Abstract
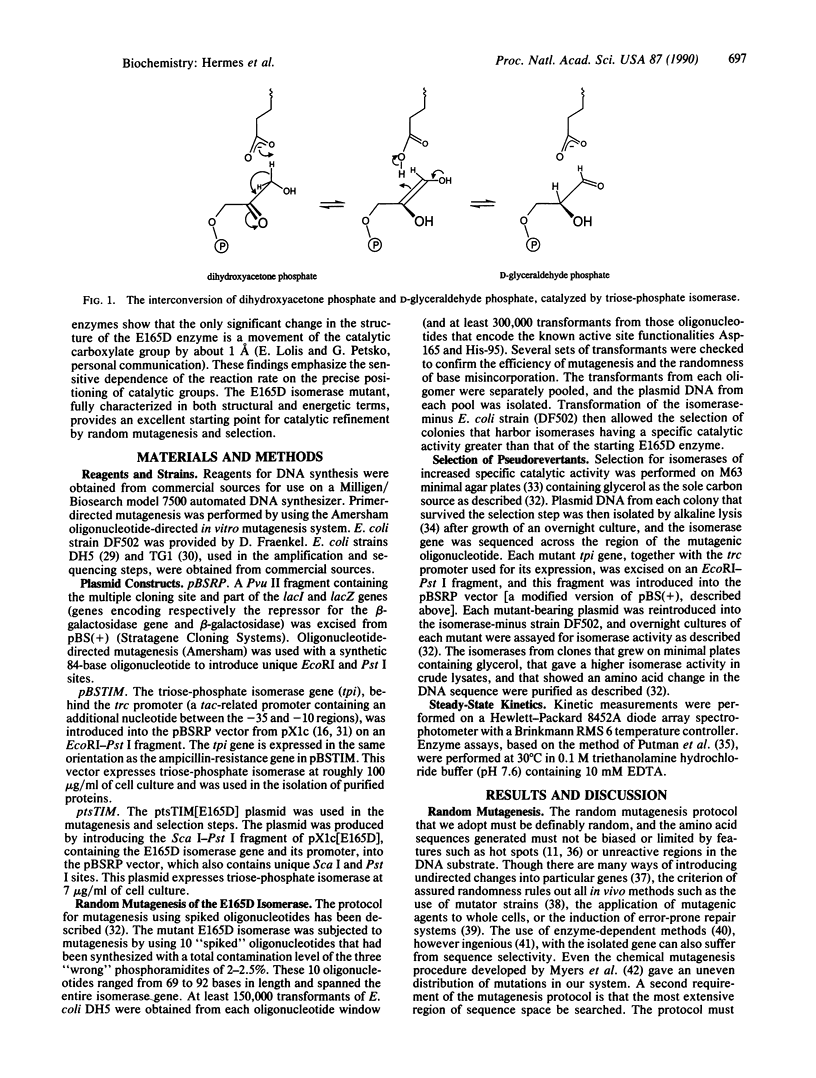
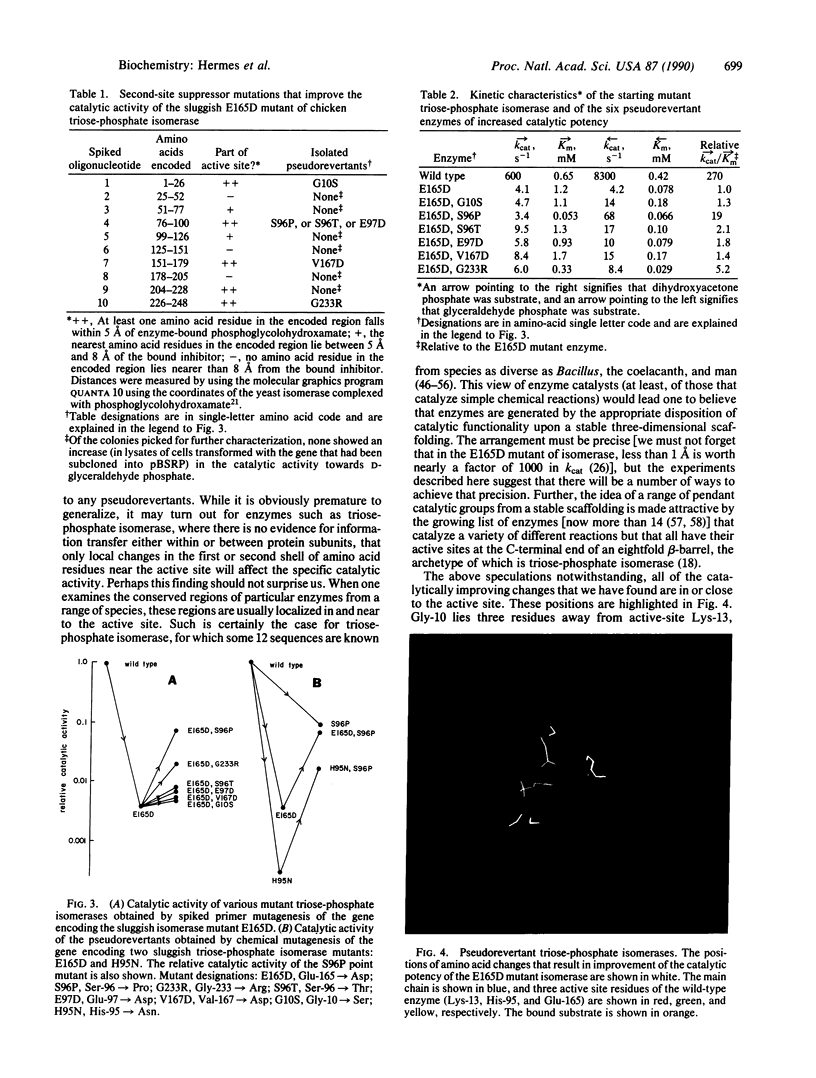
How easy is it to improve the catalytic power of an enzyme? To address this question, the gene encoding a sluggish mutant triose-phosphate isomerase (D-glyceraldehyde-3-phosphate ketol-isomerase, EC 5.3.1.1) has been subjected to random mutagenesis over its whole length by using "spiked" oligonucleotide primers. Transformation of an isomerase-minus strain of Escherichia coli was followed by selection of those colonies harboring an enzyme of higher catalytic potency. Six amino acid changes in the Glu-165----Asp mutant of triosephosphate isomerase improve the specific catalytic activity of this enzyme (from 1.3-fold to 19-fold). The suppressor sites are scattered across the sequence (at positions 10, 96, 97, 167, and 233), but each of them is very close to the active site. These experiments show both that there are relatively few single amino acid changes that increase the catalytic potency of this enzyme and that all of these improvements derive from alterations that are in, or very close to, the active site.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T. C., Davenport R. C., Jr, Giammona D. A., Lolis E., Petsko G. A., Ringe D. Crystallography and site-directed mutagenesis of yeast triosephosphate isomerase: what can we learn about catalysis from a "simple" enzyme? Cold Spring Harb Symp Quant Biol. 1987;52:603–613. doi: 10.1101/sqb.1987.052.01.069. [DOI] [PubMed] [Google Scholar]
- Alber T., Banner D. W., Bloomer A. C., Petsko G. A., Phillips D., Rivers P. S., Wilson I. A. On the three-dimensional structure and catalytic mechanism of triose phosphate isomerase. Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):159–171. doi: 10.1098/rstb.1981.0069. [DOI] [PubMed] [Google Scholar]
- Alber T., Kawasaki G. Nucleotide sequence of the triose phosphate isomerase gene of Saccharomyces cerevisiae. J Mol Appl Genet. 1982;1(5):419–434. [PubMed] [Google Scholar]
- Albery W. J., Knowles J. R. Free-energy profile of the reaction catalyzed by triosephosphate isomerase. Biochemistry. 1976 Dec 14;15(25):5627–5631. doi: 10.1021/bi00670a031. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Harris J. I. Primary structure of triosephosphate isomerase from Bacillus stearothermophilus. Eur J Biochem. 1980 Jul;108(2):599–611. doi: 10.1111/j.1432-1033.1980.tb04755.x. [DOI] [PubMed] [Google Scholar]
