; Tsai J. C.; Weissman J. S.; Dueber J. E.; Qi L. S.; Lim W. A. Engineering Complex Synthetic Transcriptional Programs With CRISPR RNA Scaffolds. Cell 2015, 160, 339–350. 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H.; Song C. Q.; Dorkin J. R.; Zhu L. J.; Li Y.; Wu Q.; Park A.; Yang J.; Suresh S.; Bizhanova A.; Gupta A.; Bolukbasi M. F.; Walsh S.; Bogorad R. L.; Gao G.; Weng Z.; Dong Y.; Koteliansky V.; Wolfe S. A.; Langer R.; et al. Therapeutic Genome Editing by Combined Viral and Non-Viral Delivery of CRISPR System Components in Vivo. Nat. Biotechnol. 2016, 34, 328–333. 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.; Marraffini L. A. CRISPR-Cas: New Tools for Genetic Manipulations From Bacterial Immunity Systems. Annu. Rev. Microbiol. 2015, 69, 209–228. 10.1146/annurev-micro-091014-104441. [DOI] [PubMed] [Google Scholar]
- Ramanan V.; Shlomai A.; Cox D. B.; Schwartz R. E.; Michailidis E.; Bhatta A.; Scott D. A.; Zhang F.; Rice C. M.; Bhatia S. N. CRISPR/Cas9 Cleavage of Viral DNA Efficiently Suppresses Hepatitis B Virus. Sci. Rep. 2015, 5, 10833. 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E. M.; Bassit L. C.; Mueller H.; Kornepati A. V.; Bogerd H. P.; Nie T.; Chatterjee P.; Javanbakht H.; Schinazi R. F.; Cullen B. R. Suppression of Hepatitis B Virus DNA Accumulation in Chronically Infected Cells Using a Bacterial CRISPR/Cas RNA-Guided DNA Endonuclease. Virology 2015, 476, 196–205. 10.1016/j.virol.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A.; Bak R. O.; Clark J. T.; Kennedy A. B.; Ryan D. E.; Roy S.; Steinfeld I.; Lunstad B. D.; Kaiser R. J.; Wilkens A. B.; Bacchetta R.; Tsalenko A.; Dellinger D.; Bruhn L.; Porteus M. H. Chemically Modified Guide RNAs Enhance CRISPR-Cas Genome Editing in Human Primary Cells. Nat. Biotechnol. 2015, 33, 985–989. 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. B.; Guthrie E. H.; Huang M. T.; Taxman D. J. Short Hairpin RNA (ShRNA): Design, Delivery, and Assessment of Gene Knockdown. Methods Mol. Biol. 2010, 629, 141–158. 10.1007/978-1-60761-657-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. J. Before and After Endosomal Escape: Roles of Stimuli-Converting SiRNA/Polymer Interactions in Determining Gene Silencing Efficiency. Acc. Chem. Res. 2012, 45, 1077–1088. 10.1021/ar200241v. [DOI] [PubMed] [Google Scholar]
- Lavergne T.; Bertrand J. R.; Vasseur J. J.; Debart F. A Base-Labile Group for 2’-OH Protection of Ribonucleosides: a Major Challenge for RNA Synthesis. Chem. - Eur. J. 2008, 14, 9135–9138. 10.1002/chem.200801392. [DOI] [PubMed] [Google Scholar]
- Beaucage S. L.; Reese C. B.. Recent Advances in the Chemical Synthesis of RNA. Current Protocols in Nucleic Acid Chemistry; John Wiley & Sons: Hoboken, NJ, 2009; Chapter 2: Unit 2.16 1–31, Unit 31.2.16.1 10.1002/0471142700.nc0216s38. [DOI] [PubMed] [Google Scholar]
- Jasinski D.; Schwartz C.; Haque F.; Guo P. Large Scale Purification of RNA Nanoparticles by Preparative Ultracentrifugation. Methods Mol. Biol. 2015, 1297, 67–82. 10.1007/978-1-4939-2562-9_5. [DOI] [PubMed] [Google Scholar]
- Flinders J.; Defina S. C.; Brackett D. M.; Baugh C.; Wilson C.; Dieckmann T. Recognition of Planar and Nonplanar Ligands in the Malachite Green-RNA Aptamer Complex. ChemBioChem 2004, 5, 62–72. 10.1002/cbic.200300701. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic Acids Res. 2003, 31, 3406–3415. 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andronescu M.; Fejes A. P.; Hutter F.; Hoos H. H.; Condon A. A New Algorithm for RNA Secondary Structure Design. J. Mol. Biol. 2004, 336, 607–624. 10.1016/j.jmb.2003.12.041. [DOI] [PubMed] [Google Scholar]
- Ding Y.; Chan C. Y.; Lawrence C. E. Sfold Web Server for Statistical Folding and Rational Design of Nucleic Acids. Nucleic Acids Res. 2004, 32, W135–W141. 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindewald E.; Afonin K.; Jaeger L.; Shapiro B. A. Multistrand RNA Secondary Structure Prediction and Nanostructure Design Including Pseudoknots. ACS Nano 2011, 5, 9542–9551. 10.1021/nn202666w. [DOI] [PMC free article] [PubMed] [Google Scholar]