- Banner D. W., Bloomer A. C., Petsko G. A., Phillips D. C., Pogson C. I., Wilson I. A., Corran P. H., Furth A. J., Milman J. D., Offord R. E. Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5 angstrom resolution using amino acid sequence data. Nature. 1975 Jun 19;255(5510):609–614. doi: 10.1038/255609a0. [DOI] [PubMed] [Google Scholar]
- Benzer S. ON THE TOPOGRAPHY OF THE GENETIC FINE STRUCTURE. Proc Natl Acad Sci U S A. 1961 Mar;47(3):403–415. doi: 10.1073/pnas.47.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklow S. C., Raines R. T., Lim W. A., Zamore P. D., Knowles J. R. Triosephosphate isomerase catalysis is diffusion controlled. Appendix: Analysis of triose phosphate equilibria in aqueous solution by 31P NMR. Biochemistry. 1988 Feb 23;27(4):1158–1167. doi: 10.1021/bi00404a013. [DOI] [PubMed] [Google Scholar]
- Botstein D., Shortle D. Strategies and applications of in vitro mutagenesis. Science. 1985 Sep 20;229(4719):1193–1201. doi: 10.1126/science.2994214. [DOI] [PubMed] [Google Scholar]
- CRICK F. H., BARNETT L., BRENNER S., WATTS-TOBIN R. J. General nature of the genetic code for proteins. Nature. 1961 Dec 30;192:1227–1232. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- Carter P., Bedouelle H., Winter G. Improved oligonucleotide site-directed mutagenesis using M13 vectors. Nucleic Acids Res. 1985 Jun 25;13(12):4431–4443. doi: 10.1093/nar/13.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Protein structure: the 14th barrel rolls out. Nature. 1988 Jun 16;333(6174):598–599. doi: 10.1038/333598a0. [DOI] [PubMed] [Google Scholar]
- Cooper R. A. Metabolism of methylglyoxal in microorganisms. Annu Rev Microbiol. 1984;38:49–68. doi: 10.1146/annurev.mi.38.100184.000405. [DOI] [PubMed] [Google Scholar]
- Corran P. H., Waley S. G. The amino acid sequence of rabbit muscle triose phosphate isomerase. Biochem J. 1975 Feb;145(2):335–344. doi: 10.1042/bj1450335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Cox E. C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- Craik C. S., Largman C., Fletcher T., Roczniak S., Barr P. J., Fletterick R., Rutter W. J. Redesigning trypsin: alteration of substrate specificity. Science. 1985 Apr 19;228(4697):291–297. doi: 10.1126/science.3838593. [DOI] [PubMed] [Google Scholar]
- Cronin C. N., Kirsch J. F. Role of arginine-292 in the substrate specificity of aspartate aminotransferase as examined by site-directed mutagenesis. Biochemistry. 1988 Jun 14;27(12):4572–4579. doi: 10.1021/bi00412a052. [DOI] [PubMed] [Google Scholar]
- De la Mare S., Coulson A. F., Knowles J. R., Priddle J. D., Offord R. E. Active-site labelling of triose phosphate isomerase. The reaction of bromohydroxyacetone phosphate with a unique glutamic acid residue and the migration of the label to tyrosine. Biochem J. 1972 Sep;129(2):321–331. doi: 10.1042/bj1290321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn I. S., Cowan R., Jennings P. A. Improved peptide function from random mutagenesis over short 'windows'. Protein Eng. 1988 Oct;2(4):283–291. doi: 10.1093/protein/2.4.283. [DOI] [PubMed] [Google Scholar]
- Fraenkel D. G. Mutants in glucose metabolism. Annu Rev Biochem. 1986;55:317–337. doi: 10.1146/annurev.bi.55.070186.001533. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hartman F. C. Irreversible inactivation of triose phosphate isomerase by 1-hydroxy-3-iodo-2-propanone phosphate. Biochem Biophys Res Commun. 1968 Dec 30;33(6):888–894. doi: 10.1016/0006-291x(68)90395-1. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Sauer R. T. Phage lambda repressor revertants. Amino acid substitutions that restore activity to mutant proteins. J Mol Biol. 1985 Nov 5;186(1):53–63. doi: 10.1016/0022-2836(85)90256-6. [DOI] [PubMed] [Google Scholar]
- Knight K. L., Sauer R. T. DNA binding specificity of the Arc and Mnt repressors is determined by a short region of N-terminal residues. Proc Natl Acad Sci U S A. 1989 Feb;86(3):797–801. doi: 10.1073/pnas.86.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles J. R. Tinkering with enzymes: what are we learning? Science. 1987 Jun 5;236(4806):1252–1258. doi: 10.1126/science.3296192. [DOI] [PubMed] [Google Scholar]
- Kolb E., Harris J. I., Bridgen J. Triose phosphate isomerase from the coelacanth. An approach to the rapid determination of an amino acid sequence with small amounts of material. Biochem J. 1974 Feb;137(2):185–197. doi: 10.1042/bj1370185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherbarrow R. J., Fersht A. R. Protein engineering. Protein Eng. 1986 Oct-Nov;1(1):7–16. doi: 10.1093/protein/1.1.7. [DOI] [PubMed] [Google Scholar]
- Lehtovaara P. M., Koivula A. K., Bamford J., Knowles J. K. A new method for random mutagenesis of complete genes: enzymatic generation of mutant libraries in vitro. Protein Eng. 1988 Apr;2(1):63–68. doi: 10.1093/protein/2.1.63. [DOI] [PubMed] [Google Scholar]
- Lu H. S., Yuan P. M., Gracy R. W. Primary structure of human triosephosphate isomerase. J Biol Chem. 1984 Oct 10;259(19):11958–11968. [PubMed] [Google Scholar]
- Maquat L. E., Chilcote R., Ryan P. M. Human triosephosphate isomerase cDNA and protein structure. Studies of triosephosphate isomerase deficiency in man. J Biol Chem. 1985 Mar 25;260(6):3748–3753. [PubMed] [Google Scholar]
- Marchionni M., Gilbert W. The triosephosphate isomerase gene from maize: introns antedate the plant-animal divergence. Cell. 1986 Jul 4;46(1):133–141. doi: 10.1016/0092-8674(86)90867-6. [DOI] [PubMed] [Google Scholar]
- McKnight G. L., O'Hara P. J., Parker M. L. Nucleotide sequence of the triosephosphate isomerase gene from Aspergillus nidulans: implications for a differential loss of introns. Cell. 1986 Jul 4;46(1):143–147. doi: 10.1016/0092-8674(86)90868-8. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Lerman L. S., Maniatis T. A general method for saturation mutagenesis of cloned DNA fragments. Science. 1985 Jul 19;229(4710):242–247. doi: 10.1126/science.2990046. [DOI] [PubMed] [Google Scholar]
- Müller W., Weber H., Meyer F., Weissmann C. Site-directed mutagenesis in DNA: generation of point mutations in cloned beta globin complementary dna at the positions corresponding to amino acids 121 to 123. J Mol Biol. 1978 Sep 15;124(2):343–358. doi: 10.1016/0022-2836(78)90303-0. [DOI] [PubMed] [Google Scholar]
- Ner S. S., Goodin D. B., Smith M. A simple and efficient procedure for generating random point mutations and for codon replacements using mixed oligodeoxynucleotides. DNA. 1988 Mar;7(2):127–134. doi: 10.1089/dna.1988.7.127. [DOI] [PubMed] [Google Scholar]
- Pichersky E., Gottlieb L. D., Hess J. F. Nucleotide sequence of the triose phosphate isomerase gene of Escherichia coli. Mol Gen Genet. 1984;195(1-2):314–320. doi: 10.1007/BF00332765. [DOI] [PubMed] [Google Scholar]
- Putman S. J., Coulson A. F., Farley I. R., Riddleston B., Knowles J. R. Specificity and kinetics of triose phosphate isomerase from chicken muscle. Biochem J. 1972 Sep;129(2):301–310. doi: 10.1042/bj1290301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIEDER S. V., ROSE I. A. The mechanism of the triosephosphate isomerase reaction. J Biol Chem. 1959 May;234(5):1007–1010. [PubMed] [Google Scholar]
- Raines R. T., Sutton E. L., Straus D. R., Gilbert W., Knowles J. R. Reaction energetics of a mutant triosephosphate isomerase in which the active-site glutamate has been changed to aspartate. Biochemistry. 1986 Nov 4;25(22):7142–7154. doi: 10.1021/bi00370a057. [DOI] [PubMed] [Google Scholar]
- Russell P. R. Transcription of the triose-phosphate-isomerase gene of Schizosaccharomyces pombe initiates from a start point different from that in Saccharomyces cerevisiae. Gene. 1985;40(1):125–130. doi: 10.1016/0378-1119(85)90031-9. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Protein engineering. The design, synthesis and characterization of factitious proteins. Biochem J. 1987 Aug 15;246(1):1–17. doi: 10.1042/bj2460001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Grisafi P., Benkovic S. J., Botstein D. Gap misrepair mutagenesis: efficient site-directed induction of transition, transversion, and frameshift mutations in vitro. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1588–1592. doi: 10.1073/pnas.79.5.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Lin B. Genetic analysis of staphylococcal nuclease: identification of three intragenic "global" suppressors of nuclease-minus mutations. Genetics. 1985 Aug;110(4):539–555. doi: 10.1093/genetics/110.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. M. Natural selection and the concept of a protein space. Nature. 1970 Feb 7;225(5232):563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- Straus D., Gilbert W. Chicken triosephosphate isomerase complements an Escherichia coli deficiency. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2014–2018. doi: 10.1073/pnas.82.7.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus D., Raines R., Kawashima E., Knowles J. R., Gilbert W. Active site of triosephosphate isomerase: in vitro mutagenesis and characterization of an altered enzyme. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2272–2276. doi: 10.1073/pnas.82.8.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels B. W., Gibson W. C., Osinga K. A., Kramer R., Veeneman G. H., van Boom J. H., Borst P. Characterization of the gene for the microbody (glycosomal) triosephosphate isomerase of Trypanosoma brucei. EMBO J. 1986 Jun;5(6):1291–1298. doi: 10.1002/j.1460-2075.1986.tb04358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton R. P., Ptashne M. Changing the binding specificity of a repressor by redesigning an alpha-helix. Nature. 1985 Aug 15;316(6029):601–605. doi: 10.1038/316601a0. [DOI] [PubMed] [Google Scholar]
- Wilks H. M., Hart K. W., Feeney R., Dunn C. R., Muirhead H., Chia W. N., Barstow D. A., Atkinson T., Clarke A. R., Holbrook J. J. A specific, highly active malate dehydrogenase by redesign of a lactate dehydrogenase framework. Science. 1988 Dec 16;242(4885):1541–1544. doi: 10.1126/science.3201242. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Tryptophan biosynthesis in Escherichia coli. Genetic determination of the proteins involved. JAMA. 1971 Nov 15;218(7):1026–1035. [PubMed] [Google Scholar]




